Abstract
In recent years interest in bacteriophages in aquatic environments has increased. Electron microscopy studies have revealed high numbers of phage particles (104 to 107 particles per ml) in the marine environment. However, the ecological role of these bacteriophages is still unknown, and the role of the phages in the control of bacterioplankton by lysis and the potential for gene transfer are disputed. Even the basic questions of the genetic relationships of the phages and the diversity of phage-host systems in aquatic environments have not been answered. We investigated the diversity of 22 phage-host systems after 85 phages were collected at one station near a German island, Helgoland, located in the North Sea. The relationships among the phages were determined by electron microscopy, DNA-DNA hybridization, and host range studies. On the basis of morphology, 11 phages were assigned to the virus family Myoviridae, 7 phages were assigned to the family Siphoviridae, and 4 phages were assigned to the family Podoviridae. DNA-DNA hybridization confirmed that there was no DNA homology between phages belonging to different families. We found that the 22 marine bacteriophages belonged to 13 different species. The host bacteria were differentiated by morphological and physiological tests and by 16S ribosomal DNA sequencing. All of the bacteria were gram negative, facultatively anaerobic, motile, and coccoid. The 16S rRNA sequences of the bacteria exhibited high levels of similarity (98 to 99%) with the sequences of organisms belonging to the genus Pseudoalteromonas, which belongs to the γ subdivision of the class Proteobacteria.
The marine bacterial community is responsible for a considerable portion of primary production and regeneration of nutrients in the microbial loop and is associated with a great variety of marine bacteriophages (5, 12). These phages are capable of infecting a large portion of the bacterioplankton (32, 34). It is assumed that as part of the marine food web, bacteriophages play important quantitative and qualitative roles in controlling marine bacterial populations (8, 24, 34, 39, 45). The phenotypic diversity and genotypic diversity of the phage populations are related to the interaction between phages and their host organisms, which provides a tool for understanding the interaction itself (13). To estimate the influence of marine bacteriophages on the diversity of bacterioplankton, we investigated phage diversity. The virus species concept proposed by Murphy et al. (37) delineates seven different families of bacteriophages based on morphological criteria and provides criteria for new phage species based on several traits, such as DNA homologies, serological data, protein profiles, and host ranges.
In this paper, we describe the diversity and genetic relationships of marine phages based on investigations of 22 representatives from 85 phage-host systems (35, 36) collected between 1988 and 1992 from waters around an island, Helgoland, located in the North Sea. All of the phages were virulent and formed plaques on their host bacteria. We assigned the phages to different virus families, species, and strains based on morphology, DNA homology, and host range. Furthermore, we characterized the phenotypic and genotypic features of the host bacteria.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
Bacterial strains and bacteriophages were kindly provided by K. Moebus, Biologische Anstalt Helgoland, Helgoland, Germany. They were isolated from North Sea water collected at one location near Helgoland, an island in the North Sea belonging to Germany. The bacterial strains were grown as described by Moebus and Nattkemper (33). Phage lysates were prepared by the overlay agar technique (31). Confluent lysis was generated, and the phages were eluted with 10 ml of SM buffer per plate after incubation for 1 h at room temperature (47). Phage stocks were stored at 4°C.
Phage-host cross-reaction test.
Two-layer agar plates containing a 10-ml bottom layer and a 3-ml soft agar upper layer were used for the phage-host cross-reaction test; the soft agar layer contained ca. 108 bacteria (34). Phage lysates were dotted in a dilution series from 100 to 106 onto the upper layer immediately after solidification in order to distinguish between a clear lysis reaction caused by plaque formation and inhibition of the bacterial lawn. After incubation overnight at 18°C in the dark, plaque formation was evaluated.
Electron microscopy of phages.
