Abstract
The distribution of Clostridium botulinum serotypes A, B, E, and F in Finnish trout farms was examined. A total of 333 samples were tested with a neurotoxin-specific PCR assay. C. botulinum type E was found in 68% of the farm sediment samples, in 15% of the fish intestinal samples, and in 5% of the fish skin samples. No other serotypes were found. The spore counts determined by the most-probable-number method were considerably higher for the sediments than for the fish intestines and skin; the average values were 2,020, 166, and 310 C. botulinum type E spores kg−1, respectively. The contamination rates in traditional freshwater ponds and marine net cages were high, but in concrete ponds equipped with sediment suction devices the contamination rates were significantly lower. Pulsed-field gel electrophoresis (PFGE) typing of 42 isolates obtained in this survey and 12 North American reference strains generated 28 pulsotypes upon visual inspection, suggesting that there was extensive genetic diversity and that the discriminatory power of PFGE typing in C. botulinum type E was high. A numerical analysis of SmaI-XmaI macrorestriction profiles confirmed these findings, as it divided the 54 isolates into 15 clusters at a similarity level of 76%. For this material, this level of similarity corresponded to a three-band difference in the macrorestriction profiles, which indicated that there is no genotypic proof of a close epidemiological relationship among the clusters.
In northern temperate regions, botulism is caused mainly by the consumption of Clostridium botulinum type E-contaminated fish or other marine meat. Although type E botulism is a persistent risk associated with northern ethnic cooking, type E botulism outbreaks have occurred rarely in developed communities consuming industrially produced foods (10). Paradoxically, however, industrially processed vacuum-packaged hot-smoked salmonoids have been responsible for a cluster of recent outbreaks in northern Europe (2, 3, 19, 22). The consumers’ demand for reduced use of sodium salts and the vacuum packaging used to prolong shelf lives have apparently created high-risk botulinogenic products that are largely dependent on refrigeration for safety (8).
Finland recently experienced its first recorded botulism outbreak (19). The incriminated food was a hot-smoked vacuum-packaged whitefish that was produced in Finland but was exported to Germany, where two persons developed botulism after they ate the fish. Although the raw material in this case had been imported from Canada, we became interested in whether farmed fish and fish farm environments in Finland harbored numbers of C. botulinum type E spores similar to the numbers harbored by the corresponding natural counterparts, which we surveyed previously. The rates of occurrence of C. botulinum type E in Finnish natural sediments (12) and nonfarmed fish (16) have been estimated to be 71 and 20%, respectively.
Aquaculture in Finland and in other Baltic Sea countries consists almost entirely of farming rainbow trout (Oncorhynchus mykiss), a Pacific Ocean salmonoid. In 1979 the aquaculture industry in Finland consisted almost entirely of freshwater pond farming, with annual production levels of 4,000 tons; since then, the industry has expanded and has become increasingly sea based. In 1996, 83% of the 18,000 tons of fish produced was grown in marine net cages (28). The Baltic Sea has the highest level of C. botulinum type E contamination detected in the world (12, 14, 17), so care should be taken to develop clean and safe farming practices.
Pulsed-field gel electrophoresis (PFGE) has been used extensively as a subtyping tool for the nosocomial pathogen Clostridium difficile (7, 18, 20, 29) and also for food-poisoning strains of Clostridium perfringens (24, 25). This technique seems to be an ideal method for typing clostridial strains, but widespread use of PFGE has been hindered by the relatively high proportion of isolates that are untypeable because of persistent DNases, which result in shearing of DNA (21, 27). We recently described a PFGE protocol for C. botulinum group II strains in which this problem was overcome to a large extent (11). In that study, 21 strains isolated from various sources and locations over a period of 60 years were typed, and marked differences were found among the macrorestriction profiles (MRPs) obtained.
The present study was performed to evaluate the prevalence and type distribution of C. botulinum in Finnish trout farms and in fish harvested from these farms. The effects of different farming practices on C. botulinum contamination levels were also considered. As very little is known about the genetic biodiversity of the different C. botulinum groups, we performed a PFGE typing and cluster analysis with the C. botulinum type E strains isolated in this study. By linking this spatially and temporally limited set of isolates with reference strains, we were able to show that PFGE is a very powerful tool for characterizing C. botulinum type E strains.
MATERIALS AND METHODS
Trout farms.
