Abstract
Lactococcus lactis subsp. cremoris AM2 was previously shown to lyse early and extensively during cheese ripening (M.-P. Chapot-Chartier, C. Deniel, M. Rousseau, L. Vassal, and J.-C. Gripon, Int. Dairy J. 4:251–269, 1994). We analyzed the bacteriolytic activities of autolytic strain AM2 by using renaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis performed with two different substrates in the gel, Micrococcus lysodeikticus and L. lactis autoclaved cells. Several lytic activities were detected in L. lactis AM2; a major lytic activity, designated A2 (46 kDa), was found only with the L. lactis cell substrate. This activity appears to be different from major peptidoglycan hydrolase AcmA characterized previously (G. Buist, J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrickman, J. Bacteriol. 177:1554–1563, 1995), which has a similar molecular mass. The two enzymes differ in substrate specificity as well as in sensitivity to pH and different chemical compounds. L. lactis AM2 is lysogenic and mitomycin C inducible. Enzyme A2 was shown to be inducible by mitomycin C and to be prophage encoded. It was identified as an enzyme similar to the lysin encoded by lactococcal small isometric temperate bacteriophages. A prophage-cured derivative of L. lactis AM2 was obtained, and this isolate exhibited different autolytic properties than AM2. After prolonged incubation in the stationary phase after growth on M17 medium, the extent of lysis of an AM2 culture was 60%, whereas over the same period there was almost no lysis in a prophage-cured derivative strain culture. These results suggest that the prophage lytic system is involved in the strain AM2 lysis observed in liquid medium and that it could also be involved in the lysis observed during cheese ripening.
Lactococci, which are widely used as starter bacteria in cheesemaking, participate in the development of cheese flavor through the actions of their enzymes (41). Autolysis of these organisms appears to be a crucial step in the release of intracytoplasmic enzymes, such as peptidases that produce free amino acids which are aroma precursors and degrading hydrophobic peptides which are responsible for bitter taste. This view is supported by the results of several experimental studies which correlated enhancement of bacterial lysis with an increase in the free amino acid production rate and a decrease in bitter taste (3, 4, 9, 10, 21). The autolytic properties of lactococci, which have been studied in buffer or liquid medium (3, 4, 23, 40) as well as in cheese (8, 43), appear to vary from strain to strain. The following factors could account for the different autolytic behaviors: the cellular cell wall hydrolase contents, regulation of the expression of the enzymes, and cell wall composition. Another possibility is that a prophage is involved, as previously proposed (12), since numerous strains of lactococci are lysogenic (11).
Bacteria synthesize cell wall hydrolases which are capable of hydrolyzing the peptidoglycan in their own cell envelopes and are also called autolysins. The following four types of enzymes are distinguished on the basis of their cleavage specificities: β-N-acetylmuramidases (lysozymes), β-N-acetylglucosaminidases, N-acetylmuramyl-l-alanine amidases, and endopeptidases (37). These enzymes, which are localized in the cell wall and are potentially lethal to the cells, are synthesized during bacterial growth, and it has been proposed that they are involved in different cellular processes, including cell division, cell wall turnover, and transformation. Cell wall hydrolases (lysins) are also encoded by bacteriophage DNA and are synthesized during the bacteriophage lytic cycle. The structural resemblance and potential evolutionary link between bacteriophage lysins and bacterial autolysins have been described previously for gram-positive bacteria, such as Streptococcus pneumoniae (14, 15) and Bacillus subtilis (13, 25). In the case of Lactococcus lactis bacteriophages, a second protein, called holin, is required for cell lysis. This protein is a small protein which is able to make holes in the cytoplasmic membrane and allows the lysin to reach the cell wall peptidoglycan (44).
In previous studies, workers started to evaluate the peptidoglycan hydrolase contents of different L. lactis strains. The renaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) technique (24, 33), which permits detection of hydrolase activity after renaturation in a substrate-containing gel, revealed activity bands in L. lactis (6, 32, 35). The gene encoding the major peptidoglycan hydrolase (AcmA) of L. lactis MG1363 has been cloned and sequenced (6). This gene encodes a 46.5-kDa protein which is probably a muramidase (30), is required for cell separation during growth, and appeared to be present in all of the strains tested.
In a previous study, we compared the autolytic behaviors of two L. lactis starter strains during ripening of pressed-curd cheese (8). We observed that L. lactis subsp. cremoris AM2 autolyzes early and extensively, whereas L. lactis subsp. lactis NCDO763 does not autolyze over a period of several months.
In this study, we used the renaturing SDS-PAGE technique to analyze the bacteriolytic activity profile of the autolytic L. lactis strain AM2 and to compare this profile with that of the nonautolytic strain NCDO763 in order to address the question of the different autolytic behaviors. In strain AM2 with a lactococcal cell substrate, we detected a major bacteriolytic activity at 46 kDa, which was not present in strain NCDO763 or in MG1363 and was different from the previously described AcmA activity. We found that this enzyme was actually encoded by prophage DNA present in lysogenic strain AM2. We propose that a prophage is involved in the lysis observed in strain AM2 during cheese ripening.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. lactis strains were obtained from the CNRZ collection (Institut National de la Recherche Agronomique, Jouy-en-Josas, France). The strains were grown at 30°C in M17 broth (39) (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% (wt/vol) lactose (M17-lac broth); the only exception was strain MG1363, which was grown in M17 broth supplemented with 0.5% (wt/vol) glucose. Growth was monitored by measuring the optical density at 650 nm (OD650) with a spectrophotometer (model Uvikon 931; Kontron Instruments Inc., Everett, Mass.).
General DNA techniques.
Restriction enzymes were obtained from Boehringer Mannheim and were used as recommended by the supplier. Molecular cloning, purification, and analysis of DNA were performed by using standard procedures (36). The pBluescript vector was obtained from Stratagene (La Jolla, Calif.).
PCR was carried out by using a model 480 DNA thermal cycler (Perkin-Elmer, Norwalk, Conn.) and Taq DNA polymerase according to the instructions of the manufacturer (Oncor, Inc., Gaithersburg, Md.). Primers LYS-1 (5′-ATGAAAAGATTAATCAAAAAATCTG; positions 463 to 487) and LYS-2 (5′-TTAATAATTTAGWGTTTGACCAG; positions 1749 to 1727) were deduced from the previously published sequence of the lysin gene (lys) of lactococcal bacteriophage Tuc2009 (2). Primers PALA-4 (5′-CTTCAACAGACAAGTCC; positions 319 to 335) and PALA-14 (5′-GATAAATGATTCCAAGC; positions 1448 to 1432) corresponded to internal sequences of the acmA gene (6).
