Abstract
A DNA-based, direct method for initial characterization of the total bacterial community in ileum and cecum of the chicken gastrointestinal (GI) tract was developed. The efficiencies of bacterial extraction and lysis were >95 and >99%, respectively, and therefore the DNA recovered should accurately reflect the bacterial communities of the ileal and cecal digesta. Total bacterial DNA samples were fractionated according to their percent G+C content. The profiles reflecting the composition of the bacterial community were reproducible within each compartment, but different between the compartments of the GI tract. This approach is independent of the culturability of the bacteria in the consortium and can be used to improve our understanding of how diet and other variables modulate the microbial communities of the GI tracts of animals.
The microbiology of the gastrointestinal (GI) tracts of farm animals has long been of interest for reasons of both food safety and animal nutrition and health. Attempts are being made to bolster host defenses by using feed ingredients which favor the growth of bacteria generally regarded as beneficial. Typical modulators of GI tract ecology are prebiotics (e.g., oligosaccharides [25]) and probiotics (e.g., lactobacilli and bifidobacteria [5, 24, 25]). Understanding of the mode of action and development of effective products would be greatly assisted by reliable tools with which to monitor the composition of the microbial community in the GI tract.
Molecular techniques have been employed to monitor for the presence of specific bacterial pathogens in chicken digesta samples (2, 6, 16, 26). In order to place findings from such single-species studies in perspective, some knowledge of the total bacterial community composition of this system must be obtained. However, direct analyses (i.e., analyses that are not based on culturability) of bacterial community structure in the various compartments of the chicken GI tract have not yet been performed. In other complex environments, DNA-based community-level molecular analyses have been used to obtain information on microbial community diversity, structure, and function (10, 12, 20, 21, 27, 30, 36). In this report, we show for the first time essentially quantitative recovery and lysis of the bacterial fraction from the distal part of broiler chicken small intestine (ileum) and cecum and also purification of total DNA representing the bacterial community, a requisite step for DNA-based community analyses. We also provide an initial depiction of the total bacterial community in these two chicken GI tract compartments obtained by DNA-based profiling.
Bacterial extraction.
Broiler chickens were raised in cages and unless otherwise indicated were fed a standard wheat-based diet with no antibiotics. The birds were killed by cervical dislocation, and the ileum and cecum were immediately removed and dissected. All digesta samples were kept on ice and processed further within 2 h. Quantitative bacterial recovery and lysis are prerequisites for all subsequent comprehensive DNA-based community analyses. Therefore, these protocols were carefully developed, and their efficiencies were demonstrated. Digesta samples from the different compartments of the broiler chicken digestive tract were found to vary in composition, the amount of material present, and bacterial density. Bacterial numbers in the ileum, the distal part of small intestine, were typically between 107 and 109 per g of digesta, and the bulk of the dry matter was undigested feed particles, which had to be eliminated prior to bacterial lysis. This was accomplished through the five-cycle differential centrifugation process. Seven grams of ileal digesta was suspended in 200 ml of wash buffer (50 mM sodium phosphate buffer [pH 8], 0.1% Tween 80). The suspensions were then shaken for 20 min on a reciprocating horizontal platform shaker at 100 oscillations/min at room temperature. Undigested feed particles were removed from the suspension by low-speed centrifugation at 200 × g for 15 min. The supernatant was carefully transferred to a clean flask and kept on ice. The digesta pellet was again suspended in wash buffer, the differential centrifugation process was repeated for a total of five rounds, and samples of both the suspended digesta and the low-speed supernatant were taken before each round for microscopic enumeration of bacteria. The bacteria in the pooled supernatants were collected by centrifugation at 30,000 × g for 15 min at room temperature. In the dual cecal compartments, solid feed particles were absent and bacterial densities were typically between 1010 and 1011 per g of digesta. For efficient bacterial recovery and lysis from this compartment, it was important to remove the viscous polysaccharides and other soluble compounds interfering with these procedures. This was achieved by a four-cycle dilution and washing process. Cecal samples (1 g) were suspended in 30 ml of wash buffer and then shaken for 10 min on a reciprocating horizontal platform shaker at moderate speed at room temperature. The resulting suspension was subjected to centrifugation at 30,000 × g for 15 min to collect the bacterial fraction. The pellet was resuspended and washed three more times in 30 ml of fresh wash buffer, and samples of the suspended bacteria were taken at each step for direct microscopic enumeration. Microscopic enumeration of bacteria was accomplished by fluorescence microscopy of DAPI (4′,6-diamidino-2-phenylindole)-stained cells essentially as described previously (32). An automated stage-manipulation system and digital camera using Image Pro Plus (Media Cybernetics, Silver Spring, Md.) and Amcas (Cultor Technology Center, Kantvik, Finland) software were used to enumerate cells.
