Abstract
A gene coding for a de novo peptide sequence containing a metal binding motif was chemically synthesized and expressed in Escherichia coli as a fusion with the maltose binding protein. Bacterial cells expressing the metal binding peptide fusion demonstrated enhanced binding of Cd2+ and Hg2+ compared to bacterial cells lacking the metal binding peptide. The potential use of genetically engineered bacteria as biosorbents for the removal of heavy metals from wastewaters is discussed.
The discharge of heavy metals into the environment due to agricultural, industrial, and military operations and the effect of this pollution on the ecosystem and human health are growing concerns. Recent research in the area of heavy metal removal from wastewaters and sediments has focused on the development of materials with increased affinity, capacity, and selectivity for target metals (7, 8, 30). The use of microorganisms to sequester, precipitate, or alter the oxidation state of various heavy metals has been extensively studied (7, 8, 15, 24, 27). Expression of metallothioneins (1, 17, 20, 24, 25) or metallopeptides (4, 28) to increase the affinity and biosorptive capabilities of bacterial cells for heavy metals is a promising technology for the development of bacterium-based biosorbents. We are studying the expression of metal binding peptides with high affinity and selectivity for target metals in Escherichia coli and the potential use of such peptides as heavy metal biosorbents. We previously reported expression of the Neurospora crassa metallothionein gene in E. coli by fusion to the maltose binding protein of expression vector pMAL (New England Biolabs) (21). This recombinant E. coli organism was shown to remove heavy metals such as Cd2+ from simulated and actual wastewater samples (3, 20). The de novo design of metal binding motifs has been described and offers the potential for the design and expression of peptides with high affinity and selectivity for various metals (5, 9, 12, 14, 23). In addition to the metallothioneins, peptides which contain an abundance of cysteine residues or Cys-Gly motifs have been shown to have a high affinity for Cd2+ and Hg2+ (5, 22). In this study, heavy metal removal by bacterial cells expressing a repetitive metal binding motif, (Cys-Gly-Cys-Cys-Gly)3, expressed as a fusion with the maltose binding protein is reported.
Synthesis of the metal binding gene.
The metal binding gene encodes 21 amino acids with the motif Cys-Gly-Cys-Cys-Gly repeated three times and linked through a three-amino-acid sequence (Fig. 1A). Four overlapping oligonucleotides were synthesized (0.2 μM scale) on an Applied Biosystems 371 DNA synthesizer. Oligonucleotides were purified and subsequently phosphorylated, hybridized, and ligated by using standard molecular biology protocols as described previously (16). The sequences for the four overlapping oligonucleotides used are as follows: SYNTOP1, 5′ GATCCTGTGGTTGCTGTGGCAAAGGTCATTGTGGCTGT 3′; SYNTOP2, 5′ TGCGGCAAAGGTCACTGCGGTTGCTGTGGTA 3′; SYNBOT1, 5′ TTTGCCGCAACAGCCACAATGACCTTTGCCACAGCA ACCACAG 3′; and SYNBOT2, 5′ AGCTTACCACAGCAACCGCAGTGACC 3′.
