Summary
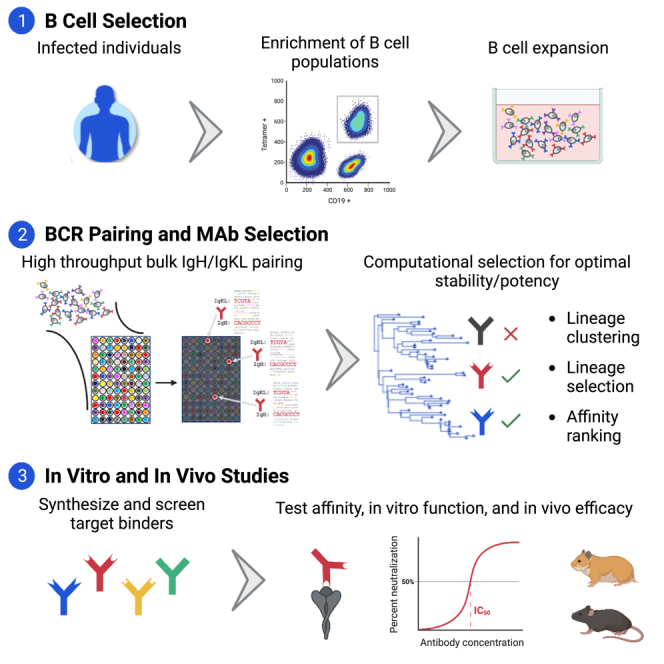
TruAB Discovery is an approach that integrates cellular immunology, high-throughput immunosequencing, bioinformatics, and computational biology in order to discover naturally occurring human antibodies for prophylactic or therapeutic use. We adapted our previously described pairSEQ technology to pair B cell receptor heavy and light chains of SARS-CoV-2 spike protein-binding antibodies derived from enriched antigen-specific memory B cells and bulk antibody-secreting cells. We identified approximately 60,000 productive, in-frame, paired antibody sequences, from which 2,093 antibodies were selected for functional evaluation based on abundance, isotype and patterns of somatic hypermutation. The exceptionally diverse antibodies included RBD-binders with broad neutralizing activity against SARS-CoV-2 variants, and S2-binders with broad specificity against betacoronaviruses and the ability to block membrane fusion. A subset of these RBD- and S2-binding antibodies demonstrated robust protection against challenge in hamster and mouse models. This high-throughput approach can accelerate discovery of diverse, multifunctional antibodies against any target of interest.
Keywords: Antibodies, SARS CoV-2, S2 antibodies, Antibody secreting cells, Cell fusion, High throughput sequencing, BCR pairSEQ, TruAB Discovery, Broadly neutralizing antibodies
Graphical abstract
Highlights
-
•
NGS-based TruAB platform enables high-throughput discovery of diverse antibodies
-
•
Paired antibodies originate from memory B cells and ASCs of donors with COVID-19
-
•
Anti-S1 and -S2 antibodies neutralize live virus and offer in vivo protection
-
•
Anti-S2 antibodies block membrane fusion and exhibit pan-betacoronavirus activity
Keitany et al. demonstrate a novel antibody discovery method yielding large numbers of spike-reactive antibodies from B cells of individuals infected with COVID19. These antibodies have diverse sequences and MOAs, including S2-binders reactive across betacoronaviruses that neutralize by blocking membrane fusion. A tested subset provides potent protection in animal models.
Introduction
Monoclonal antibodies proved to be an important drug modality during the SARS-CoV-2 pandemic and demonstrated efficacy either as prophylactic treatment for high risk immune-compromised individuals or for early therapeutic treatment following infection.1,2,3 All antibodies approved for use against COVID-19 target the S1 domain of the spike protein and have been described to neutralize primarily through the inhibition of attachment to the ACE2 receptor by the spike receptor binding domain (RBD).4,5,6,7 While antibodies targeting the S1 region can inhibit the virus from binding to the host cell receptor, this region of the spike protein is highly variable8,9 and contains the majority of mutations among SARS-CoV-2 variants.10,11,12 As a result, several of these variants pose a major challenge for antibody-based therapeutics, including the Omicron lineage which drastically reduces the effectiveness of more than 85% of tested antibodies.13,14 Therefore, diverse sets of neutralizing antibodies that bind to a variety of conserved epitopes are of clear value for combating future SARS-CoV-2 variants and for coronavirus pandemic preparedness.
A small number of antibodies that bind to relatively conserved domains in the SARS-CoV-2 RBD can broadly neutralize several sarbecovirus taxa.15 Although generally less potent than RBD-directed antibodies, the S2 domain is also a potentially viable target for broadly neutralizing antibodies.9,16,17,18 The high conservation (63–98%) of S2 domain across human coronavirus strains makes it an especially interesting antibody target.8,9 Composed of a fusion peptide (FP), heptad repeat 1 (HR1), and heptad repeat 2 (HR2) regions, the S2 domain mediates fusion of the viral envelope with the host cell membrane.19 S2 undergoes proteolytic cleavage at the S2′ site just upstream of the FP, exposing the FP and allowing for insertion and subsequent membrane fusion. A small subset of S2-binding antibodies has been tested for in vivo efficacy and can protect animals from infection through inhibition of this fusion, as well as Fc-mediated effector function.16,17,18,20 However, S2 epitopes are not well exposed on commonly used prefusion stabilized versions of the spike trimer,20,21 limiting our ability to identify S2-specific B cells.
Due to the continued growing utility of monoclonal antibody therapies, improvements in technologies for high-throughput screening of B cells are warranted. The traditional methods of antibody discovery such as hybridoma and phage display require significant investments of time and labor and are inherently low throughput, further increasing cost and limiting the diversity of discoverable antibodies.22 Refinement in techniques including the use of fluorochrome-conjugated tetramerized antigenic reagents for labeling and enrichment of antigen-specific B cells using anti-fluorochrome magnetic beads, followed by flow cytometry-based analysis of the enriched B cells improved the ability to phenotype antigen-specific B cells.23 Use of tetramers and multimers also enabled the isolation of purified populations of these cells via fluorescence-activated cell sorting (FACS) for downstream analysis including single-cell sequencing. On the other hand, modern droplet microfluidics even allows for the enrichment of antigen-specific antibody-secreting cells (ASCs) that lack surface IgG.24 While these developments have contributed to substantial increases in the speed and efficiency with which antibody therapies can be developed, antibody discovery efforts are still limited by the lack of high-throughput technologies for pairing antibody heavy and light chains at scale.
The combination of heavy (IgH) and light (IgL) chain subunits in functional antibodies determines their specificity25,26 and potential value for therapeutic use. Many high-throughput amplicon-based approaches now exist for characterizing the IgH or IgL repertoires individually27 but, given the location of these genes on separate chromosomes, identifying naturally occurring gene “pairs” and the functional antibodies they encode remains technically challenging. Single-cell sequencing, whether well-based28,29,30 or emulsion droplet-based,31,32,33 can provide large numbers of pairs with relative ease and has become the convention for antibody discovery. Despite its power, the single-cell approach has drawbacks, requiring specialized equipment and losing the ability to discover diverse antibodies in highly clonal settings where a small number of lineages make up the majority of the B cell population.
Here we validated our TruAB Discovery approach, including its use of pairSEQ, a high-throughput method previously developed for pairing the alpha and beta chains of T cell receptors34 for use in pairing and functionally characterizing naturally occurring, fully human antibody heavy and light chains from individuals infected with SARS-CoV-2 (BCR pairSEQ). Several key features distinguish BCR pairSEQ from other, single-cell approaches for natively pairing antibody sequences. First, BCR pairSEQ requires no upfront purchase of specialized lab equipment, leveraging only standard molecular biology techniques. Second, the strongest constraint on the theoretical upper limit to the number of antibodies paired in a single batch of BCR pairSEQ is the total diversity of the input cell population, rather than constraints of the technology or quantity of reagents invested. Finally, the success of single-cell approaches can be severely hampered by highly abundant B cell clones, resulting in substantial investment in the redundant recovery of a single unique pair at the expense of less abundant clones. BCR pairSEQ has no such constraint and is capable of simultaneously pairing antibodies across a huge range of abundances.
Using BCR pairSEQ, we identified more than 60,000 unique, in-frame, functional, full-length, and natively paired BCR sequences and screened over 2,000 antibodies for ability to bind multiple SARS-CoV-2 spike domains. Our pipeline yielded anti-RBD, anti-S1 (but non-RBD-specific) plus novel anti-S2 antibodies with neutralizing capacity across betacoronaviruses with demonstrated protection in vivo against SARS-CoV-2 challenge in two animal models. This study exemplifies the power of BCR pairSEQ in generating ultra-diverse antibody libraries for therapeutic applications.
Results
Thousands of antibodies isolated from BCR pairSEQ of enriched spike-specific memory B cells
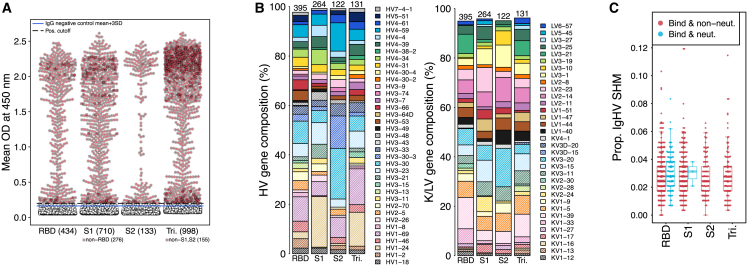
Memory B cells from 227 individuals infected with SARS-CoV-2 between April and October 2020 were enriched for antigen-specific B cells using S1/S2 tetramer or spike trimer protein. B cells were sequenced and analyzed, yielding 59,045 naturally paired, unique, functional, in-frame, and full-length antibody sequences representing 29,577 independent B cell lineages (<85% similarity in HCDR3 nucleotide sequences). We used a proprietary bioinformatics pipeline leveraging abundance, isotype, and patterns of somatic hypermutation (SHM) to select a total of 1,727 antibodies representing 1,395 clonal lineages from this set for synthesis and additional screening, of which 998 antibodies representing 912 lineages (58% and 65% binding success, respectively) bound to SARS-CoV-2 spike (Figures 1A and 1B). Of the 998 spike-binding antibodies, 434 bound to RBD, 276 bound to S1 but not RBD, 133 bound to S2, and 155 bound the full spike trimer but not to S1, RBD, or S2 alone. Our S1 non-RBD antibodies are likely N-terminal domain (NTD) specific but other regions of S1 cannot be excluded. Similarly, those identified as trimer-specific that fail to bind to RBD, S1, or S2 individually may include regions of both S1 and S2 in their epitope or may bind at the interface of spike monomers, requiring the natural trimer to bind with high affinity.
Figure 1.
Characteristics of antibodies from enriched antigen-specific memory B cells
(A) Every synthesized antibody was tested in duplicate by ELISA for binding against four spike antigens: RBD, S1 including RBD, S2, and the full spike protein in its trimeric form (Tri). Non-specific IgG controls were included in each ELISA and are denoted.
(B) Diversity of heavy (left) and light chain (right) of spike-reactive antibodies. Numbers above bars indicate the total antibodies represented. Only genes represented by at least 1.5% of antibodies in an individual category are shown. Where multiple antibodies per lineage were tested, only a representative single lineage is included.
