ABSTRACT
Bronchoalveolar lavage is usually employed for molecular diagnosis of Pneumocystis jirovecii but requires a specialized procedure. By contrast, nasopharyngeal (NP) specimens are easily obtained. In this retrospective study of 35 patients with paired NP and bronchoscopy specimens, NP specimens had a 100% negative percent agreement (95% CI 80.5–100) but only 72.2% positive percent agreement (95% CI 46.5–90.3).
KEYWORDS: Pneumocystis jirovecii pneumonia, PCR, nasopharyngeal specimens, bronchoalveolar lavage specimens
INTRODUCTION
The incidence of Pneumocystis jirovecii pneumonia (PJP) in non-HIV patients has been on the rise due to the increase of immunosuppressive therapies for the treatment of various conditions such as cancer, solid organ transplantation, hematopoietic cell transplantation, inflammatory bowel diseases, and rheumatologic and connective tissue disorders (1, 2). Infection with this pathogen can lead to significant morbidity and mortality not only due to the severity of the infection itself but also due to non-specific symptoms and radiographic presentations leading to delays in diagnosis (2, 3).
The updated EORTC/MSGERC criteria for establishing proven Pneumocystis jirovecii infection involve the detection of the organism via microscopy in tissue samples, bronchoalveolar lavage (BAL) fluid, or expectorated sputum using conventional or immunofluorescence staining (4). While polymerase chain reaction (PCR) testing of BAL or expectorated sputum is more sensitive than cytology (5, 6) and can aid in establishing probable infection (4), collecting these specimens can be challenging, particularly in non-intubated patients experiencing respiratory distress for whom the procedure could cause deterioration. Furthermore, a threshold for qPCR to differentiate between infection-causing disease and colonization is yet to be validated. Serologic markers like beta-D-glucan have limited operating characteristics, particularly specificity, in patients without HIV (7).
PCR testing for Pneumocystis jirovecii using nasopharyngeal (NP) specimens would be an appealing option due to its minimally invasive nature. However, its clinical utility has been limited to small studies of nasopharyngeal aspirates with varying testing platforms and populations (8 – 11). The Laboratoire de santé publique du Québec (LSPQ), a provincial public health and reference laboratory in Québec, Canada (serving 8.6 million citizens), has established a program for quantitative PCR (qPCR) testing for Pneumocystis (12) in respiratory samples. We sought to leverage laboratory data to report on the relative performance of NP qPCR to detect Pneumocystis jirovecii.
MATERIALS AND METHODS
We conducted a retrospective study from October 2019 to July 2023, which included all individuals who underwent Pneumocystis jirovecii testing with NP specimens at the LSPQ. NP specimens could include swabs or aspirates. Oropharyngeal, nasal, and unspecified swab specimens were excluded from the analysis. Data were extracted from the laboratory information system and included basic demographic data, qPCR results (positive, negative, or indeterminate), and qPCR quantitative results. The primary objective was to characterize the proportion of positive PCR results for Pneumocystis jirovecii NP specimens compared to BAL among paired specimens and to calculate the associated positive percentage agreement (PPA) and negative percentage agreement (NPA). We calculated PPA and NPA for paired NP and BAL specimens within 48 hours and 7 days to account for delay between collections, using BAL qPCR as the reference standard. Paired specimens were defined as clinical isolates sent within a defined period, and the analysis was limited to cases where NP specimens were collected prior to BAL sampling. As a secondary analysis, we evaluate the proportion of positive results for each specimen type and their assessed Pneumocystis burden (copies/mL) for both NP and BAL specimens.
Quantitative real-time PCR detection of Pneumocystis
Two hundred microliters of BAL or NP specimens were used to perform total nucleic acid extraction on the EMAG or NucliSens EasyMAG systems (bioMérieux, La Balme, France) and subsequently eluted in a final volume of 60 µL. When required, 200–300 µL of minimal essential medium was added to high viscosity samples and filtered through QIAshredder column by centrifugation 14,000 × g for 1 min (Qiagen, Hilden, Germany).
The Realstar Pneumocystis jirovecii PCR 1.0 Kit (Altona Diagnostics, Hamburg, Germany) was used for quantitative real-time PCR detection according to the manufacturer’s instructions on the QuantStudio 3 or 5 real-time PCR systems (Applied Biosystems, Whaltam, USA). Ten microliters of nucleic acid extract were used for qPCR amplification. Each run included four quantification controls with the kit (ranging from 1 × 101 to 1 × 104 copies/µL of reaction). These were used to generate a standard curve and calculate the organism concentration in copies per milliliter of clinical specimen in each sample. As 10 µL of 60 µL of the eluted DNA used in PCR reaction (extracted from 200 µL of input clinical specimen), a 300-fold adjustment was applied to the value obtained in copies per microliter of reaction after dilution factor adjustment to ensure that the final calculated concentration represented the organism concentration in copies per milliliter.
