Abstract
Small-subunit ribosomal DNA (SSU rDNA) from 20 phenotypically distinct strains of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria was partially sequenced, yielding 18 unique strains belonging to members of the alpha, beta, and gamma subgroups of the class Proteobacteria. To understand the origin of 2,4-D degradation in this diverse collection, the first gene in the 2,4-D pathway, tfdA, was sequenced. The sequences fell into three unique classes found in various members of the beta and gamma subgroups of Proteobacteria. None of the α-Proteobacteria yielded tfdA PCR products. A comparison of the dendrogram of the tfdA genes with that of the SSU rDNA genes demonstrated incongruency in phylogenies, and hence 2,4-D degradation must have originated from gene transfer between species. Only those strains with tfdA sequences highly similar to the tfdA sequence of strain JMP134 (tfdA class I) transferred all the 2,4-D genes and conferred the 2,4-D degradation phenotype to a Burkholderia cepacia recipient.
Bacteria capable of mineralizing 2,4-dichlorophenoxyacetic acid (2,4-D), a commonly used herbicide, are found in many different phylogenetic groups (2, 3, 7, 11, 22, 23). Evidence suggests that numerous variants of 2,4-D catabolic genes exist and that catabolic operons consist of a near-random mixing of these variants (7). Interspecies gene transfer is a well-documented phenomenon (13), and horizontal gene transfer of the 2,4-D-degrading plasmid pJP4 has been shown (3, 5). However, not all 2,4-D catabolic operons are found on plasmids (10, 11, 16, 20). The extent to which other 2,4-D genes have been exchanged in nature is unknown. The aim of this research was to assess the role of horizontal gene transfer in the evolution of 2,4-D-degrading strains. This article summarizes the results of two aspects of this work—the study of the transfer of the entire 2,4-D pathway by using standard mating experiments and a phylogenetic study of the tfdA gene. The tfdA gene codes for an α-ketoglutarate-dependent 2,4-D dioxygenase which converts 2,4-D into 2,4-dichlorophenol and glyoxylate (6). This 861-bp gene was first sequenced from Ralstonia eutropha JMP134 (19). Two more tfdA genes were cloned from chromosomal locations in Burkholderia strain RASC and Burkholderia strain TFD6 (16, 20). These proved to be identical to each other and 78.5% similar to the original. An alignment of the two variants allowed conserved areas to be identified and primers to be designed for the amplification of tfdA-like genes from other sources (24). Sequence analysis of putative tfdA fragments and the small-subunit ribosomal DNA (SSU rDNA) of the strains carrying them allowed us to construct phylogenies of the genes and their hosts and to look for congruency between them.
Mating experiments.
A collection of 2,4-D degraders containing 15 unique strains as determined by genomic fingerprinting (7) was used as a source of donors in a series of mating experiments (Table 1). Burkholderia cepacia D5, lacking the ability to grow on 2,4-D and not hybridizing to any tfd genes, was used as a recipient in mating experiments. Strain D5 contains neomycin phosphotransferase genes (nptII) carried on transposon Tn5 and is resistant to 50 μg each of kanamycin, carbenicillin, and bacitracin per ml. All of the 2,4-D strains used were sensitive to these antibiotics. Filter matings were performed with a donor-to-recipient ratio of 1:10. Colonies which grew on selective medium (500 ppm of 2,4-D in mineral salts agar [MMO] [23] including 50 μg of kanamycin, carbenicillin, and bacitracin per ml) were subjected to further tests. Their ability to catabolize 2,4-D was tested in liquid medium (same composition as that described above).
TABLE 1.