(C) Proportion of sites in heavy chain V genes in spike-reactive antibodies that have experienced somatic hypermutation relative to germline. Red dots = antibodies that bind and do not neutralize pseudovirus and blue = antibodies that bind and neutralize pseudovirus. See also Figure S1 and Table S2.
Levels of SHM among binders were relatively low (mean 2.9% in HV gene relative to germline; Figures 1C and S1), regardless of the antigen to which the antibodies reacted. On the other hand, V-genes were well-represented among the spike binders regardless of region bound, although some differences between antigen specificity were apparent (Figure 1B). For example, HV1-24 and KV3-15, both of which were previously reported to commonly bind to the NTD “supersite”, were highly represented among S1 non-RBD antibodies identified here (Figure 1B).35,36
A large subset of isolated antibodies from enriched, spike-specific memory B cells neutralize SARS-CoV-2 by blocking ACE2 binding
The majority of SARS-CoV-2 neutralizing antibodies identified thus far inhibit virus by blocking ACE2-RBD interaction,4,5,6,7 although non-ACE2-based neutralization has also been reported.16,17,18,20,35,36 Among the antibodies identified via BCR pairSEQ, a large subset blocked RBD binding to ACE2 (Figure 2A). Of 434 (130 lineages) RBD-binding antibodies, 134 (31%) demonstrated an ACE2-binding inhibition above 50%. Similar numbers were generally observed between ACE2 inhibition assays and the pseudovirus (WT,WA/2020) neutralization assay: 25% (108/434) of RBD binding antibodies from 106 different lineages achieved 70% or greater neutralization (Figure 2B). Although the majority of antibodies were RBD-specific, we also identified three non-RBD, S1 binding antibody lineages that neutralized pseudovirus. We did not observe substantial differences in levels of SHM between antigen-specific neutralizing and non-neutralizing antibodies for either RBD or non-RBD antibodies (Figure 1C). The low SHM levels are similar to what has been previously reported.37
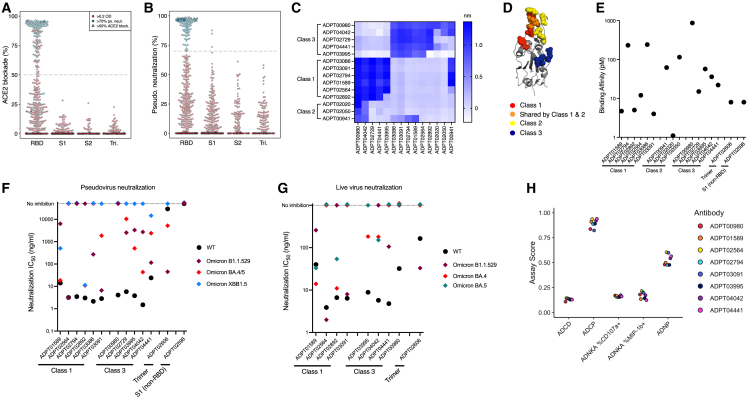
Figure 2.
Functional characterization of selected spike-specific antibodies
(A) Screening of ACE2/RBD blockade by competitive ELISA, depicted as % blockade. Antibodies that blocked this interaction by at least 50% were advanced (triangles). Only antibodies considered positive binders (OD450 > 0.2) by ELISA are shown and spike region specificity (RBD, S1, S2, and Trimer [Tri.]) is defined based on Figure 1A.
(B) Pseudovirus neutralization screening of all antibodies expressed as recombinant IgG1. Target cells used were an ACE2/TMPRSS2-expressing cell line and all antibodies were tested at 10 μg/mL. Neutralization cut-off of over 70% was used to advance antibodies (blue). Only antibodies considered positive binders to spike are shown.
(C) Heatmap showing degree of binding interference between paired antibodies as determined by Octet competition experiments. “Class” assignments38 are inferred from the epitope groups identified and critical binding residues identified by shotgun mutagenesis.
(D) Critical binding residues are highlighted on RBD structure (PDB: 7BZ5).(E) Binding affinity measured by SPR of selected antibodies to RBD (antibodies labeled with “Class”), non-RBD S1 or the full spike trimer.
(F) Pseudovirus neutralization shown by IC50 of selected antibodies against SARS-CoV-2 variants (WT = WA1). IC50 shown is the average of two independent replicates.
(G) IC50 of antibodies against live WT and Omicron variant SARS-CoV-2 viruses. IC50 values were calculated using the Plaque reduction neutralization test (PRNT) assay. IC50 shown is the average of two independent replicates.
(H) Results from bead-based effector function assay measuring complement activation (ADCD), antibody mediated phagocytosis (ADCP and ADNP), and cytotoxicity (ADNKA CD107+ and MIP1b+). The y axis shows normalized assay scores where higher scores indicate higher effector function. Also see Figures S2 and S3 and Table S1.
Antibodies with superior neutralization of pseudovirus were characterized further. Because of the large number of neutralizing antibodies identified, we narrowed down the pool by selecting highly potent antibodies from different epitope classes. RBD-binding antibodies have been classified into four different classes based on similarities in binding locations and neutralization modes of action.38 Sequence features were used to predict class assignments for our antibodies, and these were confirmed experimentally by binning antibodies using biolayer interferometry (BLI). Six antibodies were selected from class 1, three antibodies from class 2, and five antibodies from class 3 (Figure 2C). To confirm the specificities of these antibodies and delineate binding residues on the RBD protein, we performed epitope mapping via shotgun mutagenesis (Figures 2D and S2). Briefly, our antibodies were tested for binding activity against a mutation library where each residue in the SARS-CoV-2 RBD was mutated to alanine (natural alanine residues were mutated to serine). Residues which, when mutated, reduced binding by at least 75% were considered a critical part of the epitope. The binding residues were visualized on crystal structure of RBD (Figures 2D and S2).39
Antigen specificity and affinity of selected antibodies to RBD or full-length trimer were confirmed using surface plasmon resonance (SPR) assays. Antibodies bound to their respective target domains with high affinity, with most antibodies binding with KD <100 p.m. as measured by SPR (Figure 2E). All classes of antibodies neutralized both WT (WA/2020) and Alpha variant pseudovirus with IC50 values ranging from 9 to 468 ng/mL, with the majority of antibodies under 100 ng/mL (Figure S3A). Both class 1 and 3 antibodies maintained neutralization activity against Beta variant pseudovirus, but all four class 2 antibodies lost the ability to neutralize this variant. Loss of Beta variant neutralization by class 2 antibodies is consistent with previously published data that showed that antibodies in this group share a critical binding residue at RBD position 484 that is mutated from glutamate (E) to lysine (K) in the Beta variant (B.1.351)40 as we observed in our epitope mapping (Figure S2). Four of our antibodies (ADPT02564, ADPT03091, ADPT04441, and ADPT02606) maintained neutralization potency when tested against Omicron variant B.1.1.529 and BA.4/5 pseudovirus (Figure 2F). Two different antibodies (ADPT01589 and ADPT02892) were able to neutralize the latest Omicron variant (XBB1.5) as well as all other strains with the exception of B.1.1.529.
Neutralization of WT and Beta variants were confirmed using authentic live virus (Figure S3B). Class 2 and S1 antibodies did not inhibit Beta variant so we did not include them in the live virus neutralization assay. Selected RBD, trimer, and S1 binding antibodies were also tested on different variants of Omicron authentic live virus, B.1.1.529, BA.4, and BA.5 (Figure 2G). Overall, based on both pseudo and authentic live virus assays, many of our antibodies retained a level of potency against the Omicron strain tested, although the antibodies were impacted differently depending on the variant.
Previous studies have shown that some SARS-CoV-2 antibodies fail to neutralize pseudovirus, but are potent neutralizers against authentic SARS-CoV-2 virus.41 To identify these unique antibodies, we advanced all non-RBD binding antibodies to a live virus neutralization assay, including the antibodies for which pseudovirus neutralization was observed. A large number of these S1- and trimer-binding antibodies showed neutralization of live virus (Table S1). The majority of these antibodies were not characterized further because most of them were directed to the NTD, a region where many antibody-abrogating mutations were present in some of the earliest SARS-CoV-2 variants to emerge, including Alpha and Beta.11,12
We further selected 8 antibodies from among the class 1 and 3 antibodies and determined whether these RBD-specific antibodies mediated effector function. We used a multiplexed bead-based assay to measure antibody-dependent cellular phagocytosis (ADCP), antibody-dependent phagocytosis by neutrophils (ADNP), antibody-dependent cell cytotoxicity (ADCC) by measuring the % NK cell expression of CD107a, and MIP-1b and antibody-mediated complement activation.42,43,44,45 All of the selected RBD-specific antibodies mediated strong phagocytic activity as measured by both ADCP and ADNP assays (Figure 2H). To confirm phagocytic activity by RBD antibodies we performed a cell-based ADCP assay where monocytes isolated from PBMCs were used as effector cells and stable spike-expressing 293T cells were used as target cells. Similar to the bead-based assay, all RBD antibodies showed strong ADCP response (Figure S3C). These data demonstrate that in addition to neutralization, our RBD antibodies induce robust effector function that could contribute to in vivo protection.
Distinctive antibody repertoire from ASCs of acutely ill donor
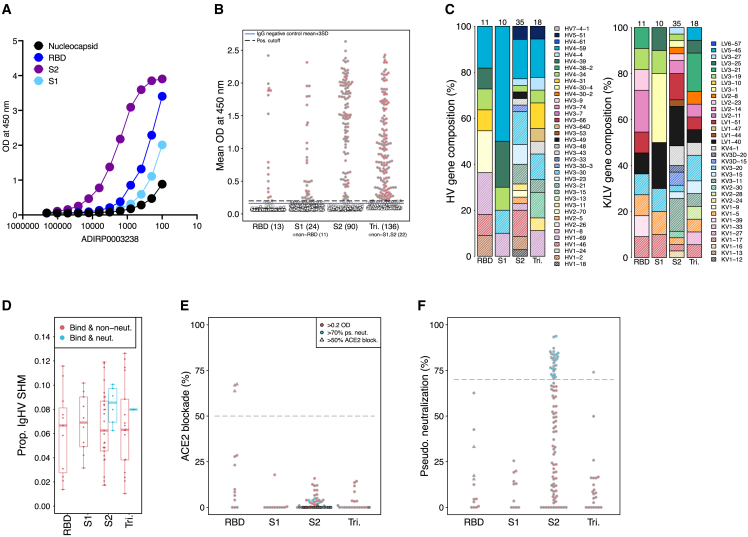
Overall, antibody discovery rates were high in memory B cells isolated from convalescent individuals due to the ability to pre-sort antigen-specific B cells. However, unlike memory B cells, ASCs cannot be enriched for antigen specificity due to lack of surface expression of BCRs. To determine whether our pairSEQ technology can be used to discover antibodies in an antigen-agnostic manner, we evaluated the ASC repertoire from one 30-year-old male sampled in July 2020 (ADIRP0003238) while still in the acute phase of illness, nine days post-symptom onset, and 3 days after last symptom. Evaluation of serum antibodies from this individual showed an unusually S2-dominant response as compared to other COVID-19-recovered individuals (Figure 3A). ASCs from this donor were isolated by cell sorting (CD19+IgD−IgM-CD27highCD38high) and sequenced on a dedicated pairSEQ plate yielding 1,460 unique, functional, in-frame, and full-length sequences from 610 clonal lineages. The ASCs were not expanded ex vivo prior to sequencing and the pairs identified were present in a median of five distinct wells. These pairs, therefore, represent naturally expanded clones. Of the antibodies identified, 366 (202 lineages) were selected using our standard bioinformatics criteria for synthesis and screening, of which 136 from 74 lineages (37% and 37% binding success, respectively) bound to SARS-CoV-2 spike (Figure 3B). The vast majority (90) of these spike-binding antibodies bound to S2, whereas only 13 bound to RBD, 11 bound to S1, and 22 bound to the spike trimer only (Figure 3B). V-genes were well-represented among those screened that bound to S2 from this donor (Figure 3C). However, patterns of SHM show that this donor’s antibodies were quite different from those found from other sources (Figures 1C, 3D, and S1). Spike-reactive antibodies had significantly greater proportions of SHM in heavy chain V-genes in these antibodies compared to those discovered from memory B cells (Wilcoxon rank-sum p = 1.9 × 10−14). Given that these antibodies were isolated from ASCs only 9 days after symptom onset early in the pandemic, the high SHM of this donor’s repertoire suggests that they likely experienced a previous infection by a related coronavirus, and B cell lineages observed were likely originally stimulated in response to that encounter.
