Abstract
Years ago, a classic textbook would define plant cell walls based on passive features. For instance, a sort of plant exoskeleton of invariable polysaccharide composition, and probably painted in green. However, currently, this view has been expanded to consider plant cell walls as active, heterogeneous, and dynamic structures with a high degree of complexity. However, what do we mean when we refer to a cell wall as a dynamic structure? How can we investigate the different implications of this dynamism? While the first question has been the subject of several recent publications, defining the ideal strategies and tools needed to address the second question has proven to be challenging due to the myriad of techniques available. In this review, we will describe the capacities of several methodologies to study cell wall composition, structure, and other aspects developed or optimized in recent years. Keeping in mind cell wall dynamism and plasticity, the advantages of performing long-term non-invasive live-imaging methods will be emphasized. We specifically focus on techniques developed for Arabidopsis thaliana primary cell walls, but the techniques could be applied to both secondary cell walls and other plant species. We believe this toolset will help researchers in expanding knowledge of these dynamic/evolving structures.
Keywords: Biophysics, cell wall composition, cell wall structure, live-imaging, mechanics, plant cell wall
This review offers a comprehensive guide to the latest techniques for understanding the dynamic, active, and diverse nature of plant cell walls, highlighting non-invasive and live-imaging strategies.
Introduction
Dynamic processes are those related to forces in action. They allow living organisms to change and adapt over time. The processes include evolution, natural selection, and learning. Thus, they are at the very core of life itself.
Plants are sessile organisms, but this does not limit their capacity to develop highly dynamic structures. One of the structures is the plant cell wall. The cell wall plays a vital role in the function and physiology of the plant cell: it provides structural support, protects the cell from internal and external damage, regulates the exchange of materials between the cell and its environment, and is involved in growth and developmental processes (Anderson and Kieber, 2020; B. Zhang et al., 2021). This is only achieved through a fine regulation of biosynthesis and remodelling, allowing the integration of internal and external cues in the plant developmental plan. The ability of plant cell walls to adapt to changing functional requirements during development and interactions with the environment can be described with the term plasticity. Plasticity is also reflected in the diverse composition of their cell walls. The specific makeup of these cell walls can vary greatly, not just between different species of plants, but also among different tissues within a single plant, at various stages of development, and in response to environmental or stress conditions (Knox, 2008; Popper et al., 2011; Bacete et al., 2018). This diversity is even observed on a microscopic scale within individual cells (Dauphin et al., 2022). Yet, this incredible versatility arises from the dynamic assembly of the same fundamental building blocks to form structures tailored to the specific needs of the plant.
Given the dynamic nature of plant cell walls and cell wall plasticity, studying the processes that govern their adaptations to various situations is crucial. To fully understand these processes, we must consider two main aspects: the underlying mechanisms driving these changes and the spatiotemporal scales at which they occur. A comprehensive approach goes beyond merely observing changes and seeks to uncover how and why they happen. Furthermore, we must focus on a temporal scale that allows the organism to adapt to the situation that triggered the change, while still being observable. To this aim, in vivo measurements, which involve investigations performed within a living organism, are especially valuable when they enable live measurements, capturing real-time data on the organism’s response to stimuli. This approach contributes significantly to our understanding of cell wall dynamics, shedding light on how plants adapt their structures and regulate their functions. However, in vivo measurements are often limited by the characteristics of plant samples since they require small samples, few cells, or cell cultures, in contrast to the larger and more complex nature of most plants. While microscopic analysis may benefit from these simplified conditions, certain procedures, such as tensile tests, usually excel with larger specimens. Particularly in the case of model plants such as Arabidopsis, their small size can be a restrictive factor. This size constraint influences the choice of suitable methods for studying dynamic processes in cell walls. Beyond these considerations, interpreting results, especially from mechanical tests, can become challenging due to the layered complexity of tissues and the variable geometries of plants.
In this review, we aim to describe the advantages and limitations of several techniques used to study cell wall dynamics in vivo. Nevertheless, a few techniques that do not allow for in vivo measurements are also mentioned because of the unmatched resolution or information obtained. Thus, we aimed to provide a full repertoire of techniques that can fully interrogate the diverse chemical and mechanical properties of plant cell walls. The current and future development of these methods will open new and exciting perspectives in the study of cell walls during dynamic processes, such as growth, development, and interaction with the environment. We have outlined in Box 1 several particularly interesting research questions and technical challenges that are associated with these methods. This knowledge will not only benefit cell wall experts but will also inform general plant biologists, biophysicists, and other researchers interested in understanding the dynamic and adaptable nature of plant cell walls.
Box 1. Research questions and technical challenges in cell wall research.
Outstanding biological questions
What is the biological reason behind the diversity of cell wall composition and structure in different tissues and developmental stages?
How do the different cell wall building blocks interact with each other?
How anisotropic are cell walls and what are the effects on cell wall mechanical properties?
How does mechanical stimulation of cell walls signal to growth and development? What are the magnitudes of the relevant forces?
How can mechanical properties measured with different techniques, which give different results, be reconciled into a unified biological trait?
How do cell wall composition and structure determine mechanical properties of plant tissues such as cell wall stiffness and viscosity?
What are the structural determinants of cell wall mechanics?
How are mechanical forces transmitted along cell wall components?
How do cell wall composition and mechanical properties translate into agronomically important traits such as yield?
How do tissue-level measurements relate to individual cell mechanisms?
Outstanding technical challenges
Precise predictions of cell wall composition from unlabelled images.
Implementation of realistic time-dependent cell wall simulations incorporating mechanical and chemical perturbations.
Measurements and quantifications in growing tissues.
An intricate network: composition and interactions of cell wall components
The plant cell wall stands as a sophisticated and complex network which is far from being completely understood. It comprises a diverse range of components, each playing a role that is as structurally essential as it is dynamic. Cellulose, hemicelluloses, pectins, and various proteins converge into a structure that is resilient, yet adaptable. Recent reviews can provide an excellent overview of primary (Cosgrove, 2022) and secondary (Zhong et al., 2019) cell wall composition and structure. Hence, here we will only provide a summarized version to refresh the advanced reader’s memory and offer an overview to newcomers to the topic.
Cellulose is the main cell wall component. This polysaccharide, composed of linear chains of β-1,4 linked d-glucose units, forms microfibrils that aggregate into larger macrofibrils. These structures lend the cell wall its incredible strength, rivalling the tensile strength of steel (Gibson, 2012). Guided by cytoskeletal elements, these cellulose microfibrils provide an architectural framework for the cell wall, largely determining its mechanical properties (recently reviewed by Chebli et al., 2021). The cytoskeleton also participates in cell wall assembly by helping to transport cell wall polysaccharides from the Golgi apparatus to the extracellular space. Regulating orientation of the cell wall material and deposition rate can produce cell wall anisotropic properties that are used by plants during their development or defence actions, as described in later sections.
Hemicelluloses, primarily composed of xyloglucan molecules in primary cell walls and of xylan and glucomannan in secondary cell walls, are believed to interact intimately with cellulose microfibrils, although the exact nature of this interaction is still under active investigation (Cosgrove, 2022; Du et al., 2022; Coen and Cosgrove, 2023). Two different models have been proposed, namely the ‘tethered network’ model—which establishes that hemicelluloses such as xyloglucan tether cellulose microfibrils together—and the ‘biomechanical hotspot’ model—according to which cellulose microfibrils form tight contacts with each other with the help of a mixture between xyloglucans and disordered cellulose (Cosgrove, 2022).
Pectins, the third critical player, not only regulate the wall’s porosity and flexibility but also can form ‘egg-box’ configurations in the presence of divalent cations such as calcium. This unique conformation contributes to the wall’s overall rigidity, adding to the importance of pectins in maintaining cell wall architecture. In addition, other cell wall elements, such as expansins, are important for controlling cell wall loosening, probably by affecting the contact between cellulose and hemicellulose—although this still needs to be experimentally validated (Cosgrove, 2016b; Samalova et al., 2022).
All plant cells have a primary cell wall composed mainly of the above-described components (Fig. 1). The thickness of the primary cell wall in growing cells varies from 50 nm to ~1 μm, depending on the cell type and developmental stage (Derbyshire et al., 2007). Secondary cell walls (Fig. 1) are thicker (≥1 μm; Taiz et al., 2022) and function as highly specialized structures that fortify cells that have ceased growing. Predominantly found in tissues responsible for mechanical support, water transport, and defence, these walls are characterized by a higher lignin content, which contributes to their rigidity.
Fig. 1.
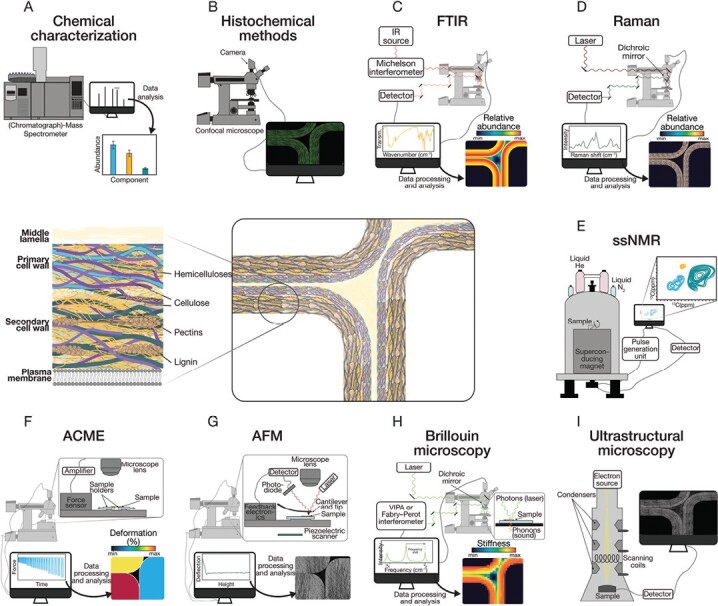
The plant cell wall is a chemically diverse structure composed mainly of polysaccharides (cellulose, hemicelluloses, and pectin), with secondary cell walls also including lignin. These molecules are arranged in a complex 3D structure that gives the cell wall its strength and rigidity. New techniques for studying plant cell walls provide insights into cell wall composition (A–E) and mechanical characteristics (F–I). (A) Chemical characterization provides accurate absolute quantification of the different cell wall components, although with no possibilities of in situ studies. (B) Histochemical methods such as using monoclonal antibodies against cell wall epitopes or click chemistry carbohydrate probes allow the location of specific components on the cell walls (e.g. hemicelluloses are marked in green on the figure). (C) Fourier-transform infrared (FTIR) spectroscopy enables relative quantifications of specific structural components (e.g. hemicelluloses) but the resolution is limited. (D) Raman spectroscopy also enables relative quantifications, in this case with higher resolution. (E) Solid-state NMR (ssNMR) allows a high level of detail in the study of chemical composition, but the data interpretation is complicated, and it is not compatible with in situ analysis. (F) An automated confocal-microextensometer (ACME) enables the measurement of extensibility and other mechanical properties in vivo in individual cells. (G) Atomic force microscopy (AFM) uses a probe (cantilever) to determine cell wall stiffness and topography, and generates an image with nanometric resolution, although it is restricted to the first cell layer. (H) Brillouin microscopy can be used to study mechanical properties in different cell layers with a micrometric resolution. The frequency shift caused by the Brillouin effect is a measurement of stiffness, whereas the amplitude of the signal can be related to viscosity. (I) Ultrastructural microscopy techniques can be used to image macromolecules at submolecular resolution, allowing the observation of cell wall microstructure.
Although we have a good understanding of cell wall composition and structure, resolving the precise interactions of the cell wall components will be an essential prerequisite to understanding global network behaviour and properties.
The plastic cell wall: tools to study the composition of an ever-changing system
From a biochemical point of view, the cell wall composition is well known for particular plant species, developmental stages, tissues, and even across a single cell wall (Somssich et al., 2016; Gigli-Bisceglia et al., 2020; Molina et al., 2021). However, there is limited holistic knowledge concerning spatio-temporal variations. For instance, a classic biochemical characterization consisting of monosaccharide and linkage analysis (Pettolino et al., 2012) is often performed using whole plants or organs of a particular developmental stage or exposed to different conditions for defined periods of time. Other biochemical characterizations such as oligosaccharide mass profiling (OLIMP), which requires enzymatic digestion followed by massspectrometry on the solubilized oligosaccharides (Günl et al., 2011), have similar drawbacks. On the other hand, histological methods (recently reviewed by Voiniciuc et al., 2018; DeVree et al., 2021) enable a more detailed analysis of cell walls at the tissue and cell levels, but they are limited by sample characteristics. For example, samples that are too thick, such as intact seedlings or plant structures, are unsuitable for most transmission-based microscopy techniques and require slicing and fixing. Other techniques, such as spectrometry-based techniques or advanced microscopy [atomic force microscopy (AFM) or electron scanning microscopy] are restricted to the epidermis and adjacent cell layers. Therefore, techniques that allow in vivo compositional analysis are of outstanding interest to detect and quantify the changes in cell wall composition during different growth stages and exposure to different environmental conditions (Fig. 1A–E; Table 1). To illustrate this, we will use three different scenarios from development, response to drought, and plant immunity. We will summarize what kind of dynamic responses occur in each case in primary cell walls, and describe the latest methods used to understand them. Finally, we will discuss which methods we think have strong potential in the future to significantly advance knowledge in the field.
