Abstract
Background
Colorectal cancer (CRC) is the third most common cancer in the world and has a high mortality rate. Colorectal adenoma (CRA) is precancerous lesions of CRC. The purpose of the present study was to construct a nomogram predictive model for CRA with low-grade intraepithelial neoplasia (LGIN) in order to identify high-risk individuals, facilitating early diagnosis and treatment, and ultimately reducing the incidence of CRC.
Methods
We conducted a single-center case-control study. Based on the results of colonoscopy and pathology, 320 participants were divided into the CRA group and the control group, the demographic and laboratory test data were collected. A development cohort (n = 223) was used for identifying the risk factors for CRA with LGIN and to develop a predictive model, followed by an internal validation. An independent validation cohort (n = 97) was used for external validation. Receiver operating characteristic curve, calibration plot and decision curve analysis were used to evaluate discrimination ability, accuracy and clinical practicability of the model.
Results
Four predictors, namely sex, age, albumin and monocyte count, were included in the predictive model. In the development cohort, internal validation and external validation cohort, the area under the curve (AUC) of this risk predictive model were 0.946 (95%CI: 0.919–0.973), 0.909 (95 % CI: 0.869–0.940) and 0.928 (95%CI: 0.876–0.980), respectively, which demonstrated the model had a good discrimination ability. The calibration plots showed a good agreement and the decision curve analysis (DCA) suggested the predictive model had a high clinical net benefit.
Conclusion
The nomogram model exhibited good performance in predicting CRA with LGIN, which can aid in the early detection of high-risk patients, improve early treatment, and ultimately reduce the incidence of CRC.
Keywords: Predictive model, Nomogram, Colorectal adenoma, Laboratory tests, Risk factor
Highlights
-
•
We developed a predictive model for colorectal adenoma based on routine laboratory index.
-
•
The predictive model involves four predictors: sex, age, albumin and monocyte count.
-
•
The primary model was displayed as a nomogram as well as a formula for clinical use.
-
•
We conducted internal validation using bootstrap technique and external validation.
-
•
The model exhibited excellent discrimination, calibration and clinical practicability.
1. Introduction
Colorectal cancer (CRC) is the third most common malignant tumor and the mortality rate associated with CRC is the second highest among cancer-related deaths worldwide [[1], [2], [3]],resulting in 935,000 deaths and more than 1.9 million new CRC in 2020 across the globe [4]. Today, CRC is the second most common cancer in China, after lung cancer [5], which brought a heavy disease burden. In recent years, the incidence of early-onset CRC (under the age of 50 years) has been increasing worldwide [1,3]. Therefore, reducing the incidence and mortality of CRC has become an urgent public health issue that needs to be addressed. Colorectal adenomatous polyp (CAP), also known as colorectal adenoma (CRA), are recognized as a precursor lesion for CRC [6,7]. We are aware that CRA may progress to CRC due to genetic and environmental factors [8], which is known as the adenoma-carcinoma sequence, and this process usually takes more than 10 years [[9], [10], [11]]. For this reason, timely identification and management of CRA are crucial. Colorectal polyps are typically asymptomatic in some cases, which poses a challenge for early detection and treatment without regular health screenings. Patients who come for voluntary examination usually have clinical symptoms (such as increased frequency of bowel movements) or abnormal test results (such as fecal occult blood), which leads to colonoscopy and the subsequent discovery of polyps. Unfortunately, some patients are diagnosed with CRC during their initial examination, which delays early treatment.
Currently, colonoscopy is the most widely used and effective procedure for detecting colorectal cancer and adenomas [8,12,13]. Colonoscopy can also perform therapeutic polypectomy, which prevents the progression of CRA to CRC. Many studies have confirmed that early diagnosis of CRA and removal of precancerous lesions by colonoscopy can significantly reduce the incidence and mortality of cancer [[14], [15], [16], [17]]. However, colonoscopy is an invasive procedure that requires complex intestinal preparation and can result in unpleasant personal experiences and complications, which limits its use for routine screening in large population. This may lead to the failure to timely detect high-risk populations with CRA. Therefore, it is crucial to identify an economical, straightforward and feasible method for the risk prediction of CRA in a given population. For the past few years, some studies have explored predictive models for CRC [[18], [19], [20], [21]], yet few have focused on predicting the risk of CRA, a precancerous lesion of CRC. Several studies have utilized clinical features to predict the risk of colorectal polyps or neoplasia with moderate accuracy and low sensitivity and specificity [22,23], and the predictors involved in the predictive model require relevant imaging examinations that are not sufficiently convenient nor economical. The diagnostic efficacy of the model in existing studies is suboptimal, and some models lack complete model validation. Therefore, there remains room for improvement in the accuracy and operability of the predictive model for CRA.
The pathology grading of CRA can be divided into low-grade intraepithelial neoplasia (LGIN) and high-grade intraepithelial neoplasia (HGIN). HGIN accounted for about 5 % of adenomas [24] and was considered a type of early cancer (intramucosal carcinoma) [25,26], which was not studied in this research. Our focus in this study was on the population of CRA with LGIN, which accounted for the majority of detected CRA and held potential clinical screening significance. The purpose of this study is to explore the risk factors for CRA with LGIN by conducting single factor analysis and multivariate logistic regression analysis based on basic demographic information and routine laboratory test results obtained during medical check-ups. Additionally, a nomogram model will be developed and validated for predicting the risk of CRA with LGIN. The predictive model is expected to facilitate the identification of high-risk individuals, guiding them towards further colonoscopy and pathological examination to determine the presence of colorectal neoplasia. Early removal of adenomas can help prevent the future incidence of colorectal cancer. (For brevity, “CRA with LGIN” in this study was abbreviated as “CRA” in the following text.)
2. Materials and methods
2.1. Study design and population
This was a single-center case-control study, which was conducted at the Department of Gastroenterology and Digestive Endoscopy Center, Beijing Chao-Yang Hospital, Capital Medical University from January 2019 to January 2023. In order to establish a reliable predictive model for CRA, our study consisted of four aspects: (1) identification of the statistically significant risk factors associated with CRA using the development cohort; (2) developing a predictive model for CRA and conducting internal validation using the development cohort; (3) conducting external validation using an independent validation cohort; (4) evaluating the discrimination, calibration, and effectiveness of clinical application in both the development cohort and validation cohort. The present study was performed according to the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) Statement [27].