Figure 3.
Distinctive antibody repertoire from the ASCs of an individual with a recent SARS-CoV-2 infection
(A) Depicts ELISA on donor (ADIRP0003238) serum showing binding to nucleocapsid, RBD, S2, and S1 proteins.
(B) ELISA binding to RBD, S2, S1, and Trimer (Tri.) proteins by antibodies selected for synthesis and screening from ADIRP0003238.
(C and D) (C) V-gene usage of selected antibodies, and (D) SHM in ADIRP0003238 donor’s ASC-derived monoclonal antibodies.
(E) Competitive ELISA screening of antibodies for ACE2/RBD blockade shown as % blockade.
(F) Pseudovirus neutralization screening of all antibodies expressed as recombinant IgG1. Target cells used were an ACE2/TMPRSS2-expressing cell line and all antibodies were tested at 10 μg/mL. Neutralizing antibodies indicated in blue. Also see Figure S1.
Functional characterization of pan-betacoronavirus reactive S2 antibodies isolated from ASCs
Unique to ADIRP0003238 donor’s ASCs were several S2-binding antibodies that, despite showing no evidence of ACE2 blockade (Figure 3E), strongly neutralized pseudovirus (Figure 3F). This was surprising given that we had previously isolated over 100 S2-binding antibodies from memory B cells from all other donors and none of these were observed to neutralize pseudovirus (Figure 2B). Memory B-cell-derived S2-binding antibodies had significantly lower SHM as compared to both S2 binders (p = 1.77 × 10−12) and S2 neutralizers (p = 4.9 × 10−5) from this donor’s ASCs (Figure 3D). These S2 neutralizers also had significantly greater SHM than all neutralizers from memory B cells (p = 4.9 × 10−5). The lack of ACE2 blockage suggests that these S2 antibodies function via an alternative mechanism.
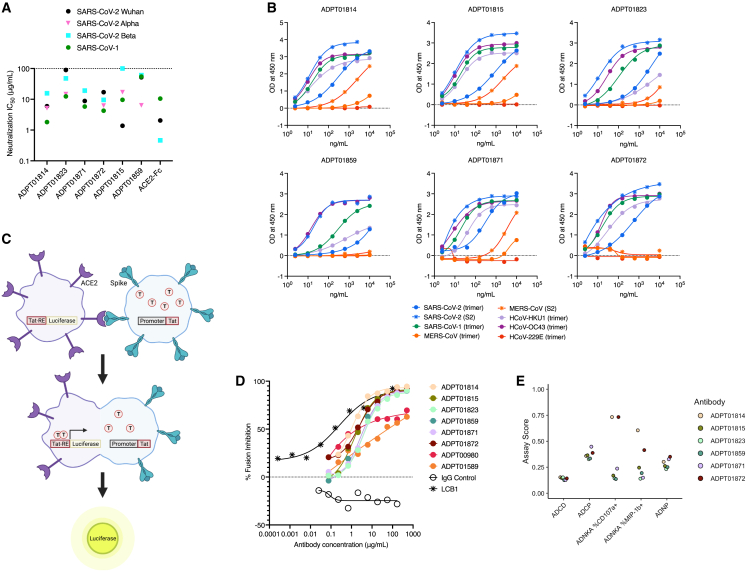
Six S2-specific antibodies that showed the highest neutralizing activity against SARS-CoV-2 during screening were chosen for further characterization. These S2 antibodies were additionally able to neutralize pseudoviruses of various SARS-CoV-2 strains as well as SARS-CoV-1 (Figure 4A), albeit at thousand-fold less potency based on IC50 values compared to the RBD antibodies we evaluated (Figure 2F). It is notable that for four out of six of these antibodies the highest potency was against SARS-CoV-1, a phenomenon not observed with RBD antibodies. The S2-specific antibodies were also tested for binding against a panel of various spike antigens across several betacoronavirus strains (SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-HKU1, and HCoV-OC43) and one alphacoronavirus strain (HCoV-229E). All six antibodies displayed binding to spike antigens from several betacoronavirus strains, although none bound to the alphacoronavirus strain HCoV-229E (Figure 4B).
Figure 4.
Neutralization, breadth, and mechanism of action for S2-binding antibodies
(A) Pseudovirus neutralization IC50 of ASC-derived S2 antibodies as described in Figure 2F. Neutralization IC50 of pseudoviruses of various SARS-CoV-2 strains as well as SARS-CoV-1 is shown.
(B) Dilution curves of antibody binding by ELISA to spike trimer and S2 protein from betacoronaviruses (SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-HKU1, and HCoV-OC43) and one alphacoronavirus (HCoV-229E).
(C) Schematic of the cell-cell fusion assay. Figure created using BioRender.
(D) Dilution curves of selected antibodies blocking cell-cell fusion. LCB1, a peptide previously shown to block fusion, was used as a positive control. Data shown is representative of 3 experiments.
(E) Results from bead-based effector function assay measuring ADCD, ADCP, ADNP, and cytotoxicity via ADNKA CD107+ and MIP1b+ measurement, as described in Figure 2H. Also see Figures S4 and S5.
Because these antibodies have the ability to neutralize virus in vitro but were not found to block ACE2 binding, we developed a cell-cell fusion inhibition assay to interrogate if these S2 antibodies were able to block cell fusion (Figure 4C). In this assay, cells expressing ACE2 on the surface and an LTR-driven luciferase reporter gene were incubated with cells expressing spike on the surface and the transcription factor Tat. Spike-binding and activation by ACE2 results in fusion of the two cells, transfer of the Tat protein, and subsequent expression of luciferase. The peptide LCB1 has been shown to block cell fusion and was used as a positive control.46 All six S2-binding antibodies blocked cell-cell fusion in this assay to similar levels (Figure 4D). Two RBD antibodies were also evaluated (ADPT1589 and ADPT00980) and both were observed to block fusion partially.
Previous efforts at S2-binding antibody epitope identification have been hindered by flexibility in the target region, partial occlusion by adjacent spike subunits, differences in prefusion and postfusion conformations, and disruption by adding stabilizing proline mutations in spike trimer.16,17,20,47,48,49 Nevertheless, S2-neutralizing antibodies have been found to predominantly target either a flexible region of the stem helix16,17,47 or the FP.20,48,49 Epitope binning experiments reveal that all of the S2-binders identified here likely share at least part of their epitope with other stem helix binders (Figure S4), namely CC40.817,47 and S2P6.16 However, given the comparatively low affinity of S2-binding antibodies combined with the difficulties listed previously, the complete epitope may also include neighboring regions of S2.
We also tested the ability of these S2-binding antibodies to mediate effector functions in a bead-based multiplex assay of ADCP, ADCC, and ADCD.44 Two antibodies (ADPT01814 and ADPT01872) demonstrated NK-mediated ADCC, shown as ADNKA scores measured by expression of both CD107a and MIP-1b on NK cells (Figure 4E). In a cell-based assay of ADCC, ADPT01814 demonstrated high fold induction of ADCC activity (Figure S5). We did not observe ADPT01872-mediated ADCC activity in this assay, a result that could be due to differences in antibody binding to antigen expressed on cells as opposed to displayed on beads.
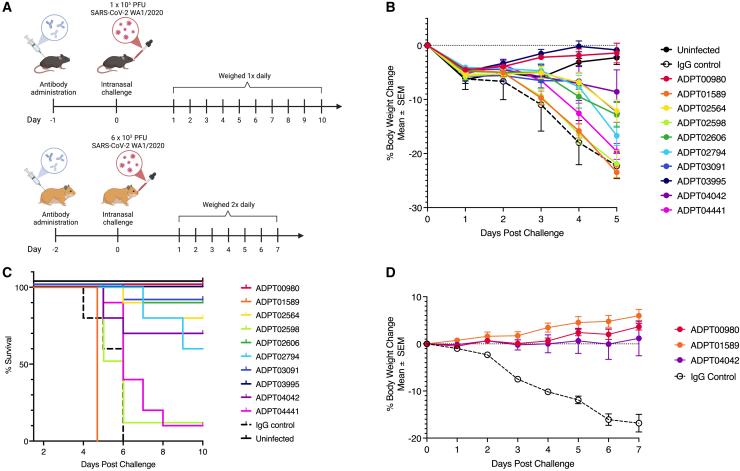
Passive transfer of RBD and NTD neutralizing antibodies protect against SARS-CoV-2 challenge in mice and Syrian hamsters
To determine whether our lead antibody candidates could elicit in vivo protection, we performed passive transfer of recombinant antibodies against RBD, NTD, and S2 into both K18h-ACE2 mice and Golden Syrian hamsters and then challenged with the original WA1 strain of SARS-CoV-2. RBD antibodies were selected based on in vitro potency and ability to neutralize multiple variants including Alpha, Beta, and Omicron (Figure 2F). With these criteria four RBD antibodies were selected from both class 1 (ADPT03091, ADPT02794, ADPT01589, and ADPT02564) and class 3 (ADPT00980, ADPT04042, ADPT03995, and ADPT04441). None of the antibodies from class 2 were selected due to lack of neutralization against the Beta variant (Figure 2F). In addition, 2 non-RBD antibodies (ADPT02606 and ADPT02598) that retained activity against the Omicron variant in pseudovirus neutralization experiments were also selected (Figure 2F). All mice were treated with 1.5 mg/kg of antibodies 24 h before intranasal infection with SARS-CoV-2 and weight was monitored (Figure 5A), with mice euthanized when they lost 25% of their body weight. Animals treated with ADPT00980 and ADPT03995 experienced no weight loss compared to control and the majority of the other antibodies (5 of 8) showed minimal weight loss 5 days after infection (Figure 5B). We calculated percent survival 10 days after virus infection and found that 7 of 10 antibodies tested show over 60% protection (Figure 5C). Class 3 antibodies showed the strongest protection, with two antibodies (ADPT00980 and ADPT03995) that protected mice 100% and another antibody (ADPT04042) with 70% protection. We observed significant protection when mice were treated with class 1 antibodies (ADPT03091 [90%], ADPT02564 [80%], and ADPT02794 [60%]). Of note, ADPT02606, an S1 specific non-RBD antibody which had the highest neutralization potency of all antibodies we tested against Omicron, showed 90% protection in mice (Figure 5C).
Figure 5.