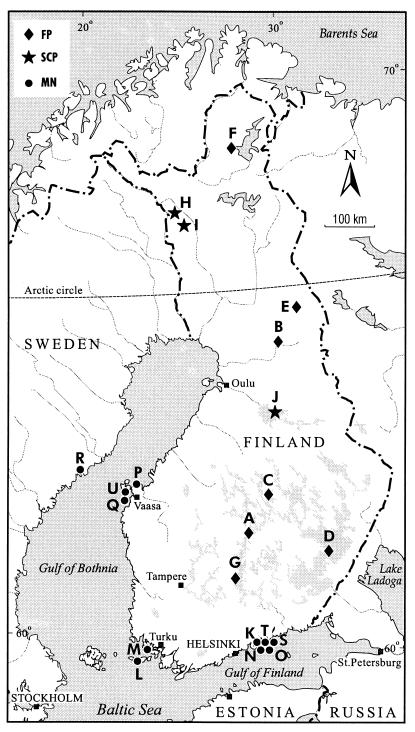
In this study, 333 sediment and fish samples were collected from 20 farms in Finland and 1 farm in Sweden. Ten of the farms (farms A to J) (Fig. 1) were traditional earth pond farms, and three of these (farms H to J) were modern with concrete walls and were equipped with self-cleaning systems for the removal of sedimented bottom matter. Eleven farms (farms K to U) were marine net cage farms with the cages anchored in shallow (depth, <10 m) littoral water. The pond farms use natural freshwater and are situated on the Finnish mainland, whereas the more abundant net cage farms are sea based in brackish water (salinity, 0.6 to 0.8%). On 17 of the farms, including all of the marine net cage farms, only rainbow trout (Oncorhynchus mykiss) was grown; on 2 of the farms lake trout (Salmo trutta lacustris) was grown; and on 2 of the farms sea trout (Salmo trutta trutta) was grown. On one of the latter farms whitefish (Coregonus lavaretus) was also grown.
FIG. 1.
Map of the study area. The 21 trout farms are indicated by capital letters. FP, freshwater pond; SCP, self-cleaning freshwater pond; MN, marine net cage.
Sediment, intestinal, and skin samples.
Sampling took place during 1995 and 1996. A total of 125 aquatic sediment samples (approximately 200 g) were collected from the sediments of the farms. A total of 165 intestinal samples were obtained after fish from the farms were stunned and eviscerated. Forty-three whole, eviscerated fish (1 to 2 kg) were collected from seven of the farms and used to obtain skin samples. The median number of each type of sample per farm was six. All samples were placed in individual plastic bags and transported chilled to the laboratory. The sediment and intestinal samples were homogenized in a blender for 30 s prior to enrichment and quantitative analysis. Each skin sample consisted of the head, skin, peritoneum, gills, and fins of a single fish, which were homogenized in a blender with 100 ml of sterile 0.1% peptone water prior to enrichment.
Reference strains.
Twelve strains from our C. botulinum type E collection (strains R-90, 31-2570, RS-1, Beluga, R-9087, C-51, C-60, C-94, 250, 36208, and KA-2, 4062) were used to obtain reference MRPs for the cluster analysis. These strains were North American and North Atlantic isolates; a more detailed description of the origin of each strain has been published previously (11).
PCR detection and isolation.
The quantitative method of Hielm et al. (13) was used to examine samples for the presence of C. botulinum types A, B, E, and F. Dynazyme II DNA polymerase (Finnzymes, Espoo, Finland) and a 96-well DNA Engine thermal cycler (MJ Research, Watertown, Mass.) were used for the PCR. The size of each amplified PCR product was determined in agarose gels by comparison with standard DNA fragments (DNA molecular weight marker VI; Boehringer, Mannheim, Germany).
Strains were isolated by streaking alcohol-treated PCR-positive cultures onto anaerobic egg yolk agar (1). Plates were incubated for 3 days, and then lipase-positive colonies from these plates were inoculated into tryptone-peptone-glucose-yeast extract (Difco Laboratories, Detroit, Mich.) broth (9). All cultures were incubated at 26°C in an anaerobic cabinet (model MK III; Don Whitley Scientific Ltd., Shipley, England) with an internal atmosphere containing 85% N2, 10% CO2, and 5% H2. The species and serotypes of 48 C. botulinum type E isolates obtained from the survey material were ascertained by performing a botulinum neurotoxin-specific PCR (13). If many isolates were obtained from a single sample, only one isolate was kept for the collection. Each isolate was given a designation consisting of (i) a prefix (K indicated that the isolate was obtained from a fish sample, and S indicated that the isolate was obtained from a sediment sample), (ii) a number, and (iii) a suffix (A to U, referring to the fish farm from which the sample was taken).