2,4-D-degrading strains, geographic origins, and GenBank accession numbers
| Strain | GenBank accession no. (SSU rDNA) | Origin | Most similar to genus and/or speciesa | Transferb | tfdA typec | GenBank accession no. (tfdA gene) | Reference or source |
|---|---|---|---|---|---|---|---|
| JMP134 | AF049542 | Australia | Ralstonia eutropha | + | I | M16730 | 3 |
| EML1549 | AF049546 | Oregon | Burkholderia sp. | + | I | 2 | |
| TFD39 | AF049539 | Saskatchewan | Burkholderia sp. | + | I | U43197 | 23 |
| K712 | AF049543 | Michigan | Burkholderia sp. | + | I | U43276 | 11 |
| TFD9 | AF049537 | Saskatchewan | Alcaligenes xylosoxidans | + | I | U43276 | 23 |
| TFD41 | AF049541 | Michigan | Ralstonia eutropha | + | I | 23 | |
| TFD38 | AF049540 | Michigan | Ralstonia eutropha | + | NDc | 23 | |
| TFD23 | AF049536 | Michigan | Rhodoferax fermentans | + | I | U43276 | 23 |
| RASC | AF049544 | Oregon | Burkholderia sp. | (+) | II | U25717 | 2 |
| TFD6 | AF049546 | Michigan | Burkholderia sp. | − | II | 23 | |
| TFD2 | AF049545 | Michigan | Burkholderia sp. | − | II | 23 | |
| TFD31 | AF049536 | Saskatchewan | Rhodoferax fermentans | − | III | 23 | |
| B6-9 | AF049538 | Ontario | Rhodoferax fermentans | ND | III | U43196 | 9 |
| I-18 | U22836 | Oregon | Halomonas sp. | ND | III | U22499 | 15 |
| K1443 | AF049531 | Michigan | Sphingomonas sp. | − | —d | 11 | |
| 2,4-D1 | AF049535 | Montana | Sphingomonas sp. | − | — | R. Sanford | |
| B6-5 | AF049533 | Ontario | Sphingomonas sp. | ND | — | 9 | |
| B6-10 | AF049534 | Ontario | Sphingomonas sp. | ND | — | 9 | |
| EML146 | AF049532 | Oregon | Sphingomonas sp. | − | — | 2 | |
| M1 | AF049530 | French Polynesia | Rhodospeudomonas sp. | ND | R. Fulthorpe |
The generus and/or species most similar to the strain is given based on similarities of SSU rDNA sequences.
Symbols: +, able to transfer 2,4-D degradation to B. cepacia D5; (+), able to transfer at very low frequency; −, no transfer detected.
ND, not determined.
—, no amplificate was obtained.
The disappearance of 2,4-D from the culture medium was monitored by high-performance liquid chromatography. Cells were removed by centrifugation, and the supernatant was filtered through 0.2-μm-pore-size filters. These samples were then analyzed on a Lichrosorb Rp-18 column (Anspec Co., Ann Arbor, Mich.) with 60% methanol–40% 0.1% H3PO4 as the eluant. 2,4-D was detected by measuring light absorption at 230 nm. The presence of tfd genes was detected by hybridizing colony blots with a DNA probe derived from the entire pJP4 plasmid. The identity of the colonies was confirmed by probing with the nptII gene of Tn5 (found in B. cepacia D5). Probes were labeled with random hexanucleotides incorporating [32P]dCTP (3,000 Ci/mmol; New England Nuclear, Boston, Mass.). Hybridizations were done under high-stringency conditions by using 50% formamide and Denhardt’s solution (18) at 42°C. Of the 15 unique strains tested, 9 transferred 2,4-D degradation abilities to D5. This transfer was confirmed by hybridization with pJP4 for eight of these strains. B. cepacia RASC could transfer degradative abilities, but neither it nor the transconjugant hybridized to the pJP4 probe. Work subsequent to this study has confirmed that the genes carried by RASC do not hybridize to those found on pJP4 under high-stringency conditions (7).
Phylogenetic analyses.
Total genomic DNA was isolated from 20 unique 2,4-D-degrading strains (including all 15 used for mating experiments) grown on 500 ppm of 2,4-D mineral salts medium amended with 50 ppm of yeast extract. SSU rDNA was amplified by using fD1 and rD1 as primers (25). Putative tfdA fragments were amplified by using primers TVU and TVL as previously described (24). PCR products were purified with a Gene Clean kit (Bio 101, La Jolla, Calif.). Sequencing was done with an Applied Biosystems model 373A automatic sequencer (Perkin-Elmer Cetus) by using fluorescently labeled dye termination at the Michigan State University Sequencing Facility. The sequencing primer used for SSU rDNA fragments was 519R (5′ GTA TTA CCG CGG CTG CTG G-3′). For tfdA fragments, the sequencing primers were the same as the amplification primers. GenBank accession numbers for these sequences are given in Table 1.
The SSU rDNA sequences were compared to sequences in GenBank by using the Basic Local Alignment Search Tool (BLAST) (1), and those strains with the highest maximal segment pair scores were retrieved from GenBank and included in the phylogenetic analysis. Sequences were aligned manually with the software SeqEd (Applied Biosystems) and with MacClade (14). Sites where nucleotides were not resolved for all sequences were deleted from the alignment, as were those nucleotides corresponding to the small loop in this region that is absent in the alpha subgroup of the class Proteobacteria. These deletions left 283 unambiguous sites for the construction of the SSU rDNA phylogenies. Phylogenetic trees were constructed by using the neighbor-joining analysis of pairwise Jukes-Cantor distances (4), and the topology was confirmed by using the maximum parsimony method PAUP (21). Desulfomonile tiedjei of the δ-Proteobacteria was used as an outgroup. Bootstrap analysis based on 100 replicates was used to place confidence estimates on the tree. Only bootstrap values of greater than 50 were used.
