Abstract
DNA helicase activity is essential for the vital DNA metabolic processes of recombination, replication, transcription, translation, and repair. Recently, an unexpected, rapid exponential ATP‐stimulated DNA unwinding rate was observed from an Archaeoglobus fulgidus helicase (AfXPB) as compared to the slower conventional helicases from Sulfolobus tokodaii, StXPB1 and StXPB2. This unusual rapid activity suggests a “molecular wrench” mechanism arising from the torque applied by AfXPB on the duplex structure in transitioning from open to closed conformations. However, much remains to be understood. Here, we investigate the concentration dependence of DNA helicase binding and ATP‐stimulated kinetics of StXPB2 and AfXPB, as well as their binding and activity in Bax1 complexes, via an electrochemical assay with redox‐active DNA monolayers. StXPB2 ATP‐stimulated activity is concentration‐independent from 8 to 200 nM. Unexpectedly, AfXPB activity is concentration‐dependent in this range, with exponential rate constants varying from seconds at concentrations greater than 20 nM to thousands of seconds at lower concentrations. At 20 nM, rapid exponential signal decay ensues, linearly reverses, and resumes with a slower exponential decay. This change in AfXPB activity as a function of its concentration is rationalized as the crossover between the fast molecular wrench and slower conventional helicase modes. AfXPB‐Bax1 inhibits rapid activity, whereas the StXPB2‐Bax1 complex induces rapid kinetics at higher concentrations. This activity is rationalized with the crystal structures of these complexes. These findings illuminate the different physical models governing molecular wrench activity for improved biological insight into a key factor in DNA repair.
Keywords: base excision repair, nucleotide excision repair, xeroderma pigmentosum type B
1. INTRODUCTION
Bolstered by their ability to unpack genetic material, helicases are widely regarded as vital to all organisms. Helicases are ubiquitous enzymes important for all RNA and DNA metabolism, including DNA replication, repair, transcription, and translation. Helicases are motor proteins that move directionally along nucleic acids to separate the strands as catalyzed by nucleoside triphosphate (NTP) hydrolysis (Raney, 2013). DNA replication, recombination, transcription, translation, and repair are initiated by helicase activity (Gorbalenya & Koonin, 1993; Patel & Picha, 2000; Pyle, 2008; Singleton et al., 2007; Sun & Wang, 2016). While strides have been made in understanding their mechanisms for DNA unzipping, given the sheer number, structural heterogeneity, diversified functionality, and cooperative activity of helicases, extensive functional and structural studies are still required (Lohman et al., 2008) for a complete physical picture of this essential activity.
Xeroderma pigmentosum group B (XPB), a DNA helicase belonging to superfamily 2, exhibits DNA repair and transcription activity (Fan et al., 2006; Schultz et al., 2000). In nucleotide excision repair (NER), XPB unwinds the DNA duplex around the damaged site. XPB also initiates RNA polymerase II transcription by dehybridizing gene promoters (Gillet & Scharer, 2006; Schaeffer et al., 1993). Disorders such as Cockayne syndrome, xeroderma pigmentosum, and trichothiodystrophy arise from XPB mutations (DiGiovanna & Kraemer, 2012). While recent biochemical and structural research (Fan & DuPrez, 2015; Fishburn et al., 2015; He et al., 2016; Kim et al., 2000; Rand et al., 2000) has led to mechanistic hypotheses for DNA unwinding by helicases, further study into the binding and activity of XPB is needed to elucidate its roles in transcription and repair.
Recently, we revealed a distinct new mode of helicase action, termed molecular wrench activity, applied by XPB to unwind DNA (Kahanda et al., 2018). As illustrated in Figure 1, a conventional helicase translocates along the DNA, sequentially breaking the hydrogen bonds between the base pairs. This process is driven by adenosine triphosphate (ATP) hydrolysis and leads to the complete dehybridization of the duplex. We studied the Sulfolobus tokodaii XPB homologs, StXPB1 and StXPB2, together with the Archaeoglobus fulgidus XPB (AfXPB) and their interactions with DNA monolayers. DNA unwinding by StXPB1 and StXPB2 took tens of minutes, in line with prior ensemble fluorescence studies (Jang et al., 2010; Tani et al., 2010). Interestingly, the AfXPB helicase activity timescale, which had not been previously reported, was mere seconds. The different slow and fast decay functions of these distinct helicases may reflect two distinct modes of their DNA unwinding activity. Concomitantly, the crystal structures of StXPB2 and AfXPB revealed key differences in their conformations when complexed with DNA. While StXPB2 did not crystalize in a stable open structure, a stable open conformation was found for AfXPB. This finding of an open structure led to the postulate that AfXPB may stay in the open conformation after DNA binding. For AfXPB to adopt its active conformation with a closed ATP‐binding groove, a 170° rotation between the two helicase domains is required, whereas StXBP2 needs only 50–80° rotations. Molecular wrench action commences when ATP binds to AfXPB and induces this open‐to‐closed domain rotation of the enzyme (Figure 1) that rapidly separates the two strands of the DNA duplex.
FIGURE 1.