In situ DNA preparation.
DNA was isolated as described previously (11). Briefly, overnight cultures were chilled on ice, resuspended in PIV (10 mM Tris [pH 7.5], 1 M NaCl) containing 3.5 to 4.0% (vol/vol) formaldehyde (Merck & Co., Darmstadt, Germany), and then left on ice for 1 h. Cell suspensions were each mixed with an equal amount of 2% (wt/vol) low-melting-temperature agarose (InCert agarose; FMC Bioproducts, Rockland, Maine), and casts were prepared with GelSyringe dispensers (New England Biolabs, Beverly, Mass.). The plugs were lysed by incubating them for 2 h in lysis solution (6 M Tris [pH 7.5], 1 M NaCl, 100 mM EDTA [pH 8.0], 0.5% Brij 58, 0.2% deoxycholate, 0.5% sodium lauroyl sarcosine, 20 μl of RNase per ml, 1 mg of lysozyme per ml, 20 U of mutanolysin per ml) with gentle shaking at 37°C, and isolation was completed by washing the preparation for 1 h with ESP (0.5 M EDTA [pH 8.0], 10% sodium lauroyl sarcosine, 100 μg of proteinase K per ml) at 50°C. Phenymethylsulfonyl fluoride inactivation of proteinase K and restriction endonuclease digestion of the agarose-embedded DNA were performed as recommended by the manufacturer.
Restriction enzyme digestion and electrophoresis.
Five rare-cutting restriction enzymes (ApaI, MluI, SmaI, XhoI, and XmaI [New England Biolabs]) were used to cleave C. botulinum DNA. All DNA extractions and digestions were repeated at least twice. The presence of potentially confounding plasmids in the DNA isolates was determined by subjecting undigested samples to PFGE prior to macrodigestion. All samples were electrophoresed with a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, Calif.) through a 1% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts) in 0.5× TBE buffer (Amresco, Solon, Ohio). Switch times were ramped from 1 to 26 s over 22 h at 14°C and 6 V/cm, unless indicated otherwise. Low Range or Lambda Ladder PFG markers (New England Biolabs) were used to determine fragment sizes. The gels were stained for 30 min in 1 liter of used running buffer containing 0.5 mg of ethidium bromide and were destained in running buffer until appropriate contrast was obtained for standard photography (26) or/and digital imaging with an Alpha Imager 2000 documentation system (Alpha Innotech, San Leandro, Calif.).
PFGE pattern analysis.
SmaI-XmaI MRPs for molecular sizes ranging from 50 to 350 kb were analyzed by using the GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). The similarity between MRPs was expressed as the Dice correlation coefficient (SD), as determined with the following equation: SD = [2nAB/(nA + nB)] × 100, where nAB is the number of matching fragments and nA and nB are the total numbers of fragments in profiles A and B (4). The position tolerance for band matching was set at 1.4% of the total length of the pattern (300 kb). The SmaI-XmaI MRPs were used to construct a dendrogram by the unweighted pair group method with arithmetic averages (4). The MRPs generated by cleavage of DNA with ApaI, MluI, and XhoI were visually inspected and compared. The genotypes resulting from the MRP analyses were clustered at a similarity level of 76%, which corresponded to a three-band difference in the MRPs and to the weakest possible level of epidemiologic relatedness according to the guidelines described by Tenover et al. (30).
Statistical analysis of the prevalence data.
In all analyses, logarithmic transformations of most-probable-number C. botulinum spore counts per kilogram were used. The data shown in Table 1 were compared by using the χ2 test (percentages) and the t test (means). Statistical analyses were done with the Statgraphics Plus software (version 2.0; Manugistics Inc., Rockville, Md.).
TABLE 1.
Prevalence and spore counts of C. botulinum type E in different types of Finnish trout farms
| Farm type | Sediment samples
|
Fish intestinal samples
|
||||
|---|---|---|---|---|---|---|
| No. of samples | % of positive samplesa | Log no. of spores kg−1b | No. of samples | % of positive samplesa | Log no. of spores kg−1b | |
| Freshwater pond | 42 | 60B | 2.11B | 54 | 19A | 1.32B |
| Self-cleaning freshwater pond | 14 | 14A | 1.22A | 17 | 6A | 1.20A |
| Marine net cage | 69 | 84B | 2.40C | 94 | 14A | 1.29B |
| Total | 125 | 68 | 2.17 | 165 | 15 | 1.29 |
Values followed by different subscripted letters are significantly different, as determined by the χ2 test (P < 0.05).