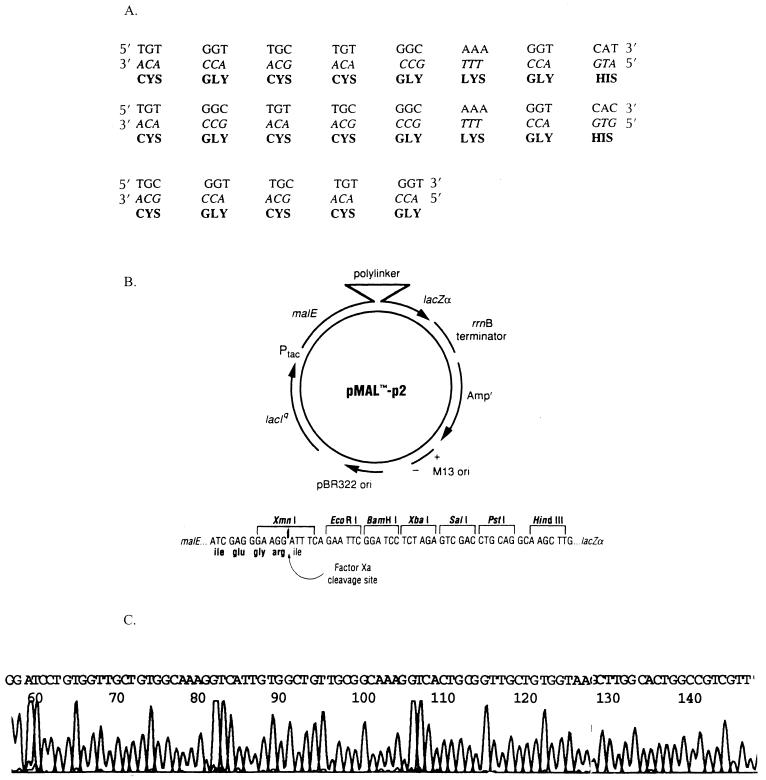
FIG. 1.
Cloning of the metal binding gene into the pMAL-p vector. (A) Nucleotide and amino acid sequence of the metal binding gene. (B) Map of vector pMAL-p. (C) DNA sequence of metal binding gene cloned in pMAL-p. Ori, origin of replication.
The constructed gene contains BamHI and HindIII overhangs at the 5′ and 3′ ends, respectively, for directional cloning into expression vector pMAL-p digested with these restriction enzymes. Following ligation, bacterial cells were transformed and screened for colonies containing the gene insert by hybridization and restriction analysis. DNA sequencing of positive clones (performed by the University of Georgia DNA Sequencing Facility) confirmed the correct in-frame cloning of the gene into pMAL-p, yielding expression vector pMAL-p-syn. Figure 1C presents the DNA sequence of pMAL-p-syn showing the metal binding gene insert flanked by the BamHI and HindIII sites of vector pMAL-p.
Expression and localization within the periplasm.
The expression of small proteins in E. coli has often been problematic due to instability and degradation (18, 21). This is especially true in cases where the protein is repetitive in nature. These problems have been minimized by expressing these proteins as fusion proteins (18, 21, 25, 26). We previously expressed the N. crassa metallothionein gene within the cytoplasm and periplasm of E. coli by using the fusion protein of vector pMAL-c and pMAL-p (encoding the maltose binding protein), respectively (21).
These and other studies have shown that bacterial cells expressing metallothionein within the cytosol are less efficient in accumulating heavy metals from solutions than cells expressing the metallothionein within the periplasm or outer membrane (4, 21, 26). In the present study only the pMAL-p vector was used to express the metal binding gene as a fusion with the maltose binding protein within the periplasm. To determine the level of expression and cellular localization of the metal binding peptide fusion, E. coli TB1 cells were transformed with either the pMAL-p-syn plasmid (containing the metal binding gene) or the parent pMAL-p plasmid as a control. Cells were grown to an optical density of 0.6 in Luria broth (LB) containing ampicillin and induced with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 h as reported previously (21). Induced cells were harvested by centrifugation at 6,000 × g and separated into cytoplasmic and periplasmic fractions utilizing the cold osmotic shock method of Neu and Heppel (19) as described previously (21). The maltose binding protein containing the metal binding peptide fusion and the maltose binding protein lacking the metal binding peptide fusion (from control cells) were purified from the cytoplasmic and periplasmic fractions by using an amylose resin affinity column under conditions recommended by the manufacturer (New England Biolabs) (21). Protein content within each fraction was determined by using the Bradford method (2) with the Bio-Rad (Richmond, Calif.) protein assay kit. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the method of Laemmli (13) as described previously (21). More than 85% of the expressed protein was found to be within the periplasm. This is consistent with results obtained from the expression of metallothionein and other proteins by using the pMAL-p vector (21). β-Galactosidase activity was used as a marker to ensure that no cytoplasmic proteins had leaked into the periplasm during fractionation (21).
