Abstract
Carbon source utilization and phospholipid fatty acid analyses were used to track the rapidly changing microbial community in composting dairy waste. Microbial abilities to utilize common plant sugars increased during composting. Community phospholipid profiles changed significantly over time. Phospholipids suggested the presence of more thermophiles and fewer bacteria with continued compost development.
New techniques to describe functional and physiological diversity are expanding our understanding of community structure and our ability to track changes in dynamic whole communities. Carbon source utilization studies, such as those done with Biolog plates, can distinguish among microbial communities in different composts (6) or soils on the basis of substrate utilization (4) or metabolic potential (17). Fatty acid profiling is used to describe microbial strains or communities or to differentiate among environmental samples by their fatty acid “fingerprint” (8). Phospholipid fatty acids (PLFAs) are rapidly turned over (15) and thus represent the current living community, both qualitatively and quantitatively (10, 14).
The objective of this study was to test the use of carbon source utilization studies and PLFA profiling to detect changes in microbial community structure in developing compost.
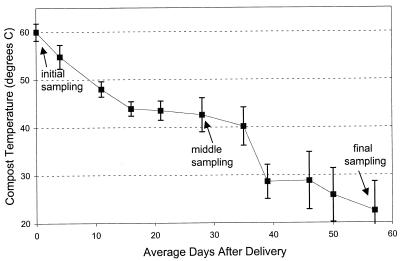
These studies were carried out with 10 compost piles made of separated solids of dairy manure and pine shaving bedding. The initial C/N ratio (determined with a model CHN-600 carbon and nitrogen analyzer [Leco Corp., St. Joseph, Mich.]) was between 55:1 and 60:1. Each pile was approximately 2 by 2.5 by 1.5 m and weighed 3.5 Mg at 65% moisture by wet weight. All piles received an inoculation of Palouse series soil (fine-silty, mixed, mesic Pachic Ultic Haploxeroll) from the surrounding field. Materials were actively composting at the initial sampling, as evidenced by the high temperature of 60°C. The compost temperature fell to near ambient air temperature after an average of 8 weeks of active composting at the experimental site (Fig. 1). The compost pH remained at an average of 8.4, and moisture content was maintained at approximately 60 to 65% water by wet weight throughout development.
FIG. 1.
Temperature at a 60-cm depth within compost piles (n = 10) of separated solids of dairy manure and bedding. Values are means ± standard errors.
Four subsamples of compost were taken from each pile at a 60-cm depth at three times during compost development. Initial samples were taken within 2 days of the material’s delivery from the dairy farm to the experimental site. Middle and final samples were taken on average 4 and 8 weeks, respectively, after initial sampling. The final sampling date was the date on which redox, nitrate, and temperature indicators signaled that composting activity had slowed significantly (18) and the C/N ratio was between 25:1 and 35:1.
Carbon source utilization trials used specially prepared microtiter plates (7) containing l-histidine, γ-aminobutyric acid, d-trehalose, d-galactose, sucrose, and d-fructose. Plate wells were filled with 150 μl of sterilized nutrient broth containing 6.25 mg of one potential carbon source and 50 μg of triphenyl tetrazolium chloride ml of broth−1. Each well was inoculated with 30 μl of a solution of 100 μg of compost ml of 0.1 M CaSO4−1. An incubation time of 72 h was used in all cases to allow growth of slower-growing microbes or of those present in low numbers in the inoculant (19). Color development was measured at 492 nm with a model 2550 microplate reader (Bio-Rad, Hercules, Calif.). No color development was detected in control plates after 72 h.
PLFAs were extracted by the procedure of Petersen and Klug (11) and were methylated by the procedure of Microbial Identification, Inc., Newark, Del. (9). Extracts were dried, resuspended in 200 μl of hexane–methyl tert-butyl ether (1:1), and injected into a gas chromatograph (5890 GC series II; Hewlett-Packard, Wilmington, Del.) equipped with a fused silica capillary column and a flame ionization detector. Extracted PLFAs were identified and quantified with software and standard solutions from Microbial Identification, Inc. Data are the percentage of the PLFA or group in the total of identified PLFAs extracted from each sample.
Principal-component (PC) analysis was used to compare multivariate data obtained from PLFA profiles (12). To enhance PC analysis and sample differentiation, those PLFAs with a loading value greater than | 0.5 | in PCs 1 through 5 of the initial analysis were used in a second iteration of PC analysis. Statistical comparisons of individual measurements or of measurements for groups of PLFAs or PCs were made with a univariate general linear model and Tukey’s honestly significant difference test and SAS software (13). Each load of raw material was considered one block in the statistical model design because the loads differed slightly in material and time of development.