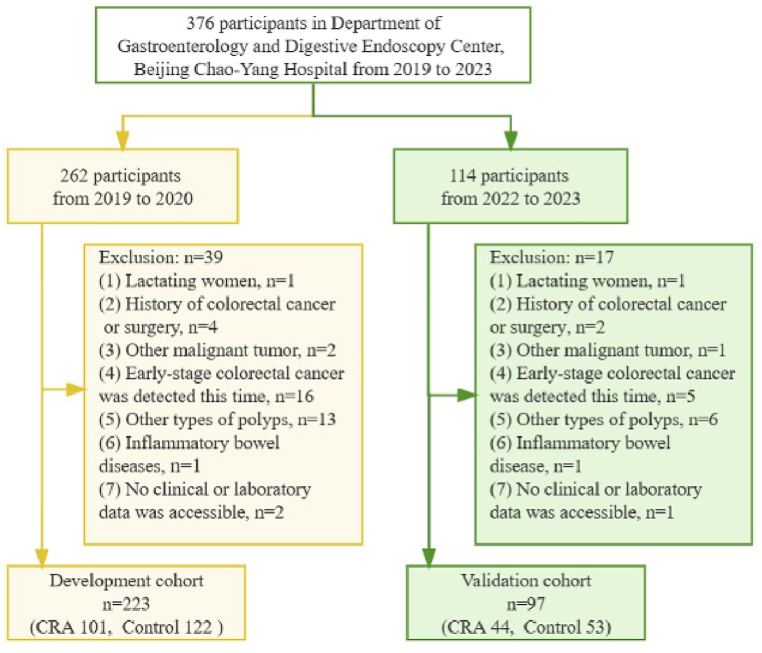
Subjects pathologically diagnosed as colorectal adenoma (CRA) with LGIN were categorized as the CRA group, while those who had no abnormalities detected during colonoscopy were classified as the control group. Inclusion criteria: (1) age over 18 years old; (2) CRA with LGIN have been confirmed by colonoscopy and biopsy or resected specimen-based pathological diagnosis; (3) accessibility to comprehensive clinical and laboratory data. Exclusion criteria: (1) pregnant women and lactating women; (2) history of colorectal cancer or surgery; (3) other malignant tumor; (4) early-stage colorectal cancer (including CRA with HGIN) detected by this colonoscopy; (5) other types of polyps, such as hyperplastic polyps, inflammatory polyps and so on; (6) inflammatory bowel diseases (IBD), such as ulcerative colitis or Crohn's disease; (7) severe metabolic abnormalities or organ failure. A total of 376 participants underwent relative examination, 320 individuals were ultimately included in the study based on predetermined criteria (Fig. 1). The participants in the development cohort (CRA 101, Control 122) were recruited from January 2019 to January 2020; the participants in the validation cohort (CRA 44, Control 53) were recruited from January 2022 to January 2023.
Fig. 1.
Flowchart of the participants selection for the development cohort and validation cohort.
2.2. Data collection
Each participant enrolled underwent a colonoscopy screening and health examination. All the clinical data were retrospectively collected from Hospital Information System (HIS), Laboratory Information Management System (LIS), Picture Archiving and Communication System (PACS) and Hospital Electronic Medical Record (HEMR). The following data were collected: (1) demographic data: sex, age, body mass index (BMI, kg/m2) and history of hypertension; (2) fasting biochemical tests: albumin (ALB, g/L), total cholesterol (TC, mmol/L), high-density lipoprotein cholesterol (HDL-C, mmol/L), low-density lipoprotein cholesterol (LDL-C, mmol/L), triglyceride (TG, mmol/L), fasting blood glucose (FBG, mmol/L); (3) blood routine tests: white blood cell count (WBC, 109/L), neutrophil count (NEUT, 109/L), lymphocyte count (LYM, 109/L), monocyte count (MONO, 108/L), red blood cell count (RBC, 1012/L) and hemoglobin (HGB, g/L), the laboratory tests were conducted within 7 days prior to a colonoscopy; (4) colonoscopy findings and pathological examination: the colonoscopy was performed by experienced endoscopists who conducted no fewer than 1000 colonoscopies annually; the resected polyps were pathologically examined; and histopathologic results were reported by experienced pathologists. The number of polyps and the maximum diameter of the polyps are collected.
2.3. Statistic analysis
Measurement data were expressed as mean ± standard deviation (SD) and counting data were expressed as frequencies and percentages. In univariate analysis, the independent sample t-test or Mann-Whitney U test was performed for measurement data and the Chi-square test was used for counting data to compare the differences between the CRA group and the control group. In single factor analysis, the independent variables with P < 0.05 were selected for the multivariate binary logistic regression analysis to determine independent risk factors associated with colorectal adenoma. We built a risk predictive model for colorectal adenoma using influencing factors selected from the development cohort and visualized it with a nomogram. Internal validation of the model was performed by the bootstrap technique (1000 resamples). Temporal validation method was used for external validation in this study, which was performed on an independent validation cohort.
We applied an area under the ROC curve (AUC) to assess the discriminative ability of the predictive model. The cut-off value corresponding to the Maximum Youden Index (YDI) was taken as the optimal cut-off value for the predictive model [28]. Under the cut-off value, sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR) and negative likelihood ratio (NLR) were calculated. Calibration plot was utilized to evaluate the accuracy of the predictive model by assessing the agreement between actual and nomogram-predicted probabilities (employing 1000 resampling bootstrap) [29]. Decision curve analysis (DCA) was conducted to evaluate the clinical practicability of the model [30,31]. The evaluation of the model performance was conducted on both the development cohort and validation cohort.
The data was statistically analyzed by SPSS 26 (IBM Corp., Chicago, IL) and R software 4.1.2. Variables with P < 0.05 (two-side test) were considered statistically significant.
The funding sources played no role in the study's design, data collection, analysis, interpretation, or manuscript writing.
3. Results
3.1. Baseline characteristics of the study
A total of 320 participants, comprising 146 males (45.63 %) and 174 females (54.37 %), were included in this study. 223 participants were enrolled in the development cohort from January 2019 to January 2020, while the validation cohort consisted of 97 participants recruited between January 2022 and January 2023. Both the development cohort and validation cohort consisted of the CRA (101 and 44) and control groups (122 and 53), respectively. No significant differences in baseline demographic characteristics were observed between the development cohort and validation cohort (Table 1).
Table 1.