High-titer phage stocks (lysates) were prepared for electron microscopy. Lysates were allowed to adsorb for 1 min to pioloform- and carbon-coated 400-mesh wide copper grids. Then the grids with the adhering phage lysates were washed three times with distilled water. Negative staining was performed with 2% (wt/vol) uranyl acetate for 40 s (19, 20). Micrographs were obtained at a primary magnification of ×40,000 by electron microscopy (Zeiss model EM 10 A microscope). The dimensions of phages were estimated by determining the mean values for 30 particles of each phage; catalase was used as an internal length calibration standard (46).
Buoyant density of phage particles.
Phage buoyant density was determined by CsCl2 gradient centrifugation by using the method of Espejo and Canelo (15).
Isolation of phage DNAs and labeling of DNA probes.
Stocks (200 ml) of phages were prepared as described above. DNAs were isolated by using the general methods described by Sambrook et al. (42). Purified phage DNAs were labeled with a nonradioactive digoxigenin labeling kit (Boehringer, Mannheim, Germany) as recommended by the manufacturer and were used as probes in subsequent DNA-DNA hybridization experiments.
DNA-DNA hybridization on nylon membranes.
For dot blot hybridization whole-phage DNA (5 to 10 μl) was dotted onto a nylon membrane and fixed with UV light (wavelength, 312 nm; 7 min). For restriction fragment DNA-DNA hybridization the phage DNA was digested with restriction enzyme HindIII (Boehringer) for 90 min at 37°C. After gel electrophoresis in 0.8% (wt/vol) agarose gels, the DNA pattern was transferred to nylon membranes (Hybond N; Amersham, Braunschweig, Germany) by Southern blotting (43), and the DNA was used as target DNA. DNA-DNA hybridization was carried out at 68°C for 16 h. Hybridization was detected as recommended by the manufacturer (Boehringer). Positive hybridization signals occurred after 1 to 4 h of incubation with 5-bromo-4-chloro-3-indolylphosphate toluidinium (salt) and nitroblue tetrazolium (salt) (70%). The levels of DNA homology between previously described phages and phages used in this work were not determined.
Determination of GC content of phage DNA.
The GC content of phage DNA was determined as described by Marmur and Doty (29).
Morphology and physiology of host bacteria.
Morphological and physiological tests were performed as described by Moaledj (30). Cell morphology (size, shape, arrangement) was determined by phase-contrast microscopy (magnification, ×1,250) after 1 to 2 days of incubation at 18°C. The media used for the physiological tests were adapted so that they fulfilled the salt requirements of marine bacteria. Gram reaction, catalase and oxidase production, and motility tests were performed with freshly prepared liquid cultures, while the cultures used to test oxidation and fermentation of glucose, saccharose, and lactose (1%, wt/vol) were incubated for up to 14 days before analysis.
PCR amplification of the 16S rRNA gene.
Bacterial DNA was isolated by the method of Anderson and McKay (2), modified for genomic DNA by omitting the NaOH step. The extracted DNA was used as target DNA in PCR (41) to amplify the 16S ribosomal RNA coding regions. The primers used for the 500-bp fragment examined were 27f (5′-AGAGTTTGATC[A/T]TGGCTCAG-3′) and 519r (5′-G[A/T]ATTACCGCGGC[G/T]GCTG-3′ (26). The sequences of the primers used for the nearly complete 16S rRNA gene (GM3F and GM4R; Escherichia coli positions 8 to 1507) have been published by Muyzer et al. (38). PCR amplification was performed with a model 480 DNA thermal cycler (Perkin-Elmer Cetus) as described by Muyzer et al. (38). To increase the specificity of amplification and to reduce the formation of spurious by-products, a “touch-down” PCR (14) was performed (65 to 55°C, 20 cycles). Aliquots (5 μl) of the amplification products were analyzed by electrophoresis in 2% (wt/vol) agarose gels, which were stained with ethidium bromide (0.5 μg/ml).
DNA sequencing of PCR products and comparative sequence analysis.