RBD-, non-RBD S1-, and trimer-directed antibodies provide effective in vivo protection
(A) Schema of the in vivo experiments. K-18-hACE2 mice and hamsters were treated with antibodies at 1.5 or 5.0 mg/kg, respectively, via intraperitoneal (IP) administration. Animals were then inoculated with SARS-CoV-2 intranasally (dose as indicated), after either 24 h (mice) or 48 h (hamsters). Mock-infected and untreated animals were used as controls.
(B) Mean ± SEM percent change in mouse body weight after inoculation of each experimental group (N = 10 mice in each group).
(C) Survival curve of mice after passive transfer of SARS-CoV-2 antibodies. Animals were euthanized when body weight was less than 75% starting weight.
(D) Percent weight change in Syrian hamsters treated with 5 mg/kg of antibody IP 48 h prior to infection with SARS-CoV-2 intranasally (N = 5 hamsters per group). Data points represent Mean ± SEM. Human IgG1 was used as isotype control.
For the Hamster model, two class 3 antibodies, ADPT0980 and ADPT04042, and one class 1 antibody, ADPT01589, were selected for treatment. Hamsters were given 5 mg/kg of antibody 48 h prior to intranasal challenge with SARS-CoV-2/USA-WA1/2020 (Figure 5A). All three RBD antibodies tested showed complete protection as observed by lack of significant weight loss compared to infected control animals treated with isotype control IgG (20% weight loss; Figure 5D). Lack of protection by ADPT01589 in the mouse model but complete protection in hamsters could be due to dosage differences or differences in the clinical manifestation of disease in the different animal models.50
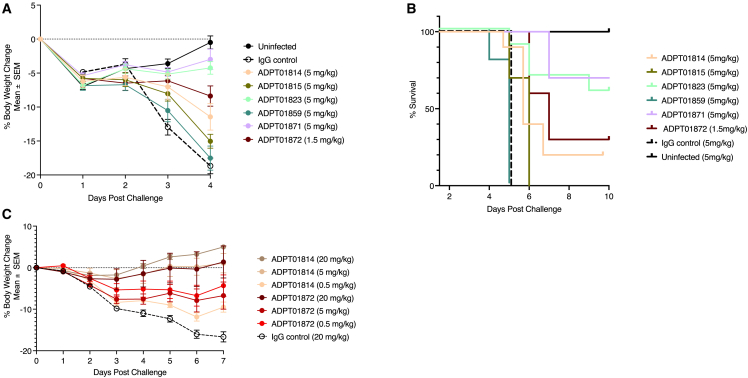
In vivo protection by membrane fusion-inhibiting S2 antibodies
To determine the protective efficacy of S2 antibodies, we similarly adopted the K18h-ACE2 mouse and Golden Syrian hamster models. Four of 6 S2 antibodies showed some level of protection compared to untreated mice (Figure 6A). Mice treated with 5 mg/kg of the S2 antibodies (ADPT01871 and ADPT01823) exhibited minimal weight loss with 70% and 60% survival 10 days after infection, respectively (Figures 6A and 6B). At 5 mg/kg, ADPT01814 also provided some degree of protection against weight loss, although survival was lower (20%). ADPT01872 was tested at a lower dose of only 1.5 mg/kg and still provided substantial protection against weight loss, although survival was only 30% (Figures 6A and 6B).
Figure 6.
Prophylactic protection of both mice and hamsters by S2-targeting antibodies at varying doses
(A–C) S2-binding antibodies also showed strong protection in K-18-hACE2 mice (A and B) and Syrian hamsters (C). Percent body weight data points represent Mean ± SEM. Experiments were conducted as detailed in Figure 5. Antibody dose varied per group as indicated in the individual plots.
Dose-response relationships were more fully explored in the hamster model for S2 antibodies ADPT01872 and ADPT01814. Each antibody was tested at 0.5, 5.0, and 20 mg/kg and a dose response was observed for both antibodies (Figure 6C). Although both ADPT01814 and ADPT01872 had minimal to moderate efficacy in mice, we observed no weight loss in hamsters at either the 20 or 5 mg/kg dose for ADPT01814 and minimal weight loss at any dose for ADPT01872. Therefore, the two S2 antibodies provided protection in both preclinical efficacy models.
Discussion
Monoclonal antibodies remain an important option in the treatment and prevention of COVID-19 and SARS-CoV-2 infections, particularly in vulnerable populations. The SARS-CoV-2 pandemic highlighted the need for methods that enable the rapid discovery of potent antibodies. Here we describe the application of an antibody discovery platform, called TruAB Discovery, which employs BCR pairSEQ for the native pairing of BCR heavy and light chain pairs at massive scale. Using TruAB Discovery, we identified a diverse set of potent and protective antibodies against SARS-CoV-2. The exceptional throughput of BCR pairSEQ allowed us to include millions of antigen-specific memory B cells and antigen-agnostic ASCs from hundreds of infected or convalescent individuals. Although single-cell technologies have enabled the pairing of naturally occurring antibody heavy and light chains, throughput remains a fundamental challenge. Following the identification and subsequent analysis of tens-of-thousands of unique, naturally paired antibodies followed by principled downsampling, our pipeline yielded more than 100 potently neutralizing antibodies. The majority of our library of antibodies bound to various, previously described regions of the RBD but, thanks to the diversity of paired antibodies identified, many retained potency to all SARS-CoV-2 variants characterized thus far, including to Omicron. The Omicron-neutralizing capabilities and crystal structures of two of these antibodies (ADPT02564 and ADPT03091, coded as COVIC-333 and COVIC-334, respectively) were independently identified by the COVIC consortium in a recent paper.51 In addition, by screening large numbers of individuals at various stages of infection, we identified a set of rare S2-binding, fusion-inhibiting antibodies from ASCs that represent promising pan-betacoronavirus broadly neutralizing candidates.
Thanks to their membrane-bound receptors, memory B cells can be enriched for antigen specificity prior to pairing via FACS. However, antibodies produced by plasma cells, which cannot be easily enriched in this way, are potentially of even greater value as therapeutics given their expected higher affinity.52,53 Therefore, we also explored an antigen-agnostic approach, applying BCR pairSEQ to the ASCs of an individual with an active SARS-CoV-2 infection. A large fraction of the resulting antibodies from ASCs were spike specific, validating the combination of plasma cells and memory B cells for BCR pairSEQ. While we observed many S2-reactive antibodies from memory B cells across individuals, exploration of ASCs from a single donor also yielded rare neutralizing antibodies that recognize the more conserved S2 region of the spike protein. Exploiting S2 as a therapeutic target confers advantages given the structural conservation of S2, extant cross-reactive neutralizing antibodies in populations due to prior exposure to common cold coronaviruses, the steric neutralization potential of antibodies against S2, and the stronger memory B cell and T cell responses.8 Characterization of our S2 antibodies showed that they can neutralize both SARS-CoV-2 and SARS-CoV-1 and recognize various other betacoronavirus taxa. Given that these antibodies were isolated from ASCs only 9 days after symptom onset early in the pandemic, the high SHM of these S2 antibodies suggests the individual (ADIRP0003238) likely experienced a previous infection by a related coronavirus and that many of the observed neutralizing antibody lineages were originally stimulated in response to that pathogen.
Interestingly, we observed S2-mediated pseudovirus neutralization was only successful in the presence of TMPRSS2 expressing cell lines, suggesting a unique mode of action that involves cleavage of spike protein by this enzyme. The mechanism of neutralization for S2 antibodies was not observed to be ACE2-dependent. Rather, our cell-cell fusion assay indicates that the antibodies block some part of the fusion machinery. Fusogenicity is just one of many characteristics that varies between strains of SARS-CoV-2,54 emphasizing the value of antibody therapies with diverse mechanisms of action that may operate with different effectiveness across strains.
S2-binding antibodies are not uncommon but few studies have demonstrated in vivo protection by an S2 antibody.16,17,18,20 Although the comparative rarity of this type of antibody seems clear, their scarcity in the literature could also be due to the widespread use of stabilized spike trimers as bait. Studies have shown that FP-directed antibodies in particular have a markedly reduced ability to bind to prefusion stabilized spike trimer protein.20,21 Enriching via these reagents could reduce or outright prevent the discovery of S2 neutralizing antibodies from memory B cells where we utilized antigen bait. Indeed, we measured weak S2-antibody affinity which was not anticipated given higher SHM observed for these antibodies. These protein-protein interactions may not be adequately measured with recombinant proteins given the different conformational states of the spike protein combined with our observations of the need of TMPRSS2 for neutralization.55 Importantly, and despite our measurement of relatively weaker affinity to recombinant protein and in vitro potency than our RBD-binders, these S2 antibodies were capable of protecting from severe disease in two preclinical models after challenge with SARS-CoV-2 virus. The robustness of the BCR pairSEQ assay means that it can be employed broadly across cell types, regardless of prior antigen enrichment allowing for the identification of these rarely reported S2-neutralizing antibodies.
Although the in vivo effector function of antibodies in treatment of infectious disease including SARS-CoV-2 has been controversial, at least one study in mice and hamsters demonstrated that monoclonal antibodies require Fc-dependent effector functions for therapeutic protection.56 Using in vitro assays, we demonstrated that selected RBD antibodies mediate effector function, specifically phagocytosis. Although these antibodies are also capable of potent neutralization in the absence of effector function, it is likely that engagement of innate immune cells played a role in the protection observed in vivo. In contrast to RBD-binders, we observed minimal phagocytosis with the S2 neutralizing antibodies, but increased NK cell mediated cytotoxicity. The different effector function of S2 also has the potential to engage NK cells without concerns of antibody-dependent enhancement (ADE). More studies are needed to fully understand the impact and potential synergy of the effector functions induced by these antibodies.
We were motivated to explore the ASC population discussed previously in part out of concern surrounding the main limitation of BCR pairSEQ: a dependence on high B cell clonality. BCR pairSEQ leverages the exceptional diversity of heavy and light chain gene sequences created during V(D)J recombination, assuming that no antibodies will converge on the identical sequence in either gene. If multiple clones of a B cell are randomly distributed across a 96-well microtiter plate, then the cooccurrence of the same pairs of heavy and light chain gene sequences across multiple wells can be leveraged to identify natural pairs of these sequences. BCR pairSEQ is thus dependent on the presence of multiple clones from a single progenitor cell. Ex vivo enrichment of memory B cell populations increases clonality sufficiently for successful pairing, but methods for expanding plasma cell populations are less well established, creating a potential hurdle for use of pairSEQ. Our ability to successfully pair nearly 1,500 antibodies from the ASCs of a single donor exploited the natural clonality induced by viral infection and enabled discovery of antibodies in an antigen-agnostic manner, and with distinct mechanism of action compared to those identified from memory B cells. These results illustrate how the TruAb Discovery approach may enhance future efforts in therapeutic antibody discovery.
In summary, over one hundred monoclonal antibodies have thus far been approved as therapeutic and diagnostic tools for infectious diseases, autoimmunity, and cancer. While technologies used for monoclonal antibody discovery have improved over time, the lengthy development times and low throughput continue to be a significant bottleneck in the discovery and development of mAbs. Here, we applied our TruAB Discovery process and BCR pairSEQ technology to discover antibodies specific for SARS-CoV-2. Our approach yielded more than 100 neutralizing antibodies with protective capacity. Our library of potent SARS-CoV-2 antibodies target diverse RBD epitopes as well as a highly conserved membrane fusion inhibiting epitope on S2. The S2 antibodies in particular may be valuable as pan-betacoronavirus prophylactic agents against current and future pandemics. The broad applicability of our TruAB Discovery approach combined with the sheer scale and throughput of our BCR pairSEQ technology makes this a notable advancement in the discovery of potent, naturally occurring antibodies with diverse modes of action, and suitable for use in various diseases.