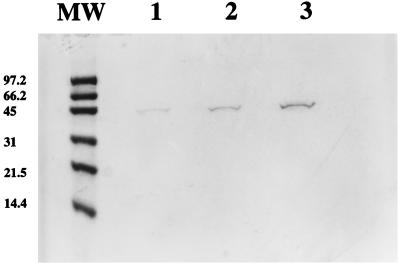
Figure 2 shows an SDS-PAGE gel of the purified metal binding peptide fusion protein isolated from the periplasmic fraction of induced cells. The purified protein appears as a 44-kDa band which is similar in size to the purified protein isolated from cells lacking the metal binding peptide (expressing only the maltose binding protein). To confirm the presence of the metal binding peptide, the purified proteins were subjected to amino acid composition analysis (performed by Baylor College of Medicine Core Protein Sequencing Facility) (21). Since the maltose binding protein does not contain any cysteine residues (6), the expression of cysteine-containing peptides by using this fusion provides a convenient and direct method for determining the authenticity of the desired product. Amino acid analysis of the purified maltose binding protein containing the putative metal binding peptide revealed that the protein contained 1.8 mol% cysteine, reflecting the addition of nine cysteines, whereas the purified maltose binding protein lacking the metal binding peptide contained 0 mol% cysteine, as expected. This is comparable to the 1.5-mol% cysteine content for the expressed metallothionein previously reported, which contains seven cysteine residues (21).
FIG. 2.
SDS-PAGE of purified metal-binding peptide fusion. The gel was stained with 0.1% Coomassie blue. Lane MW contains molecular weight markers, in thousands; lanes 1 to 3 contain 0.5, 1, and 2 μg of purified metal binding peptide, respectively.
Heavy metal removal.
The ability of cells expressing the metal binding peptide fusion to remove heavy metals from solutions was tested and compared to that of control bacteria lacking the metal binding peptide (expressing only the maltose binding protein). Induced bacterial cells (1 liter) were harvested by centrifugation and resuspended in 10 ml of LB. Several aliquots of the resuspended cells were placed in 1.5-ml centrifuge tubes and centrifuged at high speed to pellet the cells for wet weight estimation. Cells (100 mg [wet weight]) were resuspended in 30 ml of LB and then 30 μl of 5 mM stock solution of Cd2+ (CdCl2), Hg2+ (HgCl2), Pb2+ [Pb(NO3)2], or Cu2+ (Cu2SO4) was added, yielding a final concentration of 5 μM. Samples were incubated for 1 h at 37°C. Following incubation, samples were centrifuged to pellet bacterial cells and the supernatant was analyzed for heavy metal content by Accura Analytical (Norcross, Ga.). As shown in Table 1, cells expressing the metal binding peptide fusion efficiently remove Cd2+ and Hg2+ (1.1 and 1.3 nmol removed/mg of cells [wet weight]), whereas control cells lacking the metal binding peptide remove less than 0.1 nmol/mg of cells (wet weight). Removal of Pb2+ and Cu2+ was greater than in control cells, but the difference was not as great as for Cd2+ and Hg2+ (Table 1).
TABLE 1.
Heavy metal removal by cells expressing the metal binding peptide and by control cells expressing only the maltose binding protein
| Heavy metal | Total amt added (nmol)
|
Amt in supernatant after incubation
|
Calculated amt bound (nmol/mg of cells [wet wt])
|
|||
|---|---|---|---|---|---|---|
| Calculated | Measured | Cells expressing peptide | Control cells | Cells expressing peptide | Control cells | |
| Cd2+ | 150 | 149.6 ± 1.8 | 46.0 ± 8.4 | 145.2 ± 3.6 | 1.10 ± 0.12 | 0.09 ± 0.03 |
| Hg2+ | 150 | 137.3 ± 6.1 | 8.3 ± 1.2 | 142.5 ± 12.7 | 1.30 ± 0.02 | NDa |
| Pb2+ | 150 | 136.3 ± 8.9 | 40.6 ± 11.5 | 121.6 ± 6.2 | 0.95 ± 0.11 | 0.14 ± 0.06 |
| Cu2+ | 150 | 145.0 ± 2.8 | 89.3 ± 6.1 | 153.6 ± 9.5 | 0.55 ± 0.06 | ND |
ND, not determined.
