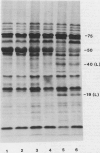
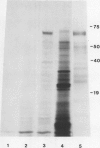
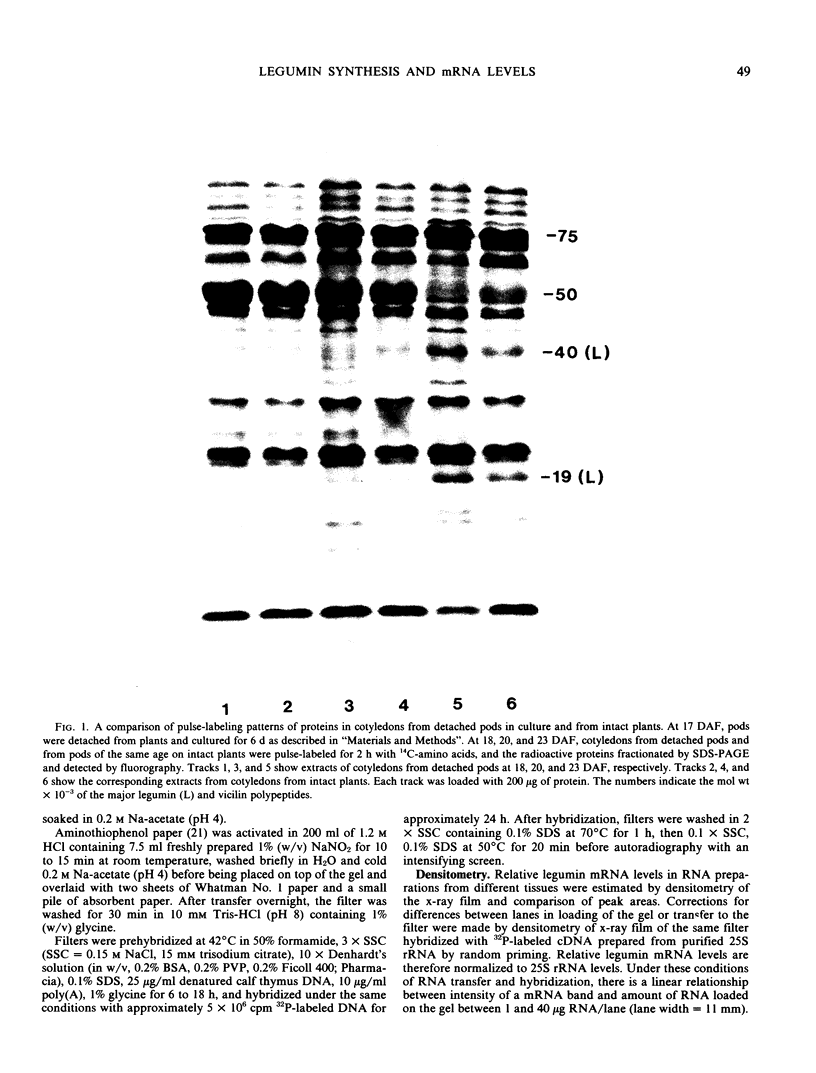
Abstract
It was shown previously that when peas (Pisum sativum L.) are grown with suboptimal sulfur supply the level of legumin (the more S-rich of the two major seed storage proteins) in the mature seed is selectively reduced (Randall, Thomson, Schroeder, 1979 Aust J Plant Physiol 6: 11-24). This paper reports a study of the cellular mechanisms involved in regulating legumin synthesis under these conditions. Pulse and pulse-chase labeling experiments were carried out with excised, immature cotyledons from normal and S-deficient plants. Legumin was isolated from cotyledon extracts by immunochromatography, and the proportion of legumin synthesis relative to total protein synthesis was determined. Results showed that reduced legumin accumulation could largely be accounted for by a greatly reduced level of legumin synthesis (80-88% reduction) rather than by a major increase in legumin breakdown.