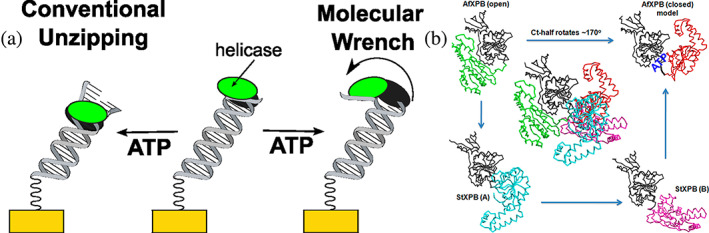
(a) An illustration of conventional and molecular wrench helicase mechanisms for Archaeoglobus fulgidus helicase (AfXPB). The domains of AfXPB are colored green and gray. (b) Structural comparison of AfXPB and helicases from Sulfolobus tokodaii (StXPB2). Center: The AfXPB structure (PDB: 2FZ4 & 2FZL) is superimposed with the two StXPB2 structures over the damage recognition domain and helicase domain1 (gray). For AfXPB, the C‐terminal halves are shown as green in the open conformation and red in the closed conformation. For StXPB2 (PDB: 5TNU), two open crystal structures are shown. The C‐terminal halves are shown as cyan for structure A and magenta for structure B. The AfXPB closed conformation is a computational model, and the ATP‐binding groove is highlighted. Reprinted (adapted) with permission from Anal Chem. 90(3):2178–2185. Copyright 2018 American Chemical Society.
Both the conventional and molecular wrench modes of XPB activity are required for DNA repair: the molecular wrench conformational change can rapidly open the duplex strands at the damage site, while the conventional DNA helicase activity by XPB promotes broader unzipping and facilitates DNA bubble extension as mediated by xeroderma pigmentosum group D (XPD), another DNA helicase within the TFIIH complex. Our results showed that XPB is a faster helicase acting as a molecular wrench than a conventional helicase. This finding harmonizes with the biological role of XPB as the primary (likely the only) helicase to instigate duplex DNA opening at the damage site and as a secondary helicase (to XPD) to extend the DNA bubble for damage incision. Furthermore, the slow kinetics of StXPBs reflect conventional DNA unzipping with XPB translocating along the DNA. In contrast, the fast decay function of AfXPB reflects the unique molecular wrench DNA unwinding activity by XPB (Figure 1; Fan & DuPrez, 2015).
The structural and kinetic signatures of this activity could profoundly influence DNA repair and transcription processes. The signature of molecular wrench activity was revealed with electrochemical DNA devices coupled with crystallography (Kahanda et al., 2018), which we will now leverage to discover the critical structural and chemical features that govern molecular wrench activity. Furthermore, AfXPB and StXPB have been shown to function with a binding partner, the endonuclease Bax1, which modulates the ATPase activity in opposite ways (Fan & DuPrez, 2015). Further study of the DNA unwinding implications would clarify their overall role and function. Here, we utilize electrochemical experiments with DNA to track the binding and unzipping dynamics of conventional (StXBP2) and molecular wrench (AfXPB) helicase activity at various concentrations. In addition, we explore the concentration‐dependent kinetics of these helicases and correlate these kinetics with their binding and structural features.
2. RESULTS AND DISCUSSION
Electrochemical devices bearing double‐stranded DNA monolayers were used to monitor DNA binding by the helicase and unwinding activity as previously described (Kahanda et al., 2018) and as illustrated in Figure 2. DNA bearing electrochemically active Nile Blue redox probes (Gorodetsky, Ebrahim, & Barton, 2008) were self‐assembled onto gold electrodes on silicon chips (Slinker et al., 2010). In this configuration, the DNA provides an electronically coupled bridge for the surface‐bound electrochemistry between the redox probes and the gold electrode (Drummond et al., 2003; Gorodetsky, Buzzeo, & Barton, 2008) as facilitated by the network of overlapping π‐orbitals of the DNA base stacking interactions (Arnold et al., 2016; Genereux & Barton, 2010). Disruption of the DNA duplex structure lowers this signal by reducing the coupling between the electrode and the redox probe. The electrochemical signal from these redox‐active DNA monolayers is recorded by square wave voltammetry (SWV), a technique sensitive to surface‐bound Faradaic/electronic reactions (Osteryoung & Osteryoung, 1985). Helicase binding disrupts the base hydrogen bonding and DNA suprastructure and lowers the peak current of the voltammetry peaks in proportion to the fraction of the monolayer bound. Our previous work showed that the DNA binding of AfXPB or StXPB2 produced a disruption energetically equivalent to breaking a single A‐T base pair (Kahanda et al., 2018). The addition of ATP initiates activity that breaks multiple base pair hydrogen bonds between the strands of the duplex, potentially fully dehybridizing the DNA and further reducing the signal. (The Nile blue strand is not tethered to the electrode once the DNA is dehybridized.) Assays were performed at room temperature.
FIGURE 2.
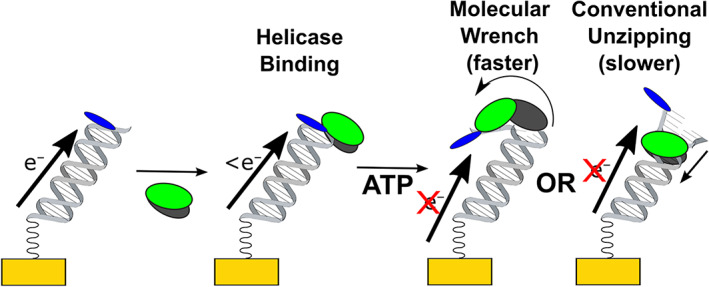
Concept figure of electrochemical detection of helicase DNA binding and unzipping activity. Electrochemistry is recorded with the reduction of a Nile blue redox probe distal to the electrode and facilitated by the DNA. Binding activity lowers the signal in proportion to the fraction of monolayer bound, and ATP treatment further lowers the signal through the loss of the duplex.