PCR products that were 500 bp long were purified with glasmilk (Bio-Rad). DNA sequencing was performed with a model ABI 377 sequencer by using a PRISM Ready Dye Deoxy terminator kit and Perkin-Elmer Taq polymerase according to the instructions of the manufacturer (ABI, Foster City, Calif.). The sequences of whole 16S ribosomal DNA fragments were determined by the method described by Buchholz-Cleven et al. (10). All sequences were aligned with sequences obtained from the Ribosomal Database Project (27) or GenBank (3). Sequence alignment was performed with the sequence editor SEQAPP (21). A phylogenetic tree was created by using the neighbor-joining algorithm and maximum likelihood as a model for evolution (PAUP test, version 6.3, developed by David Swofford). A bootstrap analysis (100 replicates) was used to validate the reproducibility of the branching pattern of the tree.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in the GenBank database under accession no. AF069653 through AF060667.
RESULTS
Selection of phages for detailed investigation.
To select a group of phages that were representative of the 85 phages isolated, phage-host cross-reaction tests were performed with the phages and 70 bacterial isolates obtained from North Sea water. The phages were assigned to sensitivity group I (SG I), SG II, and SG III on the basis of their hosts. A total of 62 (73%) of the 85 bacteriophages were highly host specific and members of SG I; these phages were found to be reproduced only by their original hosts. Sixteen phages (19%) had host ranges consisting of 2 to 10 bacteria and were members of SG II, and seven phages (8%) had broad host ranges consisting of 11 to 36 bacterial isolates and were members of SG III. A total of 22 bacteriophages were selected for further investigation. Seven of these phages belonged to SG I, nine belonged to SG II, and six were assigned to SG III.
Morphological diversity.
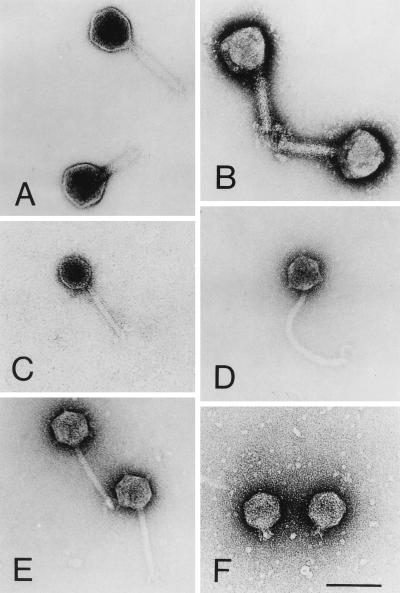
The phenotypic diversity of the 22 bacteriophages was examined by electron microscopy. The phages were identified by using morphological criteria outlined by the International Committee of Taxonomy of Viruses (37) and the species concept of Ackermann et al. (1). Morphological studies revealed that all of the phages examined had tails and thus belong to the order Caudovirales. The icosahedral heads of the phages had diameters between 50.2 and 99.3 nm. The phages could be assigned to three virus families. Eleven of the phages belonged to the family Myoviridae, which contains phages that have icosahedral heads and long contractile tails; seven phages were assigned to the family Siphoviridae, which contains phages that have icosahedral heads and long flexible tails; and four phages, which had icosahedral heads and short tails, belonged to the family Podoviridae. The phages belonging to the Myoviridae were further divided into two different morphotypes on the basis of different appendages, such as collars, antennae, or tail fibers; the four morphotype 1 phages had a collarlike structure between the head and the tail (Fig. 1B), whereas the seven morphotype 2 phages had no special appendages (Fig. 1A).
FIG. 1.
Phages belonging to three different families and their morphotypes. (A) Myoviridae, morphotype 1: head without antennae and short appendages on the tail (phage H106/1). (B) Myoviridae, morphotype 2: collarlike structure between the head and the tail and short appendages on the tail (phage H7/2; 15). (C) Siphoviridae, morphotype 1: head and tail without appendages (phage 10-77a). (D) Siphoviridae, morphotype 2: knoblike appendages on the head and tail with a hook at the end (phage 11 68c). (E) Siphoviridae, morphotype 3: knoblike appendages on the head and tail with short appendages (phage H105/1). (F) Podoviridae, morphotype 1 (phage H100/1). Bar = 100 nm.