Mean logarithmic spore count for all samples in a group. Values below the most-probable-number detection limit (30 spores kg−1) were assigned a value of 15 (log = 1.18) spores kg−1 for statistical calculations. Values followed by different subscripted letters are significantly different, as determined by the t test (P < 0.05).
RESULTS
Prevalence.
C. botulinum type E was found in 95% of the trout farms investigated in this study (all of the farms except farm I). No other serotypes were found. C. botulinum type E was found in 68% of the sediment samples, 15% of the intestinal samples, and 5% of the skin samples. For the positive samples, the mean spore counts were highest in the sediment samples and significantly lower in the intestinal and skin samples (2,020, 166, and 310 spores kg−1, respectively). The levels of C. botulinum type E contamination of sediment and fish intestinal samples were considerably lower in the self-cleaning freshwater ponds than in the other two farm types (Table 1); there was not a significant difference in the levels of contamination for the fish skin samples.
Isolated strains.
Forty-eight strains of C. botulinum type E were isolated in this study. Thirty of these strains were isolated from fish, and 18 were isolated from fish farm sediment samples. Twenty-six fish strains were isolated from rainbow trout intestines, and the remaining four fish strains were isolated from rainbow trout skin samples (strains K2-B and K3-B), lake trout intestines (K18-F), and whitefish intestines (K118-H). The fish samples were obtained from 12 of the farms in this study (farms B, C, D, F, G, H, N, O, Q, R, T, and U) (Table 2), and the sediment samples were obtained from 9 farms (farms C, D, E, F, L, M, N, P, and U).
TABLE 2.
Distribution of different C. botulinum type E pulsotypes in sediment and fish samples obtained from Finnish trout farms
| Fish farma | Farm typeb | Farmed speciesc | No. of PFGE-typeable isolates | Pulsotype(s) in sediment samplesd | Pulsotype(s) in fish samplesd |
|---|---|---|---|---|---|
| B | FP | rt | 2 | VII, XIb | |
| C | FP | rt | 3 | X | IX, XXIIIa |
| D | FP | rt | 3 | XIa, XIV | VI |
| E | FP | lt | 3 | IV(2), XIII | |
| F | FP | lt | 8 | V, XII, XIII, XVIa(2), XVIb(2) | XVIb |
| G | FP | rt | 1 | VIII | |
| H | SCP | st, wf | 1 | XXIIb | |
| M | MN | rt | 1 | XXI | |
| N | MN | rt | 4 | XXIIa(3), XXIIb | |
| O | MN | rt | 1 | XXIIa | |
| P | MN | rt | 1 | XIX | |
| Q | MN | rt | 2 | XXIV, XXIIa | |
| R | MN | rt | 1 | XIX | |
| T | MN | rt | 5 | XV(2), XXIIa(3) | |
| U | MN | rt | 6 | XXIIIb | IV, XXIIa(4) |
No C. botulinum type E strains were isolated from farms A, I, J, K, and S. The strain isolated from farm L could not be typed by PFGE due to extensive DNase activity.
FP, freshwater pond; SCP, self-cleaning freshwater pond; MN, marine net farm.
rt, rainbow trout (Oncorhynchus mykiss); lt, lake trout (Salmo trutta lacustris); st, sea trout (Salmo trutta trutta); wf, whitefish (Coregonus lavaretus).
The numbers in parentheses are the numbers of isolates representing the same pulsotype.
Plasmids.
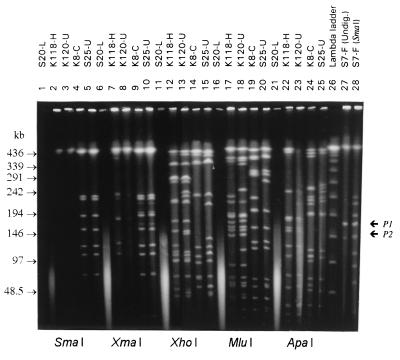
During PFGE of undigested DNA, plasmids that migrated within the analysis range (50 to 350 kb of linear DNA) were detected in 16 (30%) of the 54 isolates. Seven of these 16 isolates contained two plasmids in this range. Figure 2 shows the appearance of plasmid bands P1 and P2 in both undigested DNA and SmaI-digested DNA of strain S7-F.
FIG. 2.