Figure 3 shows the impact of ATP‐induced helicase activity on these electrochemical DNA monolayers at 8 and 200 nM concentrations of the helicases. The helicases were first added to the electrochemically active DNA monolayers, and the initial binding reaction was allowed to reach equilibrium for at least 20 min. Then, 2 mM ATP was added to the reaction mixture to initiate the unwinding reaction. In Figure 3a, the conventional StXPB2 induces a continuous monoexponential decrease in the SWV peak current observed at both concentrations. Each curve was fitted with a first‐order exponential of the form , where τ is the exponential time constant of the decay associated with separating the strands of the DNA duplexes across the complete DNA ensemble. The average exponential time constant τ ave for the population was recorded over at least three trials for each StXPB2 concentration and found to be 5000 ± 1000 and 2900 ± 500 s, respectively, for 8 and 200 nM concentrations. These reaction times are consistent with fluorescence studies of the helicase activity on ensembles of DNA (Jang et al., 2010) and RNA (Tani et al., 2010) and our prior electrochemical experiment with the StXPB2 helicase (Kahanda et al., 2018). Thus, the timescale of activity, on the order of tens of minutes, is consistent with the conventional unzipping of an ensemble of DNA. The situation is more complex for the AfXPB helicase activity in Figure 3b. For the lower 8 nM concentration, ATP‐stimulated AfXPB activity exhibited first‐order exponential decay of the DNA electrochemistry, with an exponential time constant of τ ave = 2400 ± 800 s, similar to the StXPB2 timescale associated with conventional activity. In sharp contrast, using a higher 200 nM concentration of AfXPB resulted in a much more rapid exponential decay time constant of 4 ± 2 s, nearly three orders of magnitude faster. This rapid decay is consistent with the timescale of our previous measure of AfXPB activity within experimental error (Kahanda et al., 2018) (at the lower concentration tested) and what we have ascribed as the characteristically fast kinetics of molecular wrench activity (Figures 1 and 2). Here, by testing lower and higher enzyme concentrations, we have revealed that ATP‐stimulated AfXPB activity is concentration‐dependent. These differing timescales of activity presumably alternate between conventional and molecular‐wrench modalities.
FIGURE 3.
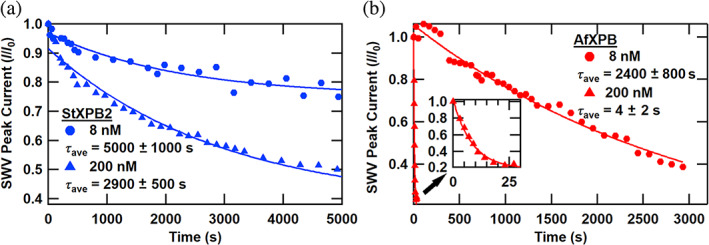
ATP‐initiated kinetics of helicases from Sulfolobus tokodaii (StXPB2) and Archaeoglobus fulgidus helicases (AfXPB). (a) Normalized square wave voltammetry (SWV) peak current versus time after ATP addition for electrochemically active DNA monolayers treated with two concentrations of StXPB2 helicase. (b) Normalized SWV peak current versus time after ATP addition for electrochemically active DNA monolayers treated with two different concentrations of AfXPB helicase. (Inset: normalized SWV peak current vs. time for the 200 nM AfXPB sample over the first 30 s.) Symbols indicate the data and solid curves are first‐order exponential decay fits to the data, and decay times are presented as average ± standard error of the mean for at least three trials.
To further understand the physical basis of these observed concentration‐dependent kinetics, we measured the steady‐state SWV peak current from electrochemical DNA monolayers versus the concentration of each helicase in the absence of ATP to measure helicase binding. As shown in Figure 4, each dataset shows sequential signal loss upon helicase addition, corresponding to the fraction of the DNA monolayer bound by the helicase. StXPB2 data were fit by a single Hill equation of the form:
| (1) |
where S[H] is the SWV signal loss at a particular concentration of helicase, S 0 is a baseline fit value, S M is a maximum signal loss value, [H] is the helicase concentration, [H 1/2] is the helicase concentration at the midpoint of the signal change, and n is the Hill coefficient. For StXPB2, H 1/2, correlating with the binding dissociation constant K D, was 158 nM, while n was 1.43, indicative of cooperative binding. Alternatively, close inspection of the AfXPB concentration dependence revealed a replicable deviation from a single binding isotherm by a dip in the data near 20 nM. These data were then fit by two simplified Langmuir binding isotherms (Esteban Fernandez de Avila et al., 2013; van de Weert & Stella, 2011) of the form:
| (2) |
where S([H]) is the square wave peak height signal at a concentration [H] of helicase, S 0 is the initial square wave peak height, S B is the square wave background signal, and K D is the binding dissociation constant associated with the system. From these fits, the lower concentration portion was fit with a tight K 1D = 25 nM, and the higher concentration portion was fit with a weaker K 2D = 334 nM. The details of this concentration‐dependent binding curve can offer insights into the physical processes responsible for the concentration‐dependent kinetics observed with ATP‐stimulated AfXPB activity.
FIGURE 4.
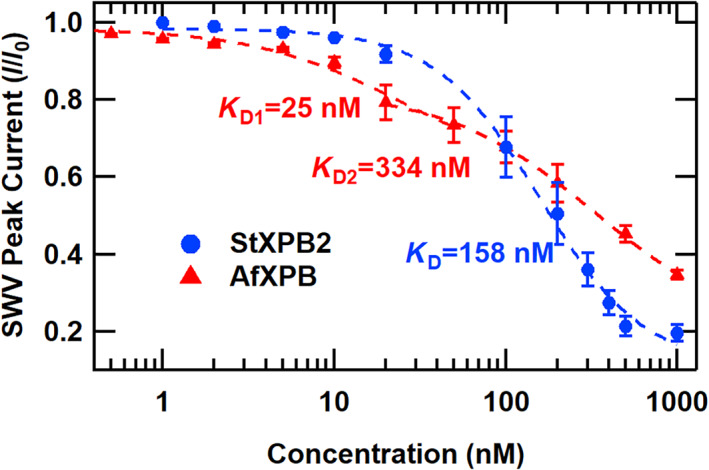
The concentration dependence of DNA binding of helicases from Sulfolobus tokodaii (StXPB2) and Archaeoglobus fulgidus helicases (AfXPB). Normalized square wave voltammetry (SWV) peak current versus helicase concentration. Symbols are data points, error bars represent the standard error of the mean for three trials, and the dashed lines represent fits to the data (no ATP was added).