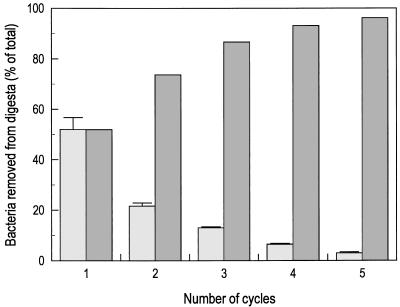
Direct microscopic counts were used to enumerate bacterial numbers in the ileal and cecal digesta for the starting samples and during the bacterial extraction. Methods that have been used for bacterial extraction from other solid matrices (9, 11) gave relatively poor recovery of bacterial cells (<50%) when applied to the digesta samples (data not shown), indicating the need to optimize these procedures for the samples being analyzed. The differential centrifugation approach resulted in extraction of approximately 96% of the cells that were present in the starting ileum digesta samples. The first cycle recovered 50% of the bacteria, whereas a minimum of four cycles were needed for 90% recovery (Fig. 1). Similarly, cell counts in the bacterial fraction from the cecum indicated quantitative recovery, with >97% of the bacterial cells present in the starting sample being recovered by the four-cycle dilution and washing process. Microscopic examination of the resuspended bacterial pellets from the high-speed centrifugation step indicated that bacteria in the supernatant pelleted quantitatively at the g forces used. Lower g forces resulted in incomplete pelleting and poor bacterial recovery, probably due to the presence of small bacterial cells in these samples (data not shown). It is worth noting that the efficiency of bacterial recovery from these samples exceeds values obtained from other types of matrices, such as soils and sediments, which typically range between 33 and 42% (11, 15). Presumably these high recoveries reflect both the importance of multiple rounds of extraction of cells from environmental samples (four or five for each protocol) and a lack of tight adhesion between bacteria and the digesta matrix.
FIG. 1.
Cell removal from digesta material ( ) and cumulative recovery (
) and cumulative recovery ( ) of bacteria from ileal digesta samples during the bacterial recovery process. The data were obtained by direct microscopic enumeration of bacterial cells in the supernatant following each round of differential, low-speed centrifugation. Percent removal is based on the number of cells present in the starting ileal digesta sample. Error bars indicate 1 standard error of the mean (n = 4).
) of bacteria from ileal digesta samples during the bacterial recovery process. The data were obtained by direct microscopic enumeration of bacterial cells in the supernatant following each round of differential, low-speed centrifugation. Percent removal is based on the number of cells present in the starting ileal digesta sample. Error bars indicate 1 standard error of the mean (n = 4).
Bacterial lysis and DNA recovery.