PFGE separation of five different macrorestriction digests of five C. botulinum type E isolates and demonstration of possible confusion between plasmids and restriction fragments in one isolate. Lanes 1, 6, 11, 16, and 21, S20-L (DNase isolate, no pulsotype); lanes 2, 7, 12, 17, and 22, K118-H (pulsotype XXIIb); lanes 3, 8, 13, 18, and 23, K120-U (pulsotype XXIIa); lanes 4, 9, 14, 19, and 24, K8-C (pulsotype XXIIIa); lanes 5, 10, 15, 20, and 25, S25-U (pulsotype XXIIIb); lane 26, lambda ladder PFG marker; lanes 27 and 28, S7-F (undigested and SmaI digested, respectively; the positions of the plasmid P1 and P2 bands are indicated). The pulse time was ramped from 1 to 26 s for 22 h at 6 V/cm and 12°C.
Macrorestriction digests.
SmaI MRPs formed the basis for PFGE typing of the 54 isolates (Fig. 3). However, the DNAs of 14 isolates were not digested by SmaI, presumably due to CG methylation of the restriction site. This resulted in one intact DNA band in the MRP, which was easily distinguished from DNase smearing (Fig. 2). The DNAs of these 14 isolates were digested with XmaI, an isoschizomer of SmaI with the same restriction site. The cleavage point of XmaI (C▾CCGGG) is different from the cleavage point of SmaI (CCC▾GGG); this makes XmaI indifferent to CG methylation while XmaI produces restriction fragments identical in appearance to the fragments produced by SmaI (Fig. 2). The 14 isolates produced identical XmaI MRPs (Fig. 4, cluster 14), but these isolates could be divided into two closely related (SD = 97%) pulsotypes on the basis of XhoI digestion data. When our DNA isolation method was used, six isolates (K9-D, K10-D, K12-D, K13-D, S20-L, and S23-N) (10%) were untypeable by PFGE due to particularly active DNases. Characterization of the other 54 isolates resulted in good ApaI, MluI, SmaI-XmaI, and XhoI MRPs. The isolates were divided into pulsotypes I to XXIV (Fig. 4) based on SmaI-XmaI MRPs, and the pulsotypes were given the subdivision suffixes a and b based on ApaI, MluI, and XhoI MRPs when appropriate.
FIG. 3.
SmaI digests of 28 isolates from Finnish trout farms. Each isolate’s designation consists of (i) a prefix (K indicates that the isolate was isolated from a fish sample, and S indicates that the isolate was isolated from sediment sample), (ii) a number, and (iii) a suffix (A to U, referring to the farm [Fig. 1] from which the sample was taken). Lanes Low Range contained the low-range PFG marker. The pulse time was ramped from 1 to 24 s for 24 h at 6 V/cm and 14°C.
FIG. 4.
Dendrogram of 54 C. botulinum type E isolates based on SmaI-XmaI macrorestriction profiles. Schematic MRPs are shown. The low-range PFG marker was included as an additional entry. A similarity analysis was performed by using the Dice coefficient, and clustering was performed by using the unweighted pair group method with arithmetic averages.
Cluster analysis and genetic diversity.
The SmaI-XmaI digests of the 54 isolates generated 24 different MRPs, which formed 15 clusters at a similarity level of 76% (Fig. 4). For these MRPs, this level of similarity corresponded roughly to a three-band difference. Analysis of MRPs generated by digestion with ApaI, MluI, and XhoI was used to distinguish between isolates that seemed to be clonal on the basis of SmaI-XmaI digestion data. The discriminative value of ApaI, MluI, and XhoI was slightly higher than that of SmaI-XmaI (ApaI, MluI, and XhoI produced 25, 27, and 28 MRPs, respectively), but since the ApaI, MluI, and XhoI profiles were not as easy to interpret as the SmaI-XmaI MRPs, they were not used for the numerical analysis. Indistinguishable pulsotypes, indicating clonality, were obtained for a wide variety of sample pairs, including sediment and fish samples, freshwater and marine samples, samples from different fish and mammal species, and European and American samples (Fig. 4). Isolates from a single farm, however, generally represented different pulsotypes (Table 2), implying that the local genetic diversity of C. botulinum type E was great.
DISCUSSION
The prevalence of C. botulinum in Finnish fish farm sediments (68%) (Table 1) corresponds well with the results of previous surveys of natural sediments (12, 13). In this study, both the spore counts and the prevalence were higher in the marine net cages than in the freshwater ponds, a finding consistent with the findings of our earlier study of Finnish natural sediments, in which C. botulinum type E spores were found in 81% of sea samples (940 spores kg−1) and 61% of freshwater samples (370 spores kg−1) (12). Studies of C. botulinum prevalence on fish farms and in farmed fish have also been conducted in several other countries, including Denmark (15), Norway (31), Great Britain (6), and Scotland (5). It seems that C. botulinum is enriched by aquaculture in areas with low levels of surrounding C. botulinum contamination (5, 6, 31), whereas this effect is not significant in environments with high levels of natural C. botulinum type E contamination (12, 15).