Clinical baseline characteristics in the development and validation cohorts.
| Development cohort |
Validation cohort |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables |
Total |
CRA group |
Control group |
P value* |
Total |
CRA group |
Control group |
P value* |
P value# |
| (n = 223) | (n = 101) | (n = 122) | (n = 97) | (n = 44) | (n = 53) | ||||
| Sex, n (%) | 0.073 | 0.839 | 0.759 | ||||||
| Male | 120 (53.81) | 61 (60.40) | 59 (48.36) | 54 (55.67) | 24 (54.55) | 30 (56.60) | |||
| Female | 103 (46.19) | 40 (39.60) | 63 (51.64) | 43 (44.33) | 20 (45.45) | 23 (43.40) | |||
| Age, years | 51.24 ± 12.65 | 59.23 ± 12.12 | 44.63 ± 8.67 | <0.001 | 54.10 ± 10.93 | 60.39 ± 12.49 | 48.89 ± 5.53 | <0.001 | 0.099 |
| BMI, kg/m2 | 24.87 ± 3.80 | 24.67 ± 3.93 | 25.03 ± 3.70 | 0.421 | 24.82 ± 4.07 | 24.81 ± 4.41 | 24.82 ± 3.80 | 0.879 | 0.590 |
| HBP, n (%) | 0.007 | 0.199 | 0.149 | ||||||
| Yes | 54 (24.22) | 33 (32.67) | 21 (17.21) | 31 (31.96) | 17 (38.64) | 14 (26.42) | |||
| No | 169 (75.78) | 68 (67.33) | 101 (82.79) | 66 (68.04) | 27 (61.36) | 39 (73.58) | |||
| ALB, g/L | 44.28 ± 4.26 | 41.58 ± 4.45 | 46.52 ± 2.39 | <0.001 | 44.71 ± 4.16 | 41.50 ± 3.86 | 47.38 ± 1.91 | <0.001 | 0.130 |
| TC, mmol/L | 4.82 ± 0.98 | 4.51 ± 0.96 | 5.09 ± 0.92 | <0.001 | 4.98 ± 0.92 | 4.71 ± 0.93 | 5.20 ± 0.86 | 0.013 | 0.185 |
| HDL-C, mmol/L | 1.31 ± 0.38 | 1.21 ± 0.34 | 1.40 ± 0.39 | <0.001 | 1.24 ± 0.32 | 1.15 ± 0.30 | 1.31 ± 0.31 | 0.008 | 0.133 |
| LDL-C, mmol/L | 3.07 ± 0.90 | 2.81 ± 0.82 | 3.29 ± 0.90 | <0.001 | 3.18 ± 0.84 | 2.96 ± 0.94 | 3.36 ± 0.72 | 0.019 | 0.307 |
| TG, mmol/L | 1.36 ± 0.79 | 1.31 ± 0.64 | 1.39 ± 0.90 | 0.713 | 1.63 ± 1.20 | 1.88 ± 1.59 | 1.43 ± 0.70 | 0.282 | 0.065 |
| FBG, mmol/L | 4.80 ± 0.86 | 4.93 ± 0.92 | 4.69 ± 0.79 | <0.001 | 5.23 ± 1.23 | 5.25 ± 1.26 | 5.22 ± 1.22 | 0.747 | 0.020 |
| WBC, 109/L | 5.96 ± 1.47 | 5.99 ± 1.57 | 5.94 ± 1.38 | 0.889 | 5.86 ± 1.47 | 5.96 ± 1.62 | 5.78 ± 1.34 | 0.747 | 0.367 |
| NEUT, 109/L | 3.48 ± 1.18 | 3.46 ± 1.33 | 3.50 ± 1.05 | 0.405 | 3.34 ± 0.98 | 3.27 ± 0.89 | 3.40 ± 1.05 | 0.712 | 0.398 |
| LYM, 109/L | 2.00 ± 0.69 | 1.93 ± 0.52 | 2.09 ± 0.85 | 0.160 | 1.99 ± 0.65 | 1.89 ± 0.54 | 2.10 ± 0.76 | 0.305 | 0.736 |
| MONO, 108/L | 3.82 ± 1.10 | 4.18 ± 1.10 | 3.53 ± 1.01 | <0.001 | 3.48 ± 1.10 | 3.90 ± 1.13 | 3.13 ± 0.09 | <0.001 | 0.011 |
| RBC, 1012/L | 4.57 ± 0.49 | 4.45 ± 0.46 | 4.68 ± 0.49 | 0.003 | 4.62 ± 0.59 | 4.37 ± 0.63 | 4.83 ± 0.46 | <0.001 | 0.506 |
| HGB, g/L | 139.78 ± 15.83 | 136.51 ± 15.21 | 142.48 ± 15.88 | 0.005 | 139.24 ± 16.94 | 134.48 ± 15.55 | 143.19 ± 17.17 | 0.005 | 0.738 |
| Number of polyps | 3.59 ± 2.48 | 2.77 ± 2.46 | 0.068 | ||||||
| Diameter max, cm | 1.23 ± 0.67 | 1.48 ± 0.87 | 0.098 | ||||||
CRA, colorectal adenoma; BMI, body mass index; HBP, history of high blood pressure; ALB, albumin; TC, total cholesterol; HDL-C, high-density lipoprotein-cholesterol; LDL-C, low-density lipoprotein-cholesterol; TG, triglyceride; FBG, fasting blood-glucose; WBC, white blood cell count; NEUT, neutrophil count; LYM, lymphocyte count; MONO, monocyte count; RBC, red blood cell count; HGB, hemoglobin. “Diameter max” refers to the maximum diameter of the polyps.
Data were expressed as n (%) or mean ± standard deviation.
P value* for difference between the CRA and Control groups in the development and validation cohorts, respectively.
P value# for difference between the development cohort and validation cohort.
3.2. Univariate logistic regression analysis for identifying the risk factors associated with CRA
In the development cohort, 10 clinical indicators were statistically significant after single factor analysis, such as age (P<0.001), history of hypertension (P<0.01), ALB (P<0.001), TC (P<0.001), HDL-C (P<0.001), LDL-C (P<0.001), FBG (P<0.05), MONO (P<0.001), RBC (P<0.01) and HGB (P<0.01) (Table 2). These indicators would be further analyzed by multivariate logistic regression. Additionally, the P value for sex comparison between the CRA group and control group in the development cohort was 0.073, which was close to 0.05. Because sex is a crucial factor in clinical diseases, it would also be included as a variable in the multivariate logistic regression model.
Table 2.