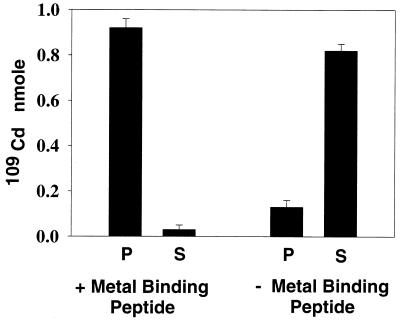
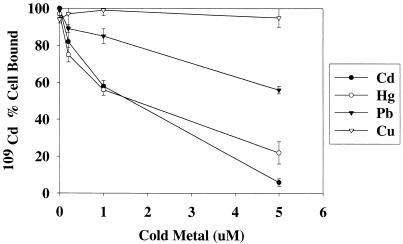
Although the level of heavy metal contamination is variable at different sites, the most promising use for a bacterium-based biosorbent may be the removal of very low levels of target metals in the presence of other metals. In this case, the bulk of the heavy metal is removed through a primary treatment, and a secondary, or polishing, application is added to further reduce the levels of target metals in the effluent (3, 17). The ability of cells expressing the metal binding peptide to remove low levels of heavy metal was tested by using part-per-billion levels of 109Cd. A stock solution was prepared by addition of 0.5 μCi of radioisotopic 109Cd (specific activity, 180 Ci/mol) to a 5-ml aliquot of unlabeled CdCl2, yielding a final specific activity of 5 μCi/mol. Bacterial cells (20 mg [wet weight]) were resuspended in 5 ml of LB and incubated with 50 μl of the 109Cd stock solution, yielding a final concentration of 0.2 μM (approximately 22 ppb). Samples were incubated for 1 h at 37°C and then centrifuged to pellet the bacterial cells. Radioactivity was measured in both the supernatant and pellets by using a Packard liquid scintillation counter as described previously (21). Figure 3 shows the total amount (in nanomoles) of 109Cd which is cell bound or remains in the supernatant after incubation. Of the 1.0 nmol of 109Cd added, cells expressing the metal binding peptide bound 0.92 nmol, whereas cells lacking the metal binding peptide bound only 0.13 nmol. The effect of coincubation with increasing concentrations of unlabeled heavy metals on the removal of 109Cd by cells was tested. In these experiments 20 mg of cells was coincubated in 5 ml of LB with 0.2 μM 109Cd and unlabeled Cd2+, Hg2+, Pb2+, or Cu2+ at a concentration of 0.2, 1, or 5 μM. Figure 4 shows the effect of increasing concentrations of the unlabeled metals on the ability of cells to remove 109Cd from solutions. Cd2+ and Hg2+ have the most inhibitory effect on 109Cd removal, indicating that these metals have a high affinity for the peptide. Pb2+ is also inhibitory at higher concentrations, but Cu2+ does not have any inhibitory effect, even at the highest concentration tested. The ability of cells expressing the metal binding peptide fusion to effectively and selectively remove Cd2+ and Hg2+ from solutions with a low initial (part-per-billion) concentration (Fig. 3 and 4) along with the data presented in Table 1 demonstrates the potential for these cells to be used at various metal-contaminated sites.
FIG. 3.
109Cd binding by cells expressing or lacking the metal binding peptide. S, supernatant; P, pellet; + metal binding peptide, cells expressing the metal binding peptide; − metal binding peptide, control cells lacking the metal binding peptide.
FIG. 4.
Effect of coincubation with cold metal on the binding of 109Cd by cells expressing the metal binding peptide. A total of 100% cell bound is equal to 1.0 nmol of 109Cd, which is the total amount added.