Effective lysis of complex mixtures of bacteria often requires a combination of various treatments known to be effective for lysing individual bacterial populations (11). To maximize the lysis efficiency for the bacterial populations present in chicken digesta, the salient steps from a variety of cell lysis protocols were combined to obtain the general bacterial lysis protocol. Samples of the bacterial fractions (0.3 g of cecal bacterial pellet and 0.7 g of ileal bacterial pellet) were resuspended in 3 ml of TE buffer (10 mM Tris [pH 8], 1 mM EDTA). These suspensions were subjected to five freeze-thaw cycles of incubation at −70°C for 60 min followed by 40°C for 15 min. Lysozyme was then added to each sample as 0.7 ml of a 200-mg/ml stock solution (in TE buffer) followed by incubation at 37°C for 3 h. After the addition of 0.2 ml of sodium dodecyl sulfate (10% [wt/vol]) and 20 μl of proteinase K solution (20 mg/ml in TE buffer), the mixture was incubated at 37°C for an additional hour. Following this incubation, 0.72 ml of 5 M NaCl, 0.6 ml of 10% CTAB (hexadecyltrimethyl ammonium bromide) in 0.7 M NaCl, and 1 g of 1,000-μm-diameter glass beads (Sigma Chemical Company, St. Louis, Mo.) were added. The mixtures were then incubated at 65°C for 20 min with vortexing for 30 s after every 5 min. Samples of the suspended bacterial fraction were taken before and after lysis to assess lysis efficiency by direct microscopic enumeration of bacteria. The cell lysate was extracted with an equal volume of chloroform-isoamyl alcohol (24:1) and then was subjected to centrifugation at 6,000 × g for 10 min at room temperature to separate the aqueous and organic phases. After transfer of the aqueous phase to a clean Corex test tube, the DNA was precipitated by addition of 0.6 volume of 100% isopropanol and incubation for 1 h at room temperature. The precipitated DNA was collected by centrifugation at 10,000 × g for 15 min at room temperature. The DNA pellet was washed briefly with 70% ethanol, vacuum dried, and dissolved in 2 ml of TE buffer. The extracted DNA was then purified by two rounds of cesium chloride-ethidium bromide equilibrium gradient centrifugation as described by Holben (9). The DNA bands from these gradients were subjected to ethidium bromide extraction, desalting, and concentration of DNA by ethanol precipitation, also as described by Holben (9). DNA concentration and purity were estimated based on A260/280.
Microscopic analysis was used to monitor the efficiency of lysis of the bacterial fractions obtained from ileal and cecal digesta. Direct microscopic counts before and after the lysis procedure indicated that >99% of the bacteria in both the ileal and cecal bacterial fractions were lysed (data not shown). Combined with the highly efficient recovery of bacteria from the digesta samples, the efficient lysis achieved with this protocol results in bacterial community DNA samples that are presumed to be representative of the bacterial populations present in the digesta from the chicken GI tract. Thus, the procedures developed here provide sound samples, not only for the community analysis described below, but also for any other DNA-based analysis (PCR, hybridizations, etc.) that might be used for the characterization of bacterial communities in the chicken GI tract.
Percent G+C profiles of the digestal communities.
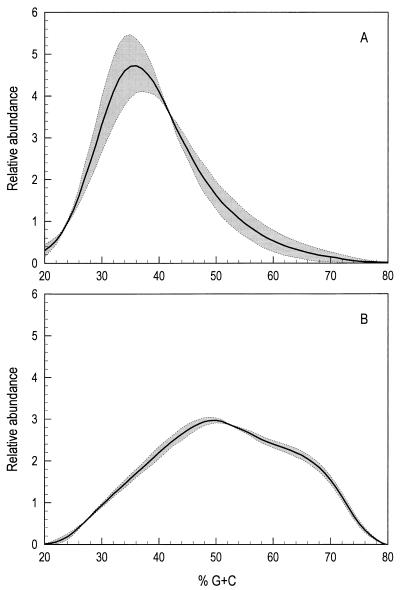
To obtain a profile of ileal and cecal digesta bacterial communities based on percent G+C content, 100 μg of each DNA sample was subjected to cesium chloride-bisbenzimidazole gradient analysis as described previously (11). Since there was no residual protein in these highly purified DNA preparations, DNA quantitation was based on A280, which minimizes background absorbance resulting from the cesium chloride gradient itself and unbound bisbenzimidazole. Determination of the percent G+C content represented by each gradient fraction was accomplished by regression analysis (r2 > 0.99) of data obtained from gradients containing standard DNA samples of known percent G+C composition (Clostridium perfringens, Escherichia coli, and Micrococcus lysodeikticus). Fractionation of total bacterial community DNA based on percent G+C content has previously been used to analyze bacterial communities and how they respond to changing conditions in soils (10), bioreactors (12), and cricket hindgut (30). The data reported here extend that analysis to studies of the bacterial communities in the GI tracts of broiler chickens. To assess variability and reproducibility in this system, replicate samples of bacterial community DNA isolated from ileal and cecal digesta of 4-week-old broiler chickens were subjected to cesium chloride-bisbenzimidazole gradient analysis. The data indicate that replicate samples from each compartment show similar profiles, while the bacterial communities in the ileal and cecal compartments were substantially different from one another (Fig. 2). While the pooling of digesta from two birds may have resulted in some averaging of differences in bacterial communities between individual birds, the replicate samples analyzed were independent replicates, and the data thus suggest that birds raised together under identical conditions have similar bacterial community compositions in their GI tracts (Fig. 2).