The fact that the contamination levels were remarkably lower in the freshwater ponds equipped with self-cleaning apparatus than in the other ponds is interesting. It seems that removal of bottom sludge effectively eliminated C. botulinum type E multiplication in the ponds with self-cleaning apparatus. Arguments that “bottomless” marine net cages should be less botulinogenic than freshwater ponds have been used to promote marine farming in the Baltic Sea. To some extent these claims are substantiated by the higher prevalence and higher spore counts in the freshwater fish samples than in the marine fish samples, while the prevalence and spore counts in the freshwater sediments were lower than the prevalence and spore counts in marine sediments (Table 1). However, although direct sediment feeding is effectively hindered in net cages, it seems that other factors, such as accumulation of biomass within the net cages and the overall higher C. botulinum type E contamination rates in the marine environment, outweigh most of this benefit. Furthermore, the widespread use of ground Baltic herring as a cheap additional feed on marine farms is likely to increase the level of contamination of farmed trout. We have found that 40% of the herring caught in the Baltic Sea contain C. botulinum type E spores (16). On some marine farms, prototypes of funnel-shaped net cages equipped with a sediment suction device are on trial in an effort to reduce nutrient escape; our findings for the self-cleaning freshwater ponds give us reason to believe that this sort of apparatus could also be used to lower C. botulinum type E contamination rates in the produce of such farms.
DNases are a persistent problem in the typing of some C. botulinum group II strains (11). In this study, we found that six isolates were untypeable by PFGE due to shearing of DNA and had to be excluded from further analysis. This unwelcome trait could not be anticipated by colony or broth appearance, and the strains produced normal, positive PCR results for the C. botulinum type E toxin sequence. To our knowledge, the CG methylation which we encountered with 14 isolates was a new phenomenon in SmaI digestion of clostridial DNA. Since the problem was sidestepped by replacing SmaI with its isoschizomer, XmaI, it did not interfere with the typing itself. There are differences in PFGE lane appearance between CG methylation and DNase lysis of agarose-imbedded DNA. Figure 2 shows that the DNA of isolate S20-L was sheared by DNases in all digests (lanes 1, 6, 11, 16, and 21), whereas the DNAs of isolates K118-H and K120-U were not digested by SmaI because of CG methylation, which resulted in a single large band of uncut DNA high in the gel (lanes 2 and 3). XmaI cut the DNAs of these isolates (lanes 7 and 8), as did the three auxiliary enzymes, and the MRPs produced were identical to (although blurrier than) the SmaI MRPs obtained for isolates in which CG methylation did not occur (lanes 4, 5, 9, and 10). It is interesting to note that the XmaI MRPs of the 14 isolates were identical to each other, although the isolates were obtained from fish from six different farms in all parts of Finland. This implies that CG methylation is actually a rarer phenomenon in C. botulinum type E strains than the first impression of the material suggested; the data suggest that CG methylation was unduly emphasized by an almost clonal lineage (Fig. 4, cluster 14) that was well dispersed throughout the study area.
Plasmids can be a source of confusion in PFGE typing, and although plasmids generally produce fainter bands than chromosomal macrorestriction fragments produce, it is advisable to take some preventive measures to avoid this problem prior to evaluation of MRPs. Olsen et al. (23) cut plasmids into insignificant fragments with XbaI; we simply electrophoresed undigested whole DNA in a PFGE gel prior to the typing runs, so that we knew whether any plasmid bands were expected in the MRPs of the isolates.
Cluster analysis (Fig. 4) of the MRPs of the C. botulinum type E isolates from Finnish trout farms confirmed that there is great diversity in strains of this serotype, as we hypothesized in our previous work (11). Compared to the origins of the reference strains (included in Fig. 4), the origins of the isolates used in this study were fairly restricted, both spatially and temporally. Even so, 18 different SmaI-XmaI MRPs were generated, and some seemingly clonal isolates were further differentiated with restriction enzymes ApaI, MluI, and XhoI. No such differentiation was observed with the clonal isolates used in our earlier study (11), which led us to propose that SmaI is the principal macrorestriction enzyme that should be used for C. botulinum group II. However, as a result of additional digestions, especially with XhoI, 22 different pulsotypes were detected in the 42 typeable isolates from Finnish trout farms (Table 2). Whether this means that XhoI is epidemiologically more relevant than SmaI for PFGE typing of C. botulinum type E isolates remains to be determined by future typing studies, preferably studies performed by using a polyphasic approach (33).