Similarly, the seven bacteriophages belonging to the Siphoviridae were subdivided into three different morphotypes. The single morphotype 1 phage had no additional appendages on its head or tail (Fig. 1C). The single morphotype 2 phage was particularly striking because it had a hook at the end of its tail (Fig. 1D). The remaining five phages, which had knoblike appendages on their heads, belonged to morphotype 3 (Fig. 1E). All four phages belonging to the Podoviridae were morphotype 1 phages with no special appendages (Fig. 1F). The buoyant densities of the phages investigated were between 1.49 and 1.54 g · cm−3.
Bacteriophage host ranges.
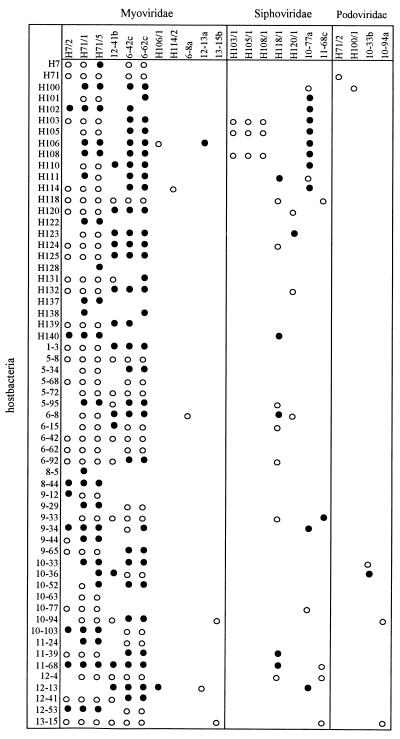
The results of our morphological characterization of the phages were related to phage host ranges (Fig. 2). Phages were arranged according to family. The following three observations were remarkable: (i) all of the phages belonging to the Myoviridae had very broad host ranges; (ii) three of the phages belonging to the Siphoviridae had identical host ranges and belonged to the same morphotype; and (iii) all four phages belonging to the Podoviridae exhibited high specificity for their host bacteria.
FIG. 2.
Bacteriophage host ranges and families. ○, phage produces plaques; •, phage inhibits growth of bacterial lawn (no plaques).
GC contents of marine bacteriophages.
All of the phages contained double-stranded DNA. The GC contents of the DNAs ranged from 33.7 to 64.7%. The GC contents of the phages belonging to the Myoviridae ranged from 33.7 to 64.7%, whereas there was less variation in the GC contents of members of the Siphoviridae (GC contents, 40.1 to 51%). The GC contents of the DNAs of members of the Podoviridae were between 38.4 and 57.6%.
DNA homologies of marine bacteriophages.