Limitations of the study
BCR pairSEQ requires a minimum degree of clonality in the B cell population of interest. While standard techniques are available for expanding these populations to induce clonality when necessary, this is an additional step not required by all BCR pairing approaches that could limit the use of BCR pairSEQ in certain cases. The antibodies discovered in this study suffer from two general issues. First, samples were collected early in the pandemic and, as a result, many of the discovered antibodies have lost potency against the now dominant Omicron variants of SARS-CoV-2. Second, some of the most promising and unusual antibodies discovered, those that neutralize via S2-binding, were difficult to fully characterize. The mechanism of action of these antibodies requires the presence of the TMPRSS2 enzyme in the cell line tested against limiting available tools. Additionally, despite efforts, we were not able to isolate the specific epitope to which they bind, information that would be valuable for understanding their function and the limitations of their binding breadth.
Significance
At least 129 antibody therapeutics have been approved for use in the US or EU against a range of indications, with 20 more under review. Despite this proliferation, hurdles remain in the development of antibody-based therapies. Key among these is the discovery of paired antibody heavy and light chain sequences. Antibodies are composed of heavy chain and light chain proteins, both of which are essential for antigen recognition and antibody stability. The sequence of each of these genes is determined through somatic V(D)J recombination including non-templated insertions and deletions, yielding the diversity that allows for recognition of potentially any antigen. However, heavy and light chain genes are encoded on separate chromosomes, complicating the native pairing of these sequences which typically requires single-cell technologies where throughput is constrained by the need to individually barcode each cell. Here, we demonstrate a combinatorial approach to antibody pairing, BCR pairSEQ, which leverages the natural diversity of these molecules to identify paired heavy and light chains from bulk B cells, circumventing the need to distinguish individual cells. We apply our approach to the identification of naturally occurring antibodies against SARS-CoV-2 derived from memory and plasma B cells of infected donors. We identify tens-of-thousands of unique antibodies and use principled computational approaches to identify ∼2,000 of the most promising therapeutic candidates for functional validation, more than half of which bound to the spike protein. We identified large numbers of neutralizing antibodies from all three canonical classes of RBD-binders and a large number of non-RBD, S1-binding neutralizers. Intriguingly, we also identified several families of S2-binders that neutralize by blocking membrane fusion and offer in vivo protection. In sum, we describe a high-throughput approach to pair native heavy and light chains from bulk B cells, increasing the rate and scale of antibody discovery.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD138 PerCP-eFluor 710 (clone MI15) | Invitrogen | Cat#46-1388-42; RRID: AB_2815146 |
| IgM BV785 (clone MHM-88) | Biolegend | Cat#314543; RRID:_AB2800831 |
| CD3 BV421 (clone SK7) | Biolegend | Cat#563798; RRID:_2744383 |
| CD14 BV421 (clone 63D3) | Biolegend | Cat#367143; RRID:_2744383 |
| CD27 PE-Cy7 (clone M-T271) | Biolegend | Cat#356411; RRID: AB_2562258 |
| CD38 FITC (clone HB-7) | Biolegend | Cat# 356609; RRID: AB_2561949 |
| CD20 APC-Cy7 (clone 2H7) | Biolegend | Cat# 302313; RRID: AB_314261 |
| IgD BV510 (clone IA6-2) | Biolegend | Cat#348219; RRID: AB_2561386 |
| CD14 PE antibody | Miltenyi | Cat#130-110-577; RRID: AB_2655050 |
| HRP Anti-human IgG | BD Biosciences | Cat#555788; RRID: AB_396123 |
| Penta-His Alexa Fluor Conjugate - AF647 | Qiagen | Cat#35370 |
| Influenza An NP Monoclonal Antibody | Thermo Fisher | Cat#MA5-29926; RRID: AB_2785735 |
| HA Tag Antibody | R&D Systems | Cat#MAB060; RRID: AB_10719128 |
| CD19 Microbeads, human | Miltenyi | Cat#130-050-301 |
| CD138 Microbeads, human | Miltenyi | Cat#130-051-301 |
| Human IgG | Equitech-Bio | Cat#SLH56 |
| Anti-S antibody (clone 6D11F2) | Genscript | Cat#A02055 |
| ADPT00980 | This manuscript | RRID: AB_2940970 |
| ADPT01589 | This manuscript | RRID: AB_2940989 |
| ADPT01814 | This manuscript | RRID: AB_2940990 |
| ADPT01815 | This manuscript | RRID: AB_2940991 |
| ADPT01823 | This manuscript | RRID: AB_2940992 |
| ADPT01859 | This manuscript | RRID: AB_2940993 |
| ADPT01871 | This manuscript | RRID: AB_2940994 |
| ADPT01872 | This manuscript | RRID: AB_2940995 |
| ADPT02564 | This manuscript | RRID: AB_2940996 |
| ADPT02598 | This manuscript | RRID: AB_2940997 |
| ADPT02606 | This manuscript | RRID: AB_2940998 |
| ADPT02794 | This manuscript | RRID: AB_2940999 |
| ADPT03091 | This manuscript | RRID: AB_2941000 |
| ADPT03995 | This manuscript | RRID: AB_2941001 |
| ADPT04042 | This manuscript | RRID: AB_2941002 |
| ADPT04441 | This manuscript | RRID: AB_2941003 |
| Bacterial and virus strains | ||
| Pseudovirus SARSCoV2 Wuhan-Hu-1 D614G | Genscript | N/A |
| Pseudovirus SARSCoV1 | Genscript | N/A |
| RVP rLuc SARSCoV2 Wuhan-Hu-1 D614G | Integral Molecular | Cat#RVP-702F |
| RVP rLuc SARSCoV2 B.1.1.529 | Integral Molecular | Cat#RVP-768L |
| RVP rLuc SARSCoV2 4/5 | Integral Molecular | Cat#RVP-774F |
| RVP rLuc SARSCoV2 XBB1.5 | Integral Molecular | Cat#RVP-786L |
| SARSCov2 WT USA-WA1/2020 | BEI Resource | Cat#NR-52281 |
| hCoV-19/South Africa/KRISP-EC-K005321/2020 | BEI Resource | Cat#NR-54008 |
| SARSCov2 WT-D614 | BEI Resource | Cat#NR- 53515 |
| SARSCov2 Omicron B.1.1.529 | Bioqual | N/A |
| SARSCov2 Omicron 4 | BEI Resource | Cat#NR-56806 |
| SARSCov2 Omicron 5 | BEI Resource | Cat#NR-58620 |
| Biological samples | ||
| PBMC | ImmuneRACE study, Adaptive | N/A |
| PBMC | Bloodworks Northwest, Seattle | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RNAlater Stabilizatoin Solution | Thermo Fisher Scientific | Cat#AM7020 |
| eBioscience 1X RBC Lysis Buffer | Thermo Fisher Scientific | Cat#00-4333-57 |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | Cat#25200056 |
| Sodium Pyruvate (100 mM) | Thermo Fisher Scientific | Cat#11360070 |
| IMDM | Thermo Fisher Scientific | Cat#12-440-053 |
| GlutaMax | Thermo Fisher Scientific | Cat#35050061 |
| Gentamicin | Thermo Fisher Scientific | Cat#15710072 |
| human AB serum | MilliporeSigma | Cat#H4522 |
| 1-Step™ Ultra TMB-ELISA Substrate Solution | Thermo Fisher Scientific | Cat#34028 |
| Stop Solution for TMB Substrates | Thermo Fisher Scientific | Cat#PIN600 |
| Tween 20 | Sigma-Aldrich | Cat#P5927 |
| Ficoll Paque Plus | Thermo Fisher Scientific | Cat#17144002 |
| PBS, pH7.4 | Thermo Fisher Scientific | Cat#10010072 |
| Blocker BSA 10X in PBS | Thermo Fisher Scientific | Cat#PI37525 |
| Human Recombinant IL-4 | BioLegend | Cat#574008 |
| CD40L, carrier free, human | R&D Systems | Cat#6420-CL-025/CF |
| IL-2 (human, carrier free) | Biolegend | Cat#589106 |
| BioGlo Luciferase | Promega | Cat#G7941 |
| Renilla-Glo luciferase | Promega | Cat#E2720 |
| PKH67 ethanolic dye solution | Sigma Adrich | Cat#P7333 |
| SARS-CoV2 Spike Trimer ECD-His | Genscript | N/A |
| SARS-CoV2 RBD-His | Genscript | Cat#Z03514 |
| SARS-CoV2 RBD-Biotin | Genscript | N/A |
| SARS-CoV2 S2-Biotin | Genscript | N/A |
| SARS-CoV2 nucleocapsid | Genscript | Cat#Z03480 |
| SARS-CoV Spike S1+S2 ECD-His (S577A, Isolate Tor2) | Sino | Cat#40634-V08B |
| MERS-CoV (NCoVSpike Protein ECD aa 1-1297, His | Sino | Cat#40069-V08B |
| MERS-CoV (NCoVSpike Protein S2 aa 726-1296, His | Sino | Cat#40070-V08B |
| HCoV-HKU1-Spike (S1+S2 ECD His) | Sino | Cat#40606-V08B |
| HCoV-229E) Spike Protein S1+S2 ECD His | Sino | Cat#40605-V08B |
| HCoV-OC43- Spike Protein S1+S2 ECD His | Sino | Cat#40607-V08B |
| HCoV-NL63-Spike Protein (S1+S2 ECD, His) | Sino | Cat#40604-V08B |
| Critical commercial assays | ||
| ADCC Reporter Bioassay | Promega | Cat#G7018 |
| Miltenyi StraightFrom® Whole Blood CD138 kit | Miltenyi | Cat#130-105-961 |
| Miltenyi StraightFrom® Whole Blood CD19 kit | Miltenyi | Cat#130-090-880 |
| Endotoxin Assay Kit | Bioendo | Cat#KC64T |
| Experimental models: Cell lines | ||
| 293T-ACE2 | Genscript | Cat#M00770 |
| 293T-Spike | Genscript | Cat#M00804 |
| TurboCHO-HT | Genscript | N/A |
| HEK293/ACE2/Tat cells | Genscript | N/A |
| CHO-K1/LTR/Spike cells | Genscript | N/A |
| Vero E6 | ATCC | Cat#CRL-1586 |
| Experimental models: Organisms/strains | ||
| Golden Syrian Hamsters:HsdHan®:AURA | Inotiv | Cat#8904M |
| Mouse: K18h-ACE2: B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Labs | RRID:IMSR_JAX:034860; Cat#034860 |
| Oligonucleotides | ||
| Provided in Table S2 | This manuscript | N/A |
| Software and algorithms | ||
| BD FACSChorus | BD Biosciences | Version 2.0 |
| FlowJo | BD Biosciences | Version10.7.2 |
| GraphPad Prism | GraphPad Software, Inc. | Version 9.0 |
| IgBLASTN | Ye et al.56 | v. 1.16.0 (ncbi.github.io/igblast/cook/How-to-set-up.html) |
| R | R Core Team57 | v. 3.6.1, 4.1.2 (cran.r-project.org) |
| Python | N/A | v. 3.8.3 (python.org) |
| Octet Analysis Studio | Sartorius | v. 12.2 |
| Protein Data Bank | RCSB PDB | rcsb.org |
| The International ImMunoGeneTics Information System | Giudicelli et al.58 | imgt.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Amy Gilbert (agilbert@adaptivebiotech.com).