Table 1.
Experimental techniques to study cell wall dynamics
| Technique | Information | In vivo analysis (temporal resolution) | Spatial resolution | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|
| Chemical characterization | Mono-/oligosaccharide composition Glycosidic linkages Decorations |
No | None | Quantitative and detailed information | Only provides global information of all cell walls in the sample. Destructive. |
Günl et al (2011); Pettolino et al. (2012) |
| Fluorescent probes | Relative amount of components Decorations Location |
Depending on the technique | 180–500 nm | Variety of tools. Defined protocols and relatively easy to use. |
Limited information (qualitative). Resolution restricted by the diffraction limit. |
Pattathil et al. (2010); Knox (2012); Bidhendi et al. (2020a) |
| Click chemistry | Relative amount of components Decorations Location Cell wall network packing density |
Depending on the technique [limited to non-Cu(I)-catalysed reactions] | 180–500 nm | Better resolution for chemical composition. Non-destructive. |
Limited number of non-toxic probes. Resolution restricted by the diffraction limit. |
Sawa et al. (2006); Anderson et al. (2012); Tobimatsu et al. (2014); Ropitaux et al. (2022) |
| FTIR | Spectra of IR absorption (determined by chemical composition and arrangement of components) | No | None | High amount of information (polysaccharides, functional groups, linkages, in muro organization). Small amount of sample. Non-destructive. Easy to use. |
Water interference Data processing is essential and can be complex. |
Alonso-Simón et al. (2011) |
| FTIR microspectroscopic imaging | No | 1–5 μm | Same as FTIR plus in situ analysis. | Resolution restricted by the diffraction limit. | Mouille et al. (2003) | |
| Raman spectroscopy microscopy | Raman spectra (determined by molecular composition and distribution) | No | 0.5–1 μm | Non-invasive. Label-free. |
Long acquisition time. Sample manipulation. Resolution limited by diffraction limit. |
Mateu et al. (2020) |
| CARS/SRS | Yes | 130–300 nm | Same as Raman but with a lower acquisition time. Allows quantitative analysis (only SRS). |
Sample manipulation. Sample fluorescence. Resolution restricted by the diffraction limit. |
Zhao et al. (2019); Xu et al. (2021) | |
| ssNMR | NMR spectra (structure of molecules, chemical composition, and dynamics) | Yes | Atomic resolution | Very high amount of information. Nanometric scale. |
Complex information. Requires specific and expensive equipment. Requires NMR-active isotopes or cryogenic temperatures. |
Zhao et al. (2020, 2021) |
| Tensile tests | Material deformation (stretching) upon force loading | Limited | Whole tissue | Measurements in living tissues. Measurement of in plane stiffness. |
Heterogeneous/non-homogeneous behaviour of cell walls. | Bidhendi and Geitmann (2018a); Suslov and Vissenberg (2018) |
| ACME | Yes | Single cell | Same as in tensile tests but allowing simultaneous imaging. | Tissues need to be accessible. | Robinson et al. (2017) | |
| AFM | Young’s modulus based on the interaction of a thin cantilever with the sample | Possible | 1–10 nm | Direct measurements of cell wall mechanics. Label-free. |
Limited to first cell layer (epidermis). Invasive. |
Krieg et al. (2018); Zhao et al. (2019) |
| Brillouin microscopy | Longitudinal elastic modulus based on Brillouin effect | Yes | 1–3 μm | 3D measurements. Non-invasive. Label-free Short acquisition times. |
Indirect measurements of cell wall mechanics. |
Caponi et al. (2020); Yang et al. (2022, Preprint) |
| FE-SEM | Ultrastructure of surfaces | No | 1–5 nm | High resolution. | Limited to surfaces. | Zheng et al. (2018, 2020) |
| Cryo-ET | Ultrastructure of surfaces in cryogenic samples | No | 1–5 nm | High resolution. Native conditions preserved. |
Imaging requires vacuum conditions and low temperatures. Limited to surfaces. |
Sarkar et al. (2018); Nicolas et al. (2022) |
| CLEM | Ultrastructure and component’s location | No | 3–5 nm | High resolution. Combined fluorescent and electron microscope imaging. |
Limited to surfaces. Fluorescence and electron imaging gap. | Chambaud et al. (2022) |
| sNano-FTIR | Spectra of IR absorption (determined by chemical composition and arrangement of components) and topography | No | 20 nm | High resolution of chemical composition. Non-destructive. Label-free. |
Limited to first cell layer. | Huth et al. (2012); Charrier et al. (2021) |
Development
The primary cell wall has multiple crucial functions in plant development. It gives shape to newly dividing cells, regulates cell growth and cell cycle progression, controls cell specialization, and responds to external stimuli (Bidhendi and Geitmann, 2016; Voxeur and Hofte, 2016; Gigli-Bisceglia and Hamann, 2018; Anderson and Kieber, 2020). Biochemical and enzymatic modifications of cell walls lead to the loosening, strengthening, extending, or softening of the structure, which is essential for plant development (Peaucelle et al., 2011; Park and Cosgrove, 2012; Cosgrove, 2016a; Coen and Cosgrove, 2023). Consequently, the connection between the composition and structure of plant cell walls is especially close for these processes. In addition to the examples presented here, we will further examine structural changes to the cell wall later in this review.
In this context, pectins are especially relevant. Both pectin composition (i.e. the relative abundance of each of the different pectins) and pectin methylesterification and de-methylesterification play important roles in plant cell growth (Peaucelle et al., 2011; Braybrook and Peaucelle, 2013; Altartouri et al., 2019). Different histological techniques such as propidium iodide staining or oligosaccharide-based probes have been used for in vivo labelling of pectins (Bidhendi et al., 2020a). These probes have several distinct advantages over immunostaining (another histological method that we discuss in the next subsection), such as smaller size, better tissue penetration, and higher resolution, the possibility to perform time-course experiments, and easier dual labelling protocols. Furthermore, these probes are suitable for studying the dynamics of (de)methylesterification because they are highly sensitive to changes in methylation and acetylation status. This sensitivity is particularly relevant when considering the ‘egg-box’ model, where different degrees of methylesterification influence the structure and functionality of the cell wall. In one prominent example, Mravec et al., (2014) demonstrated that a chitosan oligosaccharide (COS) probe coupled to Alexa Fluor 488 (COS488) binds specifically to homogalacturonan (HG)-containing pectic polysaccharides and oligogalacturonate (OGA) derived from HG. The binding of COS488 to pectic polysaccharides decreased with increasing degrees of methylation, and to oligosaccharides with increasing degrees of polymerization. Using the COS488 probe, Mravec et al. (2014) tracked HG turnover in Arabidopsis root cap cells and found that inhibiting demethylesterification and lowering HG synthesis reduced COS-binding sites and caused defects in cell adhesion. Removing methyl esters created cleavage sites for HG-degrading enzymes that promoted cell separation. This indicates that HG methylation is fine-tuned at the cellular level and may primarily contribute to cell wall stiffening rather than adhesion in mature root cap cells under mechanical stress. However, a drawback of the COS probe is its dependence on ionic strength for binding, which can be influenced by local environmental factors. In a subsequent study (Mravec et al., 2017), the authors developed oligogalacturonide (OG)-based probes (OG7–OG13) to visualize the sites of egg-box formation in situ. The OG7–OG13 probes probably do not extensively affect cell wall homeostasis at low concentrations, and their interactions do not rely exclusively on ionic interactions, making them more reliable for studying the dynamics of HG processing. Still, these probes are strongly limited by the type of interactions occurring between carbohydrates.
Another alternative to study pectin turnover is click chemistry labelling. This uses bio-orthogonal chemical reporters, which are analogues to monosaccharides (azido- or alkyne-monosaccharides). They are incorporated into the cell wall target polysaccharide using the cell’s endogenous biosynthetic machinery. Once synthesized and deposited in the cell wall, the polysaccharide containing the reporter can be coupled to an exogenous fluorescent probe through a covalent bond, allowing study of their location and relative amount (Sawa et al., 2006). In Arabidopsis, the first application of this method used a fucose analogue to study pectin organization and dynamics (Anderson et al., 2012). However, a major drawback of this approach is that it uses copper I ions as catalysers, which are toxic for Arabidopsis and damage the wall. More recently efforts have been made to improve the technique by developing copper-free labelling methods (Hoogenboom et al., 2016). To date, there are a limited number of monosaccharide analogues for click chemistry available that have no toxic effects on the cell wall. Last year, Ropitaux et al. (2022) described a method using the azido-monosaccharides Fuc-N3 and Kdo-N3 for in vivo labelling of Arabidopsis root cell walls. This method enables the labelling of the pectin polysaccharide rhamnogalacturonan-II (RG-II) and is an example of the future direction required for this field. Interestingly, the regulation of cell wall lignification in distinct cell types, which happens at specific stages during the process of cell differentiation, and its subcellular localization, can be also examined through click chemistry (Tobimatsu et al., 2014; Pandey et al., 2015; Lion et al., 2017).
Cellulose also has a critical role in plant growth, development, and morphogenesis. If cellulose microfibrils are synthesized in a preferential orientation on the cell wall, this region will be reinforced, and the anisotropy of the wall composition will facilitate cell expansion in the direction of minimum stress/stiffness (perpendicular to cellulose orientation) (Green, 1964; Wang and Jiao, 2020). Interactions between cellulose microfibrils and other cell wall components such as hemicelluloses (xyloglucan) or pectins fine-tune wall expansibility. While different models describing the interactions have been proposed (Park and Cosgrove, 2012; Altartouri et al., 2019; Coen and Cosgrove, 2023) they still need to be clarified. Lately, several studies have used solid-state nuclear magnetic resonance (ssNMR) to gain insight into the pectin–cellulose interaction. NMR is a physical phenomenon in which nuclei in a magnetic field absorb and re-emit electromagnetic radiation. This energy emitted has a specific resonance frequency, which depends on the strength of the magnetic field and the magnetic properties of the nucleus. NMR spectrometry uses the observed changes in the nuclear spin states of atoms in a molecule to determine the structure of a molecule, its chemical composition, and its dynamics. In particular, ssNMR can be used to determine the chemical structure, 3D structure, and dynamics of solids and semi-solids at an atomic level, which allows analysis of cell wall composition with high resolution. This spectroscopic method provides a plethora of information on the conformational structure and linkage of polysaccharides, cellulose packing, and water content (Zhao et al., 2020). It has been observed that mutations in genes encoding pectin methyltransferases cause an increase in highly branched arabinan, a particular type of pectin. Moreover, in these mutants, there is a closer association between pectins and cellulose. Specifically, the mutants exhibit a tighter packing between cellulose and pectic backbones (HG and RG-I), pointing to a role for these pectins in the stabilization of cellulose–pectin contacts (Kirui et al., 2021). Despite the high resolution of this technique, a strong limitation is that NMR-active isotopes (e.g. 13C and 15N) must be incorporated into cell wall polymers to obtain the spectral resolution required for resolving numerous carbon and nitrogen sites in plant cell walls. This inconvenience can be overcome by using the recently developed magic-angle spinning dynamic nuclear polarization (MAS-DNP) method, which enhances NMR sensitivity by tens to hundreds fold. This technique has been used already in poplar (Perras et al., 2017), rice (Zhang et al., 2019; Zhao et al., 2021), and cotton (Kirui et al., 2019) to study polymer composition, structural polymorphisms, and intermolecular packing. However, it cannot be used for in vivo studies because MAS-DNP is conducted at cryogenic temperature (~100 K) (Zhao et al., 2021). Other necessary considerations when using that approach include the potential dehydration of the samples due to the centrifugation induced by the spinning of the samples, the heating effect of radiofrequency pulses, and the limitations for maintaining proper physiological conditions (Ghassemi et al., 2022). Furthermore, another limitation is that current NMR methods do not allow the observation of the spatial architecture or the specific location of the different components within the cell wall. Different methods which combine microscopy techniques with NMR have been employed for the study of other extracellular matrices such as biofilms or fungal cell walls (Ghassemi et al., 2022). This is an exciting perspective to further understand interactions between different cell wall components. However, its application to plant cell walls is still to be explored.
