Abstract
The aim of this review article is to focus on the unconventional roles of epigenetic players (chromatin remodelers and long non-coding RNAs) in cell division, beyond their well-characterized functions in chromatin regulation during cell differentiation and development. In the last two decades, diverse experimental evidence has shown that subunits of SRCAP and p400/TIP60 chromatin remodeling complexes in humans relocate from interphase nuclei to centrosomes, spindle or midbody, with their depletion yielding an array of aberrant outcomes of mitosis and cytokinesis. Remarkably, this behavior is shared by orthologous subunits of the Drosophila melanogaster DOM/TIP60 complex, despite fruit flies and humans diverged over 700 million years ago. In short, the available data support the view that subunits of these complexes are a new class of moonlighting proteins, in that they lead a "double life": during the interphase, they function in chromatin regulation within the nucleus, but as the cell progresses through mitosis, they interact with established mitotic factors, thus becoming integral components of the cell division apparatus. By doing so, they contribute to ensuring the correct distribution of chromosomes in the two daughter cells and, when dysfunctional, can cause genomic instability, a condition that can trigger tumorigenesis and developmental diseases. Research over the past few years has unveiled a major contribution of long non-coding RNAs (lncRNAs) in the epigenetics regulation of gene expression which also impacts on cell division control. Here, we focus on possible structural roles of lncRNAs in the execution of cytokinesis: in particular, we suggest that specific classes of lncRNAs relocate to the midbody to form an architectural scaffold ensuring its proper assembly and function during abscission. Drawing attention to experimental evidence for non-canonical extranuclear roles of chromatin factors and lncRNAs has direct implications on important and novel aspects concerning both the epigenetic regulation and the evolutionary dynamics of cell division with a significant impact on differentiation, development, and diseases.
Keywords: Chromatin remodeling, Mitosis, Cytokinesis, Spindle, Midbody (MB)
Introduction
In eukaryotes, successful cell division requires the proper distribution of chromosomes and cytoplasmic material to daughter cells, orchestrated via coordinated cytoskeletal processes including spindle assembly, spindle positioning, chromosome segregation, and cytokinesis [1–9]. Upon entering mitosis, chromosomes condense and attach to mitotic spindle fibers to ensure that sister chromatids are pulled towards opposite sides of the cell (Fig. 1). The mitotic spindle assembles from microtubule arrays and associated proteins that orchestrate chromosome segregation during mitosis [3, 4]. The spindle is highly dynamic in nature and evolutionarily conserved, with many components shared by humans and simpler organisms. In addition to tubulins, proteins involved in spindle function include motor proteins, microtubule-associated proteins (MAPs), microtubule organizing centers, regulatory kinases and phosphatases, kinetochore protein complexes, and chromatin-associated proteins. Following chromosome segregation, the assembly of actomyosin contractile ring occurs at the cleavage furrow. The ring drives the constriction of the plasma membrane leading to abscission, the last stage of cytokinesis (Fig. 1). Before the final cut, the two newly generated daughter cells remain connected by a cytoplasmic bridge that contains the midbody (MB), an organelle first described by Walther Flemming at the end of the 1800s [5]. The MB is a complex multi-protein organelle with a tightly packed structure that forms from the bipolar microtubule array of the central spindle. It plays a pivotal role in the final step of cell division by localizing the site of abscission, and hence of physical separation of daughter cells during cytokinesis [6–12]. Several cellular and molecular pathways have been identified that localize to the MB, contributing to the proper execution of cytokinesis. Notably, various biochemical assays on MB-purified extracts have identified proteins not only related to the cytoskeleton, but also to other molecular pathways, including lipid rafts, vesicle trafficking, protein synthesis, and chromatin organization [6, 10].
Fig. 1.
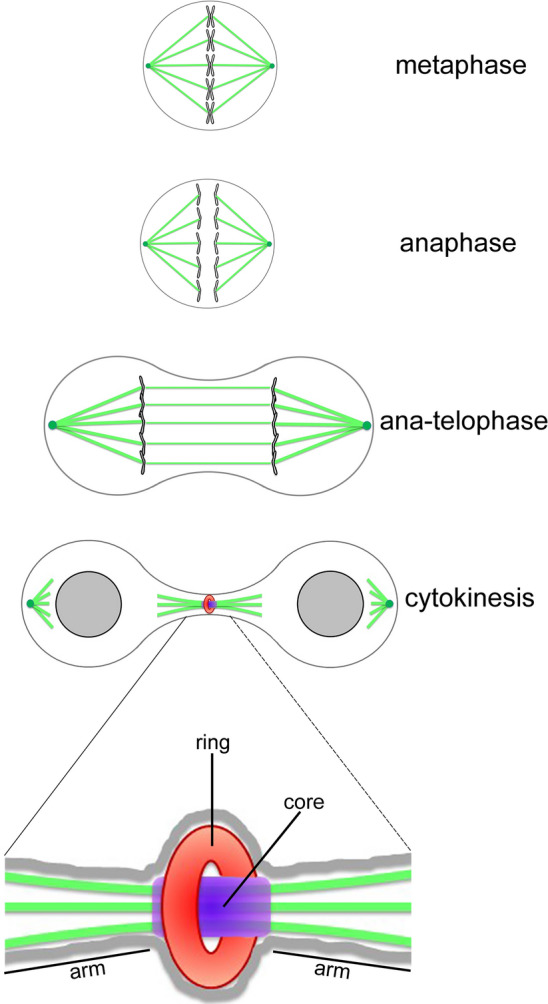
Schematic representation illustrating various stages of cell division and highlighting the three major structural regions of the MB: the ring, central body, and arm
The MB consists of three major structural regions: the ring, central core, and arms (Fig. 1) [10]. MB-proteins are generally classified in subgroups based on their location: the ring contains Anillin, Citron kinase, and other contractile ring components; the central core is marked by central spindle proteins, e.g. the Centralspindlin complex; the arms contain the Aurora B kinase and its localizing partners of the Chromosome Passenger Complex (CPC).
Studies on mitosis and cytokinesis are increasingly relevant to cancer research. Anomalies in the mitotic spindle can impact chromosome segregation, leading to aneuploidy. This phenomenon results in chromosomal instability, a significant contributor to genetic heterogeneity in cancer. Additionally, chromosomal instability plays a crucial role in clinical prognosis and the development of therapeutic resistance [13–16]. Furthermore, MB alterations can lead to cytokinesis failure, resulting in two outcomes: (i) inhibition or regression of the cleavage furrow, leading to the formation of binucleated cells, or (ii) persistent connections between daughter cells, forming long intercellular bridges and giving rise to syncytial cells. Cytokinesis failure ultimately yields tetraploid and polyploid cells with multiple centrosomes, which can further result in aneuploid daughter cells. All these dysfunctions converge to promote tumorigenic transformation [16–18]. Consequently, understanding the molecular mechanisms underlying mitosis and cytokinesis holds the potential to significantly impact both cancer prognosis and therapy.