Univariate and multivariate logistic regression analysis of risk factors for CRA based on the development cohort.
|
Variables |
Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| OR | 95 % CI | P | OR | 95 % CI | P | |
| Sex, n (%) | ||||||
| Male | 0.614 | 0.360–1.047 | 0.073 | 2.957 | 1.142–7.658 | 0.026* |
| Female | 1.000 | |||||
| Age, years | 1.149 | 1.106–1.194 | <0.001 | 1.105 | 1.052–1.161 | <0.001*** |
| BMI, kg/m2 | 0.975 | 0.909–1.046 | 0.483 | |||
| HBP, n (%) | ||||||
| Yes | 0.428 | 0.229–0.803 | 0.008 | |||
| No | 1.000 | |||||
| ALB, g/L | 0.482 | 0.399–0.582 | <0.001 | 0.525 | 0.425–0.648 | <0.001*** |
| TC, mmol/L | 0.512 | 0.370–0.709 | <0.001 | |||
| HDL-C, mmol/L | 0.231 | 0.104–0.515 | <0.001 | |||
| LDL-C, mmol/L | 0.522 | 0.373–0.729 | <0.001 | |||
| TG, mmol/L | 0.873 | 0.613–1.245 | 0.454 | |||
| FBG, mmol/L | 1.425 | 1.016–2.000 | 0.04 | |||
| WBC, 109/L | 1.023 | 0.854–1.225 | 0.805 | |||
| NEUT, 109/L | 0.975 | 0.779–1.219 | 0.823 | |||
| LYM, 109/L | 1.450 | 0.953–2.206 | 0.083 | |||
| MONO, 108/L | 1.797 | 1.364–2.369 | <0.001 | 1.903 | 1.213–2.988 | 0.005** |
| RBC, 1012/L | 0.374 | 0.210–0.666 | 0.001 | |||
| HGB, g/L | 0.976 | 0.959–0.993 | 0.006 | |||
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.3. Multivariate logistic regression analysis and construction of a nomogram model
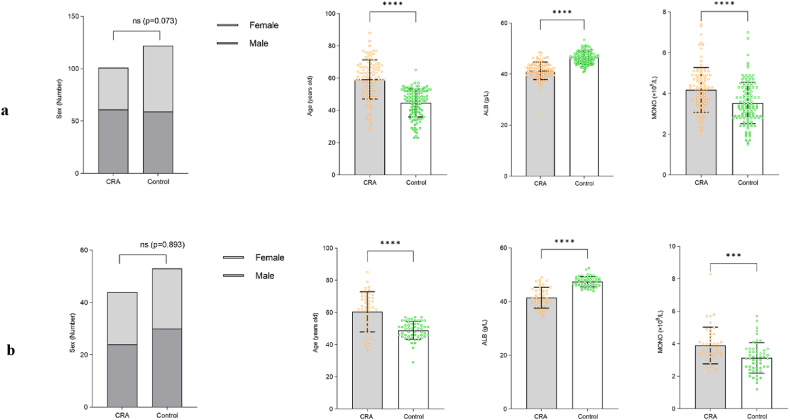
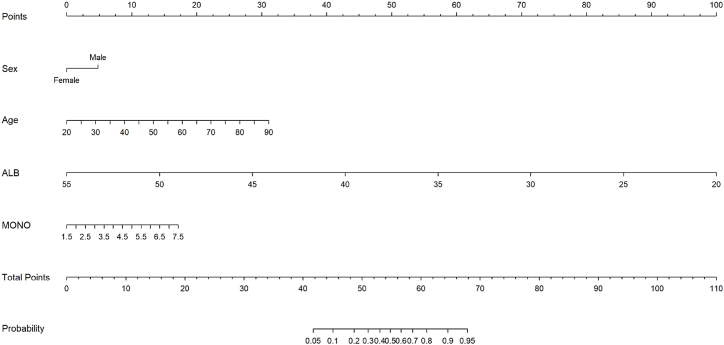
By conducting multivariate logistic regression analysis, four valuable risk factors (sex, age, ALB and MONO) were selected as predictors to establish the predictive model (Table 2), which were compared between the CRA group and control group in both the development cohort and validation cohort, and these results were illustrated in Fig. 2a and b. Hosmer-Lemeshow test result for the predictive model was good (p = 0.940). A nomogram was constructed based on the multivariate logistic regression model (Fig. 3). In order to provide more options for clinical use, we listed the formula for the predictive model here, logit(P) = 1.084*sex + 0.100*age-0.644*ALB + 0.644*MONO + 20.103; P = 1/[1+e−(1.084*sex+0.100*age−0.644*ALB+0.644*MONO +20.103)].
Fig. 2.
Comparison of four risk factors for CRA in the development and validation cohort. (a) for the development cohort; (b) for the validation cohort.
CRA, colorectal adenoma; ALB, albumin; MONO, monocyte count.
Error bar stands for standard deviation. ***P<0.001; ****P<0.0001.
Fig. 3.
Nomogram model for the risk prediction of colorectal adenoma. ALB, albumin; MONO, monocyte count.Each variable was scored on a scale of 0–100, and the total score for predicting CRA was obtained by summing up the scores corresponding to each variable. Different total score corresponded to different probability of CRA.
3.4. Evaluating the performance of the predictive model built by the development cohort
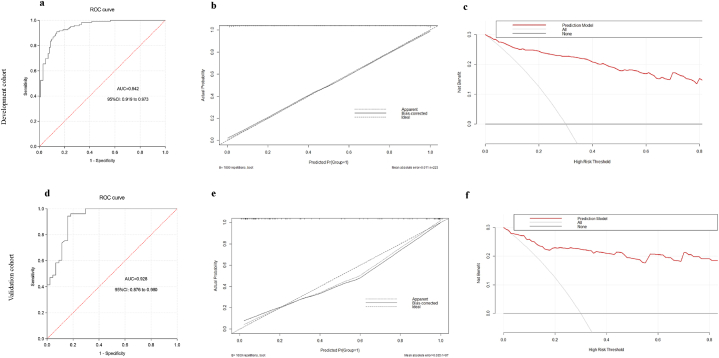
ROC curve analysis revealed the AUC of this predictive model in the development cohort was 0.946 (95%CI: 0.919–0.973), demonstrating a good discrimination of the predictive model (Fig. 4a). According to the maximum YDI (0.771), the corresponding cut-off value was 0.501, of which SEN, SPE, PLR and NLR of the predictive model were 0.861, 0.910, 9.567 and 0.153, respectively. Calibration plot demonstrated excellent concordance between the actual and nomogram-predicted probabilities in the development cohort (Fig. 4b). DCA showed the nomogram model was effective in clinical practice (Fig. 4c).