In Figure 5, we explored the ATP‐stimulated AfXPB activity at three key concentrations displaying characteristic kinetics: 8, 20, and 40 nM. At the low concentration of 8 nM, as noted in Figure 3b, exponential decay is observed with slow dynamics corresponding to τ ave = 2400 s. In contrast, at the highest concentration of 40 nM, rapid exponential decay is observed with τ ave = 7.0 ± 0.5 s. This is the fast timescale associated with molecular wrench activity. Surprisingly, at the middle concentration of 20 nM AfXPB, a unique and replicable dynamic behavior is observed throughout the enzymatic reaction. Initially, a rapid exponential drop is observed, with a τ 1ave = 6.2 ± 0.8 s. Yet, after this rapid initial decay, the SWV peak current does not drop fully to noise levels. After this, the current increases linearly with a slope m 2ave = 1.1 ± 0.2%/s. (This is one of the relatively few examples of the recovery of the electrochemical DNA signal following enzymatic activity [Boon et al., 2002; DeRosa et al., 2005; Muren & Barton, 2013].) Finally, on longer timescales, the signal again undergoes first‐order exponential decay, with τ 3ave = 1600 ± 100 s. Overall, for 20 nM AfXPB, it appears that the DNA monolayer first experiences helicase molecular wrench activity, followed by recovery of the duplex and conventional unwinding activity by the helicase. Notably, this key concentration occurs precisely where there is a change in the slope—a “kink”—in the binding curve presented in Figure 4, where we have modeled the crossover between two binding isotherms. In our previous study, molecular wrench activity was observed at 10 nM AfXPB (Kahanda et al., 2018), indicating that approximately 10–20 nM is the crossover concentration range to observe these dynamic effects.
FIGURE 5.
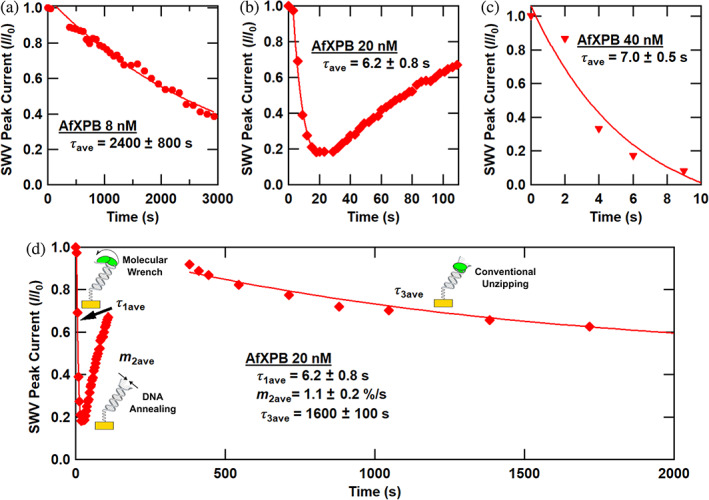
Detailed ATP‐stimulated kinetics of the Archaeoglobus fulgidus helicases (AfXPB). Normalized square wave voltammetry (SWV) peak current versus time after ATP addition for electrochemically active DNA monolayers treated with various concentrations of AfXPB helicase. (a) 8 nM AfXPB. (b) 20 nM AfXPB. (c) 40 nM AfXPB. (d) 20 nM AfXPB, longer timescale. Symbols indicate the data, solid curves are first‐order exponential decay fits to the data, and decay times are presented as average ± standard error of the mean for at least three trials.
The question arises: what fundamental biochemical features induce such concentration‐dependent AfXPB behavior and the characteristics of Figure 5d? Concerning the dynamic behavior of Figure 5d, we postulate that molecular wrench activity accounts for the rapid initial signal loss where present. We also assert that the two binding isotherms of Figure 4 correspond to two binding modes of the AfXPB helicase. Now, let us consider three distinct models, and while they are distinct, the activity may involve a combination of these views. (i) Snap‐back model: there have been several reports of a spring‐loaded, slippage, or snap‐back action of a range of DNA helicases, whereby they rebind to a previously scanned or unwound portion of the substrate with rapid DNA reannealing occurring (Le et al., 2023; Myong et al., 2005; Singh et al., 2019; Sun et al., 2011; Wu et al., 2017). This can be facilitated by multiple binding sites on the monomer helicase, giving rise to separate affinity for ssDNA, dsDNA, DNA forks, or other features. (ii) Catch‐and‐release model: we could be witnessing a modified catch‐and‐release model, whereby the helicase releases from DNA following molecular wrench activity and ATP‐to‐ADP hydrolysis and returns to DNA after rebinding to ATP apart from the DNA, thus bypassing molecular wrench activity. (iii) Cooperative model: in this view, the initial step of molecular wrench helicase activity requires two AfXPB monomer helicases to be bound to the DNA. Once unzipping is accomplished, the helicases are released, and the DNA rehybridizes. When AfXPB rebinds, it can immediately process as a conventional monomeric helicase due to the excess of ATP (2 mM) compared to AfXPB (20 nM).
