Abstract
The potential for biodegradation of aromatic hydrocarbons was evaluated in soil samples recovered along gradients of both contaminant levels and pH values existing downstream of a long-term coal pile storage basin. pH values for areas greatly impacted by runoff from the storage basin were 2.0. Even at such a reduced pH, the indigenous microbial community was metabolically active, showing the ability to oxidize more than 40% of the parent hydrocarbons, naphthalene and toluene, to carbon dioxide and water. Treatment of the soil samples with cycloheximide inhibited mineralization of the aromatic substrates. DNA hybridization analysis indicated that whole-community nucleic acids recovered from these samples did not hybridize with genes, such as nahA, nahG, nahH, todC1C2, and tomA, that encode common enzymes from neutrophilic bacteria. Since these data suggested that the degradation of aromatic compounds may involve a microbial consortium instead of individual acidophilic bacteria, experiments using microorganisms isolated from these samples were initiated. While no defined mixed cultures were able to evolve 14CO2 from labeled substrates in these mineralization experiments, an undefined mixed culture including a fungus, a yeast, and several bacteria successfully metabolized approximately 27% of supplied naphthalene after 1 week. This study shows that biodegradation of aromatic hydrocarbons can occur in environments with extremely low pH values.
Polycyclic aromatic hydrocarbons (PAHs) occur as common constituents of petroleum, coal tar, and shale oil but are most frequently formed by incomplete combustion of fossil fuels (21). These contaminants represent a class of compounds that are widely distributed in nature (31) and are generally considered to have genotoxic or carcinogenic potential (21, 31). Several species of algae, bacteria, and fungi are known to degrade PAHs (31). Lower-molecular-weight PAH compounds, including naphthalene, phenanthrene, and anthracene, have been previously shown to be mineralized by bacteria (5). The capacity of microorganisms to degrade specific PAHs in nature depends on the physical and chemical properties of the contaminants, the environment, and the activity of indigenous organisms.
In spite of the wide range of environments suffering from PAH contamination, most investigations have focused on species of microorganisms that grow on common laboratory media at room temperature (mesophiles) and at neutral pH (neutrophiles). Many environments in which PAH contamination occurs, such as mine drainage basins, are acidic and occasionally have elevated temperatures. Significant numbers of acidophilic bacteria have been found in these environments (8, 10, 15, 20, 29, 32). Many of these isolates are autotrophic or mixotrophic acidophiles, although heterotrophic acidophiles belonging to the genus Acidiphilium have been isolated from coal mine drainage (10). These bacteria are mesophilic, gram-negative, aerobic rods that utilize citrate and other simple organic compounds as energy sources. Most members of this genus grow at pH 2 to 3, and none grow above pH 6.0.
This paper describes initial investigations into the biodegradation of aromatic compounds under acidophilic conditions. Surface water and soil samples were recovered from a coal pile storage area, the downstream drainage basin, and a nearby creek. These areas provided samples with different degrees of PAH concentrations and pH values.
MATERIALS AND METHODS
Site and sample description.
Soil and surface water samples were recovered during January 1997 from a coal runoff basin at the Westinghouse Savannah River Laboratory site in Aiken, S.C. Surface runoff from coal storage piles was discharged to surface streams until 1977, when new regulatory requirements were initiated. At that time, unlined earthen containment basins were constructed to intercept, stabilize, and treat surface runoff from the coal storage area. Water leaking from these basins has contaminated nearby soil and surface water with heavy metals and PAH compounds and has threatened local groundwater supplies. Microbiological analyses were performed on samples representing three distinct levels of PAH concentrations and pH values. The samples were taken from a source coal pile (identified as source), downstream from the pile in the catch basin (identified as downstream), and from an outfall creek located downstream of the catch basin but unaffected by drainage from the coal pile (identified as creek). Each sediment sample was collected and individually sealed in sterile plastic sample bags, placed on ice, and immediately shipped overnight to the Center for Environmental Biotechnology of The University of Tennessee—Knoxville. Corresponding water samples were collected in sterile 1-liter Nalgene bottles, placed on ice, and immediately shipped to the laboratory.