Fig. 4.
ROC curves, calibration plots and clinical decision curve of the prediction model in the development cohort and validation cohort. ROC curve, receiver operating characteristic curve (a) and (d), (b) and (e), (c) and (f) represent the ROC curve, calibration curve, and clinical decision curve in the development and validation cohort, respectively.
3.5. Internal validation and external validation
The bias-corrected AUC derived from internal validation using the bootstrap method was 0.909 (95 % CI: 0.869–0.940), similar to the AUC (0.946; 95%CI: 0.919–0.973) calculated in the development cohort. In order to further validate the generalizability of the predictive model, we conducted an external validation using an independent validation cohort. ROC analysis showed that the nomogram model had an AUC of 0.928 (95%CI: 0.876–0.980), which indicated this model also has an excellent discrimination ability in the validation cohort (Fig. 4d). According to the maximum YDI (0.784), the corresponding cut-off value was 0.288, of which SEN, SPE, PLR and NLR of the predictive model were 0.841, 0.943, 14.754 and 0.169, respectively. Calibration curve demonstrated good consistency between actual observations and nomogram predictions in the external validation cohort (Fig. 4e). The decision curve of this predictive model outperformed both extreme lines across a wide range of threshold probabilities, which also demonstrated the excellent performance of the nomogram model in clinical practice in external validation. (Fig. 4f).
Calibration plots show the apparent (actual), bias-corrected (adjusted), and ideal (100 % agreement) curves with 1000 resampling bootstrap. The x-axis represents predicted probability of the colorectal adenoma model; y-axis represents actual probability of colorectal adenoma.
Clinical decision curve: The x-axis means threshold probability of colorectal adenoma, while the y-axis means net benefit. The black line stands for net benefit for the predict-none-patients as colorectal adenoma, the gray line stands for net benefit for the predict-all-patients as colorectal adenoma, and the red line represents net benefit for the model.
4. Discussion
CRC mainly originates from CRA, which may enlarge gradually or progress from atypical hyperplasia to carcinoma in situ, and eventually to invasive carcinoma [8]. Developing a simple and convenient predictive model for CRA, instead of relying on the complex and invasive colonoscopy procedure, can facilitate early intervention in disease management and effectively reduce the incidence of CRC. In this study, we have developed and validated a nomogram model to accurately predict the risk of CRA. The four predictors included in the model were sex, age, ALB and MONO.
CRA is more common in men than in women [6,22,32,33], and our findings are in line with this observation. One study reported the risk of adenomas was 1.77 times in men compared with women [34], however, in our study, the risk of CRA was about three times higher in men than in women, higher than that of the previous study. Another respective study showed that male was a predictor for colorectal polyps (OR = 3.810), which is consistent with our findings [35]. Many studies have confirmed that smoking and alcohol consumption were the risk factors for CRA [36,37], and these habits were significantly more common in men than women [38], which may be one of the reasons why CRA is more common in men. A study on the prediction for CRC also showed that male was a risk factor (OR = 2.04). Therefore, sex is a common risk factor for CRA and CRC [18]. Age is also a common risk factor closely related to the development of CRA and CRC [32,39]. Numerous studies have shown that the prevalence of adenomas increases with age, escalating by 10 %–15 % from individuals aged 50–55 years [6,40]. In the present study, the age of the CRA group was observed to be higher than that of the control group in both the development cohort and validation cohort. Of the 145 CRA patients, 77.24 % were over 50 years old, similar to the findings of the previous study. Therefore, more attention should be given to patients over 50 years old in CRC screening. Our study indicated that for each additional year of age, there is a 0.105-fold increase in the risk of CRA, which was similar to a study on CRC [18]. Serum albumin is recognized as a nutritional status indicator and a negative inflammatory marker, which is associated with acute and chronic inflammation [41,42]. The plasmatic concentration of albumin can be reduced by inflammatory conditions through the alteration of hepatic protein metabolism and induction of capillary leakage [43,44]. Numerous studies have proved that chronic inflammation is highly correlated with the pathogenesis of colon polyps and colorectal cancer [[45], [46], [47]]. Some studies demonstrated that Glasgow prognostic score (GPS), which includes albumin, can serve as a prognostic factor for advanced gastrointestinal cancers, including CRC [48], and hypoalbuminemia is associated with poor prognosis of CRC [49,50]. So far, few researches have explored the correlation between albumin and CRA. In this study, we observed that higher level of albumin was related to a decreased risk of CRA, with the odds ratio decreasing by 50 % for every 1 g/L increase in concentration. Thus, albumin serves as a protective predictor for CRA, which is a new finding of this study. There is ample evidence indicating that monocytes are closely associated with both inflammatory status and immunosuppression. Increased peripheral monocyte count (MC) could be an indicator for inflammatory diseases, and exhibit tumor-promoting functions, such as angiogenesis and invasion [[51], [52], [53], [54], [55]]. Many studies have demonstrated elevated MC is an independent prognostic factor for decreased cancer-specific survival (CSS) in CRC patients [56,57]. Monocyte chemoattractant protein 1 (MCP-1) has been demonstrated to be a potent chemotactic factor for monocytes, with higher expression levels in colonic adenoma epithelial cells compared to normal ones. It can recruit circulating monocyte to the tumor microenvironment, thereby facilitating tumor progression [58,59]. Another basic study conducted in mice also confirmed that monocytes were significantly increased in colon adenoma [60]. These studies supported our findings: MC was an independent risk factor for CRA, which could be associated with the fact that CRA is associated with inflammation and is precursor lesion for CRC. Combing with previous findings, we speculate that MC may be involved in the initiation and progression of colorectal adenoma to carcinoma, which deserves further study. A study showed that monocyte products played an important role in reducing albumin production during inflammation [54]. This study further supported this finding, as the CRA group exhibited higher levels of MC and lower concentrations of albumin.
Studies have shown that dyslipidemia, such as high triglyceride (TG) levels, increased low-density lipoprotein cholesterol (LDL-C) and decreased high-density lipoprotein cholesterol (HDL-C) levels are independent risk factors for adenomatous polyp [61,62]. However, our study did not yield similar results, possibly because of the small sample size. Additionally, although HGB and HDL-C were not found to be protective predictor in this study, their levels were lower in the CRA group compared with the control group. A study demonstrated that HGB and HDL-C were protective factors for CRC [18], which was similar to our findings in CRA. Further researches are required to investigate the roles of HGB and dyslipidemia in CRA and CRC.