Total DNA of L. lactis strains were prepared as previously described (26). They were digested with the appropriate restriction enzyme (Boehringer Mannheim), electrophoresed in a 0.7% agarose gel, and blotted onto a Hybond-N nylon membrane (Amersham International, Amersham, United Kingdom) by the Southern method as described by Sambrook et al. (36). Probes corresponding to the fragments amplified by PCR performed with the primers described above from the total DNA of L. lactis MG1363 (for acmA) or AM2 (for lys) were used as probes for detection of the lys and acmA genes in total DNA from strains. Labeling of probes and detection were performed with an ECL (enhanced chemiluminescence) gene detection system (Amersham) used according to the manufacturer’s instructions.
DNA sequencing was performed with an ABI PRISM Cycle Sequencing Ready Reaction kit (Perkin-Elmer) used according to the supplier’s recommendations. The DNA sequence was determined with an Applied Biosystems model 373A automated DNA sequencer (Perkin-Elmer).
SDS-PAGE and renaturing SDS-PAGE.
SDS-PAGE was carried out as described by Laemmli (22) with a Mini Protean II cell unit (Bio-Rad Laboratories, Inc., Hercules, Calif.) and a gel that was 75 by 55 mm.
Renaturing SDS-PAGE was performed as previously described (24, 33); 0.2% (wt/vol) Micrococcus lysodeikticus ATCC 4698 (Sigma Chemicals Co., St. Louis, Mo.) autoclaved cells or 0.4% (wt/vol) L. lactis AM2 autoclaved cells were included in 10 or 12% polyacrylamide gels for detection of bacteriolytic activities. Samples were solubilized in sample buffer (62.5 mM Tris-HCl [pH 6.8], 2.3% [wt/vol] SDS, 50 mM dithiothreitol [DTT], 10% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue) and heated for 3 min at 100°C. When necessary, the sample was centrifuged, and the soluble material was loaded onto the gel.
After electrophoresis, the gels were washed for 30 min in distilled water at room temperature with gentle shaking, and then they were transferred into renaturation buffer containing 50 mM MES (2-morpholinoethanesulfonic acid) (Sigma)–NaOH (pH 6.0) and 0.1% (wt/vol) Triton X-100. The gels were incubated with gentle shaking for 16 h at 37°C. They were rinsed with distilled water, stained with 0.1% (wt/vol) methylene blue in 0.01% (wt/vol) KOH for 2 h at room temperature with gentle shaking (20), and destained with distilled water. The bacteriolytic activities appeared as clear bands on a blue background.
Molecular masses were determined by comparison with molecular mass standards electrophoresed on the same gel and stained with Coomassie blue (Fast Stain concentrate; Zoion Biotech, Newton, Mass.). The standards were purchased from Bio-Rad Laboratories and contained phosphorylase b (molecular weight, 97,400), bovine serum albumin (66,200), ovalbumin (45,000), carbonic anhydrase (31,000), soybean trypsin inhibitor (21,500), and lysozyme (14,400).
The influence of pH and the influence of various chemical compounds on bacteriolytic activities were determined by renaturing SDS-PAGE and incubation of different gel slices in renaturation buffers containing different compounds. The following buffers were used to determine the influence of pH: 50 mM sodium acetate (pH 5.0), 50 mM MES–NaOH (pH 6.0), 50 mM Tris-HCl (pH 7.0, 7.5, and 8.0), and 50 mM TAPS [N-tris(Hydroxymethyl)methyl-3-aminopropanesulfonic acid] (Sigma)–NaOH (pH 9.0). In addition, 100 mM NaCl, 100 mM KCl, 10 mM CaCl2, 10 mM MgCl2, 10 mM ZnCl2, 10 mM CuCl2, 10 mM EDTA, 10 mM iodoacetic acid, and 10 mM DTT were tested by adding them to renaturation buffer which contained 0.1% Triton X-100.
Preparation of cell extracts and cell wall extracts.
L. lactis cells grown in M17 medium were recovered by centrifugation at 8,000 × g for 15 min at 4°C and washed with cold 50 mM Tris-HCl (pH 7.0) buffer. Various cell extracts and cell fractions were prepared as described below in order to determine bacteriolytic activities by renaturing SDS-PAGE. The culture supernatant was filtered through 0.45-μm-pore-size filters (Millex-HA; Millipore S.A., Bedford, Mass.) and tested for the presence of bacteriolytic activities.
(i) SDS cell extract.
Cells were resuspended in SDS-PAGE sample buffer and heated for 3 min at 100°C, and the insoluble material was removed by centrifugation at 13,000 × g for 10 min. The supernatant (SDS cell extract) was analyzed by renaturing SDS-PAGE.
(ii) LiCl cell extract.
The cells from a 100-ml culture were resuspended in 1 ml of a solution containing 50 mM Tris-HCl (pH 7.5), 4 M LiCl, and 1 mM phenylmethylsulfonyl fluoride. The suspension was incubated at 4°C for 30 min. The cells were then removed by centrifugation, and the supernatant was dialyzed against 100 mM LiCl–50 mM Tris-HCl (pH 7.0) and examined by renaturing SDS-PAGE.
(iii) Native cell walls.
Native cell walls were prepared after disruption of cells with glass beads. The cells were resuspended in 10 mM sodium phosphate buffer (pH 7.0) containing 1 mM phenylmethylsulfonyl fluoride and were disrupted with glass beads (diameters, 150 to 212 μm; Sigma) in a bead beater (Vibrogen). The cell walls were recovered by centrifugation at 25,000 × g for 15 min at 4°C. They were washed twice with distilled water, twice with 10 mM sodium phosphate buffer (pH 7.0), and then twice with distilled water again. The pellet was finally resuspended in distilled water.
(iv) Cell wall extract and cytoplasmic extract.