It is interesting to note that isolates having the same pulsotype were not restricted to samples from the same farm, animal species, or even country (Table 2; Fig. 4). On the dendrogram based on MRPs, the 12 North American type E reference strains characterized previously (11) fell right in the midst of the isolates used in this study (Fig. 4). The proportion of clonal isolates in the sediment and fish sample groups was larger than the proportion of clonal isolates when multiple isolates from one farm were examined. These findings imply that (i) C. botulinum type E is an organism that is adapted to northern aquatic environments and occasionally contaminates animal or human food chains at random, and (ii) there is or has been extensive spread and exchange of C. botulinum type E strains in the northern temperate regions, the vehicle of which has not been determined.
The successful use of MRPs for typing of C. botulinum group II isolates confirmed that this technique is a very powerful method when it is used for C. botulinum, with broad clinical, industrial, and perhaps even legal implications. The ability of PFGE to distinguish a majority of 42 type E isolates obtained from a very limited ecological niche, 21 trout farms, implies that in future botulism outbreaks investigators may be able to link patient isolates to incriminated foods with a certainty not possible a few years ago (32). For the fish and seafood industry, the typing of C. botulinum type E strains isolated from raw material, processing routes, and/or products is a new tool for identification of critical control points and implementation of hazard analysis critical control point systems in manufacturing processes.
ACKNOWLEDGMENTS
We thank Sirkku Ekström, Kirsi Ristkari, and Maria Stark for excellent technical assistance. The help of Timo Mäkinen in obtaining samples from the Finnish Game and Fisheries Research Institute’s research stations is gratefully acknowledged.
This research was supported by the Academy of Finland, the Walter Ehrström Foundation, and the Finnish Veterinary Foundation.
REFERENCES
- 1.American Public Health Association. Compendium of methods for the microbiological examination of foods. 3rd ed. Washington, D.C: American Public Health Association; 1992. p. 1108. [Google Scholar]
- 2.Anonymous. Botulism. Epid-aktuellt. 1991;14:9. [Google Scholar]
- 3.Anonymous. Fallbericht: Botulismus nach dem Verzehr von geräucherten Lachsforellen. Epidemiol Bull. 1998;4:20. [Google Scholar]
- 4.Applied Maths. GelCompar version 4.0 manual. Kortrijk, Belgium: Applied Maths; 1996. [Google Scholar]
- 5.Burns G F, Williams H. Clostridium botulinum in Scottish fish farms and farmed trout. J Hyg. 1975;74:1–6. doi: 10.1017/s0022172400046647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann D C, Taylor L Y, Hobbs G. The incidence of Clostridium botulinum in farmed trout raised in Great Britain. J Appl Bacteriol. 1975;39:331–336. doi: 10.1111/j.1365-2672.1975.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 7.Chachaty E, Saulnier P, Martin A, Mario N, Andremont A. Comparison of ribotyping, pulsed-field gel electrophoresis and random amplified polymorphic DNA for typing Clostridium difficile strains. FEMS Microbiol Lett. 1994;122:61–68. doi: 10.1111/j.1574-6968.1994.tb07144.x. [DOI] [PubMed] [Google Scholar]
- 8.Eklund M W. Control in fishery products. In: Hauschild A H W, Dodds K L, editors. Clostridium botulinum—ecology and control in foods. New York, N.Y: Marcel Dekker; 1992. pp. 209–232. [Google Scholar]
- 9.Food and Agricultural Organization. Manual of food quality control. Vol. 12. Rome, Italy: Food and Agricultural Organization of the United Nations; 1991. pp. 115–116. [Google Scholar]
- 10.Hauschild A H W. Epidemiology of human foodborne botulism. In: Hauschild A H W, Dodds K L, editors. Clostridium botulinum—ecology and control in foods. New York, N.Y: Marcel Dekker; 1992. pp. 68–104. [Google Scholar]