In this study, we conducted both internal and external validation to evaluate the performance of our model in terms of discrimination, calibration, and clinical practicability. The results demonstrated the model exhibited excellent performance. Additionally, there were only four predictors in the model, which are easy to obtain in clinical practice with good operability. By utilizing this predictive model, we aim to identify individuals at risk for CRA, and guide them to undergo colonoscopy as early as possible in order to detect and intervene early.
Our study has the following limitations. Firstly, because of a single-center study, the data were collected from only one medical institution, which might lead to selection bias. Secondly, as this is a retrospective study, the information extraction was mainly based on electronic medical records. Some relative information such as family history and lifestyle may not be recorded completely, so these contents were not analyzed in this study, which may weaken the discriminating ability of the model. Thirdly, the external validation in this study was temporal validation, meaning that the data was collected from the same hospital at different time with a small sample size. Therefore, the clinical applicability of this model may be lower than that derived from multi-center external validation. Fourthly, the characteristics of polyps, such as anatomical location, stratification of pathological type, etc., were not involved in this study, which will be studied in our next series of studies.
5. Conclusion
In conclusion, we have developed a reliable and accurate nomogram predictive model for CRA with LGIN in this study, which can aid in the early detection and treatment of high-risk patients, so as to reduce the incidence and mortality of CRC. In the future, this model will need to be validated in a large number of populations across multiple centers for improvement.
Funding
Not applicable.
Ethics statement
This study complied with all regulations of Helsinki Declaration. The research protocol was approved by the Ethics Committee of Beijing Chao-Yang Hospital Affiliate of Capital Medical University (Approval Number: 2023-5-25-3). Informed consent was obtained from all participants in this study.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Huaguang Wang: Writing – original draft, Validation, Software, Methodology, Formal analysis, Data curation. Xinjuan Liu: Writing – review & editing, Methodology, Investigation, Conceptualization. Jiang Long: Writing – original draft, Methodology, Formal analysis, Conceptualization. Jincan Huang: Software, Methodology, Formal analysis. Shaocheng Lyu: Validation, Methodology, Data curation, Conceptualization. Xin Zhao: Software, Methodology, Formal analysis. Baocheng Zhao: Validation, Data curation. Qiang He: Supervision, Project administration, Investigation. Zhuoling An: Supervision, Project administration, Investigation. Jianyu Hao: Supervision, Project administration, Investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20996.
Contributor Information
Qiang He, Email: heqiang@bjcyh.com.
Zhuoling An, Email: anzhuoling@bjcyh.com.
Jianyu Hao, Email: haojianyu@ccmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Patel S.G., Karlitz J.J., Yen T., Lieu C.H., Boland C.R. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 2022;7:262–274. doi: 10.1016/S2468-1253(21)00426-X. [DOI] [PubMed] [Google Scholar]
- 2.Baidoun F., Elshiwy K., Elkeraie Y., Merjaneh Z., Khoudari G., Sarmini M.T., Gad M., Al-Husseini M., Saad A. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr. Drug Targets. 2021;22:998–1009. doi: 10.2174/1389450121999201117115717. [DOI] [PubMed] [Google Scholar]
- 3.Sinicrope F.A. Increasing incidence of early-onset colorectal cancer. N. Engl. J. Med. 2022;386:1547–1558. doi: 10.1056/NEJMra2200869. [DOI] [PubMed] [Google Scholar]
- 4.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 5.Maomao C., He L., Dianqin S., Siyi H., Xinxin Y., Fan Y., Shaoli Z., Changfa X., Lin L., Ji P., Wanqing C. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19:1121–1138. doi: 10.20892/j.issn.2095-3941.2022.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sninsky J.A., Shore B.M., Lupu G.V., Crockett S.D. Risk factors for colorectal polyps and cancer. Gastrointest Endosc Clin N Am. 2022;32:195–213. doi: 10.1016/j.giec.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., Bos J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 8.Tariq H., Kamal M.U., Patel H., Patel R., Ameen M., Elona S., Khalifa M., Azam S., Zhang A., Kumar K., Baiomi A., Shaikh D., Makker J. Predicting the presence of adenomatous polyps during colonoscopy with national cancer institute colorectal cancer risk-assessment tool. World J. Gastroenterol. 2018;24:3919–3926. doi: 10.3748/wjg.v24.i34.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedlak J.C., Yilmaz Ö H., Roper J. Metabolism and colorectal cancer. Annu. Rev. Pathol. 2023;18:467–492. doi: 10.1146/annurev-pathmechdis-031521-041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad R., Singh J.K., Wunnava A., Al-Obeed O., Abdulla M., Srivastava S.K. Emerging trends in colorectal cancer: dysregulated signaling pathways. Int. J. Mol. Med. 2021;47 doi: 10.3892/ijmm.2021.4847. (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng X., Song J., Yu C., Zhou Z., Liu X., Yu J., Xu G., Yang J., He X., Bai X., Luo Y., Bao Y., Li H., Yang L., Xu M., Song N., Su X., Xu J., Ma X., Shi H. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis. Signal Transduct. Targeted Ther. 2022;7:60. doi: 10.1038/s41392-022-00881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayman C.V., Vyas D. Screening colonoscopy: the present and the future. World J. Gastroenterol. 2021;27:233–239. doi: 10.3748/wjg.v27.i3.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanwar S., Vijayalakshmi S., Sabharwal M., Kaur M., AlZubi A.A., Lee H.N. Detection and classification of colorectal polyp using deep learning. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/2805607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Xie Y.H., Gao Q.Y., Cai G.X., Sun X.M., Sun X.M., Zou T.H., Chen H.M., Yu S.Y., Qiu Y.W., Gu W.Q., Chen X.Y., Cui Y., Sun D., Liu Z.J., Cai S.J., Xu J., Chen Y.X., Fang J.Y. Fecal Clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan J., Xin L., Ma Y.F., Hu L.H., Li Z.S. Colonoscopy reduces colorectal cancer incidence and mortality in patients with non-malignant findings: a meta-analysis. Am. J. Gastroenterol. 