Response to drought
Plant cell walls are essential to how plants respond to changes in their environment. Cell walls serve as a protective barrier against environmental stressors such as temperature fluctuations, UV radiation, and mechanical stress. Moreover, cell walls have a very relevant role in another process that determines drought resistance: stomatal opening. Stomata are small openings or pores found on the epidermis layer of plants. Composed of two guard cells, they are responsible for allowing gas exchange between the plant and its environment, and regulating the water balance of the plant (Meidner and Mansfield, 1968). Fluctuations in turgor pressure within guard cells control the stomatal opening and closure. These fluctuations are, in turn, regulated by the biochemical and mechanical properties of the cell walls, which determine the extent of the morphological changes (Amsbury et al., 2016).
Different histological techniques have been used to identify the components of guard cell walls, which are different from the neighbouring epidermal cells. For example, in Arabidopsis, the guard cell wall is distinguished by a low level of methylated pectins, as detected by the differences in immunofluorescence using the monoclonal antibody LM20—binding to highly methylesterified HG epitopes (Amsbury et al., 2016). An immunostaining using the monoclonal antibodies LM6M—binding short-chain linear arabinan epitopes—and LM13—binding long-chain arabinan epitopes—revealed the relevance of short-chain linear arabinans associated with the presence of RG-I to maintain the flexibility and function of guard cell walls (Carroll et al., 2022). However, although this work was complemented with computational modelling and AFM, it did not fully evaluate the dynamics of the system or the changes in other cell wall components.
Fourier-transform infrared (FTIR) spectroscopy is a non-destructive technique used to obtain the absorption spectra of the molecules in the sample in a range of infrared wavelengths, thus generating a characteristic fingerprint determined by the composition of the sample and by the distribution of the components. When applied to plant cell walls, it can provide information about cell wall composition and in muro organization, including the orientation of cell wall polymers (Gierlinger et al., 2008; Kafle et al., 2017). Because of the large amount of data obtained and the complexity of the spectra, the data analysis step is critical (Alonso-Simón et al., 2011). Multivariate analyses are good approaches to extract the maximum amount of information from FTIR data (Gorzsás et al., 2011). An implementation of this technique, FTIR microspectroscopic imaging, permits a detailed characterization of the biochemical composition of specific cell walls in situ, although not in vivo since this approach requires dehydrated samples. It has been used, for example, to classify cell wall mutants (Mouille et al., 2003), and could be a good fit to study in depth the composition and orientation of cell wall polymers of guard cells. Using microfluidic device such as that used by Devaux et al. (2018) to follow enzyme degradation of maize cell walls could be an elegant solution.
Plant immunity
Plant cell walls are the first barrier that pathogens encounter when they infect a plant. It is for this reason that many pathogens have a plethora of enzymes that attack the cell wall and degrade it. Plants, nevertheless, learned how to recognize this damage and interpret these as a danger signal that triggers plant immunity (Bacete et al., 2018).
Intriguingly, some plants with altered cell wall composition have different resistance patterns against different pathogens. For example, in a recent study, >85% of the analysed Arabidopsis cell wall mutants were either more resistant or more susceptible than wild-type plants to at least one of the pathogens studied (Molina et al., 2021). In this work, all the mutants were examined using monosaccharide and linkage analysis and FTIR to confirm the presence of cell wall alterations, although the specific location or temporal dynamics were not addressed. There are two alternative and not incompatible interpretations for the effect of these alterations in disease resistance. First, the different composition has an effect on the cell wall’s structure, thus complicating or facilitating the access of the pathogen (passive role). Indeed, cell wall reinforcement, for example through the deposition of lignin, is a relevant plant defence response (Bacete et al., 2018). Second, the different composition is reflected in a differential production of damage-associated molecular patterns (DAMPs), which trigger plant immunity (active role). Further biochemical characterization of the differences followed by chromatographic purification has been used to isolate DAMPs present in some of these mutants, which regulate different responses to pathogen attack (Bacete et al., 2020; Mélida et al., 2020). Moreover, DAMPs may prime plants for future pathogen encounters. For example, OGs induce the accumulation of camalexin, which has antimicrobial activity (Gravino et al., 2015). To understand the dynamics of cell wall changes in plant immunity, non-destructive in vivo techniques that do not interfere with immunity-related signalling processes are necessary. An example in which these considerations are especially relevant is the use of oligosaccharide probes described above. These molecules could have immunomodulatory activity and affect cell wall homeostasis (Cabrera et al., 2010), which makes them unsuitable for these studies. However, the authors note that only a small amount is required for labelling and should not interfere with the process (Mravec et al., 2017).
Raman spectroscopy, an analysis technique based on the Raman effect or inelastic scattering, uses high-intensity laser light sources that interact with the chemical bonds in the sample. If the energy of the light matches the energy of the molecular vibrations in the sample, a small amount of the energy is scattered. The resulting photons of slightly lower or higher wavelengths can be used to identify the molecular composition of a material (Mateu et al., 2020). Raman spectrometry has been applied to plant cell walls since the 1980s (Atalla and Agarwal, 1986), although some limitations in resolution and the requirement for specialized equipment have marginalized its use. However, there is a newly acquired interest in this technique due to the recent advances in optics and detectors, the new methods to enhance the signal intensity, and the possibility to combine it with confocal microscopy enabling analyses at the micrometre level (<0.5 μm), and thus improving the spatio-temporal resolution (Mateu et al., 2020). Recent improvements have targeted signal levels over spontaneous Raman spectroscopy. For example, coherent anti-Stokes Raman scattering (CARS) focuses the excitation energy specifically onto said Raman mode, whereas stimulated Raman scattering (SRS) uses stimulated Raman gain and loss (Li et al., 2020). Both approaches reduce the scanning times, making them compatible with the observation of live processes. Moreover, SRS can provide quantitative information, although the analytes must overcome a certain threshold abundance (Zhao et al., 2019). Coherent Raman scattering (CRS) is a newly developed protocol tested in Arabidopsis which includes both CARS and SRS to improve the signal-to-noise ratio, and thus is able to avoid the use of resins, therefore reducing the sample manipulation. Interestingly, a sample’s autofluorescence does not interfere with CRS (Xu et al., 2021). Therefore, it could be used as an excellent approach to study changes in cell wall composition upon pathogen infection, following generation of DAMPs and cell wall-reinforcing events.
A dynamic architecture: tools to evaluate changes in cell wall mechanics and organization
Different techniques can be used to study plant cell wall mechanical characteristics and organization. Here, we will discuss several experimental techniques that provide information with micro- and nanoscale resolution in vivo, thus allowing the study of dynamic processes that modulate the viscoelastoplastic characteristics of plant cell walls (Fig. 1F–I; Table 1). In silico strategies, which have proven to be vital complements to experimental techniques to predict and examine diverse mechanical processes, are discussed in Box 2.
Box 2. In silico approaches to study cell walls.
Cell shapes in plants are very diverse. They depend on cell wall structure and composition and on the osmotic status of the cell. Due to their complex forms, mathematical models are often used to simulate plant cells dynamics. Modelling has become a useful tool to investigate how biophysical properties of cell walls are modulated under different conditions to produce cell and organ shapes.
Modelling tools go hand in hand with experimental techniques. By using models, complex systems can be reduced to their essential components which makes them easier to understand. Subsequently, hypotheses about the behaviour of the system can be formulated and model validation and refinement be performed. Additionally, the function of mechanisms can be isolated from global signals to understand their individual contribution to complex biological processes (Riglet et al., 2021). Models can be created to analyse different scales. Cell wall components such as cellulose can be modelled at the quantum, atomistic, and molecular level. In the following lines we describe selected approaches developed to investigate cell wall dynamics. The description of the methodologies included in this box is evidently a non-comprehensive source. Readers interested in a more detailed exploration of modelling of plant growth might wish to consult these suggested publications (Geitmann and Dyson, 2014; Morris, 2018; Forterre, 2022; Jensen, 2022).
Coarse grained molecular dynamics models have been successfully used to explore the mechanics and structural properties of cell walls after application of a stretching force (Zhang et al., 2021). Instead of having an all-atoms simulation, the main idea of coarse grained models is to have a reduced representation by grouping functional atoms or molecules into a single particle (Mehandzhiyski and Zozoulenko, 2021). Major drawbacks of these simulations are high computational power, short simulation times (fractions of a second), and the limited number of atoms or molecules that are possible to include in them. An excellent review on cell walls, which describes the general aspects of coarse grained model applications, has been recently published (Cosgrove, 2022).
The finite element method (FEM) is one approach based on continuum mechanics to solve partial differential equations arising from modelling structures and reaction to forces. In biological samples, cell walls/shapes/tissues are discretized and represented as an interconnected mesh of nodes and vertices, and their collective behaviour is modelled (Kennaway and Coen, 2019). FEM can incorporate parameters related to cell walls and mechanical forces to predict changes in cell shape or, based on cell morphology, it can predict biophysical properties. After applying boundary conditions, the effects of perturbations to the system can be calculated for each node. As a result, the displacement of each node together with strain and stress values are obtained. A recent review (Bidhendi and Geitmann, 2018b) extensively described the application of FEM in plants.
Cortical microtubule arrays guide deposition of cell wall materials which in turn determine anisotropic cell wall properties. Therefore, simulations of microtubule alignment and densities are important to understand plant growth. However, the highly dynamic behaviour of microtubule polymerization makes them a difficult subject to study (Elliott and Shaw, 2018). Cytoskeleton imaging has shown that plant cells have a homogeneous array of cortical microtubules across their plasma membrane; however, this aspect was difficult to simulate starting from nucleation sites which generated a positive feedback loop for microtubule nucleation and inhomogeneous arrays in the models. Using a recent model that incorporates a new mechanism in which nucleation was saturated in microtubule-rich regions, acting in opposition to the positive feedback loop, more homogeneous microtubule regions were obtained (Jacobs et al., 2022). Microtubule diffusion rates and the role of their interaction partners in regulating microtubule concentrations are aspects that require future investigations.
Special attention should be given to the time scales and frequencies employed by different techniques to measure dynamic properties. Properties measured in units of seconds or Hertz are practical to describe qualities in the macroscopic scale. However, the spatial scale of cell wall polymer assemblies, both microfibrils and gel-like elements, ranges between 1 μm and 100 μm (Buchanan et al., 2015). Studies on polymers and molecular condensates, which have equivalent dimensions, indicated that measuring dynamic properties in the kilo Hertz to giga Hertz frequencies and in the microseconds to picoseconds time scales are very relevant to understand the intramolecular interactions taking place in these structures (Padding and Briels, 2011; Mitrea et al., 2018; Adichtchev et al., 2019; Seifert et al., 2021; Keshmiri et al., 2023, Preprint). In the following subsections, we introduce techniques that measure dynamic properties in cell walls in these ranges. However, some of them differ in the parameter that is being measured (e.g. AFM and tensile testing compared with Brillouin microscopy, see below) and cannot be directly compared. Therefore, a full description of the method and measured parameters is required when describing sample properties. In addition, we introduce techniques that only allow for static measurements (on non-living samples) but with unrivalled resolution.
Atomic force microscopy
AFM is a high-resolution microscopy technique that can measure the mechanical properties of the sample with nanometre resolution. The interaction between the tip of a thin cantilever and the surface of the sample generates a mechanical signal that can be used to calculate Young’s modulus, one of the elastic moduli of materials used to relate stress and strain. The definition of the different elastic moduli depends on how forces are applied and how material deformations can occur (Landau et al., 1986). Young’s modulus relates to the axial deformation of a material (strain) when tensile stress is applied along the axis (Allison et al., 2010; Peaucelle et al., 2011; Routier-Kierzkowska et al., 2012; Milani et al., 2013; Caponi et al., 2019). The small size and shape (including pyramidal, conical, cylindrical, and spherical) of the cantilever’s probe is the reason for its high resolution compared, for example, with tensile tests. However, its ability to measure mechanical properties at the subcellular level implies that a significant amount of time is required to scan large areas. Additionally, the application of AFM is restricted to directly accessible/surface cell layers. The often invasive nature of the indentation requires that applied forces are controlled to avoid perturbation or damage in living samples, such as potential displacement of cell wall fibres using sharp probes (Braybrook, 2015; Bidhendi and Geitmann, 2019; Asgari et al., 2020). In addition, the contribution of turgor pressure to the measurements of elastic constants needs to be taken into account (Beauzamy et al., 2015; Braybrook, 2015; Malgat et al., 2016). Furthermore, the cell samples need to be efficiently immobilized, which limits the application of AFM for live experiments.