Spheroplasts were prepared by lysozyme treatment in sucrose-containing buffer as described previously (45). The supernatant (cell wall extract) was recovered. The spheroplast pellet was resuspended in hypotonic buffer (50 mM Tris-HCl [pH 7.0], 1 mM MgCl2, 25 U of RNase A [Sigma] per ml, 50 U of DNase I [Sigma] per ml), incubated on ice for 30 min, and centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was used as the cytoplasmic extract.
Mitomycin C induction.
A 16-h culture was used to inoculate M17-lac broth to an initial OD650 of 0.05. The culture was incubated at 30°C until the OD650 was 0.3, and mitomycin C (Sigma) was added to a final concentration of 1 μg/ml as described previously (34). The culture was incubated further at 30°C, and the OD650 was monitored regularly. A culture grown in M17-lac broth at 30°C was used as a control.
Preparation and analysis of bacteriophage DNA.
A 30-ml M17-lac broth culture of L. lactis AM2 was induced with mitomycin C as described above. After lysis was completed, the lysate was centrifuged at 12,000 × g for 10 min at 15°C to eliminate the cell debris, and the supernatant was filtered through a 0.45-μm-pore-size filter (Millex-HA; Millipore). Bacteriophage particles were precipitated with polyethylene glycol 4000, and the phage DNA was extracted as described by Sambrook et al. (36).
Electron microscopy.
Fifty milliliters of L. lactis AM2 lysate obtained after mitomycin C induction was centrifuged to eliminate the cell debris. The supernatant was filtered as described above and then centrifuged at 82,000 × g for 2 h at 4°C with an ultracentrifuge (model Centrikon T-1080; Kontron Instruments Inc.). The pellet containing the phage particles was resuspended in 10 mM Tris-HCl–10 mM MgCl2 (pH 8.0). The phage particles were stained with 2% uranyl acetate, as previously described (1), and were observed with a Zeiss model EM-10 electron microscope operating at 80 kV.
Prophage curing.
The method used for prophage curing has been described previously by Gasson and Davies (16). Different dilutions in Ringer’s solution of an exponential-phase culture (OD650, 2.0) were spread onto M17-lac agar plates and subjected to UV irradiation at a wavelength of 310 nm for 5 s with a UV table. The plates were incubated for 48 h at 30°C shielded from light. A total of 400 surviving clones were examined for prophage sensitivity. These clones were resuspended in 200 μl of 10% (vol/vol) M17 broth, and single drops (5 μl) of the suspensions were placed onto M17-lac agar plates containing 10 mM calcium chloride that previously had been inoculated with a mitomycin C lysate containing bacteriophage particles. The plates were incubated at 30°C overnight. Prophage-cured clones were tested to determine their sensitivity to the bacteriophage by determining whether clear zones of lysis were produced in the spot test. However, no sensitive clones were isolated in this way. Ten clones were then tested to determine their sensitivity to mitomycin C. One of those clones was not inducible, suggesting that it had lost the prophage. We confirmed that this clone was a prophage-cured derivative of AM2 (designated AM2-C) by Southern hybridization with a lysin lys gene probe.
Strain AM2-C was not sensitive to bacteriophage φAM2 isolated from strain AM2 after mitomycin C induction, as determined by the double-layer technique (18) or in liquid medium. This could have resulted either from a phage defect or from the presence of another integrated prophage that conferred immunity. In any case, this explains why during screening for phage sensitivity on agar plates we failed to isolate a cured AM2 derivative.
Autolysis in buffer and in M17 broth.
Exponential-phase cells (OD650, 1.0) grown in M17-lac broth were harvested by centrifugation at 8,000 × g for 10 min at 4°C. The pellet was washed in 50 mM sodium acetate buffer (pH 5.0), 50 mM MES–NaOH buffer (pH 6.0), or 50 mM sodium phosphate buffer (pH 7.0). The cells were resuspended in the same buffer to an initial OD600 of 0.7. The cell suspensions were incubated at 30°C, and autolysis was monitored by determining the decrease in OD600. The extent of autolysis was expressed as the percent decrease in OD600 after some time.
The autolysis of stationary-phase cells in M17 broth at 30°C over a several-day period was monitored by determining the change in OD650. Cell viability was determined after the cells were plated onto M17 agar plates and incubated for 48 h at 30°C. The release of X-prolyl-dipeptidyl aminopeptidase (PepX) was monitored by measuring the enzymatic activity in the culture supernatant with the Ala-Pro-paranitroanilide (Bachem, Bubendorf, Switzerland) substrate as described by Zevaco et al. (45). The activity unit used was the katal (1 kat of enzyme activity released 1 mol of paranitroaniline per s).
RESULTS
Detection of the bacteriolytic activities of the autolytic strain L. lactis subsp. cremoris AM2 by renaturing SDS-PAGE.
L. lactis subsp. cremoris AM2 was described previously as an autolytic strain during cheese ripening (8). We analyzed the bacteriolytic activities of strain AM2 by the renaturing SDS-PAGE technique and compared these activities to the activities of L. lactis subsp. lactis NCDO763, which has been described as a nonautolytic strain (8), and L. lactis subsp. cremoris MG1363, which exhibits a major AcmA autolysin cloned and sequenced previously (6). Two different substrates were included in the gel, autoclaved M. lysodeikticus and L. lactis cells. The presence of enzymatic activities was first determined with SDS cell extracts obtained from the three strains grown in M17 medium.
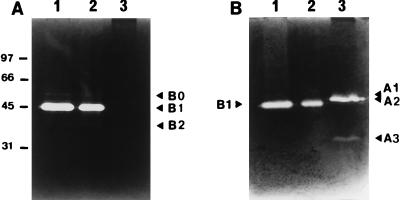
With the M. lysodeikticus substrate, no activity band was detected in the strain AM2 SDS cell extract (Fig. 1A, lane 3). Under the same conditions, the activity profiles obtained with L. lactis NCDO763 and MG1363 were consistent with the results of a previous study (6), and the following three activity bands were identified (Fig. 1A, lanes 1 and 2): band B1, a major band at 45 kDa which corresponded to major peptidoglycan hydrolase AcmA; band B0, a minor band at 50 kDa which was found to be a precursor of AcmA (6); and band B2, a minor band at 38 kDa which was probably a degradation product of AcmA (6).
FIG. 1.