2016;111:355–365. doi: 10.1038/ajg.2015.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., Chen G., Li Z., Zhang P., Li X., Gan D., Cao X., Du H., Zhang J., Zhang L., Ye Y. Colonoscopic screening is associated with reduced Colorectal Cancer incidence and mortality: a systematic review and meta-analysis. J. Cancer. 2020;11:5953–5970. doi: 10.7150/jca.46661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zauber A.G., Winawer S.J., O'Brien M.J., Lansdorp-Vogelaar I., van Ballegooijen M., Hankey B.F., Shi W., Bond J.H., Schapiro M., Panish J.F., Stewart E.T., Waye J.D. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J.Y., Wang Y.T., Li X.L., Shao Y., Han Z.Y., Zhang J., Yang L.B., Deng J., Li T., Wu T., Lu X.L., Cheng Y. Prediction model using readily available clinical data for colorectal cancer in a Chinese population. Am. J. Med. Sci. 2022;364:59–65. doi: 10.1016/j.amjms.2022.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Ma X., Dong H., Qu B., Yang T., Xu M., Sheng G., Hu J., Liu A. Construction and assessment of a joint prediction model and nomogram for colorectal cancer. J. Gastrointest. Oncol. 2022;13:2406–2414. doi: 10.21037/jgo-22-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y., Hua X., Win A.K., MacInnis R.J., Gallinger S., Marchand L.L., Lindor N.M., Baron J.A., Hopper J.L., Dowty J.G., Antoniou A.C., Zheng J., Jenkins M.A., Newcomb P.A. A new comprehensive colorectal cancer risk prediction model incorporating family history, personal characteristics, and environmental factors. Cancer Epidemiol. Biomarkers Prev. 2020;29:549–557. doi: 10.1158/1055-9965.EPI-19-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cueto-López N., García-Ordás M.T., Dávila-Batista V., Moreno V., Aragonés N., Alaiz-Rodríguez R. A comparative study on feature selection for a risk prediction model for colorectal cancer. Comput. Methods Progr. Biomed. 2019;177:219–229. doi: 10.1016/j.cmpb.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Ghajari H., Sadeghi A., Khodakarim S., Zali M., Nazari S.S.H. Designing a predictive model for colorectal neoplasia diagnosis based on clinical and laboratory findings in colonoscopy candidate patients. J. Gastrointest. Cancer. 2022;53:880–887. doi: 10.1007/s12029-021-00737-4. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y., Liu Y., Yin X., Zhang T., Hao Y., Zhang P., Yang Y., Gao Z., Liu S., Yu S., Li H., Wang G. Establishment of clinical predictive model based on the study of influence factors in patients with colorectal polyps. Front Surg. 2023;10 doi: 10.3389/fsurg.2023.1077175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bateman A.C. Pathology of colorectal polyps and cancer. Surgery. 2023;41:15–21. [Google Scholar]
- 25.Endoscopic Classification Review Group Update on the paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy. 2005;37:570–578. doi: 10.1055/s-2005-861352. [DOI] [PubMed] [Google Scholar]
- 26.Chinese Medical Association, Endoscopy Digestive, Branch Gastroenterology. Consensus on screening, diagnosis and treatment of early colorectal cancer and precancerous lesions in China. Chin. J. Intern. Med. 2015;54:375–389. (in Chinese) [Google Scholar]
- 27.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 28.Perkins N.J., Schisterman E.F. The inconsistency of "optimal" cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am. J. Epidemiol. 2006;163:670–675. doi: 10.1093/aje/kwj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg E.W., Vickers A.J., Cook N.R., Gerds T., Gonen M., Obuchowski N., Pencina M.J., Kattan M.W. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Rousson V., Lee W.C., Ferdynus C., Chen M., Qian X., Guo Y. Decision curve analysis: a technical note. Ann. Transl. Med. 2018;6:308. doi: 10.21037/atm.2018.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Symer M., Connolly J., Yeo H. Management of the malignant colorectal polyp. Curr. Probl. Surg. 2022;59 doi: 10.1016/j.cpsurg.2022.101124. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X.X., Liu M.H., Wang R.L., Tian T. Effect of gender and age on the correlation between Helicobacter pylori and colorectal adenomatous polyps in a Chinese urban population: a single center study. Gastroenterol Res Pract. 2020;2020 doi: 10.1155/2020/8596038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corley D.A., Jensen C.D., Marks A.R., Zhao W.K., de Boer J., Levin T.R., Doubeni C., Fireman B.H., Quesenberry C.P. Variation of adenoma prevalence by age, sex, race, and colon location in a large population: implications for screening and quality programs. Clin. Gastroenterol. Hepatol. 2013;11:172–180. doi: 10.1016/j.cgh.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Wang Y., Zhu K., Wang X., Yu W., Li S. Predictors for colorectal polyps in an asymptomatic population undergoing medical check-ups. Surg. Laparosc. Endosc. Percutaneous Tech. 2023;33:108–114. doi: 10.1097/SLE.0000000000001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H., Shen Z., Zhao J., Zhou Z., Xu Y. Distribution characteristics and risk factors of colorectal adenomas. Zhonghua wei chang wai ke za zhi. 2018;21:678–684. [PubMed] [Google Scholar]
- 37.Čebohin M., Samardžić S., Marjanović K., Tot Vesić M., Kralik K., Bartulić A., Hnatešen D. Adenoma characteristics and the influence of alcohol and cigarette consumption on the development of advanced colorectal adenomas. Int. J. Environ. Res. Publ. Health. 2020;17:8296. doi: 10.3390/ijerph17228296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakai K., Watari J., Tozawa K., Tamura A., Hara K., Yamasaki T., Kondo T., Kono T., Tomita T., Ohda Y., Oshima T., Fukui H., Sakurai J., Kim Y., Hayakawa Y., Fujisawa T., Morimoto T., Miwa H. Sex differences in associations among metabolic syndrome, obesity, related biomarkers, and colorectal adenomatous polyp risk in a Japanese population. J. Clin. Biochem. Nutr. 2018;63:154–163. doi: 10.3164/jcbn.18-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawwas M.F. Adenoma detection rate and risk of colorectal cancer and death. N. Engl. J. Med. 2014;370:2539–2540. doi: 10.1056/NEJMc1405329. [DOI] [PubMed] [Google Scholar]