AFM has been a relatively widely used method to evaluate mechanical properties of plant cell walls in response to developmental cues, hormonal or enzymatic treatments, turgor pressure, or pectin methylesterification (Weber et al., 2015; T. Zhang et al., 2017; Feng et al., 2018; Song et al., 2020; Wang et al., 2022). However, Young’s moduli values obtained in these experiments are usually lower than those obtained in tensile stretching experiments (see later), or osmotic pressure measurements (where a treatment is applied—typically NaCl, sorbitol or mannitol—to change the turgor pressure inside the cells and cell deformation is quantified to estimate Young’s modulus) (Sapala and Smith, 2020; Seifert et al., 2021). The last two methods allow the measurement of stiffness in the direction of cellular growth, which is presumably more relevant to growth and morphogenesis. In contrast, AFM is usually set to apply an indentation force in the normal direction.
Despite the development of novel AFM modes to study dynamic processes with little perturbation to the samples, their application in time-sensitive mechanical measurements of plant cell walls remain scarce. A recent work by Seifert et al. (2021) used multifrequency AFM for the first time on living plants—Arabidopsis hypocotyls—to obtain nanoscale maps of elasticity, viscosity, and time relaxation. The method superimposes multiple frequencies during the cantilever indentation and detects changes in its oscillation parameters, allowing calculation of the dynamic mechanical parameters of the scanned area (García and Herruzo, 2012; Cartagena-Rivera et al., 2015). An adequate model that distinguishes between reversible and irreversible deformations to provide realistic viscoelastic quantification measurements is essential. By setting the cantilever oscillation to frequencies in the kilo Hertz range (time scale of microseconds), Seifert et al. (2021) aimed to obtain mechanical measurements directly relevant for plant growth. Young’s modulus values obtained with multifrequency AFM corresponded with reported values using other macroscopic techniques such as tensile stretching and osmotic pressure experiments (hundreds of MPa), in contrast to static AFM measurements which typically produce lower Young’s modulus values (0.1–20 MPa).
Brillouin microscopy
Brillouin microscopy is based on the Brillouin light scattering effect (i.e. the interaction between incident light and the acoustic waves inside a sample), which leads to a frequency shift of a few giga Hertz in the inelastic scattered light. The sample’s mechanical properties determine the propagation characteristics of the acoustic waves. Hence, the observed frequency shift can be related to the sample’s longitudinal modulus of elasticity, which relates uniaxial stress to strain in the same axis (material is not allowed to expand laterally), allowing quantification of the material’s stiffness (Prevedel et al., 2019). Therefore, Brillouin microscopy presents important advantages in comparison with AFM (Table 1), including the possibility of non-invasive, label-free, and 3D stiffness measurements in vivo.
In recent years, this technique has gained relevance for the study of mechanobiology, although it is still relatively new in the plant sciences field. In a pioneering study, Elsayad et al. (2016) used Brillouin microscopy to examine the mechanical properties of fluorescently labelled extracellular matrices (a broad concept including plant cell walls) in onion and Arabidopsis. The results confirmed that the method was flexible enough to be applied to different organisms and showed that the hydrostatic pressure in the cell and the viscosity of the cytoplasm impact the mechanical characteristics of plant extracellular matrices. When applied to Arabidopsis, Brillouin microscopy was instrumental to demonstrate that red and far-red signalling modulates the mechanical characteristics of the extracellular matrix (Elsayad et al., 2016). In terms of sensitivity, Brillouin microscopy offered valuable quantitative insights, allowing for precise measurements of these mechanical changes. Further studies have delved into the applicability of this technique, with Altartouri et al. (2019) employing it to understand lobe formation in epidermal pavement cells. In an interesting approach, Bacete et al. (2022) applied Brillouin microscopy to explore the effects of cellulose biosynthesis inhibition and turgor alterations on root cell walls. This particular study was noteworthy because it demonstrated the technique’s ability to detect changes across the radial axis of the root, including the analysis of inner layers such as the central cylinder, in live samples. In fact, the depth at which Brillouin microscopy can effectively analyse depends only on the sample’s degree of transparency.
Due to the architecture of conventional microscopes, initial Brillouin microscope set-ups needed to rotate the sample to allow stiffness measurements in different directions (Koski et al., 2013; Eltony et al., 2022). Recently, a novel implementation to measure Brillouin scattering at different in-plane angles simultaneously, based on a light dispersive element, was used to measure mechanical properties of periclinal and anticlinal hypocotyl cell walls (Keshmiri et al., 2023, Preprint). Measurement of the anisotropic mechanical properties of cell walls with one method highlights the potential of Brillouin microscopy in plant mechanobiology.
While Brillouin microscopy has brought significant advances to plant sciences, it does carry certain limitations (Palombo and Fioretto, 2019; Prevedel et al., 2019; Antonacci et al., 2020; Zhang and Scarcelli, 2021). Firstly, the technique’s ability to probe the high-frequency longitudinal modulus within a sample is not a direct measure of the conventional Young’s modulus. This introduces an additional complexity as there is currently no theoretically established relationship between these two moduli. Nevertheless, empirical correlations have been made to relate the the two moduli with practical success, suggesting that measurements in frequencies different from the biological process time scales are still relevant (Scarcelli et al., 2015). Secondly, the acquisition time of the technique is slower than some other imaging modalities due to the intrinsic weak intensity of the Brillouin signal. Additionally, the instrument’s sensitivity is restricted by the spectral precision of the spectrometer. Despite these limitations, advancements such as confocal Brillouin (Zhang and Scarcelli, 2021) microscopy offer potential solutions, underscoring the promise of Brillouin microscopy for future plant science research.
Combined spectroscopy approaches: studying composition and structure all at once
Newly developed approaches aim to combine label-free non-destructive techniques that provide information about biophysical characteristics of materials (such as AFM or Brillouin microscopy), with others that provide information about their composition (such as FTIR and Raman spectroscopy/microscopy) (Prats-Mateu and Gierlinger, 2017; Scarponi et al., 2017). These bring a whole new perspective to the study of plant cell walls since they allow simultaneous characterization of both the biochemical composition and mechanical properties in the same sample with high temporal and spatial resolution.
Confocal Raman microscopy and AFM can achieve resolutions to probe composition, structure, and mechanics of single cell walls (<0.5 μm) (Zhao et al., 2019). A correlative application of these techniques was used to track the dynamic changes of cell walls in early wood cells and revealed how the architecture changes under wood compression (Felhofer et al., 2020). Redistribution of cell wall components in the same sample area was then observed by applying compressive forces and looking at the adhesion forces between the AFM tip and the chemical composition of the region probed. Compressed regions contained denser microfibrils and accumulation of lignin compared with open regions which were characterized by porous regions or water-rich regions and fewer denser fibres (Felhofer et al., 2020). These results agreed with Young’s modulus calculations from the slope in graphs of the repulsive force signal following tip snap. A higher Young’s modulus was observed in compressed regions (stiffer) compared with open regions (softer). In addition, this study supports cell wall models highlighting the dynamic interaction occurring at the nanodomains between lignin and other cell wall polymers (Kang et al., 2019).
The diffraction limit is a fundamental limitation of optical systems. This limit is a consequence of diffraction and the wave nature of light, resulting in a diffraction pattern known as an Airy disk (for a uniformly illuminated, circular aperture). According to the Rayleigh criterion, two points can only be resolved when the centre of one diffraction pattern is directly over the first minimum of the other. Consequently, objects smaller than half the wavelength of the incident light cannot be resolved, a phenomenon known as the Abbe limit. Several implementations of the AFM–Raman combination have tried to overcome this limitation, enabling the determination of the biochemical composition of the sample with nanometre resolution. These techniques are based in scanning near-field optical microscopy (SNOM), which places a detection tip in the near-field regime of the sample (within one wavelength) to focus and collect light. Prominent examples are near-field Raman which uses a subwavelength tip aperture to illuminate samples and can achieve resolutions between 50 nm and 150 nm (W. Zhang et al., 2017); and AFM-based tip-enhanced Raman spectroscopy (TERS), which uses an apertureless sharp metal tip positioned at the illumination focus to collect scattered light from the near field to the far field, achieving resolutions up to 1.7 nm (Zhang et al., 2016). However, there are currently no applications of near-field Raman or TERS in plant cell walls, probably because of the low Raman signal, tip manufacturing, and reproducibility issues (Prats-Mateu and Gierlinger, 2017).
In a different setup, a scattering(apertureless)-SNOM combined with an AFM (referred to as nano-FTIR) and equipped with an infrared light source can resolve infrared images, achieving resolutions that only depend on the size of the tip apex (Huth et al., 2012). This instrumentation can be used to obtain mechanical and chemical properties including topography, mechanical phase (sensitive to viscosity), optical amplitude, and phase (related to optical reflectivity and absorption) (Zancajo et al., 2020; Charrier et al., 2021). The chemical composition and the delignification process of primary cell walls in young poplar trees were monitored by measuring local values of dielectric functions of cellulose and lignin at the unprecedented resolution of 20 nm (Charrier et al., 2021). Nano-FTIR has also been used to investigate polysaccharide composition and structure in secondary cell walls and middle lamellae of three varieties of poplar hardwood with different recalcitrance levels (Bhagia et al., 2022). This method has the potential to investigate cell wall modifications induced by physical, chemical, and biological processes.
A particularly interesting emerging approach for studying plant cell wall dynamics arises from the combination of Raman and Brillouin spectroscopic methods. Indeed, both are closely related. Raman spectroscopy uses light to probe the molecules in a material, as described above. The approach is comparable with Brillouin spectroscopy, which uses light to probe vibrations in the sample. In plant cell walls, this combination has been used to examine the mechanical properties and composition of dying Populus and Geranium leaves (Rakymzhan et al., 2019). However, in this setup, Raman and Brillouin measurements are taken independently. An improved approach that combines both in one microscope, allowing simultaneous measurements, such as the one described by Traverso et al. (2015), is certainly an exciting possibility for the future combined studies of plant cell wall composition and mechanical characteristics.
Although the application of these techniques to cell walls can be technically challenging, they are undoubtedly promising strategies for the characterization of their mechanical proprieties and biochemical composition at the nanoscale.
Advanced microscopy techniques
Traditional light microscopy techniques such as those mentioned earlier do not have the necessary resolution to detect details at the nanometre scale and can only be used to observe cell wall structure or, with the methods mentioned, to study composition. However, they do not provide information to study its organization. In contrast, electron microscopy (EM) techniques, traditionally referred to as ultrastructure microscopy, allow imaging of macromolecules at submolecular resolution (Otegui and Pennington, 2019). As the wavelength of an electron is much smaller than that of a visible photon, EM techniques have better resolution than optical microscopes. In recent years, advances in optical microscopy have led to overcoming the diffraction limit obstacle, at least under some circumstances (Beyond the diffraction limit, 2009). Nevertheless, EM continues to give better resolution compared with light microscopy, although vacuum conditions and sample preparation requirements prevent measurements of live samples.
A conventional scanning electron microscope uses a heated tungsten filament to generate electrons that scan the surface of the sample. An improved version of this setup, field emission SEM (FE-SEM), uses field effect guns. These concentrate low and high energy electrons using a low electrical potential which allows for an improved resolution compared with conventional EM (Zheng et al., 2020). FE-SEM combined with high resolution AFM has been used to explore the fibrillar organization and topography of onion cell walls (T. Zhang et al., 2017; Zheng et al., 2018; Wang et al., 2020).
In electron tomography (or image reconstruction by optical sections), 2D EM projections can be used to reconstruct a 3D structure of an object by imaging at different angles (Ercius et al., 2015; Otegui and Pennington, 2019). This technique can be paired with cryofixation to examine samples in their native state, although potential drawbacks include the possibility of surface artefacts and low accuracy in sample preparation (Ercius et al., 2015). Specifically in plant sciences, cryo-electron tomography (cryo-ET) has been used recently to image the nano-architecture of plant cell walls. A study by Sarkar et al. (2018, Preprint) used this method to investigate the structure of Arabidopsis stems, revealing detailed measurements of individual microfibrils and their compositions. Their findings indicated that structural imperfections, such as buckling and twisting, in the microfibrils led to a reduction in cell wall stiffness and an increase in ductility, which they suggested could serve as a defence mechanism against rupture after extreme loading. To delve deeper into plant cell structures, Nicolas et al. (2022) used cryo-focused ion beam milling prior to cryo-ET, allowing them to access deeper layers in onion scale cells. Their research unveiled a complex interplay between regions with microfibrils and mesh-like regions, presenting interesting trends in mesh occupancy and microfibrillar orientation, with cellulose orientation patterns across successive lamellae following a bimodal and not a helicoidal pattern as previously proposed based on traditional TEM observations (Nicolas et al., 2022). The organization of the microfibrillar orientation is especially noteworthy as it holds significance for plant biomechanics and morphogenesis.