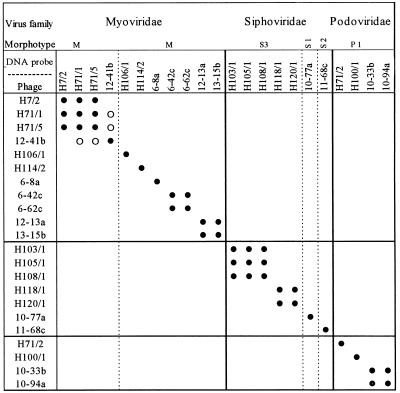
Dot blot hybridizations revealed that phages belonging to different families did not exhibit DNA homology. To obtain more detailed information, specific HindIII endonuclease restriction digestion of the DNAs and subsequent Southern hybridization were carried out with the phages that exhibited DNA homology in dot blot hybridization experiments. These experiments showed clearly that either there was a high level of homology between the DNAs of two phages belonging to the same family (i.e., DNA homology occurred in every DNA fragment) transferred to a nylon membrane or no homology was detected. Besides the fact that there was no homology between bacteriophages belonging to different families, an overview of all of the hybridization experiments revealed that within the families not all of the phages were genetically related (Fig. 3). Seven phages exhibited no DNA homology to the other phages investigated. The other 15 phages exhibited DNA homology to one or more phages belonging to the same family. In the Myoviridae, DNA homology was observed between phages 12-13a and 13-15b, as well as between phages 6-42c and 6-62c. No Southern hybridizations were performed with phages H71/1, H7/2, H71/5, and 12-41b; the nondigested DNAs of these four phages exhibited reproducible homologies. Dot blot hybridization revealed that the DNA of phage 12-41b exhibited a weak hybridization reaction to DNAs of phages H71/1 and H71/5, while there was no DNA homology between phages H7/2 and 12-41b (Fig. 3). In the Siphoviridae, phages H103/1, H105/1, and H108/1 exhibited DNA homology, while phages H118/1 and H120/1 were homologous to each other. The DNAs of two of the phages (10-33b and 10-94a) in the Podoviridae were homologous. No homology was observed for any of the other phages. According to the species concept of Ackermann et al. (1), phages belonging to the same family that exhibit DNA homology should be assigned to the same species. Within the Myoviridae six species could be differentiated; four species were differentiated within the Siphoviridae, and three species were differentiated in the Podoviridae. Altogether, 13 species were identified among the 22 phages tested.
FIG. 3.
DNA-DNA hybridization of 22 marine bacteriophages. •, strong hybridization signal; ○, weak hybridization signal. The families and morphotypes of the phages and DNA probes are indicated.
Morphology, physiology, and phylogenetic analysis of host bacteria.
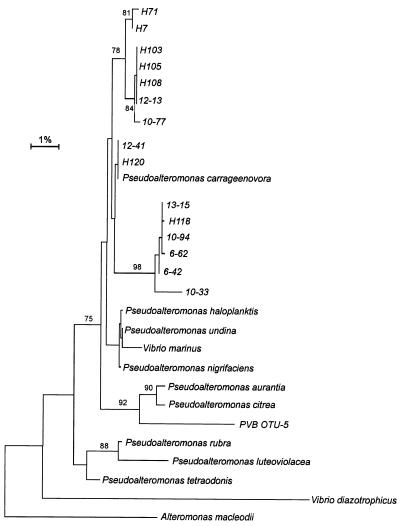
All of the bacteria examined were gram negative, motile, catalase and oxidase positive, and facultatively anaerobic. A comparative analysis of partial sequences (ca. 500 bp) of all of the isolates revealed that they belong to the γ subdivision of the class Proteobacteria and that Pseudoalteromonas species are the closest relatives (Fig. 4). The levels of sequence similarity for the host bacteria were more than 99%. The levels of similarity between the isolates and their closest known relatives were around 98%. A phylogenetic analysis performed with nearly complete 16S rRNA sequences of three isolates (H7, 12-13, and H120) gave a similar tree, which had a slightly different branching order.
FIG. 4.
Phylogenetic affiliations of host bacteria: distance tree based on the first 500 bp of 16S ribosomal DNA. The bootstrap values obtained with 100 replicates are shown. Only bootstrap values equal to or greater than 75% are shown.
DISCUSSION
Bacteriophages are ubiquitous, very abundant, and morphologically diverse in marine environments (4, 9, 16, 17). The present study provides data which reveal the great genetic diversity of marine bacteriophages, which exceeds the morphological diversity. The great genetic diversity represents a large gene pool for horizontal and vertical gene transfer, which supports the view that marine bacteriophages play an important role in controlling bacterioplankton both quantitatively and qualitatively.
Classification of marine phages.