FIG. 2.
Profiles of the bacterial community in ileal (A) and cecal (B) digesta material from 4-week-old chickens. Data were obtained as a continuous stream from the UV absorbance flow cell and are presented as percent G+C content versus relative abundance. The solid line in each panel indicates the mean of two replicate samples. The shaded areas in each panel indicate the standard error of the mean.
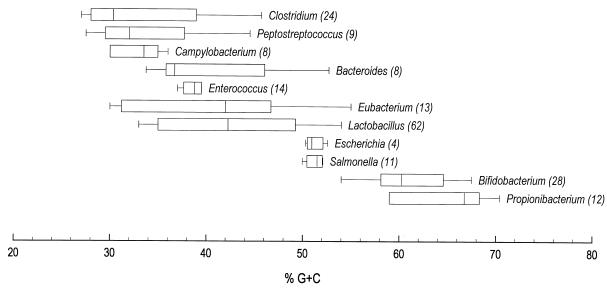
Numerous bacterial species have been characterized from the GI tract of the chicken by selective plating and subsequent identification by biochemical profiles (1, 3, 4, 7, 8, 14, 17, 23, 28, 29, 35). Such studies clearly demonstrate the presence of the selected bacterial groups, but probably do not accurately depict the total bacterial community in question. Figure 3 shows ranges of percent G+C content in the DNA of some of the genera which have been reported to be present in the GI tract of the chicken. The ileal and cecal bacterial communities illustrated by the percent G+C profiles (Fig. 2 and 4) may be, but are not necessarily, composed of the members of the bacterial genera listed. Speculation regarding the identity of organisms represented by these profiles is useful in guiding future studies, but to truly demonstrate whether a particular organism or group of organisms is present in a given region of the bacterial community profile requires additional DNA hybridization or ribosomal DNA analyses as reported previously (12, 33).
FIG. 3.
Ranges of percent G+C content in bacterial genera present in the GI tract of the chicken. Boxes indicate ranges, which accommodate 80% of the species within a given genus, and the vertical line in each box is the median of that genus. The values in parentheses show the number of species included in the survey. The figure is based on the literature data (13, 19, 22, 34, 37).
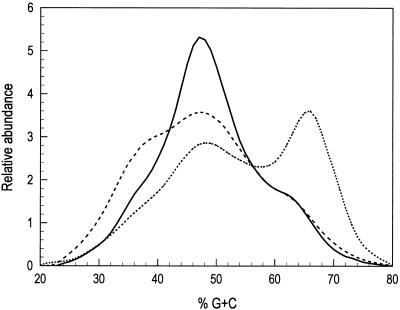
FIG. 4.