Let us consider key overall observations. At higher AfXPB concentrations (~40 nM or more), molecular wrench activity dominates and completely extinguishes the signal from the monolayer. For the dynamic intermediate concentration regime near 20 nM, the signal recovery is ~100 s. This is notably slower than the rehybridization single‐molecule experiments, which are on the order of milliseconds to seconds (Le et al., 2023; Myong et al., 2005; Singh et al., 2019; Sun et al., 2011; Wu et al., 2017). The slower timescale here follows from the recovery of the monolayer ensemble. It indicates a significant time between the end of the correlated initial unzipping activity and the initiation of the second set of unzipping events. The signal recovers to nearly 100% of the initial value, likely suggesting that the DNA strands were not completely dehybridized from the initial activity. Notably, the crossover to this dynamic intermediate regime at 20 nM correlates with a kink in the binding curve of Figure 4—the onset of the second binding isotherm. So, the rapid kinetics are initiated as an apparent second binding mode is activated, possibly corresponding to a binding partner. Carefully considered, these observations lend credence to each of the proposed models, and some combination of the models may give rise to the observed dynamic behavior.
Next, we investigated the binding and ATP‐stimulated kinetics of StXPB2‐Bax1 and AfXPB‐Bax1 complexes. The Bax1 nuclease affects the conformation and activity of StXPB2 and AfXPB differently. Crystal structure and activity analyses have shown that StXPB2‐Bax1 stabilizes StXPB2 in its closed conformation (Figure S4) and stimulates ATP hydrolysis, while AfXPB‐Bax1 stabilizes AfXPB in the open conformation consistent with the crystal structure of the complex (Figure S5) and reduces its ATPase activity (DuPrez et al., 2020). We measured the concentration dependence of initial DNA binding and ATP‐stimulated activity of StXPB2‐Bax1 and AfXPB‐Bax1 helicase‐Bax1 complexes and presented the results in Figure 6. In Figure 6a,b, forming the complex with Bax1 shifts the binding activity of both helicases. For StXPB2‐Bax1, the curve is fit with the simplified Langmuir binding isotherm of Equation (2), with K D = 640 nM. As expected, the binding of the StXPB2‐Bax1 is lowered relative to StXPB2. For AfXPB‐Bax1, the curve was fit with the Hill equation (Equation 1), with a K D of 145 nM and a Hill coefficient n of 2.2.
FIGURE 6.
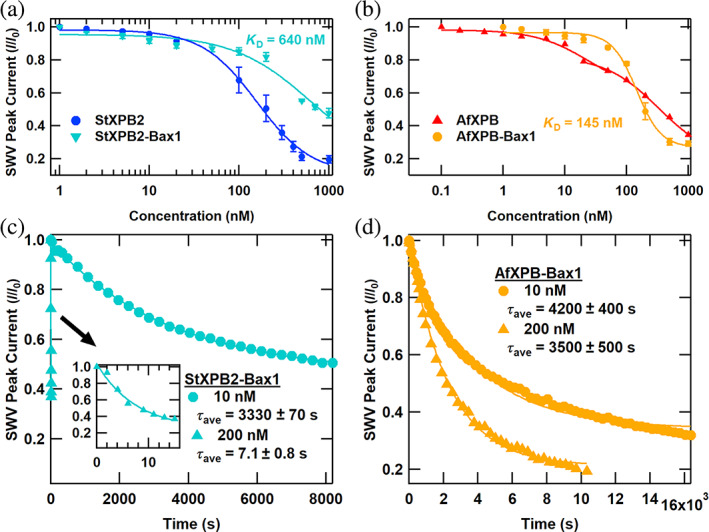
Binding and kinetics of helicase‐Bax1 complexes. (a) Normalized square wave voltammetry (SWV) peak current versus helicase concentration for helicases from Sulfolobus tokodaii (StXPB2) (Figure 4 data) and StXPB2‐Bax1 (no ATP was added). (b) Normalized SWV peak current versus helicase concentration for Archaeoglobus fulgidus helicases (AfXPB) (Figure 4 data) and AfXPB‐Bax1 (no ATP was added). (c) Normalized SWV peak current versus time after ATP addition for electrochemically active DNA monolayers treated with various concentrations of StXPB2‐Bax1 helicase. (d) Normalized SWV peak current versus time after ATP addition for electrochemically active DNA monolayers treated with various concentrations of AfXPB‐Bax1 helicase. Symbols indicate the data, solid curves are first‐order exponential decay fits to the data, and decay times are presented as average ± standard error of the mean for at least three trials.
The ATP‐stimulated kinetics of each complex at 10 and 200 nM are shown in Figure 6c,d. The kinetics of StXPB2‐Bax1 are dynamic with concentration. The exponential time constant for the lower 10 nM concentration was similar to the time constants for uncomplexed StXPB2 (3500 s). However, the higher 200 nM concentration exhibits substantially more rapid activity. A monoexponential decay with τ ave = 7.1 ± 0.8 s was observed at this concentration. In the complex, Bax1 stabilizes the active closed conformation of StXPB2 (see Figure S4) that enhances its ATPase activity (DuPrez et al., 2020). It is curious and noteworthy to see that this rapid timescale is similar to molecular wrench activity. For AfXPB‐Bax1, while exponential signal loss is observed again, the activity rate is lowered for both concentrations, with the rapid molecular wrench activity completely suppressed at 200 nM (τ ave = 3330 ± 70 s). This is consistent with the crystal structure of the complex (Figure S5) and the understanding of molecular wrench activity. An AfXPB conformational change from open to closed is requisite for rapid activity, and Bax1‐binding may lower the propensity for this conformational change to occur, limiting the ATP activity rate.