Upon receipt of the soil and water samples in the laboratory, microbial enumerations, enrichments with aromatic hydrocarbons, mineralization assays utilizing 14C-labeled compounds, and DNA extractions were immediately initiated. The pH of each soil and water sample was determined. Soil from the source area exhibited a pH of 6.1, with the pH of the corresponding water sample being 2.0. The higher pH of the source material verses source water is probably due to the buffering capacity of the large amount of organic material present. Soil from the downstream sample had a pH of 3.4, with the pH of the corresponding water sample being 2.0. Soil from the creek sample had a pH of 6.6, with the pH of the corresponding water sample being 5.4. Additional sediment and water samples were stored at 4°C.
Microbial enumerations.
Neutrophilic and acidophilic heterotrophic bacteria were enumerated by traditional spread plate analyses on solid agar media. Total viable counts for neutrophilic, heterotrophic microorganisms were determined in triplicate by adding 1.0 g of sediment to 9.0 ml of sterile 0.1% (wt/vol) sodium pyrophosphate dilution buffer (pH 7.0) (4) and then vortexing vigorously for 30 s on, 30 s off, and 30 s on. Appropriate dilutions from each series were plated on one-fourth strength YEPG (YEPG consists of [in grams per liter of dH20] yeast extract, 0.2; polypeptone, 2.0; d-glucose, 1.0; ammonium nitrate, 0.2; agar, 17.0 [pH 7.0]) and incubated at 25°C for 1 week before determinations of CFU (25). Acidophilic bacteria were enumerated by the method of Harrison (9). Serial dilutions were performed on triplicate samples, and appropriate dilutions from each dilution series were plated on acidophile medium, as described by Harrison (9). Acidophile plates were incubated for 2 weeks at 25°C before determinations of CFU.
Biodegradation analysis.
All radiochemicals were purchased from Sigma (St. Louis, Mo.). The specific activities (in millicuries per millimole) were as follows: toluene-ring-UL-14C, 60.0; naphthalene-UL-14C, 49.8; phenanthrene-9-14C, 13.3; anthracene-UL-14C, 15.0; salicylic acid-ring-UL-14C, 10.0. Mineralization assays, by the method of Sanseverino et al. (24), were performed to determine the biodegradation potentials of toluene, naphthalene, phenanthrene, and anthracene in each soil sample. Biodegradation was defined as evolution of 14CO2, as determined by liquid scintillation counting. Briefly, 2-g samples were slurried with 1 ml of the corresponding filter-sterilized (0.45-μm filter apparatus; Corning Costar Corporation, Cambridge, Mass.) water sample in 40-ml U.S. Environmental Protection Agency-certified vials (Eagle-Picher, Miami, Okla.). Sterility of the water samples used for mineralization experiments was determined by the lack of CFU on either one-fourth strength YEPG (pH 7.0) or acidophile plate medium (pH 3.0) after 1 week of incubation. Carbon dioxide traps were created by inserting an 8-ml glass vial containing 0.5 ml of a 0.5 M sodium hydroxide solution into the 40-ml mineralization vial. Approximately 100,000 dpm of the appropriate 14C-labeled substrate was added, and the vials were sealed with screw caps and teflon-lined septa. In experiments with cycloheximide treatment, a solution of 10 mg/ml of the appropriate filter-sterilized water sample was freshly prepared and 2 ml of the solution was added. Negative (killed) controls for mineralization assays were established by slurrying the soil samples in 2 ml of 37% formaldehyde (Fisher Chemicals, Fair Lawn, N.J.) and 2 ml of a filter-sterilized water sample and then boiling the vials for 15 min.