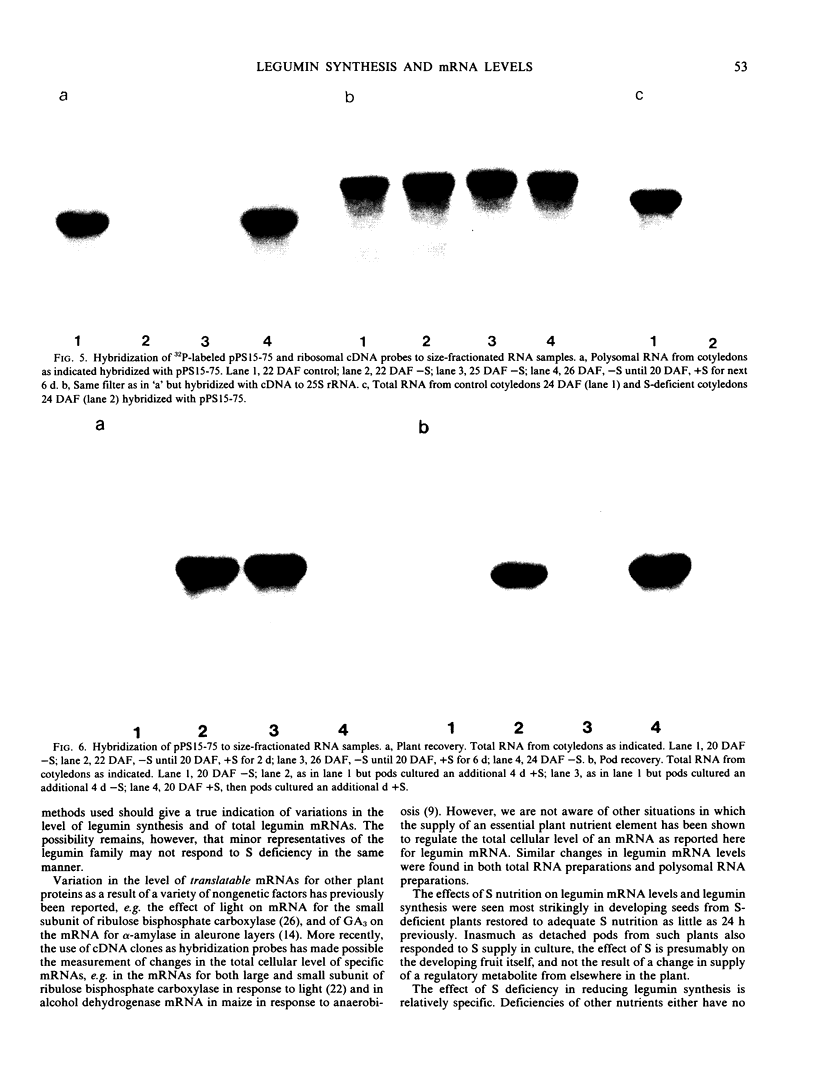
Legumin mRNA levels were assayed by two methods. In vitro translation of polysomal RNA from cotyledons of normal and S-deficient plants indicated a reduction of 60 to 70% in synthesis of legumin-related products by preparations from S-deficient plants. A legumin cDNA clone was constructed, characterized, and used to measure the levels of legumin mRNA in polysomal and total RNA preparations from developing cotyledons. Legumin mRNA levels were reduced by 90% in preparations from S-deficient plants.
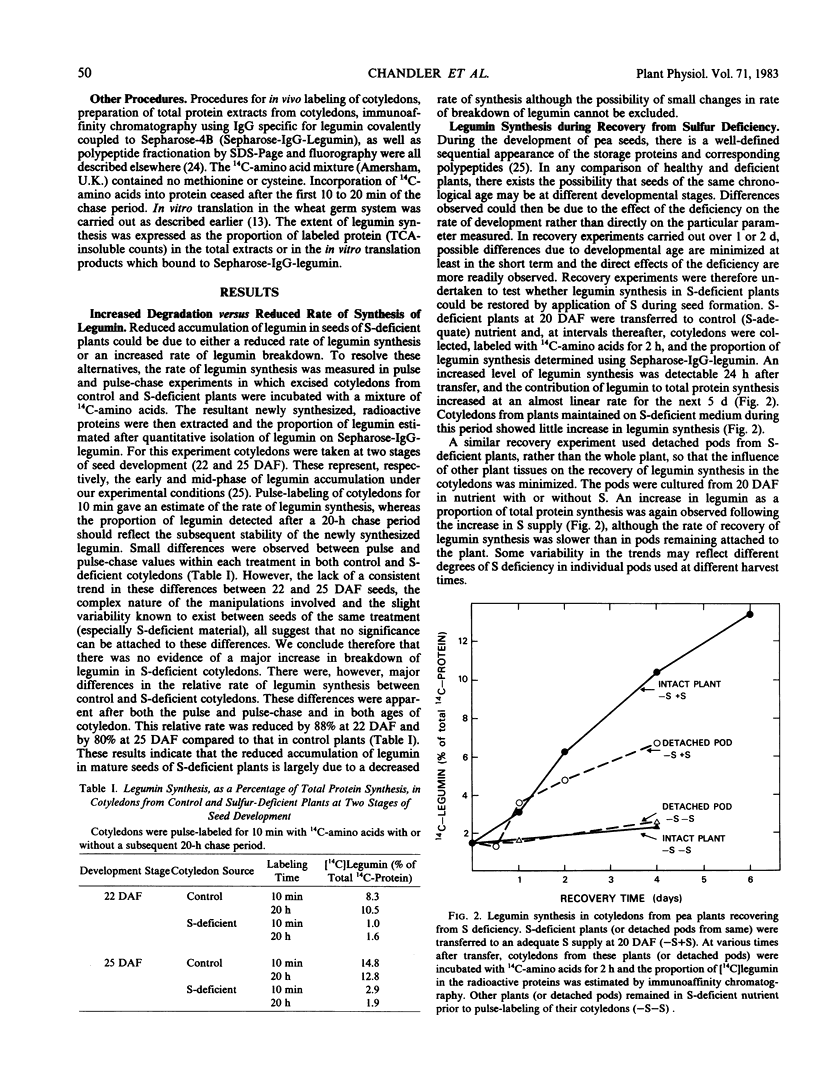
When restored to an adequate S supply, S-deficient plants (or pods taken from such plants) recovered normal levels of legumin synthesis (in vivo and in vitro) and of legumin mRNA. These results indicate that reduced legumin accumulation under conditions of S deficiency is primarily a consequence of reduced levels of legumin mRNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chandler P. M., Rimkus D., Davidson N. Gel electrophoretic fractionation of RNAs by partial denaturation with methylmercuric hydroxide. Anal Biochem. 1979 Oct 15;99(1):200–206. doi: 10.1016/0003-2697(79)90063-0. [DOI] [PubMed] [Google Scholar]
- Ereken-Tumer N., Richter J. D., Nielsen N. C. Structural characterization of the glycinin precursors. J Biol Chem. 1982 Apr 25;257(8):4016–4018. [PubMed] [Google Scholar]
- Gerlach W. L., Pryor A. J., Dennis E. S., Ferl R. J., Sachs M. M., Peacock W. J. cDNA cloning and induction of the alcohol dehydrogenase gene (Adh1) of maize. Proc Natl Acad Sci U S A. 1982 May;79(9):2981–2985. doi: 10.1073/pnas.79.9.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R., Neugebauer K., Pirkl E., Zentgraf H., Schaller H. Conversion of bacteriophage fd into an efficient single-stranded DNA vector system. Mol Gen Genet. 1980 Jan;177(2):231–242. doi: 10.1007/BF00267434. [DOI] [PubMed] [Google Scholar]