ATP-dependent chromatin remodeling complexes
Chromatin organization and remodeling are crucial aspects of development and differentiation of higher eukaryotes. In this context, ATP-dependent chromatin remodeling complexes are multi-protein machines that have been highly conserved during eukaryotic genome evolution [31]. These complexes use the energy from ATP hydrolysis to control sliding and displacement of the nucleosomes, thereby modulating histone-DNA interactions and making nucleosomal DNA more accessible to specific binding proteins during replication, transcription, and DNA repair, processes that are crucial for the proper execution of cell division.
Currently, chromatin remodeling complexes are categorized into four subfamilies based on their associated ATPase subunits [19]: (i) the mammalian switch/sucrose non-fermenting (SWI/SNF) subfamily, also called BAF complexes (Brg/Brm-associated factor); (ii) the chromodomain helicase DNA-binding (CHD) subfamily; (iii) the imitation switch (ISWI) subfamily; and (iv) the inositol requiring 80 (INO80) subfamily, which includes yeast INO80 and SWR1 complexes, as well as human p400/TIP60 and SRCAP complexes.
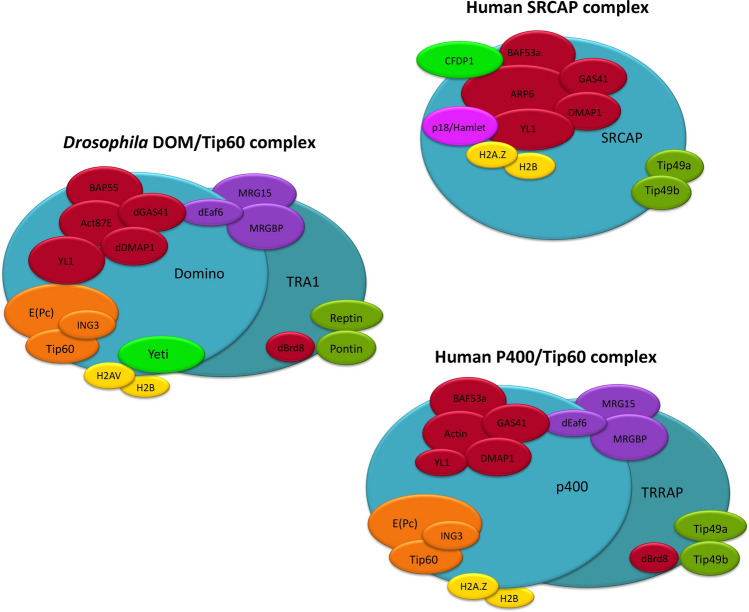
The INO80 family is responsible for exchanging the canonical histone H2A with the variant H2A.Z in various eukaryotic species [19]. The Drosophila DOM/TIP60, related to the yeast SWR1 complex shares many subunits with the p400/TIP60 and SRCAP complexes. It has been recently suggested that the subunits assigned to Drosophila DOM/Tip60 complex are indeed part of two different chromatin remodeling complexes, DOM-A and DOM-B. These complexes are analogous to the yeast SWR1 and NuA4 complexes, respectively, and are characterized by different functions and subunit compositions [20]. Overall, the subunits of these remodeling complexes exhibit strong evolutionary and functional conservation (Fig. 2; Table 1) and their dysfunction is implicated in cell cycle alterations and tumorigenesis (Table 2).
Fig. 2.
The cartoon shows the subunit composition of D. melanogaster DOM/Tip60 and related human SRCAP and p400/Tip60 complexes. The DOM/Tip60 complex consists of 17 known subunits (Act87B, BAP55, Brd8, DOM, DMAP1, E(Pc), Eaf6, GAS41, Ing3, Mrg15, MrgBP, Nipped-A, Pontin, Reptin, TRA1, YETI, and YL1). This complex is crucial for the replacement of acetylated phospho-H2A.V with unmodified H2A.V through the activity of Domino ATPase. In Drosophila, H2A.V is the only H2A variant and corresponds to mammalian H2A.X and H2A.Z [20]. Subunits are not in scale
Table 1.
Evolutionary conservation of ATP-dependent chromatin remodeling complexes
| Organism | H. sapiens | D. melanogaster | S. cerevisiae | A. thaliana | |||
|---|---|---|---|---|---|---|---|
| Complex | p400/Tip60 | SRCAP | DOM / Tip60 | NuA4 | SWR1 | AtNuA4 | AtSWR1 |
| Core | P400 | SRCAP | DOM-A | EAF1 | SWR1 | EAF1A / EAF1B | PIE1 |
| RUVBL1 | Pontin | RVB1 | TIP49a (RIN1) | ||||
| RUVBL2 | Reptin | RVB2 | RVB2A / RVB2B | ||||
| BAF53a (ACTL6A) | BAP55 | ARP4 | ARP4 | ||||
| YEATS4 | GAS41 | YAF9 | YAF9A / YAF9B | ||||
| DMAP1 | DMAP1 | SWC4 (EAF2) | SWC4 | ||||
| VPS72 | YL1 | SWC2 | SWC2 | ||||
| Actin | Act87E | ACT1 | ACT1 | ||||
| ACTR6 | ARP6 | ARP6 | ARP6 | ||||
| Tip60 (KAT5) | Tip60 | ESA1 | HAM1 / HAM2 | ||||
| MORF4L1 | MRG15 | EAF3 | MRG1 / MRG2 | ||||
| MEAF6 | dEAF6 | EAF6 | EAF6 | ||||
| MRGBP | MRGBP | EAF7 | EAF7 | ||||
| EPC1 | E(Pc) | EPL1 | EPL1A / EPL1B | ||||
| ING3 | ING3 | YNG2 | ING1 / ING2 | ||||
| TRRAP | Nipped-A (dTra1) | TRA1 | TRA1 / TRA2 | ||||
| BRD8 | Brd8 | Bdf1 | |||||
| ZNHIT1 | SWC6 (VPS71) | SEF | |||||
| CFDP1 | YETI | SWC5 | SWC5 | ||||
| Non-homologous subunits | EAF5 | SWC3, SWC7 | |||||
The D. melanogaster DOM/TIP60 complex appears to be a fusion of SWR1 and NuA4 complexes of yeast, since the TIP60 complex shares conserved subunits with either SWR1 or NuA4 complexes. Similarly, the human TIP60 complex is a fusion of SWR1 and NuA4 complexes
Table 2.