Detection of bacteriolytic activities of L. lactis MG1363, NCDO763, and AM2 by renaturing SDS-PAGE. (A) Gel containing autoclaved M. lysodeikticus cells as the substrate. (B) Gel containing autoclaved L. lactis cells as the substrate. Lane 1, MG1363 SDS cell extract; lane 2, NCDO763 SDS cell extract; lane 3, AM2 SDS cell extract. The positions of the lytic bands are indicated by arrowheads. The numbers on the left are molecular masses (in kilodaltons).
With the L. lactis substrate under the same conditions, three activity bands were detected in AM2, major band A2 (46 kDa) and minor bands A1 (50 kDa) and A3 (34 kDa) (Fig. 1B, lane 3). Detection of the minor bands varied from one experiment to another. For strains NCDO763 and MG1363 (Fig. 1B, lanes 1 and 2), band B1 was detected, but the band intensity was lower than the band intensity on the micrococcus substrate, as previously reported (6, 32).
Thus, the major bacteriolytic activity detected in L. lactis AM2, A2 activity, appears to be different from AcmA; the two activities differ in substrate specificity, and the A2 activity has a slightly higher molecular weight than AcmA.
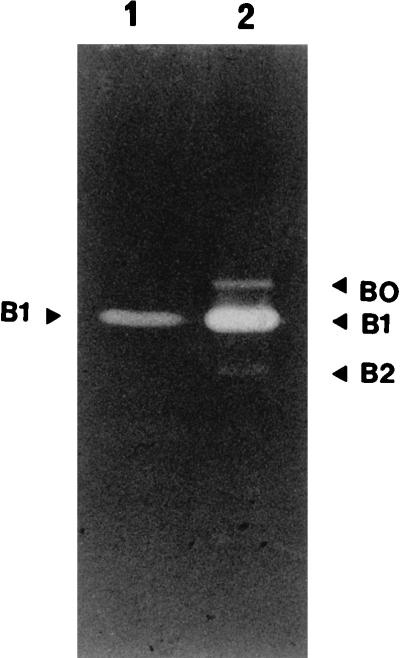
Despite the fact that AcmA activity was not detected in the SDS cell extract of L. lactis AM2, the presence of the acmA gene in AM2 total DNA was shown by the results of PCR amplification with specific primers derived from the acmA sequence and by Southern hybridization (data not shown). Total DNA digests from L. lactis AM2 and MG1363 were probed with the acmA PCR product. The acmA gene was found to reside on a 2.5-kb EcoRV fragment (data not shown). In addition, AcmA activity was detected in a native cell wall preparation obtained from L. lactis AM2 when M. lysodeikticus was the substrate (Fig. 2). These results suggest that the amount of autolysin AcmA in strain AM2 is lower than the amounts of AcmA in other strains or that it is more difficult to extract AcmA from strain AM2 than from other strains because of differences in the cell envelope structure.
FIG. 2.
Detection of bacteriolytic activities of L. lactis NCDO763 and AM2 by renaturing SDS-PAGE in a gel containing autoclaved M. lysodeikticus cells as the substrate. Lane 1, native cell wall preparation from strain AM2; lane 2, NCDO763 SDS cell extract. The positions of the lytic bands are indicated by arrowheads.
Different cell extracts and fractions (native cell wall, cell wall extract prepared with lysozyme, cytoplasmic fraction, LiCl cell extract) of strains AM2 and NCDO763 were prepared and tested to determine whether there were extra bacteriolytic activities in gels containing micrococci or lactococci, but no extra lytic activity was detected (data not shown). When the cells were harvested at different times during cell growth, no extra bands were observed in the activity profiles.
The effects of the renaturing buffer pH on the two main activities, A2 and AcmA, were studied by incubating gel slices in different buffer solutions. The optimum pH for A2 activity ranged from 5.0 to 6.0, whereas the optimum pH for AcmA activity ranged from 6.0 to 7.0. The effects of different cations and chemical compounds on the A2 and AcmA activities were also examined. The intensity of the AcmA activity band was enhanced by Ca2+, Mg2+, and Mn2+ and was decreased by Cu2+. A2 activity was inhibited by Cu2+ and was partially inhibited by Zn2+. EDTA, DTT, and iodoacetic acid had no effect on either activity.
The differences in the optimal pH values and sensitivities to some cations between the AcmA and A2 activities and the differences in substrate specificities and molecular masses strongly suggested that these activities correspond to two different bacteriolytic enzymes.
Identification of the major bacteriolytic activity detected in L. lactis subsp. cremoris AM2 as a mitomycin C-inducible lytic enzyme.
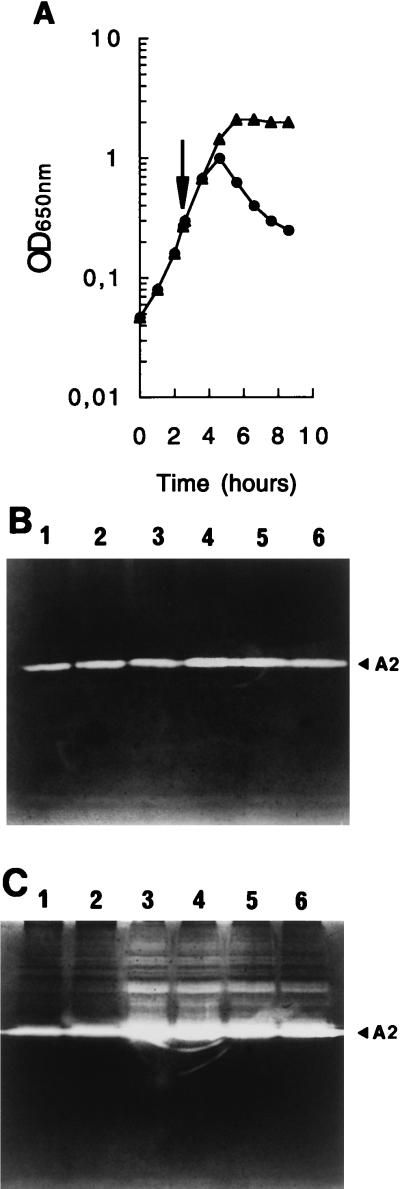
Numerous L. lactis strains have been found to be lysogenic, and the majority of them can be induced by treatment with mitomycin C (11). An exponentially growing culture of strain AM2 (OD650, 0.3) was exposed to mitomycin C (1 μg/ml). Two hours after the addition of mitomycin C, growth was arrested and the culture lysed, which resulted in a decrease in the OD650 to a value close to the initial value (Fig. 3A).