- Higgins T. J., Spencer D. Cell-free Synthesis of Pea Seed Proteins. Plant Physiol. 1977 Nov;60(5):655–661. doi: 10.1104/pp.60.5.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph T., Higgins V., Spencer D. Precursor Forms of Pea Vicilin Subunits: MODIFICATION BY MICROSOMAL MEMBRANES DURING CELL-FREE TRANSLATION. Plant Physiol. 1981 Feb;67(2):205–211. doi: 10.1104/pp.67.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moreira M. A., Hermodson M. A., Larkins B. A., Nielsen N. C. Partial characterization of the acidic and basic polypeptides of glycinin. J Biol Chem. 1979 Oct 10;254(19):9921–9926. [PubMed] [Google Scholar]
- Myers J. C., Spiegelman S. Sodium pyrophosphate inhibition of RNA.DNA hybrid degradation by reverse transcriptase. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5329–5333. doi: 10.1073/pnas.75.11.5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Jan;3(1):101–116. doi: 10.1093/nar/3.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M., Ellis R. J. Light-stimulated accumulation of transcripts of nuclear and chloroplast genes for ribulosebisphosphate carboxylase. J Mol Appl Genet. 1981;1(2):127–137. [PubMed] [Google Scholar]
- Spencer D., Higgins T. J., Button S. C., Davey R. A. Pulse-labeling Studies on Protein Synthesis in Developing Pea Seeds and Evidence of a Precursor Form of Legumin Small Subunit. Plant Physiol. 1980 Sep;66(3):510–515. doi: 10.1104/pp.66.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin E. M., Suttie J. L. Light Effects on the Synthesis of Ribulose-1,5-Bisphosphate Carboxylase in Lemna gibba L. G-3. Plant Physiol. 1980 Apr;65(4):641–647. doi: 10.1104/pp.65.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]