Subunits of SRCAP and p400/TIP60 complexes implicated in cancer biology
| Subunits | Implication in cancer biology |
|---|---|
| BAF53a | Overexpressed in bladder, cervix, myeloma, colon, and ovarian cancers [21]; aberrant expression correlated with the progression of rhabdomyosarcoma, osteosarcoma, hepatocellular carcinoma, and head and neck squamous cell carcinoma [21]; BAF53a is considered to promote cancer progression via EMT epithelial–mesenchymal transition [22] |
| EPC1 | Is mutated by chromosomal translocation in endometrial stromal sarcoma and in adult T-cell leukaemia/lymphoma SO4 cells [23–25] |
| GAS41 | Overexpressed in glioblastoma cell lines [26, 27] |
| MRGBP/MRGX/MRG15 | Overexpressed in colorectal cancer tissues [28] |
| P400 | Its siRNA-mediated decrease favors the response to 5-fluorouracil of colon cancer cells [29] |
| Pontin & Reptin | Overexpressed in bladder, colon, liver cancers, and melanoma [21, 30, 31] |
| SRCAP | Mutated in large intestine, cervix, bone, endometrium, lung, urinary tract [21]; overexpressed in pancreatic cancer [21]; interacts with the CREB binding protein (CBP), an acetyltransferase encoded by a haplo-insufficient tumor suppressor gene in B-cell lymphoma [32]. It has been identified as a physiologically relevant mediator of PSA expression in prostate cancer cells [33] |
| Tip60 | Acts as a haplo-insufficient tumor suppressor [29, 34] |
| YL1 | Overexpressed in prostate cancer [35] |
Relocation and functions of chromatin factors during cell cycle progression
The first example of versatile chromosome proteins able to change their localizations and functions during cell-cycle progression is given by the CPC, whose subunits are Aurora B, INCENP, Borealin, and Survivin [36]. In early mitosis, they associate with chromosomes, then are recruited to kinetochores to monitor their interactions with the spindle microtubules and eventually relocate to the MB. The catalytic component Aurora B, whose localization is aided by the three passenger partners, phosphorylates and activates several factors at specific times at these different locations, thus playing essential roles in cell division.
The dynamic behavior during cell-cycle progression is not restricted to the CPC proteins. In fact, diverse chromatin proteins, in addition to their role in modulating chromatin organization and gene expression, can relocate to centrosomes, spindle and MB, taking part in cell division and hence in the maintenance of genomic integrity and stability (Table 3). The chromatin proteins Skeletor and Chromator, interact with each other and redistribute during mitosis to form a molecular spindle matrix complex [37, 38]. The Nucleolar and Spindle-Associated Protein (NuSAP), a RanGTP-dependent microtubule and DNA-binding protein, is nucleolar during interphase, associates with microtubules in metaphase and with the central spindle in anaphase and its depletion results in mitosis and cytokinesis defects [39, 40]. In addition, the chromatin remodeler INO80 and three subunits of the SRCAP and p400/TIP60 chromatin remodeling complexes, Pontin (RUVBL1), Reptin (RUVBL2) and TIP60 (KAT5), were shown to relocate to the mitotic apparatus with functional relevance in ensuring the proper execution of cell division in human cells [41–48]. Notably, these proteins were found to interact with tubulins and/or with cell division regulators. INO80 binds to microtubule and was implicated in spindle assembly [41]. Pontin associates with the mitotic spindle and centrosomes via interactions with tubulins in U937 human promonocytic cells [42] and interacts with the γ-tubulin ring complex in Xenopus egg extracts [43]. Pontin and Reptin were found on the mitotic spindle [43]. In late anaphase, both Pontin and Reptin concentrate at the MB in HeLa cells [44, 45]. Accordingly, both Pontin and Reptin are found in microtubule interactome [46], and their RNAi-depletion leads to cell cycle alterations such as spindles defects, misaligned, and lagging chromosomes [43–45]. The TIP60 acetyltransferase of the p400/TIP60 chromatin remodeling complex co-localizes and physically interacts with both Plk1, a mitotic kinase, and cyclin B1, forming a ternary complex that localizes to the centrosomes and to the MB during cytokinesis [47]. Moreover, TIP60 performs Aurora B acetylation at kinetochores and is required for proper chromosome segregation [48]. Finally, an RNAi screening in Drosophila melanogaster S2 cells provided evidence for a role in cytokinesis of BAP55 [49], a subunit of the Drosophila DOM/TIP60 complex (Fig. 2; Table 1).
Table 3.
Localization of chromatin factors to sites of cell division apparatus, in human and D. melanogaster cell cultures
| Homo sapiens | ||
|---|---|---|
| Name | Localization | References |
| Aurora B | Kinetochores, spindle midzone, midbody | [36] |
| INCENP | Inner centromeres, spindle midzone, equatorial cortex | [36] |
| Borealin | Centromeres, spindle midzone, cleavage furrow | [36] |
| Survivin | Centromeres, spindle midzone, cleavage furrow | [36] |
| NuSAP | Central spindle | [39, 40] |
| Ino80 | Mitotic spindle | [41] |
| Pontin, Reptin | Mitotic spindle, centrosomes, midbody | [42, 44–46, 51] |
| BAF53a, CFDP1, GAS41, YL1 | Mitotic spindle, central spindle, midbody | [51] |
| p400 | Midbody | [51] |
| SRCAP | Centrosomes, mitotic spindle, midbody | [50] |
| TIP60 (KAT5) | Spindle poles, kinetochores, cleavage furrow, central spindle, midbody | [47, 48, 51] |
| Drosophila melanogaster | ||
|---|---|---|
| Skeletor | Skeletor defined spindle, microtubule spindle | [37] |
| Chromator | Chromator defined spindle, centrosomes | [38] |
| dTIP60 | Spindle, centrosomes, midbody | [51, 52] |
| DOM-A | Centrosomes; midbody | [50–52] |
| MRG15 | Centrosomes; midbody | [51, 52] |
| YETI | Spindle, midbody | [51, 52] |
| BAP55 | Spindle, centrosomes | [52] |
| DMAP1 | Centrosomes | [52] |
| YL1 | Centrosomes | [52] |
More recently, we studied several subunits of human SRCAP and p400/TIP60 chromatin remodeling complexes and found that they localize at the mitotic apparatus (centrosomes, spindle, and MB) in both HeLa and U2OS cell lines (Table 3; Fig. 3A), with their RNAi-induced depletion producing cell division defects at both mitosis and cytokinesis [50, 51]. These defects might be a secondary effect caused by general chromatin perturbations that in turn would result in deregulation of cell division genes. However, a direct role of chromatin remodeling subunits in cytokinesis is supported by co-IP experiments performed on chromatin-free protein extracts from telophase-synchronized HeLa cell [50, 51]. In these assays, where chromatin contribution can be excluded, SRCAP, BAF53a and TIP60 interacted at telophase with α-tubulin, Aurora B, CIT-K, and other MB-associated cytokinesis regulators. Remarkably, a similar relocation behavior during mitosis and meiosis is shared by subunits of DOM/TIP60 complex of D. melanogaster (Table 3; Fig. 3B), which are orthologous to those of SRCAP and p400/TIP60 human complexes (Table 1) [51, 52]. Since the lineages of D. melanogaster and humans separated approximately 780 million years ago, these results strongly suggested that the functional recruitment of chromatin remodelers to the mitotic apparatus is a very ancient and biologically functional process [51].
Fig. 3.