2,4-D degrader diversity.
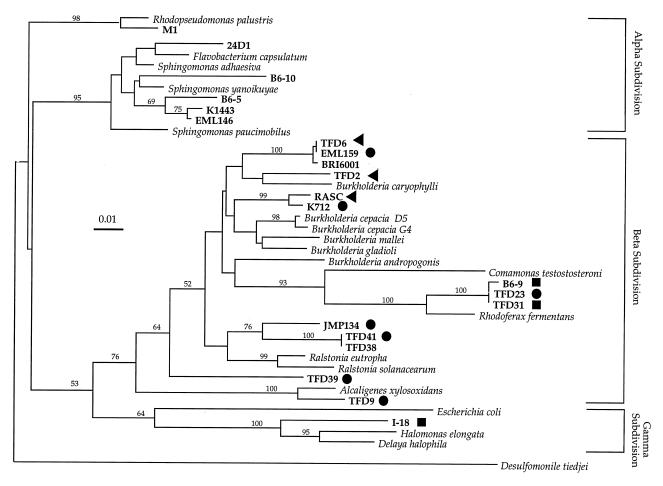
The 2,4-D degraders in this study were distributed throughout the alpha, beta, and gamma subgroups of the Proteobacteria (Fig. 1). The lack of representation of gram-positive bacteria is likely a reflection of isolation methods, not of the lack of gram-positive 2,4-D degraders. The majority of these strains were members of the beta subgroup of Proteobacteria, five of which were most closely related to the genus Burkholderia, having at least 92% sequence similarity with each other. Three were closely related to Rhodoferax fermentans (close to the class Comamonadaceae), three were related to Ralstonia eutropha, and one was related to Alcaligenes xylosoxidans. TFD39 falls outside any clear cluster. One member of the γ-Proteobacteria, strain I-18, a haloalkaliphile, was found to be closely related to the salt-loving genus Halomonas (15). The remaining six strains all clustered in the alpha branch of Proteobacteria (Fig. 1). Of this subgroup, five were most closely related to the genus Sphingomonas. One member of the α-Proteobacteria, strain M1, which is the most oligotrophic and slow growing of all the strains used in this study, is 97% similar to Rhodopseudomonas palustris. The character of strain M1 correlates well with its phylogenetic placement near the slow-growing genus Bradyrhizobium.
FIG. 1.
Neighbor-joining dendrogram (Jukes-Cantor distances) of SSU rDNA from 2,4-D-degrading bacteria (indicated in boldface type) and reference strains (indicated in italic type). Class I (•), class II (▴), and class III (■) types of tfdA genes are indicated. Bootstrap confidence limits (percentages) are indicated above each branch. Scale bar represents a Jukes-Cantor distance of 0.01.
tfdA gene fragments.
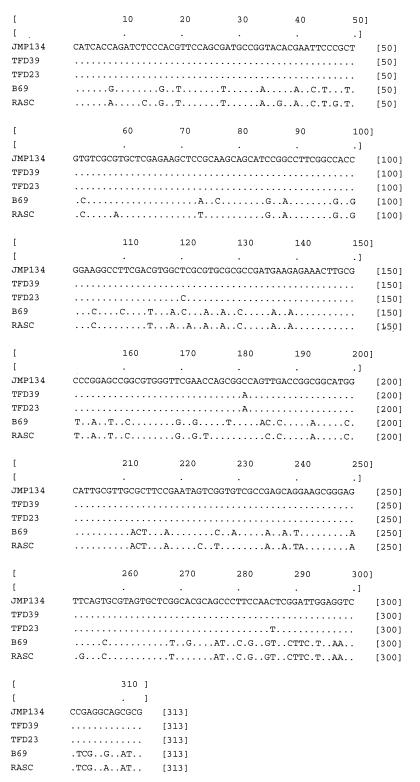
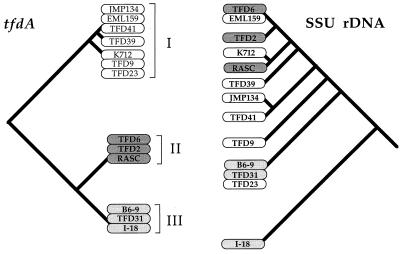
tfdA gene fragments were successfully amplified and sequenced from 10 strains of β-Proteobacteria and 1 strain of γ-Protobacteria. None of the strains from the α-Proteobacteria gave any amplificates with these primers. These 313 contiguous nucleotides were aligned with additional tfdA sequences from JMP134 and from strain RASC (Fig. 2). Three distinct classes of tfdA gene sequences with slight variations in each class were found. Class I included fragments from JMP134, TFD39, TFD23, K712, and TFD9 that differed from each other by 2 bp at the most. Class I tfdA genes are probably plasmid encoded. All strains with a class I tfdA gene examined so far contained broad-host-range, self-transmissible plasmids containing 2,4-D genes (2, 3, 11, 17). All of the strains with a class I tfdA gene were able to transfer the 2,4-D phenotype in the mating studies reported above. The class II tfdA sequences included identical fragments amplified from RASC, TFD6, and TFD2 which were 76% similar to those in class I. Class III included identical fragments from strains TFD31, B6-9, and I-18 which were 77% similar to class I genes and 80% similar to class II genes. Both class II and III tfdA genes differed from each other and from class I genes in the same nine sites corresponding to the third base pair of the codons. The tfdA phylogenetic tree is a simple one, with three distinct branches that are incongruent with the SSU rDNA-derived phylogeny (Fig. 3). Class I tfdA sequences were found in Burkholderia-like strains, in strains related to the Comamonas-Rhodoferax group, and in the Ralstonia-Acaligenes group, all in the β-Proteobacteria. Class II sequences are less widely distributed, found only in