Criteria given by the International Committee of Taxonomy of Viruses were used for taxonomic classification of the 22 marine bacteriophages investigated. According to these criteria, all of the phages belong to the order Caudovirales (tailed bacteriophages), a very common, widely distributed group. Up to 1993, 4,007 phages had been described in detail; 96% of these phages had tails and thus belonged to the Caudovirales, while only 4% had filamentous, isometric, or pleomorphic morphology (28). About 150 marine bacteriophages have been isolated and described, and only one phage, PM2 (15), has been classified as a member of the family Corticoviridae (37). The other 149 phages belong to families of tailed phages (16).
The phages investigated in this study were found to belong to all three families in the Caudovirales (Myoviridae, Siphoviridae, and Podoviridae). These phages have icosahedral heads with diameters between 50.2 and 99.3 nm. The majority of cultured marine phages studied so far have the same characteristics (6); therefore, the phages investigated belong to this major group. In contrast, estimates of diameters of phage heads based on direct counting of marine phages by electron microscopy revealed that most phages in marine environments are smaller (diameters, 30 to 60 nm) (6, 11).
Almost all of the phages isolated near Helgoland had morphological structures that have been described previously (16). The only exception was phage 11-68c, which belongs to the Siphoviridae and has a hook at the end of its tail; this morphology has not been reported previously for the marine environment. Phage H7/2 has been described previously (16), and three other phages belonging to the Myoviridae had the same collarlike structure between the head and the tail. Interestingly, the morphology of the members of the Podoviridae was less diverse than the morphology of the members of the Myoviridae and Siphoviridae.
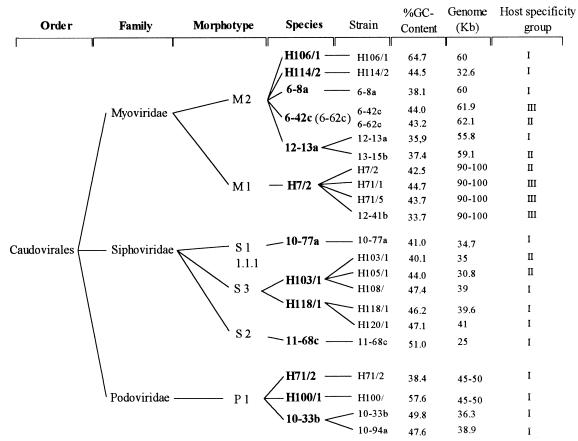
The species to which phages belong were determined in this study by using the concept of Ackermann et al. (1), which is based on the occurrence of DNA homologies in phages belonging to the same family. Like Jarvis (23) and Krylov et al. (25), Ackermann et al. did not observe DNA homology between phages belonging to different families. The present study confirmed the findings of Ackermann et al. since there was no DNA homology between phages belonging to different families, while several species were found in each family. For example, four phages belonging to the Myoviridae that had the collarlike structure were found to belong to the same species by DNA homology studies. Different phages belonging to the same species were also characterized by using several strain-specific markers (1, 28, 37), including host range, density of phage particles, and GC content of the phage DNA (Fig. 5).
FIG. 5.
Arrangement of 22 phages as members of an order, families, morphotypes, and species. Phage density data, GC contents of DNAs, and host range groups are also shown.
A total of 22 marine bacteriophages were tested, and 13 new species based on the absence of DNA homology and 22 different phage strains were differentiated, which revealed that the genetic diversity was high, much higher than the morphological diversity (Fig. 5). The very broad host ranges of phages belonging to the Myoviridae compared to the host ranges of phages belonging to the Siphoviridae and Podoviridae supports the findings of Suttle and Chan (44). In a study of cyanophages these authors found that phages belonging to the latter two families were more host specific than phages belonging to the Myoviridae.
Classification of host bacteria.