Carbon substrate utilization tests indicated that microbial utilization of γ-aminobutyric acid increased over time (P = 0.05), while histidine was utilized at similar levels at all sampling times. The compost microbial communities’ ability to utilize the common plant sugars sucrose, galactose, and fructose increased during composting (P < 0.05), particularly between the initial and middle sampling times. Trehalose utilization decreased during composting (P < 0.01). Trehalose is not as common in soil and plant material as the other sugars tested, and it is known to be utilized by a small subset of microbes, primarily yeasts (16). These data show that the functional abilities of the compost community change during composting.
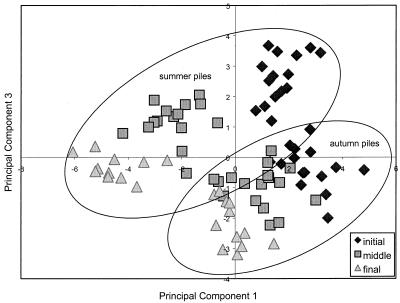
PCs 1, 2, and 3 of PLFA profiles could be used to categorize samples by stage of development (P < 0.001, P < 0.05, and P < 0.01, respectively), indicating that the community’s phospholipid makeup had shifted during composting (Fig. 2). Initial samples tended to rank higher and final samples tended to rank lower in PCs 1, 2, and 3. PCs 1 and 3 also differed by time of year in which composting had taken place. Piles created and developed in summer (May to July) ranked lower in PC 1 (P < 0.05) and higher in PC3 (P < 0.01) than autumn piles (October to December).
FIG. 2.
PCs 1 and 3 of the microbial community’s PLFA makeup in compost at a 60-cm depth.
Certain fatty acids can be used as indicators of the microbial groups that produce them. Two indicators of general bacterial growth, PLFAs 15:0 and 17:0 (20), declined over successive sampling times (P < 0.01 for both) (Table 1). An indicator of aerobic bacteria, 16:1ω7c (5), also decreased over time (P < 0.01). Indicator PLFAs gave disparate results concerning the prevalence of anaerobic organisms.
TABLE 1.
Indicator PLFAs of microbial groups in dairy waste compost at a 60-cm depth
| Organisms and indicator PLFAb | % of total PLFAs (n = 10) at sampling timea
|
||
|---|---|---|---|
| Initial | Middle | Final | |
| Eubacteria | |||
| 15:0 | 1.28 A | 1.12 A | 0.65 B |
| 17:0 | 1.34 A | 0.96 B | 0.59 C |
| Fungi | |||
| 18:2ω6c | 3.93 A | 4.61 A | 3.94 A |
| 18:3ω6c | 0.66 A | 0.94 A | 0.44 A |
| Aerobic organisms | |||
| 16:1ω7c | 4.78 A | 3.83 A | 2.03 B |
| Anaerobic organisms | |||
| 14:1ω7c DMA | 0.03 A | 0 A | 0 A |
| 16:1ω9c DMA | 0.15 A | 0 B | 0 B |
| Cyclo 17:0 | 3.69 A | 2.84 B | 2.00 C |
| Cyclo 19:0 | 4.58 B | 6.72 A | 7.59 A |
| Total cyclo compounds | 8.28 A | 9.56 A | 9.59 A |
| Thermophilic organisms | |||
| 15:0 iso | 4.82 B | 7.26 A | 7.15 A |
| 17:0 iso | 2.32 B | 5.43 A | 5.15 A |
| Iso-branched compounds | 16.6 B | 28.5 A | 28.9 A |
| Total branched compounds | 26.3 B | 35.8 A | 35.0 A |
| Nonthermophilic organisms | |||
| Anteiso | 9.57 A | 7.29 AB | 6.15 B |
| Unsaturated | 22.6 B | 29.4 A | 30.2 A |
Neither of the PLFA markers of fungi, 18:2ω6c and 18:3ω6c (2), changed significantly over time (Table 1). The proportion of fungi in the community may have varied little with compost maturation, although the types of fungi present or active at any stage may have varied significantly (1).
The temperature in the compost piles was maximal at the initial sampling time and dropped steadily over the period of active composting (Fig. 1). However, PLFA patterns indicate an increase, and then a plateau, of biomarkers of thermophilic organisms (Table 1). Although the temperatures were highest at the initial sampling, the thermophilic community may not have been well established in this fresh material. Composts maintained a thermophilic temperature (>40°C) through the middle sampling time, allowing further growth of the thermophilic community. Chang and Hudson (1) also found that thermophilic or thermotolerant fungi could increase in a compost pile even as the compost temperature decreased. In their study, numbers of thermophilic fungi increased from 0 to 107 g−1 as compost cooled from 70 to 50°C. Culturable populations of thermophilic fungi and bacteria did not decrease as compost cooled even to 15°C. More unsaturated fatty acids can be expected with falling temperatures because the half-life of the desaturase enzyme increases with decreasing incubation temperature (3).