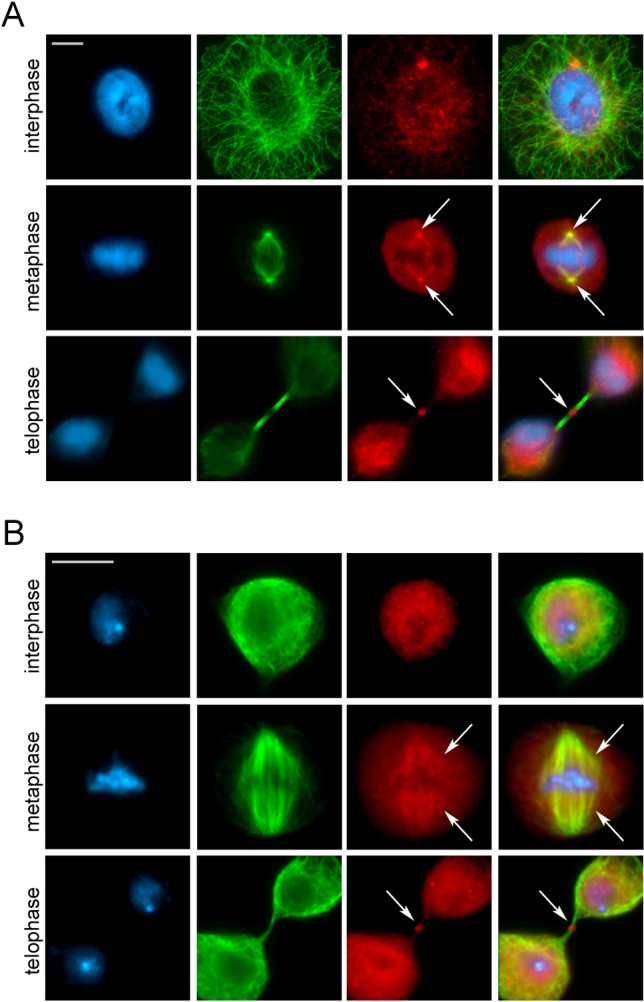
Evolutionarily conserved relocation of chromatin remodelers to the cell division apparatus: the example of TIP60 acetyl-transferase. Immunofluorescence images depict the conserved relocation of chromatin remodelers to the cell division apparatus. Human RPE-1 (A) and D. melanogaster S2 cells (B) were stained with DAPI (blue), anti-α-tubulin (green), and anti-TIP60 (red). Arrows highlight the centrosome/spindle structures (metaphase) and midbodies (telophase). Scale bars:10 μm (RPE-1) and 5 μm (S2)
Several ATPases interact with microtubules and play direct roles in mitosis and cytokinesis. Specifically, the AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis [53]. The ISWI activity is necessary for spindle maintenance, stabilizing microtubules in anaphase [54], while INO80 and CHD4 are required for spindle microtubules assembly [41, 55]. Interestingly, domain analysis of ISWI, CHD4, and INO80 revealed that they can bind microtubules through regions containing chromatin-binding domains [56]. Moreover, Spastin, a microtubule-severing ATPase ensuring the final cut at the midbody, assembles into a hexamer and recognize the C-terminal amino acids of α-tubulin [57]. Mutations in the Spastin-encoding gene are the most common causes of dominant hereditary spastic paraplegias (HSPs), a genetic motor neuron disease characterized by progressive degeneration of corticospinal tract axons [58]. Interestingly, depletion of Spastin in HeLa and MRC5 cells results in cytokinesis failure phenotypes [59], similar to those found in SRCAP-depleted HeLa cells [50]. In agreement with these results, the SRCAP ATPase interacts with Spastin and α-tubulin [50] in telophase-synchronized cells and carries putative microtubule-binding domains in its C-terminal region (Y. Prozzillo, unpublished). Thus, we hypothesized that SRCAP ATPase, as spindle and MB component, participates in two different steps of cell division: by ensuring proper chromosome segregation during mitosis and MB function during abscission at cytokinesis [50].
Dominant truncating mutations of the Srcap gene were found to trigger the onset of Floating Harbor syndrome (FHS), a rare genetic disease characterized by delayed bone mineralization and growth, skeletal, and craniofacial abnormalities, often associated with mental disability [60, 61]. Thus, in addition to gene deregulation caused by chromatin alterations, a cell division failure may contribute to the developmental defects found in FHS patients [50].
Collectively, the aforementioned studies highlighted the existence of a massive and evolutionarily conserved phenomenon where relocation of chromatin factors to the cell division apparatus has functional relevance in ensuring a faithful cell division in distantly related organisms. It is possible to speculate that the “mitotic trip” of chromatin remodelers from the interphase nucleus to the cell division apparatus takes place by exploiting interactions with microtubules and/or microtubule-associated proteins. Elucidating the molecular mechanisms underlying the moonlighting functions of chromatin proteins in cell division is also an important challenge to clarify yet poorly understood routes to tumorigenic transformation in cancer types in which these factors are aberrantly expressed and/or dysfunctional (Table 2). Gaining insight into these processes will also help to expand our understanding of the link between cell division and cancer.
Chromatin remodeling and lncRNAs
LncRNAs are crucial players controlling a plethora of biological processes [62, 64] and their deregulation is also implicated in tumorigenesis [63–66]. As the list of cancers aberrantly expressing lncRNAs is growing fast, lncRNAs have been proposed both as novel biomarkers and potential therapeutic targets for cancer [65, 66]. Increasing evidence shows that many lncRNAs are involved in chromatin regulation and gene expression and can function as scaffolds for the recruitment of chromatin factors [63]. Several lncRNAs facilitate the binding and spreading of the Polycomb repressive complex 2 (PRC2) across targeted chromatin [67–69]. Interactions between lncRNAs and subunits of different chromatin remodeling complexes such as BAF, SRCAP, NuRD, and ATRX complexes have been reported [70]. For example, the lncRNA SChLAP1 interacts with SNF5, a core subunit of the BAF complex, which is required for the proper assembly and function of the complex [71]. The well-known lncRNA XIST physically associates with the BRG1 subunit of the BAF complex and inhibits its ATPase activity in vitro [72]. The lncRNA SWINGN promotes the interaction between SWI/SNF chromatin remodeling complexes and the transcription start site of GAS6 oncogene [73]. The SWI/SNF complexes have been also identified as paraspeckle components that interact with the lncRNA NEAT1 [74]. Remarkably, the lncKdm2b, a highly expressed lncRNA in murine embryonic stem cells, interacts with SRCAP, the main subunit of the homonymous complex increasing its ATPase activity [75]. This finding is of particular interest in light of the roles played by SRCAP in mitosis and cytokinesis [50].
Regulatory roles of RNAs in cell division
Over the last two decades, several studies emerged supporting the regulatory roles of RNAs, both mRNA and lncRNAs, during cell division in evolutionary distant organisms. Evidence for non-canonical localizations of mRNAs to the mitotic apparatus suggested that they could be involved in the regulation of cell division. Centrosomally localized mRNAs were found in embryos of the mollusc Ilyanassa obsoleta [76] and in oocytes of the surf clam Spisula solidissima [77]. In Xenopus laevis egg extracts, mitotic spindle-associated RNA has been identified and suggested to play a translation-independent role in spindle assembly [78]. In mitotic extracts of both X. laevis and humans, a significant fraction of mRNAs was also identified that targets microtubule during mitosis, suggesting a conserved mechanism to regulate mitotic events and delivering translationally inactive mRNAs to daughter cells [79]. In early stages of D. melanogaster embryogenesis, several mRNAs were found associated with spindle poles, centrosomes, astral microtubules or the mitotic spindle [80], indicating that mRNA localization may play a key role in targeting various cellular machineries, including those involved in protein synthesis. The mRNA localization to subcellular structures was originally found to occur through the 3ʹ UTR regions, such as those of nanos and bicoid in early Drosophila embryos [81–83]. These and other findings [84–87] also provided evidence for a mitotic apparatus localized mRNA translation, whose initial concept emerged in the late 1950s and early 1960s [88]. Very recently, evidence have been provided showing a localized enrichment and translation of midbody associated mRNAs encoding key regulatory factors of cytokinesis [89, 90]. Remarkably, the results of Park et al. [89] suggested that the mitotic kinesin MKLP1 and the actively regulated cytoskeleton-associated protein, ARC, are necessary for the localization and translation of mRNAs in the MB dark zone, while ESCRT-III, a protein normally required for the abscission process, maintains translation levels in the MB.