The majority of all marine bacteriophages are highly host specific (6, 12, 32), and 73% lyse only the original host bacterium. However, a few phages were able to form plaques on more than 50% of the 36 bacterial strains investigated here. Therefore, the relationships of the bacterial isolates can be deduced from their phage sensitivity patterns (22, 35, 36). Moebus and Nattkemper (33) studied the taxonomy of 31 bacterial isolates obtained from phage-host systems isolated near Helgoland. These authors reported that all of their strains were members of the family Vibrionaceae and most were members of the genus Vibrio. All of the host bacteria investigated here belong to the γ subdivision of the class Proteobacteria, with levels of DNA sequence homology of more than 99%. They are all closely related to the genus Pseudoalteromonas. No phages for this genus have been described until now. However, the genus was described recently, and some species formerly classified as members of the genus Vibrio (e.g., Vibrio marinus) (18, 40) are now thought to be closely related to the Pseudoalteromonas group.
Although we investigated only a small part of the marine bacterial community, we established that there is great genetic variation in the infectious marine bacteriophages, which leads to high levels of species and strain diversity. It is likely that with in future studies increased genetic diversity among phages will be discovered, especially in groups of bacteria belonging to the “silent majority” of marine bacteria that have not been cultured yet.
ACKNOWLEDGMENTS
We are very grateful to K. Moebus (Biologische Anstalt Helgoland) for his very generous gift of the phage-host systems which we investigated in this study. We thank Bärbel Jungnickl for her help with the preparation of the photographic prints.
This investigation was supported by a grant from the Biologische Anstalt Helgoland.
REFERENCES
- 1.Ackermann H-W, DuBow M S, Jarvis A W, Jones L A, Krylov V N, Maniloff J, Rocourt J, Safferman R S, Schneider J, Seldin L, Sozzi T, Steward R, Werquin M, Wünsche L. The species concept and its application to tailed phages. Arch Virol. 1992;124:9–82. doi: 10.1007/BF01314626. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D A, Boguski M S, Lipman D J, Ostell J. GenBank. Nucleic Acids Res. 1997;25:1–6. doi: 10.1093/nar/25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergh O, Børsheim K Y, Bratbak G, Heldal M. High abundances of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 5.Boehme J, Frischer M E, Jiang S C, Kellogg C A, Pichard S, Rose J B, Steinway C, Paul J H. Viruses, bacterioplankton, and phytoplankton in the southeastern Gulf of Mexico: distribution and contribution to oceanic DNA-pools. Mar Ecol Prog Ser. 1993;97:1–10. [Google Scholar]
- 6.Børsheim K Y. Growth and mortality of bacteria in aquatic environment. Ph.D. thesis. Trondheim, Norway: University of Trondheim; 1992. [Google Scholar]
- 7.Børsheim K Y. Native marine bacteriophages. FEMS Microbiol Ecol. 1993;102:141–159. [Google Scholar]
- 8.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratbak G, Thingstad F, Heldal M. Viruses and the microbial loop. Microb Ecol. 1994;28:209–221. doi: 10.1007/BF00166811. [DOI] [PubMed] [Google Scholar]
- 10.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1996;20:301–309. [Google Scholar]
- 11.Cochlan W P, Wikner J, Steward G F, Smith D C, Azam F. Spatial distribution of viruses, bacteria and Chl a in neritic, oceanic and estuarine environments. Mar Ecol Prog Ser. 1993;92:77–87. [Google Scholar]
- 12.Coetzee J N. Bacteriophage Taxonomie. In: Goyal S M, Gerba C P, Bitton G, editors. Phage ecology. New York, N.Y: Wiley and Sons Interscience; 1987. pp. 45–86. [Google Scholar]
- 13.Cottrell M T, Suttle C A. Genetic diversity of algal viruses which lyse the photosynthetic picoflagellate Micromonas pusilla. Appl Environ Microbiol. 1995;61:3088–3091. doi: 10.1128/aem.61.8.3088-3091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. “Touch down” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espejo R T, Canelo E S. Properties of bacteriophage PM2: lipid-containing bacterial virus. Virology. 1968;34:738–747. doi: 10.1016/0042-6822(68)90094-9. [DOI] [PubMed] [Google Scholar]