- 40.Peery A.F., Crockett S.D., Murphy C.C., Lund J.L., Dellon E.S., Williams J.L., Jensen E.T., Shaheen N.J., Barritt A.S., Lieber S.R., Kochar B., Barnes E.L., Fan Y.C., Pate V., Galanko J., Baron T.H., Sandler R.S. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2018. Gastroenterology. 2019;156:254–272.e211. doi: 10.1053/j.gastro.2018.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chojkier M. Inhibition of albumin synthesis in chronic diseases: molecular mechanisms. J. Clin. Gastroenterol. 2005;39:S143–S146. doi: 10.1097/01.mcg.0000155514.17715.39. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto T., Kawada K., Hida K., Matsusue R., Itatani Y., Mizuno R., Yamaguchi T., Ikai I., Sakai Y. Combination of lymphocyte count and albumin concentration as a new prognostic biomarker for rectal cancer. Sci. Rep. 2021;11:5027. doi: 10.1038/s41598-021-84475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levitt D.G., Levitt M.D. Human serum albumin homeostasis: a new look at the roles of synthesis, catabolism, renal and gastrointestinal excretion, and the clinical value of serum albumin measurements. Int. J. Gen. Med. 2016;9:229–255. doi: 10.2147/IJGM.S102819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckart A., Struja T., Kutz A., Baumgartner A., Baumgartner T., Zurfluh S., Neeser O., Huber A., Stanga Z., Mueller B., Schuetz P. Relationship of nutritional status, inflammation, and serum albumin levels during acute illness: a prospective study. Am. J. Med. 2020;133:713–722.e7. doi: 10.1016/j.amjmed.2019.10.031. [DOI] [PubMed] [Google Scholar]
- 45.Zhang F., Qiao S. Research progress on the relationship between inflammation and colorectal cancer. Ann Gastroenterol Surg. 2022;6:204–211. doi: 10.1002/ags3.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Austin Pickens C., Yin Z., Sordillo L.M., Fenton J.I. Arachidonic acid-derived hydroxyeicosatetraenoic acids are positively associated with colon polyps in adult males: a cross-sectional study. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grahn S.W., Varma M.G. Factors that increase risk of colon polyps. Clin. Colon Rectal Surg. 2008;21:247–255. doi: 10.1055/s-0028-1089939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishizuka M., Nagata H., Takagi K., Horie T., Kubota K. Inflammation-based prognostic score is a novel predictor of postoperative outcome in patients with colorectal cancer. Ann. Surg. 2007;246:1047–1051. doi: 10.1097/SLA.0b013e3181454171. [DOI] [PubMed] [Google Scholar]
- 49.Kim J.Y., Yee J., Park T.I., Shin S.Y., Ha M.H., Gwak H.S. Risk scoring System of mortality and prediction model of hospital stay for critically ill patients receiving parenteral nutrition. Health Care. 2021;9:853. doi: 10.3390/healthcare9070853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang S.P., Wang T.J., Huang C.C., Chang S.C., Liang S.Y., Yu C.H. Influence of albumin and physical activity on postoperative recovery in patients with colorectal cancer: an observational study. Eur. J. Oncol. Nurs. 2021;54 doi: 10.1016/j.ejon.2021.102027. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto M., Saito H., Hara K., Sugezawa K., Uejima C., Tanio A., Tada Y., Kihara K., Sakamoto T., Honjo S., Fujiwara Y. Combination of C-reactive protein and monocyte count is a useful prognostic indicator for patients with colorectal cancer. In Vivo. 2020;34:299–305. doi: 10.21873/invivo.11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auffray C., Sieweke M.H., Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 53.Olingy C.E., Dinh H.Q., Hedrick C.C. Monocyte heterogeneity and functions in cancer. J. Leukoc. Biol. 2019;106:309–322. doi: 10.1002/JLB.4RI0818-311R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chittezhath M., Dhillon M.K., Lim J.Y., Laoui D., Shalova I.N., Teo Y.L., Chen J., Kamaraj R., Raman L., Lum J., Thamboo T.P., Chiong E., Zolezzi F., Yang H., Van Ginderachter J.A., Poidinger M., Wong A.S., Biswas S.K. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41:815–829. doi: 10.1016/j.immuni.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Augier S., Ciucci T., Luci C., Carle G.F., Blin-Wakkach C., Wakkach A. Inflammatory blood monocytes contribute to tumor development and represent a privileged target to improve host immunosurveillance. J. Immunol. 2010;185:7165–7173. doi: 10.4049/jimmunol.0902583. [DOI] [PubMed] [Google Scholar]
- 56.Tanio A., Saito H., Uejima C., Takaya S., Yamamoto M., Tokuyasu N., Sakamoto T., Honjo S., Ashida K., Fujiwara Y. A prognostic index for colorectal cancer based on preoperative absolute lymphocyte, monocyte, and neutrophil counts. Surg. Today. 2019;49:245–253. doi: 10.1007/s00595-018-1728-6. [DOI] [PubMed] [Google Scholar]
- 57.Wen S., Chen N., Hu Y., Huang L., Peng J., Yang M., Shen X., Song Y., Xu L. Elevated peripheral absolute monocyte count related to clinicopathological features and poor prognosis in solid tumors: systematic review, meta-analysis, and meta-regression. Cancer Med. 2021;10:1690–1714. doi: 10.1002/cam4.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu T., Guo Z., Song X., Liu L., Dong W., Wang S., Xu M., Yang C., Wang B., Cao H. High‐fat diet‐induced dysbiosis mediates MCP‐1/CCR2 axis‐dependent M2 macrophage polarization and promotes intestinal adenoma‐adenocarcinoma sequence. J. Cell Mol. Med. 2020;24:2648–2662. doi: 10.1111/jcmm.14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka S. Monocyte chemoattractant protein 1 and macrophage cyclooxygenase 2 expression in colonic adenoma. Gut. 2006;55:54–61. doi: 10.1136/gut.2004.059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soncin I., Sheng J., Chen Q., Foo S., Duan K., Lum J., Poidinger M., Zolezzi F., Karjalainen K., Ruedl C. The tumour microenvironment creates a niche for the self-renewal of tumour-promoting macrophages in colon adenoma. Nat. Commun. 2018;9:582. doi: 10.1038/s41467-018-02834-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C.S., Hsu H.S., Li C.I., Jan C.I., Li T.C., Lin W.Y., Lin T., Chen Y.C., Lee C.C., Lin C.C. Central obesity and atherogenic dyslipidemia in metabolic syndrome are associated with increased risk for colorectal adenoma in a Chinese population. BMC Gastroenterol. 2010;10:51. doi: 10.1186/1471-230X-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie C., Wen P., Su J., Li Q., Ren Y., Liu Y., Shen R., Ren J. Elevated serum triglyceride and low-density lipoprotein cholesterol promotes the formation of colorectal polyps. BMC Gastroenterol. 2019;19:195. doi: 10.1186/s12876-019-1115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.