Finally, as a particular note of caution for activity assays involving ATP, we show the impact of failing to account for the influence of ATP addition on the solution's pH. We noted that 2 mM ATP was sufficient to change the pH of our standard buffer solution to acidic conditions (pH 5–6), with catastrophic consequences for the DNA hybridization and composite self‐assembled monolayer. (To correct this, we first dissolve the ATP in pH 10 buffer for all other experiments as noted in the methods.) Figure 7 shows the impact of 2 mM ATP on DNA electrochemistry in the absence of helicase when insufficient buffer conditions yield acidic reaction conditions. In Figure 7a, successive SWV curves were recorded before and after ATP addition. Once the ATP is added, no apparent voltammetry peak can be observed, and the voltammetry background increases significantly. The impact of the acidic conditions is rapid, with activity on the order of milliseconds. To view the details of this reaction in more detail, we added ATP during a voltammetry peak and noted the changes in Figure 7b. Initially, the current drops with the loss of the DNA electrochemistry from acid‐induced DNA dehybridization. Subsequently, the signal increases dramatically as thiol reduction ensues, exposing the electrode to irreversible oxygen electrochemistry. In Figure 7c,d, we demonstrate how to precisely quantify the rapid DNA dehybridizing dynamics by subtracting a pre‐addition averaged reference curve, enabling kinetic analysis with 16.7 ms resolution corresponding to the SWV period. The resulting linear decay curve in Figure 7d reveals a signal loss rate of 18.6 pA/ms or 49% of the DNA peak current per second. We term this technique intrasweep square wave voltammetry (ISWV) and discuss this method in detail in the Supporting Information.
FIGURE 7.
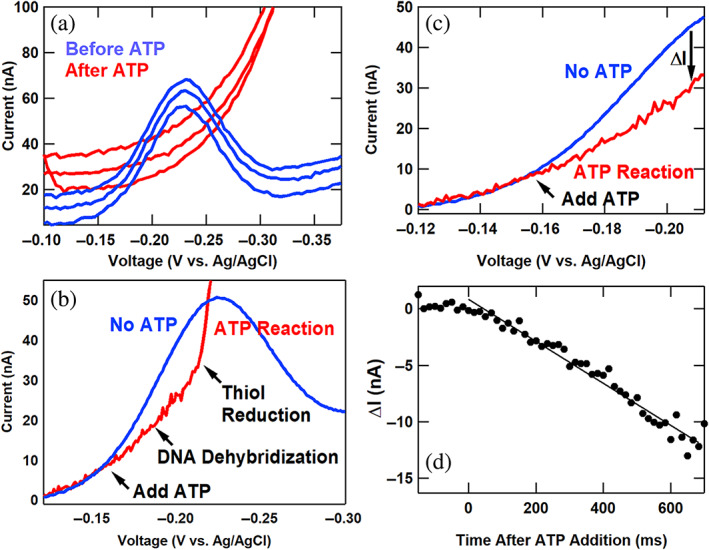
Catastrophic loss of DNA electrochemical signal under ATP‐induced acidic conditions with insufficient buffering. (a) Successive square wave voltammetry (SWV) sweeps before and after ATP addition. (b) A SWV curve from 25 SWV scans before ATP addition (blue) and a curve recorded during ATP addition (red). (c) The SWV curves of Figure 7b focused on the region between −0.120 and −0.212 V versus Ag/AgCl. (d) The difference in current between the red and blue curves of Figure 7c is plotted against time after ATP addition, showing the quantifiable rapid signal loss.
3. CONCLUSION
We investigated the concentration dependence of DNA helicase binding and ATP‐stimulated kinetics of StXPB2 and AfXPB, as well as their binding and activity in Bax1 complexes through electrochemistry with redox‐active DNA monolayers. The timescale of StXPB2 ATP‐stimulated DNA unzipping activity was concentration‐independent from 8 to 200 nM and consistent with conventional helicase unwinding. Alternatively, AfXPB activity was highly concentration dependent in this range, with exponential rate constants varying from seconds to thousands of seconds. At 20 nM, rapid exponential decay of the electrochemical signal from the reaction was followed by linear recovery and subsequent slower exponential decay. This concentration‐dependent AfXPB ATP activity is rationalized as the crossover between a rapid molecular wrench process and a slower conventional helicase mechanism. The AfXPB‐Bax1 protein complex inhibited this fast ATPase activity. In contrast, the StXPB2‐Bax1 complex induces rapid ATP‐induced kinetics at higher concentrations of the enzymes. The crystal structures of these complexes rationalize this change in activity as a function of helicase concentration: AfBax1 stabilizes AfXPB in the inactive open conformations, whereas StXPB2‐Bax1 stabilizes the active closed form of StXPB2. These findings illuminate the factors governing molecular wrench activity for improved biological insight into a key factor in DNA repair.
4. MATERIALS AND METHODS
4.1. Protein expression and purification
AfXPB and AfXPB‐Bax1 were expressed via published protocols (DuPrez et al., 2020; Fan et al., 2006). StXPB2 and StXPB2‐Bax1 expressions followed published protocols (DuPrez et al., 2020; Ma et al., 2011). The expression plasmids pET15b/StXPB2 (protein accession number: WP_010979669) and pET15b/StBax1 (protein accession number: WP_010979670), generously provided by Dr. Yulong Shen at the Shandong University of China, were transformed into Escherichia coli Rosetta (DE3) pLysS competent cells. Structures for AFXPB (PDB: 2FZ4 & 2FZL), StXPB2 (PDB: 5TNU), AfXPB‐Bax1 (PDB: 6P66), and StXPB2‐Bax1 (PDB: 6p4o & 6P4F) are deposited in the Protein Data Bank.