Analyses
Standard descriptive statistics were used for the analysis of the cohort. Continuous data were summarized as medians with interquartile ranges (IQRs). Categorical data were compared using chi-square test, while continuous data were compared using Mann-Whitney test. A P-value of <0.05 was considered statistically significant. SPSS version 25 (IBM, Chicago, Illinois) was used to perform the analyses.
RESULTS
A total of 3,428 specimens were sent to LSPQ for Pneumocystis jirovecii qPCR testing during the study period. Of these, 374 were NP, and 2,037 were BAL samples. The proportion of females was similar for both specimens (NP: 44.4% vs BAL: 45.7%), as was the median age (NP: 69 years, IQR 59–77 vs BAL: 65 years, IQR 54–73). The PCR assay infrequently yielded indeterminate results, occurring in 3 (0.8%) with NP specimens and 38 (1.9%) with BAL.
We identified 35 paired specimens within 48 hours and 58 paired specimens within a 7-day interval (Table 1). The PPA at 48 hours was 72.2% (95% CI, 46.5–90.3%), with an NPA of 100% (95% CI 80.5–100%) when comparing NP to BAL. At the 7 days interval, the PPA decreased to 60.0% (95% CI 38.7–78.9%) with an NPA of 100% (95% CI 89.4–100%).
TABLE 1.
Paired nasopharyngeal and bronchoalveolar lavage specimen analysis b
| BAL positive | BAL negative | ||
|---|---|---|---|
| Paired specimen analysis within 48 hours | |||
| NP positive | 13 | 0 | PPA = 72.2 (CI 46.5–90.3) |
| NP negative | 5 | 17 a | NPA = 100 (CI 80.5–100) |
One-paired specimen had an indeterminate BAL result with a negative nasopharyngeal result.
NP nasopharyngeal, PPA positive percent agreement, NPA negative percent agreement, BAL bronchoalveolar lavage.
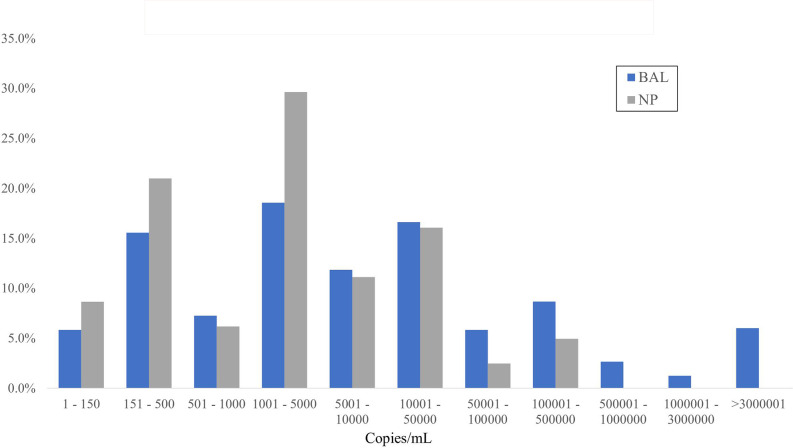
Overall, BAL specimens were more likely to be positive (29.1%; 593/2,037) as compared to NP specimens (21.7%; 81/374; P < 0.01). Of the 374 NP specimens, 283 were from swabs, and 91 were from aspirates. The proportion of positive results was similar in both cases (20.8%, for swabs vs 24.2%, for NP aspirate, P = 0.52). PCR of BAL specimens resulted in statistically higher qPCR results with a median of 6,159 copies/mL (IQR 722–51,909) compared to NP with a median of 1,662 copies/mL (IQR 362–9,150, P < 0.01). Fig. 1 illustrates the distribution of quantitative PCR results obtained from BAL and NP specimens.
Fig 1.
Distribution of positive Pneumocystis jirovecii quantitative PCR results (copies/mL) stratified by specimens.