- 16.Frank H, Moebus K. An electron microscopic study of bacteriophages from marine waters. Helgol Wiss Meeresunters. 1987;41:385–414. [Google Scholar]
- 17.Fuhrmann J A, Suttle C A. Viruses in marine planctonic systems. Oceanography. 1993;6:51–63. [Google Scholar]
- 18.Gauthier G, Gauthier M, Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 19.Gelderblom H R, Bauer H, Frank H, Wigand R. The structure of group II adenovirus. J Gen Virol. 1967;1:553–560. doi: 10.1099/0022-1317-1-4-553. [DOI] [PubMed] [Google Scholar]
- 20.Gelderblom H R, Renz H, Özel M. Negative staining in diagnostic virology. Micron Microsc Acta. 1991;22:435–447. [Google Scholar]
- 21.Gilbert D G. SeqApp—a biosequence analysis application. Bloomington: Indiana University; 1992. [Google Scholar]
- 22.Goyal S M. Methods in phage ecology. In: Goyal S M, Gerba C P, Bitton G, editors. Phage ecology. New York, N.Y: Wiley and Sons Interscience; 1987. pp. 267–288. [Google Scholar]
- 23.Jarvis A W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984;47:343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S C, Paul J H. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar Ecol Prog Ser. 1994;104:163–172. [Google Scholar]
- 25.Krylov V N, Tolmachova T O, Akhverdian V Z. DNA homology in species of bacteriophages active on Pseudomonas aeruginosa. Arch Virol. 1993;131:141–151. doi: 10.1007/BF01379086. [DOI] [PubMed] [Google Scholar]
- 26.Lane D J. 16S/23SrRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 27.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniloff J, Ackermann H-W, Jarvis A. Bacteriophage taxonomy and classification. In: Webster R G, editor. Encyclopedia of virology. New York, N.Y: Plenum Press; 1994. pp. 93–100. [Google Scholar]
- 29.Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 30.Moaledj K. Über schnelle miniaturisierte Bestimmungsverfahren für Bakterienpopulationen in aquatischen Ökosystemen. Arch Hydrobiol. 1984;100:99–121. [Google Scholar]
- 31.Moebus K. A method for the detection of bacteriophages from ocean water. Helgol Wiss Meeresunters. 1980;34:1–14. [Google Scholar]
- 32.Moebus K, Nattkemper H. Bacteriophage sensitivity patterns among bacteria isolated from marine waters. Helgol Wiss Meeresunters. 1981;34:375–385. [Google Scholar]
- 33.Moebus K, Nattkemper H. Taxonomic investigations of bacteriophage sensitive bacteria isolated from marine waters. Helgol Wiss Meeresunters. 1983;36:357–373. [Google Scholar]
- 34.Moebus K. Lytic and inhibition responses of bacteriophages among marine bacteria, with special reference to the origin of phage-host systems. Helgol Wiss Meeresunters. 1983;36:375–391. [Google Scholar]
- 35.Moebus K. Preliminary observations on the concentration of marine bacteriophages in the water around Helgoland. Helgol Wiss Meeresunters. 1992;45:411–422. [Google Scholar]
- 36.Moebus K. Further investigations on the concentration of marine bacteriophages in water around Helgoland, with references to the phage-host systems encountered. Helgol Wiss Meeresunters. 1992;46:275–292. [Google Scholar]
- 37.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of viruses. Vienna, Austria: Springer; 1995. [Google Scholar]
- 38.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:164–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 39.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. [Google Scholar]
- 40.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christen R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416.426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 41.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Ehrlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA-polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 44.Suttle C A, Chan A M. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar Ecol Prog Ser. 1993;92:99–109. [Google Scholar]
- 45.Suttle C A, Chan A M, Cottrell M T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;134:467–469. [Google Scholar]
- 46.Wrigley N G. The lattice spacing catalase as an internal standard of length in electron microscopy. J Ultrastruct Res. 1968;24:454–464. doi: 10.1016/s0022-5320(68)80048-6. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto K R, Alberts B M. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]