FIG. 3.
Effect of mitomycin C on the growth of L. lactis subsp. cremoris AM2 (A) and on its bacteriolytic activity profile (B and C). (A) Symbols: ▴, untreated culture; •, mitomycin C induction. The arrow indicates the time of mitomycin C (1 μg/ml) addition. (B and C) Samples were removed at 1-h intervals from the untreated culture (B) or from the mitomycin C-induced culture (C), and SDS cell extracts containing the same amount of proteins were analyzed by renaturing SDS-PAGE. Lanes 1 through 6, samples taken at zero time and at 1, 2, 3, 4, and 5 h after mitomycin C addition. The position of lytic band A2 is indicated by an arrowhead.
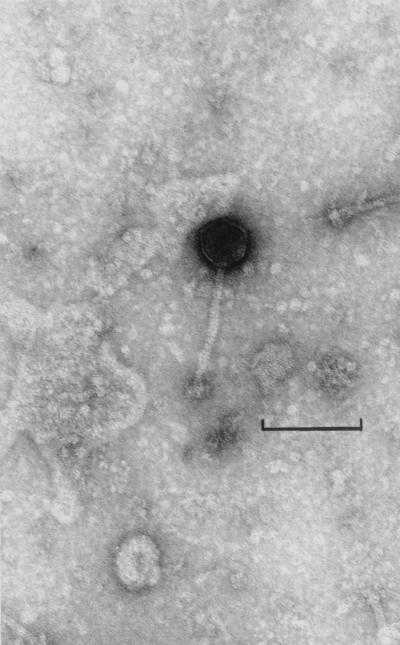
Electron microscopy of the AM2 lysate revealed the presence of a multitude of intact bacteriophage particles (Fig. 4), confirming that strain AM2 is lysogenic. A single kind of particles was present; each particle had a small isometric head, a noncontractile tail, and a base plate. The tail was about 120 nm long, and the head was about 44 nm in diameter. To characterize the bacteriophage liberated (designated φAM2), the phage DNA was extracted and digested with EcoRI (data not shown). The sum of the sizes of all of the resulting fragments was about 43 kb; these fragments probably represented the genome of a single lactococcal bacteriophage (19).
FIG. 4.
Electron micrograph of bacteriophage particles observed in an L. lactis AM2 lysate after mitomycin C induction. Bar = 50 nm.
During mitomycin C induction of L. lactis AM2, samples were removed at 1-h intervals, and the corresponding SDS cell extracts were prepared and analyzed by renaturing SDS-PAGE; the same amount of protein was placed in each well. Two hours after the addition of mitomycin C, the intensity of the A2 activity in the gel increased significantly (Fig. 3C), and it remained high over the next 3 h, during which cellular lysis took place. A concomitant increase in the A2 activity was observed in the culture supernatant (data not shown). In the control culture not treated with mitomycin C, the intensity of the A2 activity did not vary in this way during growth for the same period of time (Fig. 3B). Thus, major bacteriolytic enzyme A2 detected in AM2 appears to be mitomycin C inducible. This suggests that it is encoded by prophage DNA.
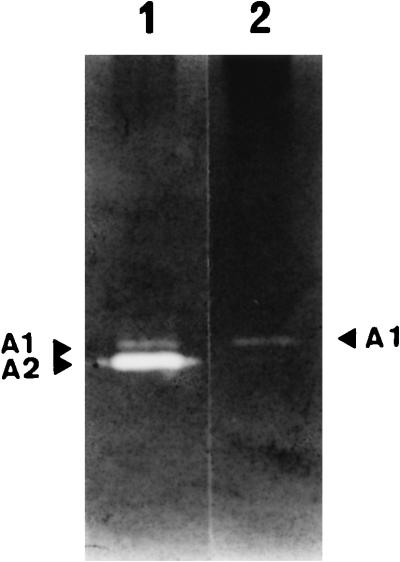
In order to confirm that the A2 enzyme was prophage encoded, a derivative of L. lactis AM2 cured of its prophage (designated strain AM2-C) was obtained as described in Materials and Methods. The bacteriolytic activities present in strain AM2-C were then determined by renaturing SDS-PAGE after preparation of an SDS cell extract. With the lactococcal substrate, A2 activity was not present in strain AM2-C, and only one of the weaker activity bands that detected in AM2, band A1 was present (Fig. 5). With the micrococcal substrate, as observed for strain AM2, no activity was detected in strain AM2-C (data not shown), although the acmA gene was detected in AM2-C total DNA by Southern hybridization with a 2.5-kb EcoRV fragment, as observed for strains AM2 and MG1363 (data not shown). These results confirmed that the major bacteriolytic activity detected in L. lactis AM2, A2 activity, is prophage encoded.
FIG. 5.
Bacteriolytic activity profile of prophage-cured derivative AM2-C of L. lactis AM2 as determined by renaturing SDS-PAGE performed with a gel containing autoclaved L. lactis cells as the substrate. Lane 1, AM2 SDS cell extract; lane 2, AM2-C SDS cell extract. The same amount of proteins was loaded into each well.
Identification of the major bacteriolytic enzyme detected in L. lactis AM2.