Our approach of using genetically engineered organisms for heavy metal removal has focused on the expression of peptides with high cysteine content within the periplasmic space of bacterial cells and the use of such bacteria as an immobilized biomass. The metal binding peptide described in this study is similar in size and cysteine content to the previously expressed N. crassa metallothionein (21), and it was designed for high affinity to Cd2+ and Hg2+ through the incorporation of Cys-Gly and Cys-Cys-Gly motifs. The potential benefits of a de novo-constructed metal binding peptide versus naturally occurring metal binding proteins include the ability to incorporate metal binding sites for several metals within one protein as well as the potential for addition of amino acid sequences for increased protein stability (11). To further increase the biosorption of heavy metals by E. coli, the expression of tandem repeat copies of the metallothionein and this peptide is currently being studied. The construction of genetically engineered organisms for heavy metal bioaccumulation has recently been reported by other investigators. Chen and Wilson (4) reported the simultaneous expression of a metallothionein gene and a mercury transport system in E. coli. This study showed that expression of the metallothionein in the cytosol without coexpression of the transport system resulted in diminished bioaccumulation of Hg2+ by the cells. These results are consistent with our previous work and other studies indicating that maximal bioaccumulation of metal occurs with expression of metal binding proteins outside the cytosol (4, 21, 25, 29). In addition, expression outside the cytosol may allow for the use of nonviable cells for metal accumulation and for efficient desorption of the bound metal (3, 7, 20). Sousa et al. reported the expression of yeast and mammalian metallothioneins within the outer membrane of E. coli when LamB was used as an anchoring domain (29). The binding of Hg2+ and Cd2+ by cells expressing the metal binding peptide fusion described in the present study is comparable to those described by Chen and Wilson (4) and Sousa et al. (28, 29). The metal binding studies described by these investigators, however, were performed under different conditions and at heavy metal concentrations of 5 μM (4) and 20 to 30 μM (28, 29), whereas our studies were performed at concentrations of 0.2 to 5 μM.
The development of an immobilized biomass from bacterial cells has been discussed as an approach for the development of heavy metal removal systems (17) and involves the immobilization of nonviable cells or cell remnants containing the metal binding peptide within a suitable matrix. The ultimate utility of such a system will depend on binding capacity, affinity, selectivity, stability, and ease of production, among other factors. The current expression system in E. coli provides several benefits for the study of metal binding peptides, including periplasmic localization, enhanced stability of the expressed peptide, and lack of cysteine in the fusion, thus providing a convenient method for analysis of cysteine-containing peptides. However, alternate cost-effective promoters (e.g., heat induced), increased levels of metallothionein expression (10), and potential for expression of metal binding peptides in other organisms need to be studied for further development. The expression and incorporation of amino acid sequences which are known to stabilize proteins against heat, pH, and salinity may further increase the utility of such a system (11).
Acknowledgments
This work was supported by a grant from the Office of Naval Research.
REFERENCES
- 1.Berka T, Shatzman A, Zimmerman J, Strickler J, Rosenberg M. Efficient expression of the yeast metallothionein gene in Escherichia coli. J Bacteriol. 1988;170:21–26. doi: 10.1128/jb.170.1.21-26.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brower J B, Ryan R L, Pazirandeh M. Comparison of ion-exchange resins and biosorbents for the removal of heavy metals from plating factory wastewater. Environ Sci Technol. 1997;31:2910–2914. [Google Scholar]