Regulatory roles of lncRNAs in controlling cell division are increasingly being demonstrated [91]. A high-content RNAi imaging screen targeting more than 2,000 human lncRNAs yielded the identification of numerous lncRNAs controlling mitotic progression, chromosome segregation and cytokinesis via regulation of key cell division players [92]. A regulatory lncRNA, termed mamRNA, was shown to be a crucial player in shaping the meiotic gene expression program in fission yeast by scaffolding the antagonistic RNA-binding proteins Mmi1 and Mei2 [93]. More recently, a widely expressed circular RNA, circZNF609, interacts with several mRNAs and increases their stability and/or translation by facilitating the recruitment of the RNA-binding protein ELAVL1 [94]. In particular, circZNF609 interacts with CKAP5 mRNA, which encodes a microtubule-stabilizing factors, and enhances its translation, thus regulating microtubule function and sustaining cell cycle progression. Importantly, circZNF609 also modulates the cancer cell response to microtubule inhibitors used in cancer chemotherapy [94].
LncRNAs as architectural components of the midbody: a working hypothesis
A large fraction of lncRNAs is exported to the cytosol, where they could be assigned to specific organelles or distributed in the cytoplasm associating with RNA-binding proteins [63]. Moreover, lncRNAs are emerging players functioning as phase separation anchors in different subcellular localizations and in the formation of biomolecular condensates [95–98]. Well-known examples are given by NEAT1 and NORAD lncRNAs [98–100].
In light of these functions, it is an intriguing possibility that ncRNAs, in addition to their well-established regulatory functions in controlling of cell division, also play structural roles at sites of the mitotic apparatus [101]. A direct physical role of a human centromeric 1.3 kb long lncRNA in maintaining centromere integrity has been proposed [102]. By targeting CENP-A and its chaperone HJURP to the centromere, this lncRNA ensures proper chromosome dynamics during mitosis and its knockdown results in the formation of multipolar spindles and lagging chromosomes [102].
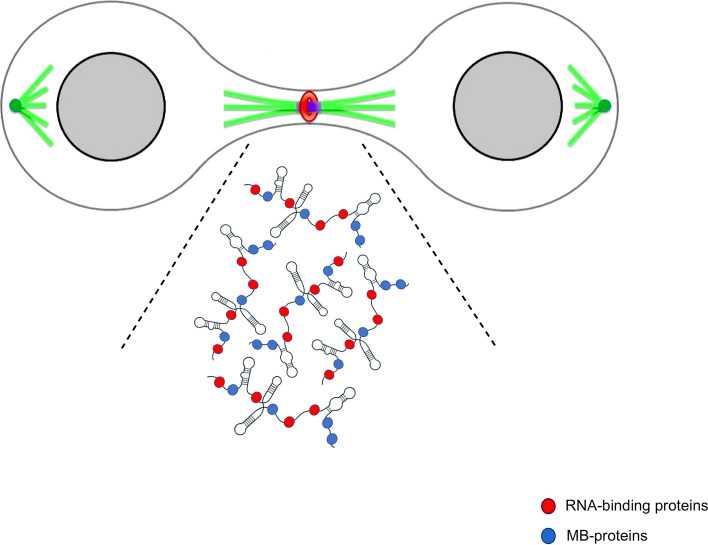
Several cellular and molecular pathways and categories of proteins have been assigned to the MB, contributing to the proper execution of cytokinesis (Fig. 1) [6, 10]. Moreover, MB-associated mRNAs have been recently reported [89, 90] but little is known about the presence of architectural lncRNAs on this organelle.
So far, only few examples suggesting a recruitment of lncRNAs to the MB have been reported. First, Moulton Clemson et al., using FISH, described MB localization of XIST RNA during cytokinesis in human female fibroblasts [103]. XIST RNA, the first long non-coding RNA to be identified, is a major epigenetic effector triggering the X-chromosome inactivation in mammalian female cells [104]. This result would also imply a role of XIST in female cell division which was not further investigated. Intriguingly, the Aurora B kinase, a key component of the MB, interacts with XIST RNA controlling its binding to chromatin [105]. However, the FISH signals found with the XIST probe at the cleavage plane [103] appeared to be somewhat dispersed and not precisely aligned with the MB/central spindle structure. Recently, Chu et al., have identified 81 XIST RNA-binding proteins in mouse cell lines [106]. Interestingly, we discovered that 56 orthologs of these XIST RNA-binding proteins are present in the human MB proteome and interactome datasets (Fig. 4) [10]. These findings collectively provide support for the recruitment of XIST at MB. Finally, in mouse 3T3 cells, the GAA repeat-containing RNAs (GRC-RNAs), a polypurine triplet repeat-rich lncRNA, was found to localize at the midzone area in early telophase and at the MB in late telophase [107]. Finally, the lncKdm2b interacts with the SRCAP ATPase [75], which is recruited to spindle and MB in HeLa and MRC5 cells [51]. However, it is unknown whether the interaction between lncKdm2b and SRCAP is nucleus-specific or also occurs at the MB.
Fig. 4.

Venn diagrams illustrating overlapping protein sets. The Venn diagrams present an analysis of protein sets to reveal commonalities between mouse XIST-binding proteins [106] and orthologous proteins detected in the human MB-proteome and interactome. The overlapping between circles shows the number of proteins in common between the groups. Notably, among the 81 XIST RNA-binding proteins found in mouse, 56 have orthologs in both the human proteome and the interactome of MB. [Y. Prozzillo, unpublished]
Together, the sparse evidence recalled above hints at the fascinating possibility that lncRNAs serve as structural components of the MB. In other words, specific classes of lncRNAs might have the ability to interact with RNA-binding proteins, cytokinesis regulators and other factors, providing an architectural platform for MB assembly (Fig. 5), thus contributing to the proper execution of cytokinesis. Since MB dysfunctions cause abscission failure, leading to genetically unstable states that would promote tumorigenic transformation, investigating in depth the roles of lncRNAs in MB assembly and function can have a strong impact on cancer biology.
Fig. 5.