- 11.Hielm S, Björkroth J, Hyytiä E, Korkeala H. Genomic analysis of Clostridium botulinum group II by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1998;64:703–708. doi: 10.1128/aem.64.2.703-708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hielm S, Hyytiä E, Andersin A-B, Korkeala H. A high prevalence of Clostridium botulinum type E in Finnish freshwater and Baltic Sea sediment samples. J Appl Microbiol. 1998;84:133–137. doi: 10.1046/j.1365-2672.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- 13.Hielm S, Hyytiä E, Ridell J, Korkeala H. Detection of Clostridium botulinum in fish and environmental samples using polymerase chain reaction. Int J Food Microbiol. 1996;31:357–365. doi: 10.1016/0168-1605(96)00984-1. [DOI] [PubMed] [Google Scholar]
- 14.Huss H. Distribution of Clostridium botulinum. Appl Environ Microbiol. 1980;39:764–769. doi: 10.1128/aem.39.4.764-769.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huss H H, Pedersen A, Cann D C. The incidence of Clostridium botulinum in Danish trout farms. I. Distribution in fish and their environment. J Food Technol. 1974;9:445–450. [Google Scholar]
- 16.Hyytiä E, Hielm S, Korkeala H. Prevalence of Clostridium botulinum type E in Finnish fish and fishery products. Epidemiol Infect. 1998;120:245–250. doi: 10.1017/s0950268898008693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johanssen A. Clostridium botulinum in Sweden and the adjacent waters. J Appl Bacteriol. 1963;26:43–47. [Google Scholar]
- 18.Kato H, Kato N, Watanabe K, Ueno K, Ushijima H, Hashira S, Abe T. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficile in a neonatal intensive care unit. J Clin Microbiol. 1994;32:2067–2070. doi: 10.1128/jcm.32.9.2067-2070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korkeala H, Wihlman H, Hielm S, Hyytiä E. Tyhjiöpakatun savusiian aiheuttama botulismi-ruokamyrkytys Saksassa. Suom Elainlaakaril. 1997;103:280–281. [Google Scholar]
- 20.Kristjánsson M, Samore M H, Gerding D N, DeGirolami P C, Bettin K M, Karchmer A W, Arbeit R D. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin W-J, Johnson E A. Genome analysis of Clostridium botulinum type A by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1995;61:4441–4447. doi: 10.1128/aem.61.12.4441-4447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Öberg S. Botulism. Epid-aktuellt. 1994;17:5. [Google Scholar]
- 23.Olsen J E, Skov M N, Angen Ø, Threlfall E J, Bisgaard M. Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype typhimurium defined by ribotyping, IS200 typing and PFGE. Microbiology. 1997;143:1471–1479. doi: 10.1099/00221287-143-4-1471. [DOI] [PubMed] [Google Scholar]
- 24.Pyhälä S, Lyytikäinen O, Björkroth J, Korkeala H. Clostridium perfringens—bakteeri kuokkavieraana pitopöydässä. Suom Elainlaakaril. 1998;104:336–341. [Google Scholar]
- 25.Ridell J, Björkroth J, Eisgrüber H, Schalch B, Stolle A, Korkeala H. Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food poisoning outbreaks. J Food Prot. 1997;61:240–243. doi: 10.4315/0362-028x-61.2.240. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 6.18–6.19. [Google Scholar]
- 27.Samore M H, Kristjánsson M, Venkataraman L, DeGirolami P C, Arbeit R D. Comparison of arbitrarily-primed polymerase chain reaction, restriction enzyme analysis and pulsed-field gel electrophoresis for typing Clostridium difficile. J Microbiol Methods. 1996;25:215–224. [Google Scholar]
- 28.Statistics Finland. Statistical yearbook of Finland 1997. Helsinki, Finland: Statistics Finland; 1998. p. 140. [Google Scholar]
- 29.Talon D, Bailly P, Delmée M, Thouverez M, Mulin B, Iehl-Robert M, Cailleaux V, Michel-Briand Y. Use of pulsed-field gel electrophoresis for investigation of an outbreak of Clostridium difficile infection among geriatric patients. Eur J Clin Microbiol Infect Dis. 1995;14:987–993. doi: 10.1007/BF01691381. [DOI] [PubMed] [Google Scholar]
- 30.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tjaberg T B, Håstein T. Utbredelse av Clostridium botulinum i norske fiskoppdrettsanlegg. Nor Veterinaertidsskr. 1975;87:718–720. [Google Scholar]
- 32.Townes J M, Cieslak P R, Hatheway C L, Solomon H M, Holloway J T, Baker M P, Keller C F, McCroskey L M, Griffin P M. An outbreak of type A botulism associated with a commercial cheese sauce. Ann Intern Med. 1996;125:558–563. doi: 10.7326/0003-4819-125-7-199610010-00004. [DOI] [PubMed] [Google Scholar]
- 33.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]