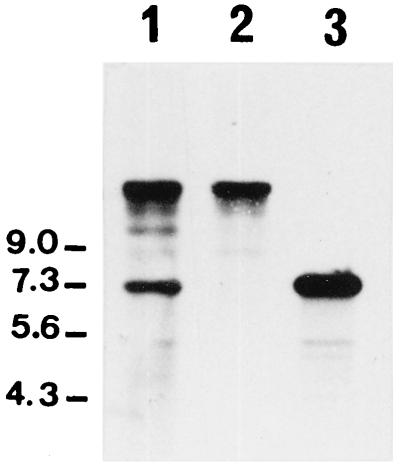
Several lysin genes of bacteriophages that infect L. lactis have been cloned and sequenced previously. Four different types of lysins, which do not exhibit sequence similarity to each other, have been characterized (17). One of these lysins, the lysin encoded by small isometric temperate bacteriophages Tuc2009 (2) and φLC3 (5), has a molecular mass of 46,000 Da, which is close to the molecular mass of the A2 lytic activity described above. Thus, we examined whether A2 could be identical to this enzyme. Using oligonucleotide primers corresponding to the 5′ and 3′ ends of the lysin structural gene (lys), we performed tests to determine the presence of the lysin gene in φAM2 bacteriophage DNA and in AM2 total DNA by PCR amplification. A DNA fragment that was 1.2 kb long (the expected size of the lysin gene) was amplified from both DNA templates. As a control for bacteriophage DNA purity, we verified that the lactococcal pepX gene (28, 31) could be amplified from AM2 total DNA but not from the bacteriophage DNA (data not shown). The lysin PCR amplification product was used to probe total DNA digests from both L. lactis AM2 and AM2-C and bacteriophage φAM2. The lys gene was found to reside on a 7.3-kb ClaI fragment in φAM2 and AM2 DNA but was not detected in AM2-C DNA, confirming that AM2-C was a bacteriophage φAM2-cured derivative of AM2 (Fig. 6). In addition, the probe hybridized with a second fragment which was larger than 9 kb, indicating that strains AM2 and AM2-C could contain another prophage that had a homologous lysin gene and was not inducible by mitomycin C. After being cloned in the E. coli pBluescript plasmid vector, the PCR amplification product was sequenced. The nucleotide sequence confirmed that the amplified gene was identical to the lysin lys gene.
FIG. 6.
Southern blot hybridization analysis of total DNA from different strains and bacteriophage φAM2. Lane 1, L. lactis subsp. cremoris AM2; lane 2, L. lactis subsp. cremoris AM2-C; lane 3, bacteriophage φAM2. DNA was cut with ClaI and hybridized with the 1.2-kb lys PCR product as the probe. The numbers on the left indicate molecular sizes (in kilobases).
Thus, the major bacteriolytic activity detected in strain AM2, A2 activity, is prophage encoded and is similar to the 46-kDa lysin previously identified in small isometric temperate bacteriophages Tuc2009 and φLC3.
Growth characteristics and temperature sensitivity of L. lactis AM2 and its prophage-cured derivative.
L. lactis AM2 was previously described as a temperature-sensitive strain, and it was postulated that the thermosensitivity of this organism results from induction of a thermoinducible prophage (12). Therefore, we examined whether the prophage-cured derivative obtained in this study, strain AM2-C, was sensitive to temperature.
In M17-lac broth, strains AM2 and AM2-C grew at the same rate at each temperature tested (30, 33, and 35°C) and did not grow at 37 and 40°C (data not shown).
The effect of a temperature shift to 40°C during growth at 30°C was then tested. The strains were grown in M17-lac broth at 30°C to an OD650 of 0.25, 0.3, or 0.5, and the cultures were subsequently shifted to 40°C. The two strains exhibited the same initial growth rate, followed by growth inhibition about 2 h after the temperature shift. A portion of the culture (about 25%) lysed after the growth inhibition (data not shown). Thus, prophage-cured derivative strain AM2-C was still temperature sensitive. The temperature sensitivity of the two strains could be attributed to the presence of a second prophage.
The only difference which was observed between the two strains grown in M17 medium was the fact that a strain AM2-C culture sedimented more rapidly than strain AM2 and produced cell aggregates, although the chains were not longer.
Autolytic properties of L. lactis AM2 and its prophage-cured derivative.
Exponential-phase cells of L. lactis AM2 and AM2-C were resuspended in buffers at different pH values between 5 and 7 and incubated at 30°C. Then lysis was monitored by determining the decrease in OD600 during a 24-h period. No differences in the autolytic properties of AM2 and AM2-C were detected (results not shown).
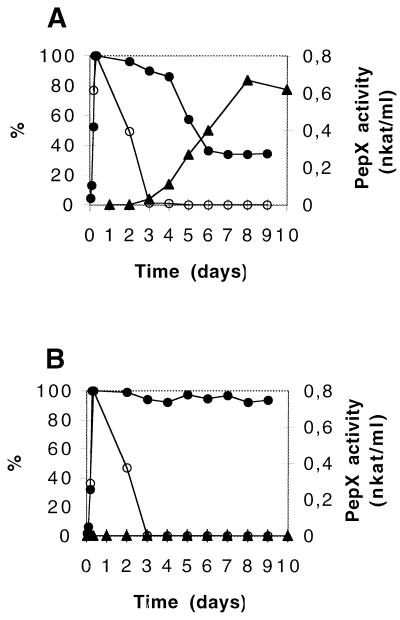
Strains AM2 and AM2-C were grown in M17-lac medium and then were incubated after they reached the stationary phase for several days at 30°C. Lysis was monitored by determining the change in the OD650 of each culture, as well as the release into the culture supernatant of PepX, which was used as an intracytoplasmic marker (38); cell viability was determined on agar plates at 1-day intervals (Fig. 7). In both cases, the viable cell population decreased rapidly between the first day and the third day. In contrast, only the OD650 of the AM2 culture decreased rapidly between the fourth day and the sixth day before it stabilized at a value which was around 40% of the initial OD650 value. The OD650 of the AM2-C culture remained constant over a 9-day period. For strain AM2, we observed that the decrease in OD650 was accompanied by a concomitant release of PepX activity into the culture supernatant. In contrast, no PepX activity was detected in the strain AM2-C culture supernatant. These results confirmed that the decrease in OD650 reflected cell lysis and the release of the intracytoplasmic contents.
FIG. 7.
Autolysis after prolonged incubation in the stationary phase after growth in M17-lac broth at 30°C of L. lactis subsp. cremoris AM2 (A) and L. lactis subsp. cremoris AM2-C (B). Bacterial lysis was monitored by determining the changes in broth turbidity (•) (expressed as percentages of the maximum OD650), in cell viability (○) (expressed as percentages of the maximum number of CFU per milliliter), and in the PepX activity released into the culture supernatant (▴) (expressed in nanokatals per milliliter of culture supernatant). PepX activity was measured with Ala-Pro-paranitroanilide as the substrate. Similar results were obtained in three independent experiments.
Previously, the lysis of an L. lactis culture observed after it reaches the stationary phase has been attributed to activity of the major autolysin, AcmA (7). Thus, the slow rate of autolysis during the beginning the stationary phase observed for strains AM2 and AM2-C could be related to the low amounts of AcmA detected in the two strains. In contrast, the lysis of the AM2 culture, but not the AM2-C culture, observed after several days could be related to the presence of the A2 bacteriolytic activity in the AM2 culture.