The high-resolution images acquired with EM techniques can be correlated with other techniques which allow for in vivo visualization of cellulose microfibril orientation. The fluorescent dyes Calcofluor White, Congo Red, and Pontamine Fast Scarlet 4B target have been used to stain cellulose and observed with polarized fluorescence microscopy (although Calcofluor White also binds to other cell wall polysaccharides and Congo Red also binds proteins) (Prentø, 2009; Anderson et al., 2010). Polarized light is usually not produced by common wide-field microscopes, but most confocal microscopes have plane polarized laser light (Gratton and vande Ven, 2006). These dyes can absorb light and fluoresce depending on light polarization. This phenomenon is called difluorescence or fluorescence dichroism, which has been exploited to observe cellulose microfibrils bound to the dye and oriented parallel to the direction of polarization in root hairs, pollen tubes, and leaf epidermal cells (Thomas et al., 2017; Altartouri et al., 2019; Bidhendi and Geitmann, 2019; Bidhendi et al., 2020a).
Correlative light and electron microscopy (CLEM) is a microscopy technique that combines light (fluorescence) and electron microscope imaging (van den Dries et al., 2022). Although it suffers from the resolution gap between conventional fluorescence microscopy (200 nm) and EM (1 nm), technical advances reduced this difference. CLEM was used to study the formation of plasmodesmata at graft interfaces in Arabidopsis between scion and rootstock hypocotyls of frozen and fixed materials (Chambaud et al., 2022). Using endoplasmic reticulum-retained yellow fluorescent protein (YFP) and red fluorescent protein (RFP) fluorophores, the origin of the observed plasmodesmata was established.
Tensile testing
The classical test to probe the mechanical properties of materials is tensile testing. The idea is to deform a material by applying a force (stretching), performed by extensometers, which provide information about the mechanical characteristics of materials (Bidhendi and Geitmann, 2019). Uniaxial or biaxial experiments are usually performed by fixing one or two ends of the sample, respectively, and stretching the opposite ends. Two approaches have been commonly used for plants in tensile testing: (i) stretching the material until mechanical failure occurs, allowing the extraction of mechanical properties such as stiffness and strength (Bidhendi and Geitmann, 2018a); and (ii) the creep method, where a constant load is used to stretch the sample and irreversible time-dependent mechanical properties can be detected (Suslov and Vissenberg, 2018). The steps of mounting and clamping or gluing the sample onto the extensometer should be done carefully to avoid building stress before the start of the experiment (Bidhendi et al., 2020b). This limitation applies especially to small samples (as they are more easily damaged) during in vivo studies. In comparison with other approaches that measure mechanical characteristics in the normal direction to the surface, tensile testing allows in-plane stiffness and elastic modulus determination (Park and Cosgrove, 2012). These parameters are more relevant for plant cell expansion and directional growth as they reflect the direction of deformation by turgor pressure (Bidhendi et al., 2020b; Majda et al., 2022).
Diverse setups have been used to measure creep and mechanical failure of plant materials at cellular and tissue level to confirm and predict mechanical properties. The automated confocal-microextensometer (ACME) methodology has been used to quantify physical properties in Arabidopsis seedlings with cellular and temporal resolution (Robinson et al., 2017). An interesting outcome of this methodology was the observation of gibberellin treatments increasing cell wall elasticity (Robinson et al., 2017), a key insight for understanding plant growth. Intriguingly, a strain gradient was also discovered along the hypocotyl, hinting at complex, yet to be identified, cell wall properties shaping plant mechanics.
A pitfall in extensometer experiments is assuming that plant tissues are homogeneous (Robinson et al., 2017; Bidhendi et al., 2020b). Plant cells are comprised of cell walls, aqueous cytoplasm, ions, and other macromolecules. In addition, cell wall composition is expected to be heterogeneous, potentially resulting in anisotropic mechanical properties. Intriguingly, extensometer results also show that the longitudinal direction of plant organs is stiffer than the transversal direction (Vanstreels et al., 2005; Bidhendi et al., 2020b). This is contrary to what would be expected for materials expanding, where softer tissues are expected in the direction of principal growth. It was suggested that extensometer experiments quantify tissue-level mechanical properties which differ from cellular properties including cell wall stiffness, cellular geometry, and patterning strength (Majda et al., 2022). In addition, non-linear behaviour has been observed in plant tissues when measuring typical strain–stress curves. It is therefore challenging to calculate Young’s modulus from such curves, as it is typically calculated using the slope of the linear region in strain–stress curves (Bidhendi et al., 2020b). A more sophisticated model would be required to extract mechanical parameters from non-linear strain–stress curves.
Beyond the wall
During their life, plant cells are exposed to a wide range of mechanical forces. As plant cells are connected to their neighbours via their middle lamella and cell walls, they experience mechanical tension arising in the neighbouring cells during growth and division. In addition, external factors such as wind, dehydration, pathogens, and herbivores create changes in the mechanical environment of cells, which they perceive. How plants sense mechanical forces, transduce them into chemical signals, and produce adequate responses to shape their growth is an area of intense research (Hamant and Haswell, 2017; Bacete and Hamann, 2020; Codjoe et al., 2021; Trinh et al., 2021). In this section, we describe tools to visualize and track how cell walls respond to such forces.
Molecular probes
Mechanical forces fundamentally influence plant morphogenesis (Trinh et al., 2021). Therefore, to understand how forces are integrated in plant tissues and organs, methods to measure tension changes in cells are necessary. Probes are attractive tools for visualization of specific features as they can be readily applied to the samples before or during experiments. Therefore, probes have great potential to test mechanical forces in vivo. A limitation of using probe methods is the need for an independent technique to calibrate the magnitude of forces observed; otherwise, the measurements remain purely qualitative.
Flipper-TR, an emergent tool to measure tension changes in different cellular membranes (plasma membrane, endoplasmic reticulum, lysosomes), has been developed recently (Colom et al., 2018; Assies et al., 2021). It is a probe that adapts the chemistry of push–pull fluorophores, molecules that can be planarized by lateral or axial forces, resulting in measurable changes in the lifetime (Fin et al., 2012; Doval et al., 2014; Dal Molin et al., 2015). Flipper-TR consist of two dithienothiophene flipper molecules, which are twisted with respect to each other due to repulsion forces around the connecting rotatable bond. Flipper-TR signal has been shown to be dependent on both lipid packing and cellular tensions, and was used to detect changes in fluorescent lifetime upon osmotic shocks (Colom et al., 2018). It was also used in a study that investigated the coupling between membrane tension and cell volume during changes in osmotic pressure (Roffay et al., 2021). We are not aware of any reports where Flipper-TR has been used to answer a biological question in plants. If the probe can be efficiently incorporated into the plasma membranes of plant cells, it could be a valuable tool to study the membrane tension in a time-dependent setting.
Microviscosity in different plant cell compartments, including the cell wall and plasma membrane, has been mapped using molecular rotors (Michels et al., 2020). Upon photoexcitation, chemical groups rotate with respect to the rest of the molecule, with the degree of rotation dependent on their interactions with the immediate environment. Rotation causes a detectable change in fluorescence lifetime and intensity of the probe. Molecular rotors were used to detect changes in free volume and lipid packing in the plasma membrane and changes in the network mesh size in the cell wall. Temporal tracking of localized microviscosity during changing environmental conditions is thus possible, as exemplified by experiments where suspension cells were treated with osmoticum at different concentrations. Hyperosmotic shocks (by the addition of mannitol) caused increases in the lifetime of plasma membrane-localized probes, indicating compression forces. In addition, differences in microviscosity were detected in vivo using the plasma membrane probe in different root cell types (epidermis root hair-growing and non-growing cells) and using the cell wall-targeted probe in leaf pavement cells in the wild type and mutants with defective cells walls (Michels et al., 2020).
Microfluidics
Live-imaging methods allow observation and quantifiation of the evolution of cellular processes over time. Depending on the duration and the conditions of the experiment, live imaging of certain tissues can be performed with very simple setups (Rahni and Birnbaum, 2019). However, to study dynamic responses to environmental stimuli, a tighly controlled system that allows the growing conditions to be changed, to apply specific external cues, and to quantify the induced responses is required. Microfluidics devices (Fig. 2) have been used in plant biology since more than a decade ago due to their capacity to perform live imaging on multiple samples under different conditions (Yanagisawa et al., 2021). Microfluidics devices have been particularly useful to study cell growth dynamics and the behaviour of fluorescent reporters. They have provided insights into how roots respond to sugar levels, hormones, and pathogenic interactions (Grossmann et al., 2011, 2012; Jones et al., 2014; Stanley et al., 2014; von Wangenheim et al., 2017; Fendrych et al., 2018; Liu et al., 2022). However, they have also been employed to study embryo, pollen tube, root hair, leaf, and whole seedling development (Gooh et al., 2015; Vang et al., 2018; Bertrand-Rakusová et al., 2020; Ueda et al., 2020).
Fig. 2.
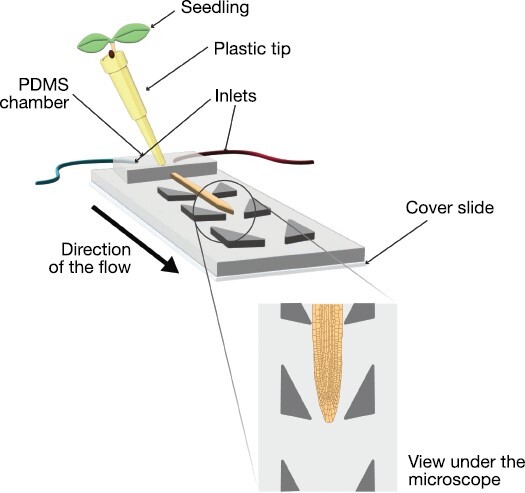
Microfluidics devices allow the study of dynamic responses. The schema represents a basic implementation for a microfluidics device with two inlets, which permits changing of the growing conditions and/or the delivery of specific treatments. The growth of the root tip can be observed under a microscope.
Microfluidics devices, offering a fresh perspective on cell wall dynamics, have been instrumental in recent investigations. Root hairs provide an excellent example of a well-established model system to study cellular polarization and coordination of tip cell growth (Ramalho et al., 2022). With microfluidics, Denninger et al. (2019) managed to follow root hair tip development over time, discerning a two-phase growth mechanism and the molecular domino effect that triggers root hair formation. Similarly, empirical data obtained from microfluidics, in combination with computational modelling, have been used to answer a long-standing question in biology: the mechanism that signals a cell to halt its growth (Liu et al., 2022). In this work, the authors pinpointed a location in the elongation zone where growth rates begin to slow down, dubbing it the start of growth cessation. To infer cell wall stiffness, cell shrinkage in hyperosmotic solutions was observed using a microfluidics device (drawing on methods by Sapala and Smith, 2020, and devices from Grossmann et al., 2011; Denninger et al., 2019). The study saw the recorded shrinkage rates as evidence of increasing cell wall stiffness along the root’s longitudinal axis, a perspective diverging from previous models attributing these changes to alterations in cell shape (Sapala et al., 2018).
Conclusions
Traditional techniques to study cell walls have uncovered useful information about cell wall composition and structure, but have fallen short in assessing their dynamic behaviour. The supramolecular network formed among cellulose, hemicellulose, and pectin in primary cell walls has received a lot of attention aiming to understand cell morphology, growth, and responses to environmental signals. In addition, lignin reinforcement in secondary cell walls of certain cell types after differentiation has been the focus of structural and biomass production studies. Nevertheless, it has proven difficult to resolve the organization of cell walls and their chemical and mechanical properties at the nanoscale without modifying the native chemical structure. Quantifying and understanding how fibrils and matrix polysaccharides are produced, interact chemically, and stretch or bend mechanically will enable us to connect the nanoscale behaviour to macroscopic properties, including stiffness, material properties, defence mechanisms, and features optimizing enzyme biomass digestibility.