Materials availability
Biotinylated RBD/S1 and spike tetramers and decoy reagents were generated as previously described.23 All antibody discovery procedures and characterization details are provided in the text STAR methods section. Primer information has been provided in Table S2. Information concerning particular antibodies can be requested through the Coronavirus Immunotherapeutics Consortium https://covic.lji.org. Where applicable and feasible, additional information may be shared by the lead contact. Antibodies, reagents, cell lines and animals used for biological studies were obtained from commercial or internal sources described in the key resources table. There are restrictions to the availability of all materials from Adaptive Biotechnologies due to the need for a Materials Transfer Agreement.
Experimental model and study participant details
Mouse model
Five-week-old female, transgenic mice with an average body weight of 20 g were used in this study. Experiments were performed at Texas Biomedical Research Institute (TBRI), in compliance with local, state, and federal regulations and were approved by the TBRI Institutional Animal Care and Use Committees (IACUC).
Hamster model
The hamster studies were carried out at Bioqual Inc. (Rockville, MD), in compliance with local, state, and federal regulations and were approved by Bioqual IACUC.
Human samples
Both frozen PBMC and fresh blood were used in this study. Samples were collected based on each institution’s study protocol as reviewed by their institution review board. Fresh blood samples and PBMCs used in this study were collected either from ImmuneRace study or Bloodworks Northwest (Seattle, WA), with sample details as described previously.59
Cell lines
HEK293T ACE2/TMPRSS2 expressing cell line (female) was used as target cells in pseudovirus neutralization assay. Cell-based ADCP and ADCC assay was performed using SARS-CoV-2 spike glycoprotein stable HEK293T cell line as target cells. ADCC Bioassay Effector Cells (Promega) was used as effector cells. HEK293/ACE2/Tat cell and CHO-K1/LTR/Spike (female) cell lines were used in fusion assay. VeroE6 cells (female) were used for live virus neutralization assays.
Method details
PBMC and blood samples
Briefly, 10-50 mL was obtained from COVID-19 patients who were either in the acute phase of infection or had clinically cleared and recovered from the SARS-CoV-2 virus. B cells were either isolated from fresh blood using Miltenyi StraightFrom® Whole Blood CD138 or CD19 or from PBMC using CD138 or CD19 microbeads following manufacturer’s instructions.
Tetramer generation, cell sorting and B cell expansion
Biotinylated RBD/S1 and spike tetramers and decoy reagents were generated as previously described.23 Briefly, biotinylated proteins were incubated with streptavidin-PE at a ratio of 1:4 at room temperature for 30 minutes. Isolated CD19/CD138 cells were stained with fluorescent markers CD3-BV421, CD20-APCcy7, IgM-BV510, IgD-BV785, CD27-PEcy7, CD38-FITC. S1/RBD PE tetramer or His-tagged spike trimer were used for binding antigen-specific cells. For trimer-specific cells, anti-His-Alexa647 was used as secondary antibody (at 1:100 dilution). Samples were sorted to select antibody-secreting B cells (ASCs) using relevant markers (CD3-/CD19+/IgD-IgM-CD27hi/CD38hi) or antigen-specific memory B cells (CD3–/CD19+/IgD-/IgM-Decoy-S1/RBD or Trimer+). All cells were run on the Melody sorter (BD) and analyzed using FlowJo software (TreeStar). ASCs were not expanded while sorted antigen-specific cells were cultured in 96-well plates at a max of 5,000 cells per well in B-cell media (IMDM, 1% GlutaMax, 0.1% Gentamicin 1:1000, 10% human AB serum and 1 mM sodium pyruvate) containing 40 ng/mL IL-4, 100 ng/mL recombinant HA-tagged CD40L and 250 ng/mL anti-HA for 7 days.
pairSEQ to identify BCR-paired sequences
We performed pairSEQ on expanded memory B cells and non-expanded ASCs as previously described34 with BCR-specific primers (Table S2). Briefly, B cells were allocated to each well in 96-well plates. Next, mRNA was extracted, converted to cDNA and amplified by BCR-specific primers. Well-specific barcodes were attached to the sequences, and the BCR molecules pooled for sequencing. Computational demultiplexing followed to map each BCR sequence back to the well in which it originated, after which pairSEQ leveraged the natural diversity of BCRs to identify pairs of heavy and light chains. The probability that two or more BCR clones occupy the same set of wells by chance is extremely low. Thus, a pair of BCR heavy and light chain sequences that uniquely share a set of wells are very likely to represent the same BCR clone. Additional details of the pairSEQ computational approach to identifying statistically significant pairs of heavy and light chains have been published previously.34
Antibody sequence analysis and synthesis
Germline genes for both heavy and light chain sequences were inferred using IgBLASTN60 version 1.16.0 against IMGT58 database release 202011-3. Somatic hypermutation was measured as a proportion of sites mutated relative to the inferred germline genes, as performed by IgBLASTN. Antibodies were selected for synthesis based on observed patterns of SHM and the absence of structural features (e.g., free cysteines, degradation hotspots, sites susceptible to deamidation and oxidation) that may cause complications in downstream antibody development. Selected antibodies were cloned onto an IgG1 backbone and transfected into GenScript proprietary TurboCHO-HT expression system. The cells were maintained at 37°C with 5% CO2 on an orbital shaker. Filtered cell culture supernatant was loaded onto an affinity purification column at an appropriate flowrate. After washing and elution with appropriate buffers, the eluted fractions were pooled and buffer exchanged to the final formulation buffer. The purified protein was analyzed by SDS-PAGE and SEC-HPLC analysis to determine the molecular weight and purity. The concentration was determined by A280 method. Endotoxin removal was performed by treating protein A column with 0.2 M NaOH solution incubated for 2 hours. Endotoxin Analysis was done using LAL Endotoxin Assay Kit (Bioendo, Cat. No. KC64T).
ELISA
Antigen specificity for synthesized antibodies was initially determined using an Enzyme-linked, immunosorbent assay (ELISA) targeting the spike protein of SARS-CoV-2. The RBD, S1 domain, S2 domain, nucleocapsid and the trimeric form of the spike protein were used as target proteins. Reactivity of S2 antibodies to other coronaviruses was tested by immobilization spike proteins form SARS-Cov1, MERS-CoV, HCoV-HKU1, HCoV-229E and HCoV-OC43. Proteins were diluted in PBS to 1 μg/mL and 50 μL added per well to 96-well plate and incubated with shaking for 1 hour at room temperature, or overnight at 4°C. Plates were washed three times using 200 μL of Wash Buffer (PBST, 0.05% Tween). Plates were blocked with 200 μL Blocking Buffer for 1 hour at room temperature with PBST with 5% BSA. Primary antibodies were diluted in blocking buffer starting at 1 or 10 μg/mL during screening and with a serial dilution of 1:3 for positive clones. Diluted proteins were added to the plate well and incubated for 1 hour at room temperature. The secondary antibody, mouse anti-human IgG conjugated to horseradish peroxidase, was added at 1:1000 dilution in blocking buffer and incubated for 1 hour at room temperature. Plates were washed three times and developed with 50 μL of TMB (3,3=,5,5=-tetramethylbenzidine) liquid substrate for 5-10 min before adding 50 μL of stopping reagent. Absorbance was measured at 450 nm using a plate reader. All ELISAs included a non-specific IgG control tested at 1 ug/mL that had a mean OD of 0.070 with a standard deviation of 0.031.
Spike subunit specificity was determined hierarchically. Antibodies that bound RBD with a mean OD at 450 nm of at least 0.2 when tested at a concentration of 1 μg/mL were considered RBD-specific. The same classification procedure was performed for S2. Antibodies that bound to S1 and not RBD were classified as NTD-binders. Those that bound to the full trimer and none of the individual subunits were classified as trimer-binders.
Surface plasmon resonance affinity measurement
The confirmation of affinity of selected mAbs was determined by Biacore 8K at a third-party vendor as per manufacturer’s instructions. Briefly, RBD, S1 or Trimer were immobilized at 25°C while HBS-EP was used as the running buffer. The sensor chip surface of flow cells 1 and 2 were activated. Antigens were diluted in NaAC and injected into the flow cell 2 to achieve conjugation and while flow cell 1 was set as blank. Antibodies were injected over the surface as association phase, followed by injecting running buffer as dissociation phase. All the data were processed using the Biacore 8K Evaluation software version 1.1. Flow cell 1 and blank injection of buffer in each cycle were used as double reference for Response Units subtraction.
ACE2 blockade
Antibodies with confirmed antigen specificity to one or more proteins of SARS-CoV-2 were assessed for blockade of ACE2 via a competitive ELISA. The ability of candidate antibodies to block the interaction between viral RBD protein and human ACE2 surface receptor was measured by ELISA. In initial screening, antibodies with mean ACE2 blockade of at least 50% were considered successful blockers. IC50 was calculated based on performing inhibition assay with serial dilution of mAbs starting from 30 μg/mL and three-fold dilution for a total of 10 points in duplicate. Anti-S antibody, Clone 6D11F2 and human IgG were included as positive and negative controls, respectively. Non-RBD antibodies (S1, S2, trimer alone or nucleocapsid) that did not block ACE2 were advanced to both live and pseudovirus assays to evaluate if these antibodies were able to use alternative mechanisms for blocking the virus.
Epitope binning
Unidirectional binning assays in traditional sandwich format were performed in 16-channel mode (Octet RH16). The assay running buffer contains 1xPBS+1%. BSA, 0.03% Tween20. All the samples were diluted in the assay running buffer before the experiment. Each binning assay consisted of the following steps: (1) AHC (Anti-Human IgG Fc Capture) sensors were saturated with 10 μg/mL of the 1st antibody for 10 min; (2) a buffer baseline was established for 30 sec; (3) the sensors were soaked in an irrelevant human IgG1 solution (10 μg/mL) for 5 min to block the unoccupied Fc binding sites; (4) a buffer baseline was established for 30 sec; (5) an antigen (SARS-CoV-2-spike Trimer, RBD or S2 in 125 nM) association phase for 5 min; (6) a buffer baseline was established for 30 sec; (7) a binning step, in which sensors were exposed to competing antibody (125 nM) for 5 min; (8) the capture surfaces were regenerated for 40 sec. All data analysis was performed by using Octet Analysis Studio 12.2.
Shotgun mutagenesis and specificity
Epitope mapping services were provided by Integral Molecular (Philadelphia, PA) as described previously.61 Briefly, a mutation library of the target protein expressed on human cells was created by high-throughput, site-directed mutagenesis. Each residue was individually mutated to alanine, with alanine codons mutated to serine. The mutant library was arrayed in 384-well microplates and transiently transfected into HEK-293T cells. Following transfection, cells were incubated with the indicated antibodies at concentrations pre-determined using an independent immunofluorescence titration curve on wild type protein. Monoclonal antibodies were detected using an Alexa Fluor 488-conjugated secondary antibody and mean cellular fluorescence was determined using IntelliCyt iQue flow cytometry platform. Mutated residues were identified as being critical to the mAb epitope if they did not support the reactivity of the test mAb but did support the reactivity of the reference mAb. This counter screen strategy facilitates the exclusion of mutants that are locally misfolded or that have an expression defect.