Profiles of the cecal bacterial community in 10-day-old chickens. Data were obtained as a continuous stream from the UV absorbance flow cell and are presented as percent G+C content versus relative abundance. Each line represents the profile obtained from pooled cecal samples of six broiler chickens. Solid line (——), uninoculated control; dotted line (........), inoculation with 108 cecal bacteria from commercially raised, free-range chickens; dashed line (–––), inoculation with 105 C. perfringens cells.
Changing digestal communities by live-fed bacteria.
To determine whether it is possible to alter the community in the GI tract and detect such changes in community structure by using this DNA-based community analysis, newly hatched chicks were inoculated with a single dose either of bacteria from the cecal contents of commercially raised, free-range chickens or C. perfringens. C. perfringens (1:1 mixture of ATCC 13124 and ATCC 3626) was grown overnight anaerobically at 37°C in RCM medium (Difco, Detroit, Mich.). The bacterial cells were collected by centrifugation at 20,000 × g and then resuspended in anaerobic 0.9% NaCl solution. Approximately 105 bacteria in 0.1 ml of solution were introduced into the crop of the broiler chicks with a 1-ml syringe. In another treatment, broiler ceca were dissected from 30 5-week-old broiler chickens, and the contents (digesta) were pooled in an anaerobic glove box and then stored frozen at −70°C in 1-ml aliquots. An aliquot was used to inoculate 50 ml of anaerobic brain heart infusion medium (LAB M; Bury, United Kingdom) in a tightly stoppered serum bottle. The culture was grown static overnight at 37°C. The bacteria were harvested by centrifugation at 20,000 × g and then resuspended in anaerobic 0.9% NaCl solution. Approximately 108 bacteria were introduced as 0.1 ml of an appropriately diluted bacterial suspension directly into the crop of the broiler chicks as described above. Bacterial numbers in the inocula were determined by direct microscopy. Following bacterial inoculation, the birds were raised under identical conditions for 10 days on a standard, antibiotic-free, corn-based diet, and then the cecal bacterial community DNA was recovered and analyzed. The bacterial community profiles from the ceca (cecal digesta of six chickens were pooled) of differentially inoculated broiler chickens were different from each other and from those of the uninoculated control birds (Fig. 4).
Compared to the percent G+C profile from the uninoculated broiler chickens, those inoculated with C. perfringens showed an increase in the relative abundance of DNA, and hence bacteria, having 25 to 40% G+C content, and a decrease in the relative abundance of bacteria whose DNA is in the 42 to 55% G+C range (Fig. 4). Although no subsequent confirming analyses were performed, this is consistent with an increase in the abundance of clostridia, campylobacteria, and enterococci or perhaps unculturable bacterial populations having the indicated percent G+C content. The cecal bacterial community from chickens inoculated with the mixed bacterial populations from commercially raised birds showed a relatively large increase in bacterial populations having 55 to 75% G+C content and a corresponding decrease in the relative abundance of bacterial populations having 30 to 55% G+C content. These data are consistent with an increase in bifidobacteria and propionibacteria (Fig. 3 and 4), although no confirming analysis was performed.
Profiling of the DNA from the total GI tract bacterial community according to its percent G+C content as described here is totally independent of the culturability of the component bacteria. Further studies employing DNA hybridization or quantitative PCR with specific primers would help confirm the presence and abundance of specific bacterial groups in the total microbial community of the chicken GI tract (11, 18, 31, 33). However, these approaches are dependent on reference bacteria and therefore on previous strain isolation under laboratory conditions. Assessment of the relative contribution of uncultured organisms to the total bacterial community is an arduous task requiring extensive molecular analyses, including primer and probe design, PCRs, cloning, sequencing, and nucleic acid hybridization experiments. With the approaches described here, we have at hand a relatively rapid community-level analysis which generates data in the form of DNA abundance versus percent G+C content. This technique provides a window on population dynamics in the community to guide future experiments related to the microbial ecology of the GI tracts of chickens.
Acknowledgments
This work was financially supported by the Cultor Corporation, Helsinki, Finland, and Finnfeeds International, Ltd., Marlborough, United Kingdom.