LncRNA–MB-proteins aggregates triggering MB assembly. LncRNAs promote the formation of phase separation, functioning as architectural scaffolds for diverse RNAs and proteins interaction giving rise to biomolecular condensates in different subcellular localizations [95–98]. The cartoon shows a hypothetical network of interactions between lncRNAs and proteins (RNA-binding proteins, MB-proteins including cytokinesis regulators, chromatin remodelers, and other MB-associated factors) driving the formation of molecular aggregates that trigger the proper MB assembly during cytokinesis
Conclusions: an upcoming challenge
To comprehensively explore and dissect the phase-specific roles of epigenetic players in cell division, their degradation could be performed in a time-controlled way using different approaches. One effective method is the immuno-depletion technique originally developed by Gergely et al., [108], which allows for the inactivation of a given protein of interest (POI) through the injection of specific interfering antibody. This approach has recently proven successful in providing evidence for the mitotic roles of the splicing factors Sf3A2 and Prp31 in Drosophila melanogaster embryos [109]. Over the years, several tools have been devised to achieve protein degradation via proteasome recruitment. Among those, the PROTACs (PROteolysis Targeting Chimeras) systems rely on chimeric molecules composed of a specific ligand for the POI and an E3 ubiquitin ligase. Advances of this system include the use of light-responsive degraders with photocaged or photoswitchable molecules [110]. Targeted protein inactivation can be also achieved by the adding a specific tag fused with the POI. In such cases, a small heterobifunctional molecule can bind the tagged protein, inducing its inactivation through dysfunctional mislocalization or proteosomal degradation [111]. These systems have been widely employed to dissect protein functions in diverse model systems [112, 113].
The dynamic relocation of lncRNAs during cell division can be examined using the CasFAS system, a recently developed imaging method for visualization of endogenous RNAs in living cells [114]. Additionally, optogenetics systems based on light-responsive molecules can be refined and integrated with other techniques, such as the recombinant codon-optimized Cas9 (rCas9), to provide valuable tools for investigating potential cell division phase-specific structural roles of lncRNAs [115–119].
In conclusion, unraveling the unexpected roles of epigenetic regulators during mitosis and cytokinesis in different organisms presents a significant challenge in the field of cell biology that can be met through the combined application of the aforementioned approaches.
Acknowledgements
We are grateful to Patrizia Lavia for her helpful suggestions.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by YP, MVS, GM, and PD. The first draft of the manuscript was written by PD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement. This research was funded by grants from MUR-PRIN 2017, project number 2017FNZRN3 (PD), and “Teresa Ariaudo Research Program 2018”, Pasteur Institute of Italy—Fondazione Cenci Bolognetti (GM).
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
No ethical approval is required.
Consent to publish
This manuscript does not contain any individual person’s data in any form.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuri Prozzillo and Maria Virginia Santopietro contributed equally to this work.
Contributor Information
Giovanni Messina, Email: giovanni.messina@unimib.it.
Patrizio Dimitri, Email: patrizio.dimitri@uniroma1.it.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112(4):407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD. Nine unanswered questions about cytokinesis. J Cell Biol. 2017;216(10):3007–163. doi: 10.1083/jcb.201612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18(3):187–201. doi: 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- 4.Forth S, Kapoor TM. The mechanics of microtubule networks in cell division. J Cell Biol. 2017;216(6):1525–1531. doi: 10.1083/jcb.201612064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flemming W. Neue Beiträge zur Kenntnis der Zelle. Arch Mikrosk Anat. 1891;37:685–751. doi: 10.1007/BF02954311. [DOI] [Google Scholar]
- 6.Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305(5680):61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307(5716):1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 8.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131(5):847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capalbo L, Bassi ZI, Geymonat M, Todesca S, Copoiu L, Enright AJ, et al. The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis. Nat Commun. 2019;10(1):4513. doi: 10.1038/s41467-019-12507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell. 2012;23(6):1024–1034. doi: 10.1091/mbc.E11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petsalaki E, Zachos G. The abscission checkpoint: a guardian of chromosomal stability. Cells. 2021;10(12):3350. doi: 10.3390/cells10123350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 14.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 15.Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet. 2020;21(1):44–62. doi: 10.1038/s41576-019-0171-x. [DOI] [PubMed] [Google Scholar]
- 16.Lens SMA, Medema RH. Cytokinesis defects and cancer. Nat Rev Cancer. 2019;19(1):32–45. doi: 10.1038/s41568-018-0084-6. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 18.Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid and tetraploid genotypes in Min mice. J Cell Biol. 2007;178(7):1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 20.Scacchetti A, Schauer T, Reim A, Apostolou Z, Campos Sparr A, Krause S, Heun P, Wierer M, Becker PB. Drosophila SWR1 and NuA4 complexes are defined by DOMINO isoforms. Elife. 2020;9:e56325. doi: 10.7554/eLife.56325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayes K, Qiu Z, Alhazmi A, Landry JW. ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy. Adv Cancer Res. 2014;121:183–233. doi: 10.1016/B978-0-12-800249-0.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng L, Wang X, Liao W, Liu J, Liao Y, He Q. BAF53a is a potential prognostic biomarker and promotes invasion and epithelial-mesenchymal transition of glioma cells. Oncol Rep. 2017;38(6):3327–3334. doi: 10.3892/or.2017.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Can Res. 2006;66(1):107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 24.Nakahata S, Saito Y, Hamasaki M, Hidaka T, Arai Y, Taki T, Taniwaki M, Morishita K. Alteration of enhancer of polycomb 1 at 10p11.2 is one of the genetic events leading to development of adult T-cell leukemia/lymphoma. Genes Chromosomes Cancer. 2009;48(9):768–776. doi: 10.1002/gcc.20681. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Spencer GJ, Lynch JT, Ciceri F, Somerville TD, Somervaille TC. Enhancers of Polycomb EPC1 and EPC2 sustain the oncogenic potential of MLL leukemia stem cells. Leukemia. 2014;28(5):1081–1091. doi: 10.1038/leu.2013.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munnia A, Schutz N, Romeike BF, Maldener E, Glass B, Maas R, Nastainczyk W, Feiden W, Fischer U, Meese E. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene. 2001;20(35):4853–4863. doi: 10.1038/sj.onc.1204650. [DOI] [PubMed] [Google Scholar]
- 27.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol Cell Biol. 2006;26(11):4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi K, Sakai M, Shimokawa T, Yamada Y, Nakamura Y, Furukawa Y. C20orf20 (MRG-binding protein) as a potential therapeutic target for colorectal cancer. Br J Cancer. 2010;102(2):325–331. doi: 10.1038/sj.bjc.6605500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, Gourraud PA, Chevillard-Briet M, Cazaux C, Trouche D. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene. 2009;28(12):1506–1517. doi: 10.1038/onc.2008.499. [DOI] [PubMed] [Google Scholar]
- 30.Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J. Pontin and reptin, two related ATPases with multiple roles in cancer. Can Res. 2008;68(17):6873–6876. doi: 10.1158/0008-5472.CAN-08-0547. [DOI] [PubMed] [Google Scholar]