DISCUSSION
Our analysis of paired specimens using qPCR for Pneumocystis jirovecii showed excellent NPA (100% at both 48 hours and 7 days) with more limited PPA (72.2% at 48 hours and 60.0% at 7 days) when comparing NP to BAL specimens. This was substantially lower PPA than reported in prior studies of NP samples (9, 10, 13). One retrospective study from Hong Kong showed excellent PPA (100%) when comparing NP aspirate PCR to BAL methenamine silver stain; however, microscopy itself has lower sensitivity compared to PCR technology (9, 14). A prospective South African study also showed high sensitivity (86.4%) when using NP aspirate compared to low respiratory tract samples, including induced sputum and BAL (13). However, this study focused primarily on HIV-infected children who are more likely to have higher fungal loads. Thus, our PPA may have been lower for a variety of reasons, including the comparator and the population tested. Our study highlights the potential of NP specimens for Pneumocystis jirovecii PCR testing, given the excellent NPA. In the right clinical context, a positive result may potentially avoid a BAL, particularly if the patients have contraindications for the invasive procedure. However, despite being an active area of research, the lack of a validated threshold for differentiating between disease and colonization makes interpretation challenging, particularly given the lower quantitative PCR results compared to BAL (15).
We also observed a lower positivity proportion with NP compared to BAL specimens (21.7% vs 29.1%), along with lower quantitative PCR results. The qPCR for BAL yielded 3.7-fold higher overall median results than NP. These findings align with the current understanding of the physiopathology of Pneumocystis jirovecii, as it mainly resides in the alveoli, suggesting that lower respiratory tract sampling should have better sensitivity (16, 17). However, it also likely reflects that patients who undergo bronchoscopy, by virtue of the barrier to testing, are clinically different than those who do not, and these differences are likely also related to overall yield.
Our study has several limitations. The clinical data obtained were limited to basic demographics, limiting our ability to assess the population which could benefit the most from this type of specimen. This absence of extensive clinical data limited our ability to evaluate the test’s clinical performance set by the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium consensus definition. Another area for improvement is the retrospective nature of this study, leading to selection bias. Patients who underwent PJP testing with NP specimens were more likely to be severely ill or have underlying contraindications for BAL. Additionally, in the paired specimen analysis, selection bias could have been introduced as patients with positive NP qPCR results may have been less likely to obtain a BAL. In contrast, patients with negative NP qPCR results but persistent symptoms were more likely to undergo a BAL for diagnostic purposes, and fungal burdens may have increased in the time interval. To address these limitations, a prospective paired study evaluating the test performance of NP qPCR or other alternative non-invasive samples, such as oropharyngeal wash, would represent a substantial improvement.
In summary, we provide valuable insight into the real-world microbiological performance of qPCR of NP specimens for PJP. While the PPA of NP qPCR may diminish over time or with ongoing therapy, the retained excellent NPA may preclude the need for invasive sampling in those which PJP already highly suspected. Nevertheless, the lower PPA and lower quantitative values, combined with the absence of a validated clinical threshold, argue for judicious use as part of an overall diagnostic strategy. Our results underscore the need for prospective studies to validate and further assess a non-invasive testing approach.
Contributor Information
Philippe J. Dufresne, Email: philippe.dufresne@inspq.qc.ca.
Kimberly E. Hanson, University of Utah, Salt Lake City, Utah, USA
REFERENCES
- 1. Wickramasekaran RN, Jewell MP, Sorvillo F, Kuo T. 2017. The changing trends and profile of pneumocystosis mortality in the United States, 1999-2014. Mycoses 60:607–615. doi: 10.1111/myc.12636 [DOI] [PubMed] [Google Scholar]