- 4.Chen S, Wilson D B. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2+-contaminated environments. Appl Environ Microbiol. 1997;63:2442–2445. doi: 10.1128/aem.63.6.2442-2445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieckmann G R, McRorie D K, Tierney D L, Utschig L M, Singer C P, O’Halloran T V, Penner-Hahn J E, DeGrado W F, Pecoraro V L. De novo design of mercury-binding two- and three-helical bundles. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 6.Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 7.Gadd G M. Metal tolerance. In: Edwards C, editor. Microbiology of extreme environments. New York, N.Y: McGraw-Hill; 1990. pp. 178–210. [Google Scholar]
- 8.Gadd G M, White C. Microbial treatment of metal pollution—a working biotechnology? Trends Biochem Technol. 1993;11:353–359. doi: 10.1016/0167-7799(93)90158-6. [DOI] [PubMed] [Google Scholar]
- 9.Gregory D S, Martin A C R, Cheetham J C, Rees A R. The prediction and characterization of metal binding sites in proteins. Protein Eng. 1993;6:29–35. doi: 10.1093/protein/6.1.29. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman B J, Broadwater J A, Johnson P, Fox B G, Kenealy W R. Lactose fed-batch overexpression of recombinant metalloproteins in Escherichia coli BL21 (DE3): process control yielding high levels of metal-incorporated, soluble protein. Protein Expr Purif. 1995;5:646–654. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 11.Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem. 1991;202:715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- 12.Klemba M, Gardner K H, Marino S, Clarke N D, Regan L. Novel metal-binding proteins by design. Nat Struct Biol. 1995;2:368–373. doi: 10.1038/nsb0595-368. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Valentine J S. Engineering metal-binding sites in proteins. Curr Opin Struct Biol. 1997;7:495–500. doi: 10.1016/s0959-440x(97)80112-1. [DOI] [PubMed] [Google Scholar]
- 15.Macaskie L E. An immobilized cell bioprocess for the removal of heavy metals from aqueous flows. J Chem Technol Biotechnol. 1990;49:357–379. doi: 10.1002/jctb.280490408. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Mattison P L. Bioremediation of metals—putting it to work. Santa Rosa, Calif: Cognis, Inc.; 1992. [Google Scholar]
- 18.Murooka Y, Nagaoka T. Expression of cloned monkey metallothionein in Escherichia coli. Appl Environ Microbiol. 1987;53:204–207. doi: 10.1128/aem.53.1.204-207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 20.Pazirandeh M. Development of a metallothionein based heavy metal biosorbent. Biochem Mol Biol Int. 1996;39:789–795. doi: 10.1080/15216549600201881. [DOI] [PubMed] [Google Scholar]
- 21.Pazirandeh M, Chrisey L A, Mauro J M, Campbell J R, Gaber B P. Expression of the Neurospora crassa metallothionein gene in Escherichia coli and its effect on heavy-metal uptake. Appl Microbiol Biotechnol. 1995;43:1112–1117. doi: 10.1007/BF00166934. [DOI] [PubMed] [Google Scholar]
- 22.Pettit L D, Gregor J E, Kozlowski H. Complex formation between metal ions and peptides. Perspect Bioinorg Chem. 1991;1:1–41. [Google Scholar]
- 23.Regan L. Protein design: novel metal-binding sites. Trends Biochem Sci. 1995;20:280–285. doi: 10.1016/s0968-0004(00)89044-1. [DOI] [PubMed] [Google Scholar]
- 24.Rittle K A, Drever J L, Colberg P J S. Precipitation of arsenic during bacterial sulfate reduction. Geomicrobiol J. 1995;13:1–11. [Google Scholar]
- 25.Romeyer F M, Jacobs F A, Masson L, Hanna Z, Brousseau R. Bioaccumulation of heavy metals in Escherichia coli expressing an inducible synthetic human metallothionein gene. J Biotechnol. 1988;8:207–220. [Google Scholar]
- 26.Romeyer F M, Jacobs F A, Brousseau R. Expression of a Neurospora crassa metallothionein and its variants in Escherichia coli. Appl Environ Microbiol. 1990;56:2748–2754. doi: 10.1128/aem.56.9.2748-2754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen H, Wang Y-T. Characterization of enzymatic reduction of hexavalent chromium by Escherichia coli ATCC 33456. Appl Environ Microbiol. 1993;59:3771–3777. doi: 10.1128/aem.59.11.3771-3777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sousa C, Cebolla A, de Lorenzo V. Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol. 1996;14:1017–1020. doi: 10.1038/nbt0896-1017. [DOI] [PubMed] [Google Scholar]
- 29.Sousa C, Kotrba P, Ruml T, Cebolla A, De Lorenzo V. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J Bacteriol. 1998;180:2280–2284. doi: 10.1128/jb.180.9.2280-2284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Totura G. Innovative uses of specialty ion exchange resins provide new cost-effective options for metals removal. Environ Prog. 1996;15:208–212. [Google Scholar]