We thank Andrew Morgan and Mike Bedford for useful comments and stimulating discussions. We also thank Osmo Siikanen, Hilkka Heikkinen, Liisa Leppä, and Esa Wainio for excellent technical assistance; Hanna Jatila for preparing the starter cultures; and Hannele Kettunen for animal maintenance and recovery of intestinal specimens.
REFERENCES
- 1.Adami A, Cavazzoni V. An experimental standard pattern of assessing the caecal microflora of chicken. Ann Microbiol Enzimol. 1993;43:329–336. [Google Scholar]
- 2.Arun K B, Kumar M C, York M D, Pomeroy B S. Direct immunofluorescent technique in diagnosis of experimental salmonellosis of turkeys. Avian Dis. 1974;19:59–65. [PubMed] [Google Scholar]
- 3.Barnes E. The intestinal microflora of poultry and game birds during life and after storage. J Appl Bacteriol. 1979;46:407–419. doi: 10.1111/j.1365-2672.1979.tb00838.x. [DOI] [PubMed] [Google Scholar]
- 4.Barnes E M, Mead G C, Barnum D A. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br Poult Sci. 1972;13:311–325. doi: 10.1080/00071667208415953. [DOI] [PubMed] [Google Scholar]
- 5.Barrow P. Probiotics for chickens. In: Fuller R, editor. Probiotics: the scientific basis. London, United Kingdom: Chapman & Hall; 1992. pp. 225–257. [Google Scholar]
- 6.Chuma T, Yamada T, Okamoto K, Yugi H, Ohya T. Application of a DNA-DNA hybridization method for detection of Campylobacter jejuni in chicken feces. J Vet Med Sci. 1993;55:1027–1029. doi: 10.1292/jvms.55.1027. [DOI] [PubMed] [Google Scholar]
- 7.Devriese L A, Hommez J, Wijfels R, Haesebrouck F. Composition of the enterococcal and streptococcal intestinal flora of poultry. J Appl Bacteriol. 1991;71:46–50. [PubMed] [Google Scholar]
- 8.Fuller R M, Coates M E, Harrison G F. The influence of specific bacteria and a filterable agent on the growth of gnotobiotic chicks. J Appl Bacteriol. 1979;46:335–342. [Google Scholar]
- 9.Holben W E. Isolation and purification of bacterial community DNA from environmental samples. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: American Society for Microbiology; 1997. pp. 431–436. [Google Scholar]
- 10.Holben W E, Harris D. DNA-based monitoring of total bacterial community structure in environmental samples. Mol Ecol. 1995;4:627–631. doi: 10.1111/j.1365-294x.1995.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 11.Holben W E, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holben W E, Noto K, Sumino T, Suwa Y. Molecular analysis of bacterial communities in a three-compartment granular activated sludge system indicates community-level control by incompatible nitrification processes. Appl Environ Microbiol. 1998;64:2528–2532. doi: 10.1128/aem.64.7.2528-2532.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdeman Moore L V, Johnson J L, Moore W E C. Genus Peptostreptococcus Klyver and van Niel 1936, 406. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1083–1092. [Google Scholar]
- 14.Impey C S, Mead G C, George S M. Competitive exclusion of salmonellas from the chick caecum using a defined mixture of bacterial isolates from the caecal microflora of adult bird. J Hyg. 1982;89:479–490. doi: 10.1017/s0022172400071047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobsen C S, Rasmussen O F. Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol. 1992;58:2458–2462. doi: 10.1128/aem.58.8.2458-2462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakoyiannis C K, Winter P J, Marshall R B. Identification of Campylobacter coli isolates from animals and humans by bacterial restriction endonuclease DNA analysis. Appl Environ Microbiol. 1984;48:545–549. doi: 10.1128/aem.48.3.545-549.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley R W, Moore W E C. Abstracts of the 82nd Annual Meeting of the American Society for Microbiology 1982. Washington, D.C: American Society for Microbiology; 1982. The small intestinal flora of poultry, abstr. Q 131; p. 231. [Google Scholar]