- 31.Grigoletto A, Lestienne P, Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochem Biophys Acta. 2011;1815(2):147–157. doi: 10.1016/j.bbcan.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, Meyer SN, Mo T, Basso K, Brindle PK, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7(3):322–337. doi: 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slupianek A, Yerrum S, Safadi FF, Monroy MA. The chromatin remodeling factor SRCAP modulates expression of prostate specific antigen and cellular proliferation in prostate cancer cells. J Cell Physiol. 2010;224(2):369–375. doi: 10.1002/jcp.22132. [DOI] [PubMed] [Google Scholar]
- 34.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 35.Barboro P, Rubagotti A, Boccardo F, Carnemolla B, Darrigo C, Patrone E, Balbi C. Nuclear matrix protein expression in prostate cancer: possible prognostic and diagnostic applications. Anticancer Res. 2005;25(6B):3999–4004. [PubMed] [Google Scholar]
- 36.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 37.Walker DL, Wang D, Jin Y, Rath U, Wang Y, Johansen J, Johansen KM. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J Cell Biol. 2000;151(7):1401–1412. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rath U, Ding Y, Deng H, Qi H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. The chromodomain protein, Chromator, interacts with JIL-1 kinase and regulates the structure of Drosophila polytene chromosomes. J Cell Sci. 2006;119:2332–2341. doi: 10.1242/jcs.02960. [DOI] [PubMed] [Google Scholar]
- 39.Raemaekers T, Ribbeck K, Beaudouin J, Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg J, Carmeliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162(6):1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou HY, Wang TH, Lee SC, Hsu PH, Tsai MD, Chang CL, Jeng YM. Phosphorylation of NuSAP by Cdk1 regulates its interaction with microtubules in mitosis. Cell Cycle. 2011;10(23):4083–4089. doi: 10.4161/cc.10.23.18200. [DOI] [PubMed] [Google Scholar]
- 41.Park EJ, Hur SK, Lee HS, Lee SA, Kwon J. The human Ino80 binds to microtubule via the E-hook of tubulin: implications for the role in spindle assembly. Biochem Biophys Res Commun. 2011;416(3–4):416–420. doi: 10.1016/j.bbrc.2011.11.069. [DOI] [PubMed] [Google Scholar]
- 42.Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, et al. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskelet. 2003;56(2):79–93. doi: 10.1002/cm.10136. [DOI] [PubMed] [Google Scholar]
- 43.Ducat D, Kawaguchi S, Liu H, Yates JR, 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell. 2008;19(7):3097–3110. doi: 10.1091/mbc.e07-11-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sigala B, Edwards M, Puri T, Tsaneva IR. Relocalization of human chromatin remodeling cofactor TIP48 in mitosis. Exp Cell Res. 2005;310(2):357–369. doi: 10.1016/j.yexcr.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 45.Gentili C, Castor D, Kaden S, Lauterbach D, Gysi M, Steigemann P, et al. Chromosome missegregation associated with RUVBL1 deficiency. PLoS ONE. 2015;10(7):e0133576. doi: 10.1371/journal.pone.0133576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, Wainman A, Zitzmann N, Deane C, Ohkura H, Wakefield JG. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 2008;6(4):e98. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang SM, Song M, Yang TY, Fan R, Liu XD, Zhou PK. HIV-1 Tat impairs cell cycle control by targeting the Tip60, Plk1 and cyclin B1 ternary complex. Cell Cycle. 2012;11(6):1217–1234. doi: 10.4161/cc.11.6.19664. [DOI] [PubMed] [Google Scholar]
- 48.Mo F, Zhuang X, Liu X, Yao PY, Qin B, Su Z, et al. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol. 2016;12(4):226–232. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Echard A, Hickson GR, Foley E, O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14(18):1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messina G, Prozzillo Y, Delle Monache F, Santopietro MV, Atterrato MT, Dimitri P. The ATPase SRCAP is associated with the mitotic apparatus, uncovering novel molecular aspects of Floating-Harbor syndrome. BMC Biol. 2021;19(1):184. doi: 10.1186/s12915-021-01109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messina G, Prozzillo Y, Monache FD, Santopietro MV, Dimitri P. Evolutionary conserved relocation of chromatin remodeling complexes to the mitotic apparatus. BMC Biol. 2022;20(1):172. doi: 10.1186/s12915-022-01365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prozzillo Y, Fattorini G, Ferreri D, Leo M, Dimitri P, Messina G. Knockdown of DOM/Tip60 complex subunits impairs male meiosis of drosophila melanogaster. Cells. 2023;12(10):1348. doi: 10.3390/cells12101348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115(3):355–367. doi: 10.1016/S0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 54.Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol. 2009;187(6):813–829. doi: 10.1083/jcb.200906020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama H, Nakos K, Santarella-Mellwig R, Rybina S, Krijgsveld J, Koffa MD, Mattaj IW. CHD4 is a RanGTP-dependent MAP that stabilizes microtubules and regulates bipolar spindle formation. Curr Biol. 2013;23(24):2443–2451. doi: 10.1016/j.cub.2013.09.062. [DOI] [PubMed] [Google Scholar]
- 56.Yokoyama H. Chromatin-binding proteins moonlight as mitotic microtubule regulators. Trends Cell Biol. 2016;26(3):161–164. doi: 10.1016/j.tcb.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 57.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176(7):995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Errico A, Ballabio A, Rugarli E. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet. 2002;11(2):153–163. doi: 10.1093/hmg/11.2.153. [DOI] [PubMed] [Google Scholar]
- 59.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10(1):42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, Allanson J, Kim CA, Wieczorek D, Moilanen JS, Lacombe D, Gillessen-Kaesbach G, Whiteford ML, Quaio CR, Gomy I, Bertola DR, Albrecht B, Platzer K, McGillivray G, Zou R, McLeod DR, Chudley AE, Chodirker BN, Marcadier J, Majewski J, Bulman DE, White SM, Boycott KM, FORGE Canada Consortium Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Hum Genet. 2012;90(2):308–13. doi: 10.1016/j.ajhg.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Messina G, Atterrato MT, Dimitri P. When chromatin organisation floats astray: the Srcap gene and Floating-Harbor syndrome. J Med Genet. 2016;53(12):793–797. doi: 10.1136/jmedgenet-2016-103842. [DOI] [PubMed] [Google Scholar]
- 62.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 63.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543(7644):199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, Harman CCD, Virtue AT, Zhu S, Bailis W, Stein J, Hughes C, Raj A, Wherry EJ, Goff LA, Klion AD, Rinn JL, Williams A, Flavell RA, Henao-Mejia J. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM, Verjovski-Almeida S. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9(8):e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marín-Béjar O, Marchese FP, Athie A, Sánchez Y, González J, Segura V, Huang L, Moreno I, Navarro A, Monzó M, García-Foncillas J, Rinn JL, Guo S, Huarte M. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patty BJ, Hainer SJ. Non-coding RNAs and nucleosome remodeling complexes: an intricate regulatory relationship. Biology (Basel) 2020;9(8):213. doi: 10.3390/biology9080213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y, Yan W, Chen X. SNF5, a core component of the SWI/SNF complex, is necessary for p53 expression and cell survival, in part through eIF4E. Oncogene. 2010;29(28):4090–100. doi: 10.1038/onc.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jégu T, Blum R, Cochrane JC, Yang L, Wang CY, Gilles ME, Colognori D, Szanto A, Marr SK, Kingston RE, Lee JT. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat Struct Mol Biol. 2019;26(2):96–109. doi: 10.1038/s41594-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grossi E, Raimondi I, Goñi E, González J, Marchese FP, Chapaprieta V, Martín-Subero JI, Guo S, Huarte M. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat Commun. 2020;11(1):936. doi: 10.1038/s41467-020-14623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci USA. 2015;112(14):4304–9. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W, Zhu P, Wang Y, Wang S, Xia P, Du Y, Meng S, Huang G, Wu J, Chen R, Tian Y, Fan Z. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018;37(8):e97174. doi: 10.15252/embj.201797174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420(6916):682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 77.Alliegro MC, Alliegro MA, Palazzo RE. Centrosome-associated RNA in surf clam oocytes. Proc Natl Acad Sci USA. 2006;103(24):9034–8. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121(2):223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 79.Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179(7):1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 81.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3’ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67(5):927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 82.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71(2):301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 83.Macdonald PM, Struhl G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336(6199):595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 84.Eliscovich C, Peset I, Vernos I, Méndez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10(7):858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- 85.Romasko EJ, Amarnath D, Midic U, Latham KE. Association ofmaternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: localized translational control supporting female meiosis in mammals. Genetics. 2013;195:349–358. doi: 10.1534/genetics.113.154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pascual R, Segura-Morales C, Omerzu M, Bellora N, Belloc E, Castellazzi CL, Reina O, Eyras E, Maurice MM, Millanes-Romero A, Méndez R. mRNA spindle localization and mitotic translational regulation by CPEB1 and CPEB4. RNA. 2020;27(3):291–302. doi: 10.1261/rna.077552.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fernandez-Nicolas A, Uchida A, Poon J, Yajima M. Vasa nucleates asymmetric translation along the mitotic spindle during unequal cell divisions. Nat Commun. 2022;13(1):2145. doi: 10.1038/s41467-022-29855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waldron A, Yajima M. Localized translation on the mitotic apparatus: a history and perspective. Dev Biol. 2020;468(1–2):55–58. doi: 10.1016/j.ydbio.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park S, Dahn R, Kurt E, Presle A, VanDenHeuvel K, Moravec C, Jambhekar A, Olukoga O, Shepherd J, Echard A, Blower M, Skop AR. The mammalian midbody and midbody remnant are assembly sites for RNA and localized translation. Dev Cell. 2023 doi: 10.1016/j.devcel.2023.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farmer T, Han KJ, Vaeth KF, Taliaferro JM, Prekeris R. The novel role of midbody-associated mRNAs in regulating abscission. bioRxiv. 2022 doi: 10.1101/2022.10.27.514111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guiducci G, Stojic L. Long noncoding RNAs at the crossroads of cell cycle and genome integrity. Trends Genet. 2021;37(6):528–546. doi: 10.1016/j.tig.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 92.Stojic L, Lun ATL, Mascalchi P, Ernst C, Redmond AM, Mangei J, Barr AR, Bousgouni V, Bakal C, Marioni JC, Odom DT, Gergely F. A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat Commun. 2020;11(1):1851. doi: 10.1038/s41467-020-14978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andric V, Nevers A, Hazra D, Auxilien S, Menant A, Graille M, Palancade B, Rougemaille M. A scaffold lncRNA shapes the mitosis to meiosis switch. Nat Commun. 2021;12(1):770. doi: 10.1038/s41467-021-21032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rossi F, Beltran M, Damizia M, Grelloni C, Colantoni A, Setti A, Di Timoteo G, Dattilo D, Centrón-Broco A, Nicoletti C, Fanciulli M, Lavia P, Bozzoni I. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol Cell. 2022;82(1):75–89.e9. doi: 10.1016/j.molcel.2021.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22(3):183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo J, Qu L, Gao F, Lin J, Liu J, Lin A. LncRNAs: architectural scaffolds or more potential roles in phase separation. Front Genet. 2021;12:626234. doi: 10.3389/fgene.2021.626234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Somasundaram K, Gupta B, Jain N, Jana S. LncRNAs divide and rule: the master regulators of phase separation. Front Genet. 2022;13:930792. doi: 10.3389/fgene.2022.930792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70(6):1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 100.Elguindy MM, Mendell JT. NORAD-induced Pumilio phase separation is required for genome stability. Nature. 2021;595(7866):303–308. doi: 10.1038/s41586-021-03633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ito KK, Watanabe K, Kitagawa D. The emerging role of ncRNAs and RNA-Binding proteins in mitotic apparatus formation. Noncoding RNA. 2020;6(1):13. doi: 10.3390/ncrna6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quénet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife. 2014;3:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 105.Hall LL, Byron M, Pageau G, Lawrence JB. AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol. 2009;186(4):491–507. doi: 10.1083/jcb.200811143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–16. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng R, Shen Z, Tripathi V, Xuan Z, Freier SM, Bennett CF, Prasanth SG, Prasanth KV. Polypurinerepeat- containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci. 2010;123(Pt 21):3734–44. doi: 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000;19(2):241–252. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pellacani C, Bucciarelli E, Renda F, Hayward D, Palena A, Chen J, Bonaccorsi S, Wakefield JG, Gatti M, Somma MP. Splicing factors Sf3A2 and Prp31 have direct roles in mitotic chromosome segregation. Elife. 2018;7:e40325. doi: 10.7554/eLife.40325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reynders M, Trauner D. Optical control of targeted protein degradation. Cell Chem Biol. 2021;28(7):969–986. doi: 10.1016/j.chembiol.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 111.Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, Scott TG, Vittori S, Perry JA, Qi J, Winter GE, Wong KK, Gray NS, Bradner JE. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. 2018;14(5):431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prozzillo Y, Messina G. droTAG: Adapting dTAG toolkit to drosophila melanogaster. Am J Biomed Sci Res. 2021 doi: 10.34297/AJBSR.2021.11.001662. [DOI] [Google Scholar]
- 113.Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21(3):181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang H, Peng J, Peng S, Wang Q, Jiang X, Xue X, Tao Y, Xiang L, Ji Q, Liu SM, Weng X, Zhou X. Live-cell RNA imaging using the CRISPR-dCas13 system with modified sgRNAs appended with fluorescent RNA aptamers. Chem Sci. 2022;13(47):14032–14040. doi: 10.1039/d2sc04656c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nelles DA, Fang MY, Aigner S, Yeo GW. Applications of Cas9 as an RNA-programmed RNA-binding protein. Bioessays. 2015;37(7):732–9. doi: 10.1002/bies.201500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat Methods. 2013;10(9):873–5. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335(6073):1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.You M, Jaffrey SR. Designing optogenetically controlled RNA for regulating biological systems. Ann N Y Acad Sci. 2015;1352(1):13–9. doi: 10.1111/nyas.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Childs-Disney JL, Yang X, Gibaut QMR, Tong Y, Batey RT, Disney MD. Targeting RNA structures with small molecules. Nat Rev Drug Discov. 2022;21(10):736–762. doi: 10.1038/s41573-022-00521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.