It has long been surmised that the microbial community in compost undergoes successional changes as the compost develops. This study shows that carbon source utilization tests and PLFA analysis can be used to track changes in dynamic microbial communities as their environments change.
Acknowledgments
This research was supported by funds from the National Science Foundation Graduate Fellows Program and the Washington State University Department of Crop & Soil Sciences. The USDA-ARS Land Management and Water Conservation Unit provided laboratory equipment and assistance.
REFERENCES
- 1.Chang Y, Hudson H J. The fungi of wheat straw compost. Trans Br Mycol Soc. 1967;50:649–666. [Google Scholar]
- 2.Federle T W. Microbial distribution in soil—new techniques. In: Megusar F, Gantar M, editors. Perspectives in microbial ecology. Proceedings of the Fourth International Symposium on Microbial Ecology. Ljubljana, Slovenia: Slovene Society for Microbiology; 1986. pp. 493–498. [Google Scholar]
- 3.Fulco A J, Fujii D K. Adaptive regulation of membrane lipid biosynthesis in Bacilli by environmental temperature. In: Kates M, Kuksis A, editors. Membrane fluidity: biophysical techniques and cellular regulation. Totowa, N.J: Humana Press; 1980. pp. 77–98. [Google Scholar]
- 4.Garland J L, Mills A L. A community-level physiological approach for studying microbial communities. In: Ritz K, et al., editors. Beyond the biomass: compositional and functional analysis of soil microbial communities. New York, N.Y: John Wiley; 1994. pp. 77–83. [Google Scholar]
- 5.Guckert J B, Antworth C P, Nichols P D, White D C. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol Ecol. 1985;31:147–158. [Google Scholar]
- 6.Insam H, Amor K, Renner M, Crepaz C. Changes in functional abilities of the microbial community during composting of manure. Microb Ecol. 1996;31:77–87. doi: 10.1007/BF00175077. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy A C. Carbon utilization and fatty acid profiles for characterization of bacteria. In: Weaver R W, et al., editors. Methods of soil analysis, part 2: microbiological and biochemical properties. Madison, Wis: Soil Science Society of America, Inc.; 1994. pp. 543–556. [Google Scholar]
- 8.Kennedy A C, Busacca A J. Particulate matter: health and regulatory issues. Proceedings of the Air and Waste Management Association. Pittsburgh, Pa: Air and Waste Management Association; 1995. Microbial analysis to identify source of PM-10 material; pp. 670–675. [Google Scholar]
- 9.Microbial Identification, Inc. Microbial identification system. Newark, N.J: Microbial Identification, Inc.; 1993. [Google Scholar]
- 10.Petersen S O, Henriksen K, Blackburn T H, King G M. A comparison of phospholipid and chloroform fumigation analyses for biomass in soil: potentials and limitations. FEMS Microbiol Ecol. 1991;85:257–268. [Google Scholar]
- 11.Petersen S O, Klug M J. Effects of sieving, storage, and incubation temperature on the phospholipid fatty acid profile of a soil microbial community. Appl Environ Microbiol. 1994;60:2421–2430. doi: 10.1128/aem.60.7.2421-2430.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pielou E C. The interpretation of ecological data. New York, N.Y: John Wiley; 1984. [Google Scholar]
- 13.SAS Institute. SAS/STAT user’s guide, version 6. Vol. 2. Cary, N.C: SAS Institute; 1988. [Google Scholar]
- 14.Vestal J R, White D C. Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. BioScience. 1989;39:535–541. [PubMed] [Google Scholar]
- 15.White D C, Davis W M, Nickels J S, King J D, Bobbie R J. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 16.Windholz M, Budavari S, Blumett R F, Otterbein E S, editors. The Merck index. 10th ed. Rahway, N.J: Merck and Co., Inc.; 1983. [Google Scholar]
- 17.Winding A. Fingerprinting bacterial soil communities using Biolog microtiter plates. In: Ritz K, et al., editors. Beyond the biomass: compositional and functional analysis of soil microbial communities. New York, N.Y: John Wiley; 1994. pp. 85–94. [Google Scholar]
- 18.Woods End Research Laboratory. Interpretation guideline for microbiological and biochemical tests. Mt. Vernon, Maine: Woods End Research Laboratory; 1992. [Google Scholar]
- 19.Wünsche L, Brüggemann L, Babel W. Determination of substrate utilization patterns of soil microbial communities: an approach to assess population changes after hydrocarbon pollution. FEMS Microbiol Ecol. 1995;17:295–306. [Google Scholar]
- 20.Zelles L, Rackwitz R, Bai Q Y, Beck T, Beese F. Discrimination of microbial diversity by fatty acid profiles of phospholipids and lipopolysaccharides in differently cultivated soils. In: Collins H P, et al., editors. The significance and regulation of soil biodiversity. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 115–122. [Google Scholar]