Burkholderia-like branches. However, even in this subgroup, this tfdA variant is found in strains that differ by 7% at the SSU rDNA level (RASC and TFD2). However, the class III sequences were most interesting, being found both in the Comamonas-Rhodoferax group and in a strain of the γ-Proteobacteria, I-18, strains that differ by 24% at the SSU rDNA level. Class III genes have since been found in a collection of randomly isolated non-2,4-D degraders, including gram-positive bacilli, as well as in various gram-negative bacteria, even though the gene is not expressed (10).
FIG. 2.
Alignment of 313 nucleotides of internal fragments of tfdA genes from representative strains. Nucleotides identical to tfdA from pJP4 are represented by periods.
FIG. 3.
Phylogenetic incongruency of tfdA genes and SSU rDNA from diverse 2,4-D-degrading bacteria. Dendrograms for tfdA and SSU rDNA are indicated. Shading indicates the type of tfdA sequence, either class I, II, or III. Note that branch lengths are not drawn to scale.
An interesting result was the detection of two different tfdA gene variants in sibling strains. TFD23 and TFD31 are identical at the ribosomal gene level, but one harbors a class I gene and the other harbors a class III gene. Similarly, TFD6 and EML159 are rRNA siblings that carry a class II and class I gene, respectively.
None of the α-Proteobacteria yielded a PCR product when amplified with the conserved tfdA primers. This finding complements our observation that none of these bacteria hybridized to the tfdA gene, even under conditions of low stringency, indicating that any tfdA-like genes in the α-Proteobacteria are likely to be more divergent from the ones sequenced here (7, 11). In addition, none of the Sphingomonas strains in the study hybridized with a whole pJP4 probe, and similarly, no Sphingomonas strains scored positive for transfer of 2,4-D-degrading ability to recipient B. cepacia D5. Together these results suggest a reduced gene flow between members of the α- and β- or γ-Proteobacteria or poor gene expression of β- or γ-derived genes by α-Proteobacteria. Although plasmid pJP4 is a broad-host-range plasmid and has been known to transfer to α-Proteobacteria such as Rhizobium and Agrobacterium species and to γ-Proteobacteria such as Pseudomonas putida, Pseudomonas fluorescens, and Pseudomonas aeruginosa, the 2,4-D pathway is not expressed in these strains of the α- or γ-Proteobacteria (3). Phylogenetically limited expression of plasmid-borne 3-chlorobenzoate-degradative genes has also been noted for the pseudomonads (8). Subsequent studies have found divergent but related sequences for the tfdB and tfdC genes in 2,4-D-degrading Sphingomonas strains (7, 12, 24).
With the exceptions of the minor differences within the class I pJP4-like tfdA sequences, there were no intermediate tfdA sequences. The most likely explanation of this is that the rate of horizontal transfer of the tfd genes is high relative to the rate at which mutations can accumulate. Examination of sequences of tfdA genes from a greater variety of organisms may turn up more intermediate variation.
ACKNOWLEDGMENTS
We thank Arturo Massol-Deya and Jizhong Zhou for primers and Tom Schmidt, Bonnie Bratina, and Jim Smith for advice and assistance with sequence analysis. We thank William Holben, Jong-Ok Ka, Nancy Tonso, Penny Amy, and Charles Greer for the use of their strains and Richard Lenski for valuable discussions in comparing phylogenies of tfdA and SSU rDNA. We also thank the people in the Research on Microbial Evolution Laboratory for their support.