DISCUSSION
Using the renaturing SDS-PAGE technique, we evaluated the bacteriolytic activities of L. lactis subsp. cremoris AM2, a strain previously reported to autolyze during cheese ripening when it was used as a starter strain (8). The enzymatic activity profile of strain AM2 varied according to the substrate included in the gel (that is, M. lysodeikticus or L. lactis cells). Previously, M. lysodeikticus has been reported to allow more sensitive detection of lytic activities by several authors (24, 32). However, in the case of strain AM2, the use of L. lactis cells as the substrate allowed detection of three bacteriolytic activities which were not detected with micrococci, including a major activity, the A2 activity (46 kDa). All of these activities differ from the previously detected activities, as determined by renaturing SDS-PAGE. The AcmA major peptidoglycan hydrolase (6) appears to be present in strain AM2 but at a lower level than in the other strains tested. Major bacteriolytic enzyme A2 detected in strain AM2 was clearly distinct from AcmA on the basis of differences in substrate specificity, molecular mass, optimal pH, and sensitivity to different divalent cations.
According to our results, the major A2 enzyme detected in L. lactis AM2 by renaturing SDS-PAGE is not an autolysin but is encoded by prophage DNA present in lysogenic strain AM2. We identified the A2 enzyme as an enzyme which was similar to the lysin of Tuc2009 (2) and φLC3 (5), which are small isometric temperate bacteriophages whose lys gene was previously cloned and sequenced. This lysin is a 429-amino acid protein which has an N-terminal catalytic domain and a C-terminal domain containing two highly homologous sequence repeats of 43 amino acids which are presumably involved in binding and recognition of the cell wall substrate (5).
Detection of prophage-associated lytic activities by the renaturing SDS-PAGE technique was reported previously for Bacillus subtilis prophages (13, 25). However, in contrast to the A2 activity, these lytic activities produced weak bands compared to the bands corresponding to autolysins.
The presence of the A2 activity in the AM2 activity electrophoretic profile did not result from induction of the prophage after cells were harvested and washed during preparation of cell extracts, since when the washing step was omitted, the band was still detected and it had the same intensity. This indicates that the A2 enzyme is present in the cells or at least a portion of the cells during the growth in M17 medium. Spontaneous induction of bacteriophages from lysogenic L. lactis strains grown on M17 medium has been described previously (34), and this could account for the detection of the A2 activity band.
We compared the autolytic properties of L. lactis AM2 and its prophage-cured derivative, AM2-C. These two strains differ in the ability to lyse during prolonged incubation in the stationary phase after growth on M17 medium, suggesting that the prophage lytic enzyme A2 may play a role in the lysis observed. Since prophage induction is likely to happen only in growing cells, lysis probably results from the A2 activity that accumulates in a portion of the cells. This enzyme, which is synthesized without a signal sequence (2, 5), cannot be transported through the cytoplasmic membrane. Thus, it requires the action of a holin also encoded by the prophage to make holes in the cell membrane or permeabilization of the cytoplasmic membrane under starvation conditions.
It has been proposed previously that a prophage is involved in the lysis of L. lactis starter strains observed in cheese (12, 29). The previous studies described temperature-sensitive strains that exhibited thermoinducible lysis when they were subjected to a temperature shift similar to the one used in the manufacture of cheddar cheese, and the authors attributed this property to induction of a prophage. The prophage identified in the present study does not seem to be involved in the thermosensitivity of L. lactis AM2. In addition, we previously observed lysis of L. lactis AM2 in pressed-curd cheese manufactured without a temperature shift (8). Nevertheless, the possibility that the bacteriophage lytic enzyme which we identified is involved in the early lysis of L. lactis AM2 in cheese is supported by several observations. First, there appears to be a lower amount of the AcmA major autolysin in strain AM2 than in other L. lactis strains, especially strain NCDO763, which was found to be nonautolytic in cheese. Second, strain AM2 and its prophage-cured derivative differ in the autolytic properties studied after prolonged incubation in M17 medium in the stationary phase. Third, A2 activity was detected especially in strains that were previously described as nonbitter strains or strains in which there was a rapid decrease in cell viability in cheese trials (12, 27, 42, 43) (unpublished data). Further comparisons in cheese assays of strain AM2 and its prophage-cured derivative when they are used as starter strains should allow us to clarify the role of the prophage lytic proteins in the lysis of AM2 observed in cheese.
ACKNOWLEDGMENTS
We are very grateful to M.-C. Chopin for helpful advice and discussions and to J.-C. Gripon for his continued interest in this work. We warmly thank B. Cesselin for the electron microscopy observations and B. Nicolas and P. Regent for photographic work.