The specificity of antibodies to SARS-CoV-2 was evaluated using a protein membrane array consisting of a library of over 4000 cell surface proteins. Epitope mapping was performed using shotgun mutagenesis consisting of a library of cells engineered to express SARS-CoV-2 containing single amino acid substitutions of alanine at each position. Critical amino acids for antibody binding to SARS-CoV-2 were identified using flow cytometry.
Pseudovirus neutralization assay
A lentiviral pseudotype bearing spike viral protein expressing a firefly luciferase read-out was used to screen for functional antibody responses against SARS-CoV-2 under biosafety level 2 laboratory conditions. Antibodies with mean pseudovirus inhibition of at least 70% were classified as neutralizers. HEK293T ACE2/TMPRSS2 expressing cell line was used as target cells. Luciferase activity was read-out using the Renilla-Glo Luciferase Assay System. IC50 of selected mAbs was determined by performing a serial dilution starting from 100 μg/mL and six or four fold dilution for a total of 10 dilutions in triplicate. Anti-S antibody, clone 6D11F2 (Genscript) and human IgG were included as positive and negative controls, respectively. IC50 Calculations were performed using GraphPad Prism, with four parameter log-logistic regression model.
Live virus microneutralization assay
Neutralization of SARS-CoV-2 was determined using microneutralization (MN) assay performed in a biosafety level 3 facility (at Battelle). A SARS-CoV-2 viral stock generated from in vitro passaging in VERO E6 cells of strain USA-WA1/2020 (BEI Resources Lot No. 70035360 or equivalent) and hCoV-19/South Africa/KRISP-EC-K005321/2020 (BEI Resource lot No.NR-54008) from a qualified lot was used in the MN assay. The candidate antibodies were analyzed with seven-point four-fold serial dilution from a defined starting concentration. The primary assay endpoint was IC50, which is the antibody concentration that neutralizes 50% of the input virus and was calculated by fitting a four-parameter log-logistic regression model using the drc v3.0-1 package in R.57,62 was performed by Battelle Standard Operating Procedures. Compared to the “no virus” control and “virus only” controls within the assay, the viral infectivity post antibody neutralization was quantified using an in situ ELISA readout performed by following Battelle SOP. A neutralizing monoclonal antibody (mAb) specifically targeting the SARS-CoV-2 spike protein was used as PC and a non-neutralizing antibody was used as NC in the MN assay. Antibodies that inhibited pseudovirus but not live virus were still selected because it suggested that mechanism of action might be present in the specific cell type used to replicate the virus.
Plaque reduction neutralization test
A PRNT was performed in a biosafety level 3 facility (at BIOQUAL, Inc.) using 24-well plates. Purified IgG was prepared starting from 1ug/ml and performed 4 fold dilution for a total of 6 dilutions. Diluted samples were then incubated with 30 plaque-forming units of SARS-CoV-2 WT-D614, BEI Resources NR-53515) and Omicron variants, B.1.1.529 (Bioqual), 4 (BEI Resources NR-56806) and 5 (BEI Resources NR-58620) in an equal volume of culture media (DMEM-10% FBS with gentamicin) for 1 h at 37°C. The serum-virus mixtures were added to a monolayer of confluent TMPRSS2 cells and incubated for 1 h at 37°C in 5% CO2. Each well was then overlaid with 1 ml of culture media containing 0.5% methylcellulose and incubated for 3 days at 37°C in 5% CO2. The plates were then fixed with methanol at -20°C for 30 min and stained with 0.2% crystal violet for 30 min at room temperature. Neutralization titers were defined as the highest IgG concentration that resulted in 50% (PRNT50) reduction in the number of plaques.
Cell fusion assay
To characterize the antibody potential in blocking the ACE2-spike mediated fusion, a cell-based fusion assay was developed by Genscript. This assay comprises two genetically engineered cell pools: a) HEK293/ACE2/Tat cell pool was established by introduction of human ACE2 and human immunodeficiency virus type 1 (HIV) Tat gene, followed by selection with 1 μg/mL Puromycin and 5 μg/mL Blasticidin; b) CHO-K1 cells were engineered to express human spike (WT) and tat inducible HIV long terminal repeat (LTR)-firefly luciferase gene and selected with 8 μg/mL Puromycin and 400 μg/mL Hygromycin B to obtain a stable cell pool. The CHO-K1/LTR/Spike cells were plated in a 96-well plate prior to 1-hour incubation with diluted antibody solutions diluted three-fold starting at 100 μg/mL. Then HEK293/ACE2/Tat cells were added and incubated for an additional 4 hours before luminescence readout. LCB1 peptide previously shown to block fusion was used as a positive control.46
Multiplex bead-based effector function
Bead-based ADCP, ADNP, ADCD (antibody-dependent complement deposition) and ADNKA (antibody dependent natural killer cell assay) were performed as described previously.42,43,44,45 Briefly SARS-CoV-2 spike D614 protein was coupled to beads carrying fluorescent tags with distinct emission wavelengths. Biotinylated protein antigens were coupled to fluorescent, carboxylate-modified 1 μm beads, yellow-green (505/515 nm, F8823) for ADCP and ADNP and red (580/605 nm, F8821) for ADCD. Antigen-coupled beads were resuspended in 0.1% BSA in PBS. Immune complexes were formed by combining mixed fluorescently coupled beads and antibodies diluted to 5 μg/mL and plates were incubated at 37°C for 2 hours. For ADCP, Immune complexes were washed with PBS prior to adding THP-1 cells (ATCC) and incubating with immune complexes for 16-18 hours at 37°C before cells were fixed with 4% PFA. For ADNP, fresh PBMC from a healthy control donor were incubated with immune complexes for 1 hour and neutrophils were stained with CD66b and fixed with 4% PFA. For ADCD, lyophilized guinea pig complement was diluted in gelatin veronal buffer supplemented with calcium and magnesium. Diluted complement was mixed with immune complexes and incubated at 37°C for 20 minutes, complement deposition was detected using anti-C3 flourescein-conjugated goat IgG. Uptake of immune complexes by cells was determined by flow cytometry and phagocytic score was calculated using the formula: (%Bead + Cells x Geomean fluorescence of Bead + Cells)/100.
For ADNKA, spike protein was immobilized on ELISA plates for 2hrs at 37°C, washed and blocked with BSA overnight at 4°C. Immune complexes were formed by adding monoclonal antibodies to the ELISA plate and incubating for 2hrs at 37°C before adding NK cells isolated from fresh blood and incubating for 5 hours at 37°C in media supplemented with GolgiStop, Brefeldin A and anti-CD107a PE-Cy5. Cells were stained with cell surface markers anti-CD3 PacBlue (BD), anti-CD16 APC-Cy5, and anti-CD56 PE-Cy7. To measure MIP-1b, NK cells were permeabilized with Perm B and stained with anti-MIP-1b PE antibody.
Cell-based phagocytosis assay
A cell-based ADCP assay was performed using SARS-CoV-2 spike glycoprotein stable 293T cell line as target cells and CD14+ cells isolated from PBMC as per manufacturer’s protocol (Miltenyi) as effector cells. Target cells were fluorescently labeled with PKH67 ethanolic dye solution (Catalog # P7333) for 1-5 minutes before being stopped with 1% BSA and washed with complete media to remove excess dye. Target cells were then incubated with monoclonal antibodies at different concentrations for 5-10 minutes. Monocytes were isolated from healthy donors and fluorescently labeled with Cell Trace Violet. A 4:1 effector:target ratio was used, and cells were incubated overnight at 37°C. Anti-human CD14-PE antibody was used to stain monocytes. Antibody-mediated phagocytosis was determined by flow cytometry, gating on CD14+ cells that were double-positive for cell trace violet and PKH67.
Cell-based antibody-dependent cell cytotoxicity assay
A cell-based ADCC assay was performed using SARS-CoV-2 S glycoprotein stable 293T cell line as target cells and ADCC Bioassay Effector Cells (Promega) as effector cells. Antibody dilutions were made in assay buffer (RPMI 1640 with 4% low IgG serum). Target cells were plated at 1.25 x 104 cells per well in a white clear bottom 96-well plate in assay buffer. Antibody dilutions were added to cells and incubated for 15 minutes at 37°C. Effector cells were then added at a concentration of 6.25 x 104 cells per well (E:T ratio of 5:1) and the plate was incubated overnight at 37°C. After overnight incubation the plate was equilibrated to room temperature and an equivalent amount of Bio-Glo Luciferase Assay Reagent (Promega) was added to each well, and after 5 minutes luminescence was read on a plate reader (Bio-Tek). Fold induction of ADCC activity above background wells containing cells but no antibody was calculated.
In vivo viral challenge studies
In vivo studies were performed with Golden Syrian Hamsters and mice. Hamsters were divided into 5 animals per group. Animals were treated intraperitoneally (IP) with different concentration of antibodies 48 hours before intranasal challenge with 6x103 PFU of SARS-CoV-2/USA-WA1/2020 (NR-52281; BEI Resources). The hamster studies were carried out at Bioqual Inc. (Rockville, MD), in compliance with local, state, and federal regulations and were approved by Bioqual IACUC. Hamsters were acclimated for 10 days upon arrival and divided into 5 animals per group. Animals were treated intraperitoneally (IP) with different concentration of antibodies 48 hours before intranasal challenge with 6x103 PFU of SARS-CoV-2/USA-WA1/2020 (NR-52281; BEI Resources). Throughout the study, animals were weighed daily and clinically observed prior to challenge, and twice daily post-challenge for a period of 7 days. Throughout the study, animals were weighed daily and clinically observed prior to challenge, and twice daily post-challenge for a period of 7 days.
Efficacy of anonymized mAbs was tested in 5-week-old female, transgenic mice (n=10/antibody) with an average body weight of 20 g at TBRI. These mice are engineered to express the human ACE2 under the K18 promoter (K18h-ACE2; The Jackson Labs Cat No. 034860.63 Antibodies are delivered IP at 1.5 mg/kg 24 hr before intranasal (25 μL each nostril) challenge with 1 x 105 PFU SARS-CoV-2/human/USA/WA-CDC-WA-1 (GenBank MN985325) at passage six. Virus stocks were confirmed by next generation sequencing to be 100% identical to the BEI original P4 stock. The Bristol deletion and other deletions/mutations were also confirmed to be absent from working stocks. Mice were observed for a 10-day experimental period. During this time, the animals were weighed daily and sacrificed if ≥25% body weight loss occurred. Blood samples were collected prior to virus challenge. The single dose was selected based on a dose titration study with CC12.3, a monoclonal antibody isolated from a convalescent COVID-19 patient. The selected dose of 1.5 mg/kg was based on the dose at which 40-60% of animals survived at the end of the 10-day period.
Quantification and statistical analysis
Wilcoxon rank-sum tests were done using R software. Live virus neutralization IC50 values were calculated using the drc package in R and pseudovirus neutralization IC50 values were evaluated using Graphpad Prism. Descriptive statistics pertaining to SHM were determined using either R or python software. Mean and standard error of the mean (SEM) for in vivo results were calculated using Graphpad Prism. Other details of statistical calculations can be found in the detailed STAR methods, results and figure legends for each experiment/analysis.
Additional resources
Human PBMC samples were collected as part of the ImmuneRACE study, ClinicalTrials.gov Identifier: NCT04494893.59
Acknowledgments
We would like to acknowledge all the individuals who donated blood for this study, and Shannon Sexson for help in design and development of the BCR pairSEQ assay. We thank The Coronavirus Immunotherapeutic Consortium (CoVIC) for funding the mouse efficacy studies. CoVIC was in turn supported by the COVID-19 Therapeutics Accelerator (INV-006133), the Bill and Melinda Gates Foundation (OPP1210938), a supplement to NIH/NIAID grant U19 AI142790-S1. Finally, we thank Shahin Shafiani for editorial support.