Helicase samples for electrochemistry studies were thawed and passed through a PD10 column (GE) for buffer exchange into 10 mM TrisCl pH 8, 200 mM NaCl, and 10 mM MgCl2. Protein concentrations were determined by absorbance measurements at 280 nm before freezing in liquid nitrogen and shipment on dry ice.
4.2. Synthesis of DNA
Double‐stranded DNA was prepared using the 17mer sequence 3′‐CTCTATATTTCGTGCGTNB‐5′ and its fully complementary sequence 5′‐(C6 thiol)‐GAGATATAAAGCACGCA‐3′. These oligonucleotides were synthesized on an H‐2 DNA Synthesizer from K & A Labs (Germany). TNB denotes the position of the thymine modified with a Nile Blue redox probe. This probe‐labeled base was formed by coupling Nile Blue perchlorate (Sigma Aldrich) with the 5‐[3‐acrylate NHS ester] deoxyuridine phosphoramidite from Glen Research. The dye was covalently coupled and processed under UltraMild conditions according to established procedures (Gorodetsky, Ebrahim, & Barton, 2008). The C6 thiol linker was coupled to the oligo as the Glen Research thiol‐modifier C6 S‐S phosphoramidite, and the dithiol was reduced for self‐assembly.
4.3. Purification of DNA
All oligonucleotides were purified via two rounds of high‐performance liquid chromatography on a Shimadzu LC‐20 AD instrument outfitted with a SIL‐20A autosampler and an SPD‐M20A diode array detector as previously described (McWilliams et al., 2015; Wohlgamuth et al., 2014). The identity of the desired products was confirmed by matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry on a Shimadzu Axima Confidence mass spectrometer.
4.4. Preparation of the double‐stranded DNA
The oligonucleotides were quantified via UV–visible spectroscopy on a Beckman DU‐800 UV–visible spectrophotometer. The formation of duplex DNA was verified by temperature‐dependent absorbance measurements with melting temperature analysis.
4.5. Fabrication of devices
The chips/substrates featuring multiplexed gold electrodes for DNA self‐assembly and electrochemical experiments were prepared as previously described (McWilliams et al., 2015; Slinker et al., 2010).
4.6. Self‐assembly of DNA monolayers
The DNA monolayers were self‐assembled onto gold electrode pads from a solution with 25 μM of the duplex DNA, 5 mM phosphate, 50 mM sodium chloride, pH = 7 buffer solution over 12–18 h. The substrates were backfilled with mercaptohexanol for 1 h to remove nonspecifically bound DNA and then thoroughly rinsed with buffer to remove residual mercaptohexanol.
4.7. Electrochemical measurements on DNA monolayers
The multiplexed substrates were placed in a custom mount and connected to electrochemical testing hardware (a CH Instruments CHI730D Electrochemical Analyzer and a CHI 684 Multiplexer). SWV was generally performed at 40 Hz with a 0.025 mV amplitude and 4 mV increment. The electrochemical measurements were performed in pH 7.9 buffer containing 50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, and 4 mM spermidine. For helicase binding experiments, helicases were added to this solution and allowed to equilibrate for at least 20 min, and SWV was recorded under the helicase incubation solution. For kinetics experiments, helicases were first added to the solution and allowed to equilibrate for at least 20 min. Then, ATP was added to this helicase incubation solution, and the SWV signal was recorded over time with ATP and helicase present. The ATP solution was prepared at pH 10 buffer as above for ATP experiments to ensure a pH of ~7.9 after ATP addition. See Figures S1–S3 for example electrochemistry and controls of these data. For details of the ISWV experiment (Figure 7), please see the Supporting Information.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Data S1. Example SWV curves, control experiment for ATP addition, control experiment for DNA unzipping, summarized time constants, summarized binding dissociation constants, investigation by intrasweep square wave voltammetry (ISWV), illustration of DNA and kinetic rates, ATP reaction by conventional SWV, ATP reaction by ISWV.
ACKNOWLEDGMENTS
This work was funded by NIH grant number R01GM108893, Office of Naval Research grant number N00014‐16‐1‐2741, and Welch Foundation grant number AT‐2079‐20210327.
Wettasinghe AP, Seifi MO, Bravo M, Adams AC, Patel A, Lou M, et al. Molecular wrench activity of DNA helicases: Keys to modulation of rapid kinetics in DNA repair. Protein Science. 2023;32(12):e4815. 10.1002/pro.4815
Review Editor: John Kuriyan
Contributor Information
Li Fan, Email: li.fan@ucr.edu.
Jason D. Slinker, Email: slinker@utdallas.edu.
DATA AVAILABILITY STATEMENT
The data are available from the authors on reasonable request.