An intriguing and still largely unexplored question in plant cell wall research is how composition and structure change with developmental stage progression. Changes occurring on the time scale of minutes to days have recently been investigated due to advances in live-imaging technologies, such as microfluidics, and non-invasive techniques, such as Brillouin microscopy and molecular probes, which allow observation and tracking of the evolution of single cell walls within individual organisms. A limitation has been that each technique can test one or few properties of the sample, thus requiring multiple measurements using different methods to fully characterize the walls. Technologies allowing real-time characterization of chemical and mechanical properties within the same measurement would be beneficial to study cell wall dynamics. Another caveat is that most of the current techniques only allow measurements to be made on sample surfaces (epidermis) or on whole tissues. Specific contributions of subepidermal tissues, whose composition is likely to differ from that of the epidermis, to the structural properties of cell walls remain uncharacterized. Enhancing the resolution and performance of non-destructive, contactless techniques will help to uncover the contributions of the different cell wall components to specific plant tissues/cell types. Computational approaches have provided insightful knowledge to predict mechanical properties and cellular morphologies, interpret unlabelled microscopic images, and incorporate results from different sources to guide new experimental steps (Box 2). Simulations could thus play a vital role in integrating knowledge about cell wall mechanics, composition, and structural changes over time, which could help us to understand the mechanisms responsible for cell wall plasticity.
We believe the methods described here form a common toolbox for cell wall studies that could be used to answer challenging questions in the near future including why cell wall composition and structure are so diverse among cell types and how mechanical forces are sensed and transduced in plant cells (Box 1). Furthermore, many of them allow in vivo measurements. This opens the door to move from static measurements to study cell walls by following changes occurring in the observed organism induced during growth or in response to environmental cues (during biotic and abiotic stresses).
Acknowledgements
We apologize to colleagues whose work has not been highlighted due to space constraints. We would like to thank Daniela Sueldo for her critical reading and helpful comments on the manuscript, and Anamika Chatterjee for checking the grammar of this manuscript—despite not speaking the ‘Cell wall speak’. We also thank Thorsten Hamann for his critical reading, guidance, and support during the writing of this manuscript, and for his advice on career development, which encouraged us to write this review.
Contributor Information
Luis Alonso Baez, Institute for Biology, Faculty of Natural Sciences, Norwegian University of Science and Technology, 5 Høgskoleringen, Trondheim, 7491, Norway.
Laura Bacete, Institute for Biology, Faculty of Natural Sciences, Norwegian University of Science and Technology, 5 Høgskoleringen, Trondheim, 7491, Norway; Umeå Plant Science Centre (UPSC), Department of Plant Physiology, Umeå University, 901 87 Umeå, Sweden.
Anja Geitmann, McGill University, Canada.
Author contributions
LAB and LB: writing the manuscript; LB: designing the figures with input from LAB.
Conflict of interest
The authors have reviewed the potential for conflicts of interest and declare that no financial, personal, or academic affiliations exist that could influence the objective discussion of techniques or the scientific discussions presented in this paper.
Funding
This work was supported by the Faculty of Natural Sciences of the Norwegian University of Science and Technology (grant no. 70443343 to LB) and the Research Council of Norway (NFR, grant no. 315325).
References
- Adichtchev SV, Karpegina YA, Okotrub KA, Surovtseva MA, Zykova VA, Surovtsev NV.. 2019. Brillouin spectroscopy of biorelevant fluids in relation to viscosity and solute concentration. Physical Review E 99, 062410. [DOI] [PubMed] [Google Scholar]
- Allison DP, Mortensen NP, Sullivan CJ, Doktycz MJ.. 2010. Atomic force microscopy of biological samples. WIREs Nanomedicine and Nanobiotechnology 2, 618–634. [DOI] [PubMed] [Google Scholar]
- Alonso-Simón A, Garcia-Angulo P, Mélida H, Encina A, Alvarez JM, Acebes JL.. 2011. The use of FTIR spectroscopy to monitor modifications in plant cell wall architecture caused by cellulose biosynthesis inhibitors. Plant Signaling and Behaviour 6, 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altartouri B, Bidhendi AJ, Tani T, Suzuki J, Conrad C, Chebli Y, Liu N, Karunakaran C, Scarcelli G, Geitmann A.. 2019. Pectin chemistry and cellulose crystallinity govern pavement cell morphogenesis in a multi-step mechanism. Plant Physiology 181, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsbury S, Hunt L, Elhaddad N, Baillie A, Lundgren M, Verhertbruggen Y, Scheller HV, Knox JP, Fleming AJ, Gray JE.. 2016. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Current Biology 26, 2899–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Carrol A, Akhmetova L, Somerville C.. 2010. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiology 152, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Kieber JJ.. 2020. Dynamic construction, perception, and remodeling of plant cell walls. Annual Reviews Plant Biology 71, 39–69. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS, Somerville CR.. 2012. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proceedings of the National Academy of Sciences, USA 109, 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci G, Beck T, Bilenca A, et al. 2020. Recent progress and current opinions in Brillouin microscopy for life science applications. Biophysical Reviews 12, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari M, Brulé V, Western TL, Pasini D.. 2020. Nano-indentation reveals a potential role for gradients of cell wall stiffness in directional movement of the resurrection plant Selaginella lepidophylla. Scientific Reports 10, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assies L, García-Calvo J, Piazzolla F, et al. 2021. Flipper probes for the community. Chimia 75, 1004–1011. [DOI] [PubMed] [Google Scholar]
- Atalla RH, Agarwal UP.. 1986. Recording Raman spectra from plant cell walls. Journal of Raman Spectroscopy 17, 229–231. [Google Scholar]
- Bacete L, Hamann T.. 2020. The role of mechanoperception in plant cell wall integrity maintenance. Plants 9, 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacete L, Mélida H, López G, Dabos P, Tremousaygue D, Denancé N, Miedes E, Bulone V, Goffner D, Molina A.. 2020. Arabidopsis response regulator 6 (ARR6) modulates plant cell-wall composition and disease resistance. Molecular Plant-Microbe Interactions 33, 767–780. [DOI] [PubMed] [Google Scholar]
- Bacete L, Mélida H, Miedes E, Molina A.. 2018. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. The Plant Journal 93, 614–636. [DOI] [PubMed] [Google Scholar]
- Bacete L, Schulz J, Engelsdorf T, et al. 2022. THESEUS1 modulates cell wall stiffness and abscisic acid production in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 119, e2119258119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauzamy L, Derr J, Boudaoud A.. 2015. Quantifying hydrostatic pressure in plant cells by using indentation with an atomic force microscope. Biophysics Journal 108, 2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand-Rakusová H, Chebli Y, Geitmann A.. 2020. Silicone chambers for pollen tube imaging in microstructured in vitro environments. Methods in Molecular Biology 2160, 211–221. [DOI] [PubMed] [Google Scholar]
- Beyond the diffraction limit. 2009. Nature Photonics 3, 361. [Google Scholar]
- Bhagia S, Ďurkovič J, Lagaňa R, Kardošová M, Kačík F, Cernescu A, Schäfer P, Yoo CG, Ragauskas AJ.. 2022. Nanoscale FTIR and mechanical mapping of plant cell walls for understanding biomass deconstruction. ACS Sustainable Chemistry & Engineering 10, 3016–3026. [Google Scholar]
- Bidhendi AJ, Chebli Y, Geitmann A.. 2020a. Fluorescence visualization of cellulose and pectin in the primary plant cell wall. Journal of Microscopy 278, 164–181. [DOI] [PubMed] [Google Scholar]
- Bidhendi AJ, Geitmann A.. 2016. Relating the mechanics of the primary plant cell wall to morphogenesis. Journal of Experimental Botany 67, 449–461. [DOI] [PubMed] [Google Scholar]
- Bidhendi AJ, Geitmann A.. 2018a. Tensile testing of primary plant cells and tissues. In: Geitmann A, Gril J, eds. Plant biomechanics: from structure to function at multiple scales. Cham: Springer International Publishing, 321–347. [Google Scholar]
- Bidhendi AJ, Geitmann A.. 2018b. Finite element modeling of shape changes in plant cells. Plant Physiology 176, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidhendi AJ, Geitmann A.. 2019. Methods to quantify primary plant cell wall mechanics. Journal of Experimental Botany 70, 3615–3648. [DOI] [PubMed] [Google Scholar]
- Bidhendi AJ, Li H, Geitmann A.. 2020b. Modeling the nonlinear elastic behavior of plant epidermis. Botany 98, 49–64. [Google Scholar]
- Braybrook SA. 2015. Measuring the elasticity of plant cells with atomic force microscopy. Methods in Cell Biology 125, 237–254. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Peaucelle A.. 2013. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: the relationship between auxin and pectin. PLoS One 8, e57813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL.. 2015. Biochemistry and molecular biology of plants, 2nd edn. Chichester: Wiley. [Google Scholar]
- Cabrera JC, Boland A, Cambier P, Frettinger P, Van Cutsem P.. 2010. Chitosan oligosaccharides modulate the supramolecular conformation and the biological activity of oligogalacturonides in Arabidopsis. Glycobiology 20, 775–786. [DOI] [PubMed] [Google Scholar]
- Caponi S, Canale C, Cavalleri O, Vassalli M.. 2019. Characterization tools for mechanical probing of biomimetic materials. In: Kumar C, ed. Nanotechnology characterization tools for tissue engineering and medical therapy. Berlin Heidelberg: Springer, 69–111. [Google Scholar]
- Caponi S, Fioretto D, Mattarelli M.. 2020. On the actual spatial resolution of Brillouin imaging. Optics Letters 45, 1063–1066. [DOI] [PubMed] [Google Scholar]
- Carroll S, Amsbury S, Durney CH, Smith RS, Morris RJ, Gray JE, Fleming AJ.. 2022. Altering arabinans increases Arabidopsis guard cell flexibility and stomatal opening. Current Biology 32, 3170–3179.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartagena-Rivera AX, Wang W-H, Geahlen RL, Raman A.. 2015. Fast, multi-frequency and quantitative nanomechanical mapping of live cells using the atomic force microscope. Scientific Reports 5, 11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambaud C, Cookson SJ, Ollat N, Bayer E, Brocard L.. 2022. A correlative light electron microscopy approach reveals plasmodesmata ultrastructure at the graft interface. Plant Physiology 188, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier AM, Normand AC, Passian A, Schaefer P, Lereu AL.. 2021. In situ plant materials hyperspectral imaging by multimodal scattering near-field optical microscopy. Communications Materials 2, 59. [Google Scholar]
- Chebli Y, Bidhendi AJ, Kapoor K, Geitmann A.. 2021. Cytoskeletal regulation of primary plant cell wall assembly. Current Biology 31, R681–R695. [DOI] [PubMed] [Google Scholar]
- Codjoe JM, Miller K, Haswell ES.. 2021. Plant cell mechanobiology: greater than the sum of its parts. The Plant Cell 34, 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E, Cosgrove DJ.. 2023. The mechanics of plant morphogenesis. Science 379, eade8055. [DOI] [PubMed] [Google Scholar]
- Colom A, Derivery E, Soleimanpour S, Tomba C, Molin MD, Sakai N, González-Gaitán M, Matile S, Roux A.. 2018. A fluorescent membrane tension probe. Nature Chemistry 10, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2016a. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. Journal of Experimental Botany 67, 463–476. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2016b. Catalysts of plant cell wall loosening. F1000Research 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2022. Building an extensible cell wall. Plant Physiology 189, 1246–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Molin M, Verolet Q, Colom A, Letrun R, Derivery E, Gonzalez-Gaitan M, Vauthey E, Roux A, Sakai N, Matile S.. 2015. Fluorescent flippers for mechanosensitive membrane probes. Journal of the American Chemical Society 137, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauphin BG, Ranocha P, Dunand C, Burlat V.. 2022. Cell-wall microdomain remodeling controls crucial developmental processes. Trends in Plant Science 27, 1033–1048. [DOI] [PubMed] [Google Scholar]
- Denninger P, Reichelt A, Schmidt VAF, Mehlhorn DG, Asseck LY, Stanley CE, Keinath NF, Evers J-F, Grefen C, Grossmann G.. 2019. Distinct RopGEFs successively drive polarization and outgrowth of root hairs. Current Biology 29, 1854–1865.e5. [DOI] [PubMed] [Google Scholar]