This work was supported by the National Science Foundation (grant DEB9120006), a part of the Joint Project on Microbial Evolution with the Research and Development Corporation of Japan (JRDC).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amy P S, Schulke J W, Frazier L M, Seidler R J. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1985;49:1237–1245. doi: 10.1128/aem.49.5.1237-1245.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich B, Meyer M, Schlegel H. Transfer and expression of the herbicide-degrading plasmid pJP4 in aerobic autotrophic bacteria. Arch Microbiol. 1983;134:92–97. doi: 10.1007/BF00407938. [DOI] [PubMed] [Google Scholar]
- 6.Fukumori F, Hausinger R P. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J Bacteriol. 1993;175:2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulthorpe R R, McGowan C, Maltseva O V, Holben W H, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulthorpe R R, Wyndham R C. Transfer and expression of the catabolic plasmid pBRC60 in wild bacterial recipients in a freshwater ecosystem. Appl Environ Microbiol. 1991;57:1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulthorpe R R, Wyndham R C. Involvement of a chlorobenzoate catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl Environ Microbiol. 1992;58:314–325. doi: 10.1128/aem.58.1.314-325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogan D A, Buckley D H, Nakatsu C H, Schmidt T M, Hausinger R P. Distribution of the tfdA gene in soil bacteria that do not degrade 2,4-dichlorophenoxyacetic acid, 2,4-D. Microb Ecol. 1997;34:90–96. doi: 10.1007/s002489900038. [DOI] [PubMed] [Google Scholar]
- 11.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leander M, Vallaeys T, Fulthorpe R R. Amplification of putative chlorocatechol dioxygenase fragments from alpha and beta proteobacteria. Can J Microbiol. 1998;44:482–486. doi: 10.1139/cjm-44-5-482. [DOI] [PubMed] [Google Scholar]
- 13.Levy S B, Miller R V. Gene transfer in the environment. New York, N.Y: McGraw Hill Publishing Company; 1989. [Google Scholar]
- 14.Maddison W P, Maddison D R. MacClade: analysis of phylogeny and character evolution, version 3.0. Sunderland, Mass: Sinauer Associates; 1992. [Google Scholar]
- 15.Maltseva O, McGowan C, Fulthorpe R, Oriel P. Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology. 1996;142:1115–1122. doi: 10.1099/13500872-142-5-1115. [DOI] [PubMed] [Google Scholar]
- 16.Matheson V G, Forney L J, Suwa Y, Nakatsu C, Sexstone A J, Holben W H. Evidence for acquisition in nature of a chromosomal 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase gene by different Burkholderia spp. Appl Environ Microbiol. 1996;62:2457–2463. doi: 10.1128/aem.62.7.2457-2463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatsu C H, Fukumori F, Fulthorpe R, Hausinger R, Holben W, Kamagata Y, Korona R, Lenski R, Maltseva O, Matheson V G, McGowan C, Suwa Y, Tiedje J, Tonso N, Top E, Wright A, Forney L. Genetic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D) catabolic genes. In: Horikoshi K, Fukuda M, Kudo T, editors. Microbial diversity and genetics of biodegradation. Tokyo, Japan: Japan Scientific Societies Press; 1997. pp. 197–206. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Streber W R, Timmis K N, Zenk M H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suwa Y, Wright A D, Fukumori F, Nummy K A, Hausinger R P, Holben W E, Forney L J. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol. 1996;62:2464–2469. doi: 10.1128/aem.62.7.2464-2469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford D L. PAUP: Phylogenetic Analysis Using Parsimony, version 3.1. Computer; 1993. program distributed by the Illinois Natural History Survey, Champaign, Ill. [Google Scholar]
- 22.Tiedje J M, Duxbury J M, Alexander M, Dawson J E. 2,4-D metabolism: pathway of degradation of chlorocatechols by Arthrobacter sp. J Agric Food Chem. 1969;17:1021–1025. doi: 10.1021/jf60165a037. [DOI] [PubMed] [Google Scholar]
- 23.Tonso N L, Matheson V G, Holben W E. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb Ecol. 1995;30:3–24. doi: 10.1007/BF00184510. [DOI] [PubMed] [Google Scholar]
- 24.Vallaeys T, Fulthorpe R R, Wright A M, Soulas G. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microb Ecol. 1996;20:163–172. [Google Scholar]
- 25.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]