REFERENCES
- 1.Accolas J-P, Spillmann H. The morphology of six bacteriophages of Streptococcus thermophilus. J Appl Bacteriol. 1979;47:135–144. [Google Scholar]
- 2.Arendt E-K, Daly C, Fitzgerald G F, Van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bie R, Sjöström G. Autolytic properties of some lactic acid bacteria used in cheese production. Part I. Material and methods. Milchwissenschaft. 1975;30:653–657. [Google Scholar]
- 4.Bie R, Sjöström G. Autolytic properties of some lactic acid bacteria used in cheese production. Part II. Experiments with fluid substrates and cheese. Milchwissenschaft. 1975;30:739–747. [Google Scholar]
- 5.Birkeland N-K. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage φLC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- 6.Buist G, Kok J, Leenhouts J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buist G, Karsens H, Nauta A, van Sinderen D, Venema G, Kok J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol. 1997;63:2722–2728. doi: 10.1128/aem.63.7.2722-2728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapot-Chartier M-P, Deniel C, Rousseau M, Vassal L, Gripon J-C. Autolysis of two strains of Lactococcus lactis during cheese ripening. Int Dairy J. 1994;4:251–269. [Google Scholar]
- 9.Crow V L, Martley F G, Coolbear T, Roundhill S J. The influence of phage-assisted lysis of Lactococcus lactis subsp. lactis ML8 on Cheddar cheese ripening. Int Dairy J. 1995;5:451–472. [Google Scholar]
- 10.Crow V L, Coolbear T, Gopal F G, Martley F G, McKay L L, Riepe H. The role of autolysis of lactic acid bacteria in the ripening of cheese. Int Dairy J. 1995;5:855–875. [Google Scholar]
- 11.Davidson B E, Powell I B, Hillier A J. Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol Rev. 1990;87:79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x. [DOI] [PubMed] [Google Scholar]
- 12.Feirtag J M, McKay L. Thermoinducible lysis of temperature-sensitive Streptococcus cremoris strains. J Dairy Sci. 1987;70:1779–1784. [Google Scholar]
- 13.Foster S J. Analysis of Bacillus subtilis 168 prophage-associated lytic enzymes: identification and characterization of CWLA-related prophage proteins. J Gen Microbiol. 1993;139:3177–3184. doi: 10.1099/00221287-139-12-3177. [DOI] [PubMed] [Google Scholar]
- 14.Garcia E, Garcia J L, Garcia P, Arraras A, Sanchez-Puelles J M, Lopez R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci USA. 1988;85:914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia P, Garcia J L, Garcia E, Sanchez-Puelles J M, Lopez R. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene. 1990;86:81–88. doi: 10.1016/0378-1119(90)90116-9. [DOI] [PubMed] [Google Scholar]
- 16.Gasson M J, Davies F L. Prophage-cured derivatives of Streptococcus lactis and Streptococcus cremoris. Appl Environ Microbiol. 1980;40:964–966. doi: 10.1128/aem.40.5.964-966.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasson M J. Lytic systems in lactic acid bacteria and their bacteriophages. Antonie Leewenhoek. 1996;70:147–159. doi: 10.1007/BF00395931. [DOI] [PubMed] [Google Scholar]
- 18.Hull R R. Methods for monitoring bacteriophage in cheese factories. Aust J Dairy Technol. 1977;32:63–64. [Google Scholar]
- 19.Jarvis A W. Bacteriophages of lactic acid bacteria. J Dairy Sci. 1989;72:3406–3428. [Google Scholar]
- 20.Jayaswal R K, Lee Y, Wilkinson B J. Cloning and expression of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. J Bacteriol. 1990;172:5783–5788. doi: 10.1128/jb.172.10.5783-5788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawabata S, Vassal L, Le Bars D, Cesselin B, Nardi M, Gripon J-C, Chapot-Chartier M-P. Phage-induced lysis of Lactococcus lactis during Saint-Paulin cheese ripening and its impact on proteolysis. Lait. 1997;77:229–239. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Langsrud T, Landaas A, Castberg H B. Autolytic properties of different strains of group N streptococci. Milchwissenschaft. 1987;42:556–560. [Google Scholar]
- 24.Leclerc D, Asselin A. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can J Microbiol. 1989;35:749–753. doi: 10.1139/m89-125. [DOI] [PubMed] [Google Scholar]
- 25.Longchamps P F, Mauël C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of the prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 26.Loureiro Dos Santos A, Chopin A. Shotgun cloning in Streptococcus lactis. FEMS Microbiol Lett. 1987;42:209–212. [Google Scholar]
- 27.Martley F G, Lawrence R C. Cheddar cheese flavour. II. Characteristics of single strain starters associated with good or poor flavour development. N Z J Dairy Sci Technol. 1972;7:38–44. [Google Scholar]
- 28.Mayo B, Kok J, Venema K, Bockelmann W, Teuber M, Reinke H, Venema G. Molecular cloning and sequence analysis of the X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:38–44. doi: 10.1128/aem.57.1.38-44.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer W. Expression and release of proteolytic enzymes of Lactococcus lactis. Ph.D. thesis. Wageningen, The Netherlands: University of Wageningen; 1996. [Google Scholar]
- 30.Mou L, Sullivan J J, Jago G R. Autolysis of Streptococcus cremoris. J Dairy Res. 1976;43:275–282. doi: 10.1017/s0022029900015831. [DOI] [PubMed] [Google Scholar]
- 31.Nardi M, Chopin M-C, Chopin A, Cals M-M, Gripon J-C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO763. Appl Environ Microbiol. 1991;57:45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Østlie M H, Vegarud G, Langsrud T. Autolysis of lactococci: detection of lytic enzymes by polyacrylamide gel electrophoresis and characterization in buffer systems. Appl Environ Microbiol. 1995;61:3598–3603. doi: 10.1128/aem.61.10.3598-3603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214:241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 34.Reyrolles D, Chopin M-C, Letellier F, Novel G. Lysogenic strains of lactic acid streptococci and lytic spectra of their temperate bacteriophages. Appl Environ Microbiol. 1982;43:349–356. doi: 10.1128/aem.43.2.349-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riepe H R, Pillidge C J, Gopal P K, McKay L L. Characterization of the highly autolytic Lactococcus lactis subsp. cremoris strains CO and 2250. Appl Environ Microbiol. 1997;63:3757–3763. doi: 10.1128/aem.63.10.3757-3763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hackenbeck R, editors. New comprehensive biochemistry. 27. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 131–167. [Google Scholar]
- 38.Tan P S T, Chapot-Chartier M-P, Pos K M, Rousseau M, Boquien C-Y, Gripon J-C, Konings W. Localization of peptidases in lactococci. Appl Environ Microbiol. 1992;58:285–290. doi: 10.1128/aem.58.1.285-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vegarud G, Castberg H B, Langsrud T. Autolysis of group N streptococci. Effect of media composition modifications and temperature. J Dairy Sci. 1983;66:2294–2303. [Google Scholar]
- 41.Visser S. Proteolytic enzymes and their relation to cheese ripening and flavor: an overview. J Dairy Sci. 1993;76:329–350. [Google Scholar]
- 42.Wiederholt K M, Steele J L. Prophage curing and partial characterization of temperate bacteriophages from thermolytic strains of Lactococcus lactis subsp. cremoris. J Dairy Sci. 1993;76:921–930. [Google Scholar]
- 43.Wilkinson M G, Guinee T P, O’Callaghan D M, Fox P F. Autolysis and proteolysis in different strains of starter bacteria during Cheddar cheese ripening. J Dairy Sci. 1994;61:249–262. [Google Scholar]
- 44.Young R Y. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zevaco C, Monnet V, Gripon J-C. Intracellular X-prolyl dipeptidyl peptidase from Lactococcus lactis subsp. lactis: purification and properties. J Appl Bacteriol. 1990;68:357–366. [Google Scholar]