- Derbyshire P, Findlay K, McCann MC, Roberts K.. 2007. Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. Journal of Experimental Botany 58, 2079–2089. [DOI] [PubMed] [Google Scholar]
- Devaux M-F, Jamme F, André W, Bouchet B, Alvarado C, Durand S, Robert P, Saulnier L, Bonnin E, Guillon F.. 2018. Synchrotron time-lapse imaging of lignocellulosic biomass hydrolysis: tracking enzyme localization by protein autofluorescence and biochemical modification of cell walls by microfluidic infrared microspectroscopy. Frontiers in Plant Science 9, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVree BT, Steiner LM, Głazowska S, Ruhnow F, Herburger K, Persson S, Mravec J.. 2021. Current and future advances in fluorescence-based visualization of plant cell wall components and cell wall biosynthetic machineries. Biotechnology for Biofuels 14, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doval DA, Molin MD, Ward S, Fin A, Sakai N, Matile S.. 2014. Planarizable push–pull oligothiophenes: in search of the perfect twist. Chemical Science 5, 2819–2825. [Google Scholar]
- Du J, Anderson CT, Xiao C.. 2022. Dynamics of pectic homogalacturonan in cellular morphogenesis and adhesion, wall integrity sensing and plant development. Nature Plants 8, 332–340. [DOI] [PubMed] [Google Scholar]
- Elliott A, Shaw SL.. 2018. Update: plant cortical microtubule arrays. Plant Physiology 176, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayad K, Werner S, Gallemi M, Kong J, Sanchez Guajardo ER, Zhang L, Jaillais Y, Greb T, Belkhadir Y.. 2016. Mapping the subcellular mechanical properties of live cells in tissues with fluorescence emission-Brillouin imaging. Science Signaling 9, rs5. [DOI] [PubMed] [Google Scholar]
- Eltony AM, Shao P, Yun SH.. 2022. Measuring mechanical anisotropy of the cornea with Brillouin microscopy. Nature Communications 13, 1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercius P, Alaidi O, Rames MJ, Ren G.. 2015. Electron tomography: a three-dimensional analytic tool for hard and soft materials research. Advanced Materials 27, 5638–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felhofer M, Bock P, Singh A, Prats-Mateu B, Zirbs R, Gierlinger N.. 2020. Wood deformation leads to rearrangement of molecules at the nanoscale. Nano Letters 20, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J.. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nature Plants 4, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, et al. 2018. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Current Biology 28, 666–675.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fin A, Vargas Jentzsch A, Sakai N, Matile S.. 2012. Oligothiophene amphiphiles as planarizable and polarizable fluorescent membrane probes. Angewandte Chemie International Edition 51, 12736–12739. [DOI] [PubMed] [Google Scholar]
- Forterre Y. 2022. Basic soft matter for plants. In: Jensen KH, Forterre Y, eds. Soft matter in plants. London: The Royal Society of Chemistry, 1–65. [Google Scholar]
- García R, Herruzo ET.. 2012. The emergence of multifrequency force microscopy. Nature Nanotechnology 7, 217–226. [DOI] [PubMed] [Google Scholar]
- Geitmann A, Dyson R.. 2014. Modeling of the primary plant cell wall in the context of plant development. In: Assmann S, Liu B, eds. Cell biology. New York: Springer, 1–17. [Google Scholar]
- Ghassemi N, Poulhazan A, Deligey F, Mentink-Vigier F, Marcotte I, Wang T.. 2022. Solid-state NMR investigations of extracellular matrixes and cell walls of algae, bacteria, fungi, and plants. Chemical Reviews 122, 10036–10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson LJ. 2012. The hierarchical structure and mechanics of plant materials. Journal of the Royal Society Interface 9, 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierlinger N, Goswami L, Schmidt M, Burgert I, Coutand C, Rogge T, Schwanninger M.. 2008. In situ FT-IR microscopic study on enzymatic treatment of poplar wood cross-sections. Biomacromolecules 9, 2194–2201. [DOI] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, Engelsdorf T, Hamann T.. 2020. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cellular and Molecular Life Sciences 77, 2049–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, Hamann T.. 2018. Outside-in control—does plant cell wall integrity regulate cell cycle progression? Physiologia Plantarum 164, 82–94. [DOI] [PubMed] [Google Scholar]
- Gooh K, Ueda M, Aruga K, Park J, Arata H, Higashiyama T, Kurihara D.. 2015. Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Developmental Cell 34, 242–251. [DOI] [PubMed] [Google Scholar]
- Gorzsás A, Stenlund H, Persson P, Trygg J, Sundberg B.. 2011. Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem: the chemical landscape of secondary xylem. The Plant Journal 66, 903–914. [DOI] [PubMed] [Google Scholar]
- Gratton E, vande Ven MJ.. 2006. Laser sources for confocal microscopy. In: Pawley JB, ed. Handbook of biological confocal microscopy, 3rd edn. New York: Springer, 80–125. [Google Scholar]
- Gravino M, Savatin DV, Macone A, De Lorenzo G.. 2015. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. The Plant Journal 84, 1073–1086. [DOI] [PubMed] [Google Scholar]
- Green PB. 1964. Cell walls and the geometry of plant growth. Brookhaven Symposia in Biology 16, 203–217. [PubMed] [Google Scholar]
- Grossmann G, Guo W-J, Ehrhardt DW, Frommer WB, Sit RV, Quake SR, Meier M.. 2011. The RootChip: an integrated microfluidic chip for plant science. The Plant Cell 23, 4234–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Meier M, Cartwright HN, Sosso D, Quake SR, Ehrhardt DW, Frommer WB.. 2012. Time-lapse fluorescence imaging of Arabidopsis root growth with rapid manipulation of the root environment using the RootChip. Journal of Visualized Experiments (65), 4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Kraemer F, Pauly M.. 2011. Oligosaccharide mass profiling (OLIMP) of cell wall polysaccharides by MALDI-TOF/MS. Methods in Molecular Biology 715, 43–54. [DOI] [PubMed] [Google Scholar]
- Hamant, O, Haswell ES.. 2017. Life behind the wall: sensing mechanical cues in plants. BMC Biology 15, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom J, Berghuis N, Cramer D, Geurts R, Zuilhof H, Wennekes T.. 2016. Direct imaging of glycans in Arabidopsis roots via click labeling of metabolically incorporated azido-monosaccharides. BMC Plant Biology 16, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth F, Govyadinov A, Amarie S, Nuansing W, Keilmann F, Hillenbrand R.. 2012. Nano-FTIR absorption spectroscopy of molecular fingerprints at 20 nm spatial resolution. Nano Letters 12, 3973–3978. [DOI] [PubMed] [Google Scholar]
- Jacobs B, Schneider R, Molenaar J, Filion L, Deinum EE.. 2022. Microtubule nucleation complex behavior is critical for cortical array homogeneity and xylem wall patterning. Proceedings of the National Academy of Sciences, USA 119, e2203900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen OE. 2022. Theoretical tools and concepts for modelling growing plant tissues. In: Jensen KH, Forterre Y, eds. Soft matter in plants. London: The Royal Society of Chemistry, 85–118. [Google Scholar]
- Jones AM, Danielson JA, ManojKumar SN, Lanquar V, Grossmann G, Frommer WB.. 2014. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3, e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafle K, Park YB, Lee CM, Stapleton JJ, Kiemle SN, Cosgrove DJ, Kim SH.. 2017. Effects of mechanical stretching on average orientation of cellulose and pectin in onion epidermis cell wall: a polarized FT-IR study. Cellulose 24, 3145–3154. [Google Scholar]
- Kang X, Kirui A, Dickwella Widanage MC, Mentink-Vigier F, Cosgrove DJ, Wang T.. 2019. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nature Communications 10, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennaway R, Coen E.. 2019. Volumetric finite-element modelling of biological growth. Open Biology 9, 190057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshmiri H, Cikes D, Samalova M, et al. 2023. Imaging the microscopic viscoelastic anisotropy in living cells. BioRxiv 10.1101/2023.05.28.542585 [Preprint]. [DOI]
- Kirui A, Du J, Zhao W, Barnes W, Kang X, Anderson CT, Xiao C, Wang T.. 2021. A pectin methyltransferase modulates polysaccharide dynamics and interactions in Arabidopsis primary cell walls: evidence from solid-state NMR. Carbohydrate Polymers 270, 118370. [DOI] [PubMed] [Google Scholar]
- Kirui A, Ling Z, Kang X, Dickwella Widanage MC, Mentink-Vigier F, French AD, Wang T.. 2019. Atomic resolution of cotton cellulose structure enabled by dynamic nuclear polarization solid-state NMR. Cellulose 26, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J. 2008. Revealing the structural and functional diversity of plant cell walls. Current Opinion in Plant Biology 11, 308–313. [DOI] [PubMed] [Google Scholar]
- Knox J. 2012. In situ detection of cellulose with carbohydrate-binding modules. Methods in Enzymology 510, 233–245. [DOI] [PubMed] [Google Scholar]
- Koski KJ, Akhenblit P, McKiernan K, Yarger JL.. 2013. Non-invasive determination of the complete elastic moduli of spider silks. Nature Materials 12, 262–267. [DOI] [PubMed] [Google Scholar]
- Krieg M, Fläschner G, Alsteens D, Gaub BM, Roos WH, Wuite GJL, Gaub HE, Gerber C, Dufrêne YF, Müller DJ.. 2018. Atomic force microscopy-based mechanobiology. Nature Reviews. Physics 1, 41–57. [Google Scholar]
- Landau LD, Pitaevskii LP, Kosevich AM, Lifshitz EM.. 1986. Theory of elasticity. Volume 7 (Theoretical physics), 3rd edn. Oxford: Butterworth-Heinemann. [Google Scholar]
- Li S, Li Y, Yi R, Liu L, Qu J.. 2020. Coherent anti-Stokes Raman scattering microscopy and its applications. Frontiers in Physics 8, 598420. [Google Scholar]
- Lion C, Simon C, Huss B, et al. 2017. BLISS: a bioorthogonal dual-labeling strategy to unravel lignification dynamics in plants. Cell Chemical Biology 24, 326–338. [DOI] [PubMed] [Google Scholar]
- Liu S, Strauss S, Adibi M, et al. 2022. Cytokinin promotes growth cessation in the Arabidopsis root. Current Biology 32, 1974–1985.e3. [DOI] [PubMed] [Google Scholar]
- Majda M, Trozzi N, Mosca G, Smith RS.. 2022. How cell geometry and cellular patterning influence tissue stiffness. International Journal of Molecular Sciences 23, 5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgat R, François F, Boudaoud A.. 2016. A mechanical model to interpret cell-scale indentation experiments on plant tissues in terms of cell wall elasticity and turgor pressure. Frontiers in Plant Science 7, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu BP, Bock P, Gierlinger N.. 2020. Raman imaging of plant cell walls. Methods in Molecular Biology 2149, 251–295. [DOI] [PubMed] [Google Scholar]
- Mehandzhiyski AY, Zozoulenko I.. 2021. A review of cellulose coarse-grained models and their applications. Polysaccharides 2, 257–270. [Google Scholar]
- Meidner H, Mansfield TA.. 1968. Physiology of stomata. London: McGraw-Hill. [Google Scholar]
- Mélida H, Bacete L, Ruprecht C, Rebaque D, del Hierro I, López G, Brunner F, Pfrengle F, Molina A.. 2020. Arabinoxylan-oligosaccharides act as damage associated molecular patterns in plants regulating disease resistance. Frontiers in Plant Science 11, 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels L, Gorelova V, Harnvanichvech Y, Borst JW, Albada B, Weijers D, Sprakel J.. 2020. Complete microviscosity maps of living plant cells and tissues with a toolbox of targeting mechanoprobes. Proceedings of the National Academy of Sciences, USA 117, 18110–18118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani P, Braybrook SA, Boudaoud A.. 2013. Shrinking the hammer: micromechanical approaches to morphogenesis. Journal of Experimental Botany 64, 4651–4662. [DOI] [PubMed] [Google Scholar]
- Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, Kriwacki RW.. 2018. Methods for physical characterization of phase-separated bodies and membrane-less organelles. Journal of Molecular Biology 430, 4773–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A, Miedes E, Bacete L, et al. 2021. Arabidopsis cell wall composition determines disease resistance specificity and fitness. Proceedings of the National Academy of Sciences, USA 118, e2010243118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ. 2018. Mathematical modelling in plant biology. Cham: Springer. [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Hofte H.. 2003. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. The Plant Journal 35, 393–404. [DOI] [PubMed] [Google Scholar]
- Mravec J, Kračun SK, Rydahl MG, et al. 2014. Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141, 4841–4850. [DOI] [PubMed] [Google Scholar]
- Mravec J, Kračun SK, Rydahl MG, Westereng B, Pontiggia D, De Lorenzo G, Domozych DS, Willats WGT.. 2017. An oligogalacturonide‐derived molecular probe demonstrates the dynamics of calcium‐mediated pectin complexation in cell walls of tip‐growing structures. The Plant Journal 91, 534–546. [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Fäßler F, Dutka P, Schur FKM, Jensen G, Meyerowitz E.. 2022. Cryo-electron tomography of the onion cell wall shows bimodally oriented cellulose fibers and reticulated homogalacturonan networks. Current Biology 32, 2375–2389.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Pennington JG.. 2019. Electron tomography in plant cell biology. Microscopy 68, 69–79. [DOI] [PubMed] [Google Scholar]