Author contributions
Conceptualization, B.E.R.R., G.J.K., A.E.G., L.B., H.S.R., and S.B.; Methodology, P.E., A.S., B.H., M.K., E.Y., G.J.K., B.E.R.R., M.E.G., A.J.M., J.T., Y.L., A.M., J.G., E.E., N.L., R.B., L.L., J.W., C.L., L.C., C.A., G.A., and J.N.D.; Investigation, G.J.K., B.E.R.R., M.E.G., A.J.M., A.M., J.T., Y.L., E.E., N.L., L.L., J.W., C.L., L.C., and C.A.; Visualization, G.J.K., B.E.R.R., and M.E.G.; Analysis and Software, B.E.R.R., B.H., H.A., T.C., A.D., R.H., C.L., D.P., and C.W.; Resources, A.C. and J.N.D.; Writing – Original Draft, G.J.K., B.E.R.R., M.E.G., A.E.G., and Y.L.; Writing – Review & Editing, G.J.K., B.E.R.R., M.E.G., and A.E.G.
Declaration of interests
At the time of execution of this work, G.J.K, B.E.R.R., M.E.G., A.J.M., J.T., Y.L., P.E., A.M., J.G., E.E., N.L., R.B., A.C., H.A., T.C., A.D., R.H., C.L., D.P., C.W., J.N.D., L.B., A.S., B.H., M.K., E.Y., H.S.R., S.B., and A.E.G. were employees and held equity in Adaptive Biotechnologies. L.L., J.W., C.L., and L.C. declare employment and equity at GenScript. G.A. is a founder of Seromyx Systems and C.A. declares no conflict of interest. Patent applications relating to SARS COV-2 antibodies are pending.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2023.07.011.
Supplemental information
Data and code availability
-
•
Section 1: Data. Details of the pairSEQ computational approach to identifying statistically significant pairs of heavy and light chains have been published previously.34 Antibody sequence data for ADPT00980, ADPT01589, ADPT01814, ADPT01815, ADPT01823, ADPT01859, ADPT01871, ADPT01872, ADPT02564, ADPT02598, ADPT02606, ADPT02794, ADPT03091, ADPT03995, ADPT04042 and ADPT04441 are listed in Table S3.
-
•
Section 2: Code. This paper does not report original code.
-
•
Section 3: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Levin M.J., Ustianowski A., De Wit S., Launay O., Avila M., Templeton A., Yuan Y., Seegobin S., Ellery A., Levinson D.J., et al. Intramuscular AZD7442 (tixagevimab–cilgavimab) for prevention of Covid-19. N. Engl. J. Med. 2022;386:2188–2200. doi: 10.1056/NEJMoa2116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 3.Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., Hebert C., Perry R., Boscia J., Heller B., et al. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 7.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah P., Canziani G.A., Carter E.P., Chaiken I. The case for S2: the potential benefits of the S2 subunit of the SARS-CoV-2 spike protein as an immunogen in fighting the COVID-19 pandemic. Front. Immunol. 2021;12:637651. doi: 10.3389/fimmu.2021.637651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva R.P., Huang Y., Nguyen A.W., Hsieh C.-L., Olaluwoye O.S., Kaoud T.S., Wilen R.E., Qerqez A.N., Park J.-G., Khalil A.M., et al. Identification of a conserved S2 epitope present on spike proteins from all highly pathogenic coronaviruses. Elife. 2023;12:e83710. doi: 10.7554/eLife.83710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakur S., Sasi S., Pillai S.G., Nag A., Shukla D., Singhal R., Phalke S., Velu G.S.K. SARS-CoV-2 mutations and their impact on diagnostics, therapeutics and vaccines. Front. Med. 2022;9:815389. doi: 10.3389/fmed.2022.815389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodcroft E.B., Aksamentov I., Neher R., Bedford T., Hadfield J., Zuber M., Scott-Brown J., Sanderson T., Bloom J., Roemer C., et al. CoVariants: SARS-CoV-2 mutations and variants of interest. https://covariants.org/
- 12.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Sempowski G.D., Saunders K.O., Acharya P., Haynes B.F. SARS-CoV-2 neutralizing antibodies for COVID-19 prevention and treatment. Annu. Rev. Med. 2022;73:1–16. doi: 10.1146/annurev-med-042420-113838. [DOI] [PubMed] [Google Scholar]
- 16.Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Yuan M., Song G., Beutler N., Shaabani N., Huang D., He W.T., Zhu X., Callaghan S., Yong P., et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl. Med. 2022;14:eabi9215. doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Song G., Liu H., Yuan M., He W.T., Beutler N., Zhu X., Tse L.V., Martinez D.R., Schäfer A., et al. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease. Immunity. 2023;56:669–686.e7. doi: 10.1016/j.immuni.2023.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacon C., Tucker C., Peng L., Lee C.-C.D., Lin T.-H., Yuan M., Cong Y., Wang L., Purser L., Williams J.K., et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022;377:728–735. doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobeil S.M.-C., Henderson R., Stalls V., Janowska K., Huang X., May A., Speakman M., Beaudoin E., Manne K., Li D., et al. Structural diversity of the SARS-CoV-2 Omicron spike. Mol. Cell. 2022;82:2050–2068.e6. doi: 10.1016/j.molcel.2022.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedrioli A., Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42:1143–1158. doi: 10.1016/j.it.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Taylor J.J., Martinez R.J., Titcombe P.J., Barsness L.O., Thomas S.R., Zhang N., Katzman S.D., Jenkins M.K., Mueller D.L. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J. Exp. Med. 2012;209:2065–2077. doi: 10.1084/jem.20112272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shembekar N., Hu H., Eustace D., Merten C.A. Single-cell droplet microfluidic screening for antibodies specifically binding to target cells. Cell Rep. 2018;22:2206–2215. doi: 10.1016/j.celrep.2018.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunik V., Peters B., Ofran Y. Structural consensus among antibodies defines the antigen binding site. PLoS Comput. Biol. 2012;8:e1002388. doi: 10.1371/journal.pcbi.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sela-Culang I., Kunik V., Ofran Y. The structural basis of antibody-antigen recognition. Front. Immunol. 2013;4:302. doi: 10.3389/fimmu.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vázquez Bernat N., Corcoran M., Hardt U., Kaduk M., Phad G.E., Martin M., Karlsson Hedestam G.B. High-quality library preparation for NGS-based immunoglobulin germline gene inference and repertoire expression analysis. Front. Immunol. 2019;10:660. doi: 10.3389/fimmu.2019.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Boehmer L., Liu C., Ackerman S., Gitlin A.D., Wang Q., Gazumyan A., Nussenzweig M.C. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeKosky B.J., Kojima T., Rodin A., Charab W., Ippolito G.C., Ellington A.D., Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein L.D., Chen Y.-J.J., Wu J., Chaudhuri S., Hsiao Y.-C., Schneider K., Hoi K.H., Lin Z., Guerrero S., Jaiswal B.S., et al. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2019;2:304. doi: 10.1038/s42003-019-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B., Sherwood A.M., Berkebile A.D., Berka J., Emerson R.O., Williamson D.W., Kirsch I., Vignali M., Rieder M.J., Carlson C.S., Robins H.S. High-throughput pairing of T cell receptor a and b sequences. Sci. Transl. Med. 2015;7:301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 35.Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science. 2021;372:1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 40.Yang W.-T., Huang W.-H., Liao T.-L., Hsiao T.-H., Chuang H.-N., Liu P.-Y. SARS-CoV-2 E484K mutation narrative review: epidemiology, immune escape, clinical implications, and future considerations. Infect. Drug Resist. 2022;15:373–385. doi: 10.2147/IDR.S344099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudreau C.M., Yu W.-H., Suscovich T.J., Talbot H.K., Edwards K.M., Alter G. Selective induction of antibody effector functional responses using MF59-adjuvanted vaccination. J. Clin. Invest. 2020;130:662–672. doi: 10.1172/JCI129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atyeo C., Pullen K.M., Bordt E.A., Fischinger S., Burke J., Michell A., Slein M.D., Loos C., Shook L.L., Boatin A.A., et al. Compromised SARS-CoV-2-specific placental antibody transfer. Cell. 2021;184:628–642.e10. doi: 10.1016/j.cell.2020.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackerman M.E., Moldt B., Wyatt R.T., Dugast A.-S., McAndrew E., Tsoukas S., Jost S., Berger C.T., Sciaranghella G., Liu Q., et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473:112630. doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao M., Su P.-Y., Castro D.A., Tripler T.N., Hu Y., Cook M., Ko A.I., Farhadian S.F., Israelow B., Dela Cruz C.S., et al. Rapid, reliable, and reproducible cell fusion assay to quantify SARS-Cov-2 spike interaction with hACE2. PLoS Pathog. 2021;17:e1009683. doi: 10.1371/journal.ppat.1009683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X., Yi C., Zhu Y., Ding L., Xia S., Chen X., Liu M., Gu C., Lu X., Fu Y., et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microbiol. 2022;7:1063–1074. doi: 10.1038/s41564-022-01155-3. [DOI] [PubMed] [Google Scholar]
- 49.Low J.S., Jerak J., Tortorici M.A., McCallum M., Pinto D., Cassotta A., Foglierini M., Mele F., Abdelnabi R., Weynand B., et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu H., Chan J.F.-W., Yuen K.-Y. Animal models in SARS-CoV-2 research. Nat. Methods. 2022;19:392–394. doi: 10.1038/s41592-022-01447-w. [DOI] [PubMed] [Google Scholar]
- 51.Callaway H.M., Hastie K.M., Schendel S.L., Li H., Yu X., Shek J., Buck T., Hui S., Bedinger D., Troup C., et al. Bivalent intra-spike binding provides durability against emergent Omicron lineages: results from a global consortium. Cell Rep. 2023;42:112014. doi: 10.1016/j.celrep.2023.112014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith K.G., Light A., Nossal G.J., Tarlinton D.M. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phan T.G., Paus D., Chan T.D., Turner M.L., Nutt S.L., Basten A., Brink R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J. Exp. Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimura I., Yamasoba D., Tamura T., Nao N., Suzuki T., Oda Y., Mitoma S., Ito J., Nasser H., Zahradnik J., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell. 2022;185:3992–4007.e16. doi: 10.1016/j.cell.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R Core Team . 2019. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 58.Giudicelli V., Chaume D., Lefranc M.-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adaptive Biotechnologies, and Covance ImmuneRACE - Immune Response Action to COVID-19 Events. https://clinicaltrials.gov/ct2/show/NCT04494893
- 60.Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davidson E., Doranz B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritz C., Baty F., Streibig J.C., Gerhard D. Dose-response analysis using R. PLoS One. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCray P.B., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Section 1: Data. Details of the pairSEQ computational approach to identifying statistically significant pairs of heavy and light chains have been published previously.34 Antibody sequence data for ADPT00980, ADPT01589, ADPT01814, ADPT01815, ADPT01823, ADPT01859, ADPT01871, ADPT01872, ADPT02564, ADPT02598, ADPT02606, ADPT02794, ADPT03091, ADPT03995, ADPT04042 and ADPT04441 are listed in Table S3.
-
•
Section 2: Code. This paper does not report original code.
-
•
Section 3: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.