REFERENCES
- Arnold AR, Grodick MA, Barton JK. DNA charge transport: from chemical principles to the cell. Cell Chem Biol. 2016;23(1):183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon EM, Salas JE, Barton JK. An electrical probe of protein‐DNA interactions on DNA‐modified surfaces. Nat Biotechnol. 2002;20(3):282–286. [DOI] [PubMed] [Google Scholar]
- DeRosa MC, Sancar A, Barton JK. Electrically monitoring DNA repair by photolyase. Proc Natl Acad Sci U S A. 2005;102(31):10788–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3):785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21(10):1192–1199. [DOI] [PubMed] [Google Scholar]
- DuPrez K, He F, Chen Z, Hilario E, Fan L. Structural basis of the XPB–Bax1 complex as a dynamic helicase–nuclease machinery for DNA repair. Nucleic Acids Res. 2020;48(11):6326–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban Fernandez de Avila B, Watkins HM, Pingarron JM, Plaxco KW, Palleschi G, Ricci F. Determinants of the detection limit and specificity of surface‐based biosensors. Anal Chem. 2013;85(14):6593–6597. [DOI] [PubMed] [Google Scholar]
- Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22(1):27–37. [DOI] [PubMed] [Google Scholar]
- Fan L, DuPrez KT. XPB: an unconventional SF2 DNA helicase. Prog Biophys Mol Biol. 2015;117(2–3):174–181. [DOI] [PubMed] [Google Scholar]
- Fishburn J, Tomko E, Galburt E, Hahn S. Double‐stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A. 2015;112(13):3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem Rev. 2010;110(3):1642–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LCJ, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106(2):253–276. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure‐function relationships. Curr Opin Struct Biol. 1993;3(3):419–429. [Google Scholar]
- Gorodetsky AA, Buzzeo MC, Barton JK. DNA‐mediated electrochemistry. Bioconjugate Chem. 2008;19(12):2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodetsky AA, Ebrahim A, Barton JK. Electrical detection of TATA binding protein at DNA‐modified microelectrodes. J Am Chem Soc. 2008;130(10):2924–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yan CL, Fang J, Inouye C, Tjian R, Ivanov I, et al. Near‐atomic resolution visualization of human transcription promoter opening. Nature. 2016;533(7603):359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim YK, Kwon HM, Yeo WS, Kim DE, Min DH. A graphene‐based platform for the assay of duplex‐DNA unwinding by helicase. Angew Chem Int Ed. 2010;49(33):5703–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahanda D, DuPrez KT, Hilario E, McWilliams MA, Wohlgamuth CH, Fan L, et al. Application of electrochemical devices to characterize the dynamic actions of helicases on DNA. Anal Chem. 2018;90(3):2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP‐dependent promoter melting by transcription factor IIH. Science. 2000;288(5470):1418–1421. [DOI] [PubMed] [Google Scholar]
- Le ST, Choi S, Lee S‐W, Kim H, Ahn B. ssDNA reeling is an intermediate step in the reiterative DNA unwinding activity of the WRN‐1 helicase. J Biol Chem. 2023;299:105081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non‐hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9(5):391–401. [DOI] [PubMed] [Google Scholar]
- Ma XQ, Hong Y, Han WY, Sheng DH, Ni JF, Hou GH, et al. Single‐stranded DNA binding activity of XPBI, but not XPBII, from Sulfolobus tokodaii causes double‐stranded DNA melting. Extremophiles. 2011;15(1):67–76. [DOI] [PubMed] [Google Scholar]
- McWilliams MA, Bhui R, Taylor DW, Slinker JD. The electronic influence of abasic sites in DNA. J Am Chem Soc. 2015;137:11150–11155. [DOI] [PubMed] [Google Scholar]
- Muren NB, Barton JK. Electrochemical assay for the signal‐on detection of human DNA methyltransferase activity. J Am Chem Soc. 2013;135(44):16632–16640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. [DOI] [PubMed] [Google Scholar]
- Osteryoung JG, Osteryoung RA. Square‐wave voltammetry. Anal Chem. 1985;57(1):101–110. [Google Scholar]
- Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. [DOI] [PubMed] [Google Scholar]
- Rand MD, Grimm LM, Artavanis‐Tsakonas S, Patriub V, Blacklow SC, Sklar J, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20(5):1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney KD. Erratum to: structure and mechanisms of SF1 DNA helicases. DNA helicases and DNA motor proteins. 1st ed. New York: Springer‐Verlag; 2013. [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JHJ, et al. DNA‐repair helicase – a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260(5104):58–63. [DOI] [PubMed] [Google Scholar]
- Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly JM. Molecular structure of human TFIIH. Cell. 2000;102(5):599–607. [DOI] [PubMed] [Google Scholar]
- Singh SP, Soranno A, Sparks MA, Galletto R. Branched unwinding mechanism of the Pif1 family of DNA helicases. Proc Natl Acad Sci U S A. 2019;116:24533–24541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. [DOI] [PubMed] [Google Scholar]
- Slinker JD, Muren NB, Gorodetsky AA, Barton JK. Multiplexed DNA‐modified electrodes. J Am Chem Soc. 2010;132(8):2769–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Johnson DS, Patel G, Smith BY, Pandey M, Patel SS, et al. ATP‐induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Wang MD. Single‐molecule perspectives on helicase mechanisms and functions. Crit Rev Biochem Mol Biol. 2016;51(1):15–25. [DOI] [PubMed] [Google Scholar]
- Tani H, Fujita O, Furuta A, Matsuda Y, Miyata R, Akimitsu N, et al. Real‐time monitoring of rna helicase activity using fluorescence resonance energy transfer in vitro. Biochem Biophys Res Commun. 2010;393(1):131–136. [DOI] [PubMed] [Google Scholar]
- van de Weert M, Stella L. Fluorescence quenching and ligand binding: a critical discussion of a popular methodology. J Mol Struct. 2011;998(1–3):144–150. [Google Scholar]
- Wohlgamuth CH, McWilliams MA, Mazaheripour A, Burke AM, Lin KY, Doan L, et al. Electrochemistry of DNA mono layers modified with a perylenediimide base surrogate. J Phys Chem C. 2014;118(50):29084–29090. [Google Scholar]
- Wu WQ, Hou XM, Zhang B, Fossé P, René B, Mauffret O, et al. Single‐molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci Rep. 2017;7:43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Example SWV curves, control experiment for ATP addition, control experiment for DNA unzipping, summarized time constants, summarized binding dissociation constants, investigation by intrasweep square wave voltammetry (ISWV), illustration of DNA and kinetic rates, ATP reaction by conventional SWV, ATP reaction by ISWV.
Data Availability Statement
The data are available from the authors on reasonable request.