- 2. Liu CJ, Lee TF, Ruan SY, Yu CJ, Chien JY, Hsueh PR. 2019. Clinical characteristics, treatment outcomes, and prognostic factors of pneumocystis pneumonia in non-HIV-infected patients. Infect Drug Resist 12:1457–1467. doi: 10.2147/IDR.S199761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsu JM, Hass A, Gingras M-A, Chong J, Costiniuk C, Ezer N, Fraser RS, McDonald EG, Lee TC. 2020. Radiographic features in investigated for Pneumocystis jirovecii pneumonia: a nested case-control study. BMC Infect Dis 20:492. doi: 10.1186/s12879-020-05217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lagrou K, Chen S, Masur H, Viscoli C, Decker CF, Pagano L, Groll AH. 2021. Pneumocystis jirovecii disease: basis for the revised EORTC/MSGERC invasive fungal disease definitions in individuals without human immunodeficiency virus. Clin Infect Dis 72:S114–S120. doi: 10.1093/cid/ciaa1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Senécal J, Smyth E, Del Corpo O, Hsu JM, Amar-Zifkin A, Bergeron A, Cheng MP, Butler-Laporte G, McDonald EG, Lee TC. 2022. Non-invasive diagnosis of Pneumocystis jirovecii pneumonia: a systematic review and meta-analysis. Clin Microbiol Infect 28:23–30. doi: 10.1016/j.cmi.2021.08.017 [DOI] [PubMed] [Google Scholar]
- 6. Fan L-C, Lu H-W, Cheng K-B, Li H-P, Xu J-F. 2013. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of Pneumocystis jirovecii pneumonia: a bivariate meta-analysis and systematic review. PLoS One 8:e73099. doi: 10.1371/journal.pone.0073099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Del Corpo O, Butler-Laporte G, Sheppard DC, Cheng MP, McDonald EG, Lee TC. 2020. Diagnostic accuracy of serum (1-3)-β-D-glucan for Pneumocystis jirovecii pneumonia: a systematic review and meta-analysis. Clin Microbiol Infect 26:1137–1143. doi: 10.1016/j.cmi.2020.05.024 [DOI] [PubMed] [Google Scholar]
- 8. Desoubeaux G, Chesnay A, Mercier V, Bras-Cachinho J, Moshiri P, Eymieux S, De Kyvon M-A, Lemaignen A, Goudeau A, Bailly É. 2019. Combination of β-(1, 3)-D-glucan testing in serum and qPCR in nasopharyngeal aspirate for facilitated diagnosis of Pneumocystis jirovecii pneumonia. Mycoses 62:1015–1022. doi: 10.1111/myc.12997 [DOI] [PubMed] [Google Scholar]
- 9. To KKW, Wong SCY, Xu T, Poon RWS, Mok K-Y, Chan JFW, Cheng VCC, Chan K-H, Hung IFN, Yuen K-Y. 2013. Use of nasopharyngeal aspirate for diagnosis of pneumocystis pneumonia. J Clin Microbiol 51:1570–1574. doi: 10.1128/JCM.03264-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sivaraj V, Cliff P, Douthwaite S, Smith M, Kulasegaram R. 2021. Pneumocystis jirovecii pneumonia PCR test on upper respiratory tract swab. HIV Med 22:321–324. doi: 10.1111/hiv.13014 [DOI] [PubMed] [Google Scholar]
- 11. Guigue N, Alanio A, Menotti J, Castro ND, Hamane S, Peyrony O, LeGoff J, Bretagne S. 2015. Utility of adding Pneumocystis jirovecii DNA detection in nasopharyngeal aspirates in immunocompromised adult patients with febrile pneumonia. Med Mycol 53:241–247. doi: 10.1093/mmy/myu087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Population by age group, Canada and regions, July 1, 2022 (in French only). Available from: https://statistique.quebec.ca/en/produit/tableau/population-by-age-group-canada-and-regions. Accessed 27 March 2023
- 13. Samuel CM, Whitelaw A, Corcoran C, Morrow B, Hsiao N-Y, Zampoli M, Zar HJ. 2011. Improved detection of Pneumocystis jirovecii in upper and lower respiratory tract specimens from children with suspected pneumocystis pneumonia using real-time PCR: a prospective study. BMC Infect Dis 11:329. doi: 10.1186/1471-2334-11-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson JW, Limper AH, Grys TE, Karre T, Wengenack NL, Binnicker MJ. 2011. Pneumocystis jirovecii testing by real-time polymerase chain reaction and direct examination among immunocompetent and immunosuppressed patient groups and correlation to disease specificity. Diagn Microbiol Infect Dis 69:145–152. doi: 10.1016/j.diagmicrobio.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sarasombath PT, Thongpiya J, Chulanetra M, Wijit S, Chinabut P, Ongrotchanakun J, Jitmuang A, Wanachiwanawin D. 2021. Quantitative PCR to discriminate between pneumocystis pneumonia and colonization in HIV and non-HIV immunocompromised patients. Front Microbiol 12:729193. doi: 10.3389/fmicb.2021.729193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cushion MT. 2010. Are members of the fungal genus Pneumocystis (a) commensals; (B) opportunists; (C) pathogens; or (d) all of the above? PLoS Pathog 6:e1001009. doi: 10.1371/journal.ppat.1001009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maillet M, Maubon D, Brion JP, François P, Molina L, Stahl JP, Epaulard O, Bosseray A, Pavese P. 2014. Pneumocystis jirovecii (Pj) quantitative PCR to differentiate Pj pneumonia from Pj colonization in immunocompromised patients. Eur J Clin Microbiol Infect Dis 33:331–336. doi: 10.1007/s10096-013-1960-3 [DOI] [PMC free article] [PubMed] [Google Scholar]