- Padding JT, Briels WJ.. 2011. Systematic coarse-graining of the dynamics of entangled polymer melts: the road from chemistry to rheology. Journal of Physics: Condensed Matter 23, 233101. [DOI] [PubMed] [Google Scholar]
- Palombo F, Fioretto D.. 2019. Brillouin light scattering: applications in biomedical sciences. Chemical Reviews 119, 7833–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JL, Wang B, Diehl BG, Richard TL, Chen G, Anderson CT.. 2015. A versatile click-compatible monolignol probe to study lignin deposition in plant cell walls. PLoS One 10, e0121334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ.. 2012. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiology 158, 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, et al. 2010. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiology 153, 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H.. 2011. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Current Biology 21, 1720–1726. [DOI] [PubMed] [Google Scholar]
- Perras FA, Luo H, Zhang X, Mosier NS, Pruski M, Abu-Omar MM.. 2017. Atomic-level structure characterization of biomass pre- and post-lignin treatment by dynamic nuclear polarization-enhanced solid-state NMR. Journal of Physical Chemistry. A 121, 623–630. [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A.. 2012. Determining the polysaccharide composition of plant cell walls. Nature Protocols 7, 1590–1607. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB.. 2011. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual Review of Plant Biology 62, 567–590. [DOI] [PubMed] [Google Scholar]
- Prats-Mateu B, Gierlinger N.. 2017. Tip in–light on: advantages, challenges, and applications of combining AFM and Raman microscopy on biological samples. Microscopy Research and Technique 80, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentø P. 2009. Staining of macromolecules: possible mechanisms and examples. Biotechnic and Histochemistry 84, 139–158. [DOI] [PubMed] [Google Scholar]
- Prevedel R, Diz-Muñoz A, Ruocco G, Antonacci G.. 2019. Brillouin microscopy: an emerging tool for mechanobiology. Nature Methods 16, 969–977. [DOI] [PubMed] [Google Scholar]
- Rahni R, Birnbaum KD.. 2019. Week-long imaging of cell divisions in the Arabidopsis root meristem. Plant Methods 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakymzhan A, Yakupov T, Yelemessova Z, Bukasov R, Yakovlev VV, Utegulov ZN.. 2019. Time‐resolved assessment of drying plants by Brillouin and Raman spectroscopies. Journal of Raman Spectroscopy 50, 1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho JJ, Jones VAS, Mutte S, Weijers D.. 2022. Pole position: how plant cells polarize along the axes. The Plant Cell 34, 174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglet L, Gatti S, Moyroud E.. 2021. Sculpting the surface: structural patterning of plant epidermis. iScience 24, 103346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S, Huflejt M, Barbier de Reuille P, Braybrook SA, Schorderet M, Reinhardt D, Kuhlemeier C.. 2017. An automated confocal micro-extensometer enables in vivo quantification of mechanical properties with cellular resolution. The Plant Cell 29, 2959–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffay C, Molinard G, Kim K, et al. 2021. Passive coupling of membrane tension and cell volume during active response of cells to osmosis. Proceedings of the National Academy of Sciences, USA 118, e2103228118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropitaux M, Hays Q, Baron A, Fourmois L, Boulogne I, Vauzeilles B, Lerouge P, Mollet J, Lehner A.. 2022. Dynamic imaging of cell wall polysaccharides by metabolic click‐mediated labeling of pectins in living elongating cells. The Plant Journal 110, 916–924. [DOI] [PubMed] [Google Scholar]
- Routier-Kierzkowska AL, Weber A, Kochova P, Felekis D, Nelson BJ, Kuhlemeier C, Smith RS.. 2012. Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiology 158, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M, Gahurova E, Hejatko J.. 2022. Expansin-mediated developmental and adaptive responses: a matter of cell wall biomechanics? Quantitative Plant Biology 3, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapala A, Runions A, Routier-Kierzkowska A-L, et al. 2018. Why plants make puzzle cells, and how their shape emerges. eLife 7, e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapala A, Smith RS.. 2020. Osmotic treatment for quantifying cell wall elasticity in the sepal of Arabidopsis thaliana. Methods in Molecular Biology 2094, 101–112. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Kowalczyk M, Apte S, Yap EG, Das J, Adams PD, Bajaj C, Guindos P, Auer M.. 2018. Cryo-electron tomography 3D structure and nanoscale model of Arabidopsis thaliana cell wall. bioRxiv 10.1101/492140. [Preprint]. [DOI]
- Sawa M, Hsu T-L, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong C-H.. 2006. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proceedings of the National Academy of Sciences, USA 103, 12371–12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli G, Polacheck WJ, Nia HT, Patel K, Grodzinsky AJ, Kamm RD, Yun SH.. 2015. Noncontact three-dimensional mapping of intracellular hydromechanical properties by Brillouin microscopy. Nature Methods 12, 1132–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarponi F, Mattana S, Corezzi S, et al. 2017. High-performance versatile setup for simultaneous Brillouin–Raman microspectroscopy. Physical Review X 7, 031015. [Google Scholar]
- Seifert J, Kirchhelle C, Moore I, Contera S.. 2021. Mapping cellular nanoscale viscoelasticity and relaxation times relevant to growth of living Arabidopsis thaliana plants using multifrequency AFM. Acta Biomaterialia 121, 371–382. [DOI] [PubMed] [Google Scholar]
- Somssich M, Khan GA, Persson S.. 2016. Cell wall heterogeneity in root development of Arabidopsis. Frontiers in Plant Science 7, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Zhao S, Shen W, Collings C, Ding S-Y.. 2020. Direct measurement of plant cellulose microfibril and bundles in native cell walls. Frontiers in Plant Science 11, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CE, Stöckli M, van Swaay D, Sabotič J, Kallio PT, Künzler M, deMello AJ, Aebi M.. 2014. Probing bacterial–fungal interactions at the single cell level. Integrative Biology 6, 935–945. [DOI] [PubMed] [Google Scholar]
- Suslov D, Vissenberg K.. 2018. Cell wall expansion as viewed by the creep method. In: Geitmann A, Gril J, eds. Plant biomechanics: from structure to function at multiple scales. Cham: Springer International Publishing, 305–320. [Google Scholar]
- Taiz L, Møller IM, Murphy A, Zeiger E.. 2022. Plant Physiology and Development, 7th edn. Oxford: Oxford University Press. [Google Scholar]
- Thomas J, Idris NA, Collings DA.. 2017. Pontamine fast scarlet 4B bifluorescence and measurements of cellulose microfibril angles. Journal of Microscopy 268, 13–27. [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Wouwer DV de, Allen E, Kumpf R, Vanholme B, Boerjan W, Ralph J.. 2014. A click chemistry strategy for visualization of plant cell wall lignification. Chemical Communications 50, 12262–12265. [DOI] [PubMed] [Google Scholar]
- Traverso AJ, Thompson JV, Steelman ZA, Meng Z, Scully MO, Yakovlev VV.. 2015. Dual Raman–Brillouin microscope for chemical and mechanical characterization and imaging. Analytical Chemistry 87, 7519–7523. [DOI] [PubMed] [Google Scholar]
- Trinh D-C, Alonso-Serra J, Asaoka M, et al. 2021. How mechanical forces shape plant organs. Current Biology 31, R143–R159. [DOI] [PubMed] [Google Scholar]
- Ueda M, Kimata Y, Kurihara D.. 2020. Live-cell imaging of zygotic intracellular structures and early embryo pattern formation in Arabidopsis thaliana. Methods in Molecular Biology 2122, 37–47. [DOI] [PubMed] [Google Scholar]
- van den Dries K, Fransen J, Cambi A.. 2022. Fluorescence CLEM in biology: historic developments and current super-resolution applications. FEBS Letters 596, 2486–2496. [DOI] [PubMed] [Google Scholar]
- Vang S, Seitz K, Krysan PJ.. 2018. A simple microfluidic device for live cell imaging of Arabidopsis cotyledons, leaves, and seedlings. Biotechniques 64, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstreels E, Alamar MC, Verlinden BE, Enninghorst A, Loodts JKA, Tijskens E, Ramon H, Nicolaï BM.. 2005. Micromechanical behaviour of onion epidermal tissue. Postharvest Biology and Technology 37, 163–173. [Google Scholar]
- Voiniciuc C, Pauly M, Usadel B.. 2018. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiology 176, 2590–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Hauschild R, Fendrych M, Barone V, Benková E, Friml J.. 2017. Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife 6, e26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Hofte H.. 2016. Cell wall integrity signaling in plants: ‘To grow or not to grow that’s the question’. Glycobiology 26, 950–960. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Wen Z, et al. 2022. Xylan-based nanocompartments orchestrate plant vessel wall patterning. Nature Plants 8, 295–306. [DOI] [PubMed] [Google Scholar]
- Wang X, Wilson L, Cosgrove DJ.. 2020. Pectin methylesterase selectively softens the onion epidermal wall yet reduces acid-induced creep. Journal of Experimental Botany 71, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiao Y.. 2020. Cellulose microfibril-mediated directional plant cell expansion: gas and brake. Molecular Plant 13, 1670–1672. [DOI] [PubMed] [Google Scholar]
- Weber A, Braybrook S, Huflejt M, Mosca G, Routier-Kierzkowska A-L, Smith RS.. 2015. Measuring the mechanical properties of plant cells by combining micro-indentation with osmotic treatments. Journal of Experimental Botany 66, 3229–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhao Y, Suo Y, Guo Y, Man Y, Jing Y, He X, Lin J.. 2021. A label-free, fast and high-specificity technique for plant cell wall imaging and composition analysis. Plant Methods 17, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa N, Kozgunova E, Grossmann G, Geitmann A, Higashiyama T.. 2021. Microfluidics-based bioassays and imaging of plant cells. Plant and Cell Physiology 62, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bevilacqua C, Hambura S, et al. 2022. Pulsed stimulated Brillouin microscopy enables high-sensitivity mechanical imaging of live and fragile biological specimens. bioRxiv 10.1101/2022.11.10.515835. [Preprint]. [DOI] [PMC free article] [PubMed]
- Zancajo VMR, Lindtner T, Eisele M, Huber AJ, Elbaum R, Kneipp J.. 2020. FTIR nanospectroscopy shows molecular structures of plant biominerals and cell walls. Analytical Chemistry 92, 13694–13701. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gao Y, Zhang L, Zhou Y.. 2021. The plant cell wall: biosynthesis, construction, and functions. Journal of Integrative Plant Biology 63, 251–272. [DOI] [PubMed] [Google Scholar]
- Zhang J, Scarcelli G.. 2021. Mapping mechanical properties of biological materials via an add-on Brillouin module to confocal microscopes. Nature Protocols 16, 1251–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao C, Mentink-Vigier F, et al. 2019. Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. The Plant Cell 31, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Vavylonis D, Durachko DM, Cosgrove DJ.. 2017. Nanoscale movements of cellulose microfibrils in primary cell walls. Nature Plants 3, 17056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fang Z, Zhu X.. 2017. Near-field Raman spectroscopy with aperture tips. Chemical Reviews 117, 5095–5109. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu J, Wang X, Durachko DM, Zhang S, Cosgrove DJ.. 2021. Molecular insights into the complex mechanics of plant epidermal cell walls. Science 372, 706–711. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sheng S, Wang R, Sun M.. 2016. Tip-enhanced Raman spectroscopy. Analytical Chemistry 88, 9328–9346. [DOI] [PubMed] [Google Scholar]
- Zhao W, Fernando LD, Kirui A, Deligey F, Wang T.. 2020. Solid-state NMR of plant and fungal cell walls: a critical review. Solid State Nuclear Magnetic Resonance 107, 101660. [DOI] [PubMed] [Google Scholar]
- Zhao W, Kirui A, Deligey F, Mentink-Vigier F, Zhou Y, Zhang B, Wang T.. 2021. Solid-state NMR of unlabeled plant cell walls: high-resolution structural analysis without isotopic enrichment. Biotechnology for Biofuels 14, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Man Y, Wen J, Guo Y, Lin J.. 2019. Advances in imaging plant cell walls. Trends in Plant Science 24, 867–878. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ning G, Cosgrove DJ.. 2020. High-resolution imaging of cellulose organization in cell walls by field emission scanning electron microscopy. Methods in Molecular Biology 2149, 225–237. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang X, Chen Y, Wagner E, Cosgrove DJ.. 2018. Xyloglucan in the primary cell wall: assessment by FESEM, selective enzyme digestions and nanogold affinity tags. The Plant Journal 93, 211–226. [DOI] [PubMed] [Google Scholar]
- Zhong R, Cui D, Ye Z.. 2019. Secondary cell wall biosynthesis. New Phytologist 221, 1703–1723. [DOI] [PubMed] [Google Scholar]