- 18.Knight I T, Holben W E, Tiedje J M, Colwell R R. Nucleic acid hybridization techniques for detection, identification, and enumeration of microorganisms in the environment. In: Levin M A, Seidler R J, Rogul M, editors. Microbial ecology: principles, methods, and applications. New York, N.Y: McGraw-Hill, Inc.; 1992. pp. 65–91. [Google Scholar]
- 19.Laskin A I, Lechevalier H A. Handbook of microbiology. 2nd ed. Vol. 3. Boca Raton, Fla: CRC Press, Inc.; 1981. pp. 559–678. [Google Scholar]
- 20.Lee S H, Fuhrman J A. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- 21.Lee S H, Fuhrman J A. Species composition shift of confined bacterioplankton studied at the level of community DNA. Mar Ecol Prog Ser. 1991;79:195–201. [Google Scholar]
- 22.Moore W E C, Holdeman Moore L V. Genus Eubacterium Prevot 1938, 294. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1353–1373. [Google Scholar]
- 23.Morishita T, Lam K M, McCapes R H. The microbiologic ecology of the turkey poult jejunum. Prev Vet Med. 1992;14:233–240. [Google Scholar]
- 24.Nguyen H T, Eckenfelder B, Levesque A. Growth promoting efficiency of two probiotics, TOYCERIN® and PACIFLOR®, in broiler diets. Arch Gefluegelkd. 1988;52:240–245. [Google Scholar]
- 25.Oyarzabal O A, Conner D E. Application of direct fed bacteria and fructooligosaccharides for Salmonella control in broilers during feed withdrawal. Poult Sci. 1996;76:186–190. doi: 10.3382/ps.0750186. [DOI] [PubMed] [Google Scholar]
- 26.Rigby C E. Enzyme-linked immunosorbent assay for detection of Salmonella lipopolysaccharide in poultry specimens. Appl Environ Microbiol. 1984;47:1327–1330. doi: 10.1128/aem.47.6.1327-1330.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritz K, Griffiths B S, Torsvik V L, Hendriksen N B. Analysis of soil and bacterioplankton community DNA by melting profiles and reassociation kinetics. FEMS Microbiol Lett. 1997;149:151–156. [Google Scholar]
- 28.Saikia P K, Devriese L A, Kalita C C, Haesebrouck F, Dutta G N. Composition of the enterococcal intestinal flora of chickens in a tropical area. Lett Appl Microbiol. 1994;19:436–437. [Google Scholar]
- 29.Salanitro J P, Blake I G, Muirhead P A, Maglio M, Goodman J R. Bacteria isolated from the duodenum, ileum, and cecum of young chicks. Appl Environ Microbiol. 1978;35:782–790. doi: 10.1128/aem.35.4.782-790.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santo-Domingo J W, Kaufman M G, Klug M J, Holben W E, Harris D, Tiedje J M. Influence of diet on the structure and function of the bacterial hindgut community of crickets. Mol Ecol, 1998;7:761–767. [Google Scholar]
- 31.Sayler G S, Layton A C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- 32.Schallenberg M, Kalff J, Rasmussen J B. Solutions to problems in enumerating sediment bacteria by direct counts. Appl Environ Microbiol. 1989;55:1214–1219. doi: 10.1128/aem.55.5.1214-1219.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleper C, Holben W, Klenk H-P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smibert R M. Genus Campylobacter Sebald and Veron, 1963, 907. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1986. pp. 111–118. [Google Scholar]
- 35.Snoeyenbos G H, Soerjadi A S, Weinack O M. Gastrointestinal colonization by Salmonellae and pathogenic Escherichia coli in monoxenic and halogenic chicks and poults. Avian Dis. 1982;26:566–575. [PubMed] [Google Scholar]
- 36.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood B J B, Holzapfel W H. The genera of lactic acid bacteria. Vol. 2. London, United Kingdom: Blackie Academic and Professional; 1995. [Google Scholar]