For mineralization experiments with pure cultures, isolates were grown for 48 h in 1 liter of liquid acidophile medium (9). Individual organisms were concentrated by centrifugation at 5,500 × g, the resulting cell pellet was washed twice in minimal salts medium (pH 3.0), and the final cell pellet was suspended in 50 ml of fresh minimal salts medium (resulting optical density of 1.8 to 2.0 at 543 nm). Each mineralization vial received 1 ml of culture and 4 ml of either acidophile medium, minimal salts medium, or a filter-sterilized water sample from the downstream area. Radiolabeled salicylic acid, toluene, naphthalene, and phenanthrene were used. Negative (killed) controls were established by the same protocol as described above for soil samples. In all mineralization experiments, active assays were halted by the addition of 1 ml of 37% formaldehyde (Fisher Chemicals).
Molecular diagnostics.
Duplicate 50-g samples were used for DNA extraction studies by a modification of the method of Ogram et al. (14, 19, 28). Cell lysis was initiated by heat and sodium dodecyl sulfate treatment and completed by using bead-mill homogenization. After concentration of nucleic acids from the aqueous phase by isopropanol precipitation, the samples were dialyzed against TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) overnight. The samples were then extracted once with Tris-saturated phenol (pH 7.5), followed by extraction once with chloroform-isoamyl alcohol (24:1). The final aqueous phase was recovered and ethanol precipitated overnight at −20°C. DNA was collected by centrifugation and dried under vacuum. The final pellet was suspended in 1 ml of sterile TE buffer (pH 7.5) and stored at −20°C until being used for hybridization studies.
DNA recovered from each soil sample was vacuum blotted onto 0.22-μm Biotrans nylon membranes (ICN Biomedical, Inc., Costa Mesa, Calif.) and fixed by baking at 80°C for 1 h and then rinsing in 2× SSC buffer (SSC consists of [in grams per liter of dH2O] sodium chloride, 17.53; sodium citrate, 8.82 [pH 7.0]), followed by baking at 80°C for another hour. Membranes were prehybridized for 12 to 18 h and then hybridized for an additional 12 to 18 h with the appropriate probe, as previously described (28). The membranes were washed three times with high-stringency wash buffer (in grams per liter, sodium chloride, 0.59; Tris base, 2.42; EDTA, 0.37; sodium dodecyl sulfate, 5.00 [pH 7.5]), and positive hybridization signals were determined by autoradiography. DNA was recovered from the isolated organisms by the method of Marmur (18).
PCR, cloning, and sequencing of 16S rDNA from isolates.
PCR amplification of 16S ribosomal DNA (rDNA) from soil isolates was performed with primers 27f and 1492r (Escherichia coli numbering system), as previously described (16, 26). The 1.5-kb PCR products were cloned with a TA cloning kit (Invitrogen, San Diego, Calif.) according to the manufacturer’s instructions. Bacterial colonies were screened for plasmids containing the correct insert by the rapid-boiling plasmid minipreparation technique of Holmes and Quigley (12), followed by restriction digestion with EcoRI. Plasmid DNA containing the correct insert for DNA sequence analysis was prepared (22), and the amount of plasmid DNA recovered was determined with a DyNA Quant 2 fluorometer (Hoefer, San Francisco, Calif.). DNA sequencing was conducted with an automated DNA sequencer (Applied Biosystems Division of Perkin-Elmer, Foster City, Calif.) by using the sequencing primer 27f (16). Tentative phylogenetic identification of microorganisms by DNA sequence analysis of cloned small-subunit rRNA was performed with the BLAST program (1) (National Center for Biotechnology Information).
Nucleotide sequence accession numbers.
The DNA sequences of strains SRS 1 and SRS 2 have been deposited in the GenBank database under accession no. AF082659 and AF082660, respectively.
RESULTS
Enumeration of microorganisms from acidic soil samples was performed by traditional agar spread plate analysis. Neutrophilic heterotrophs were present at values of 1.1 × 104 (± 4.1 × 103) CFU/g of source soil, 1.0 × 103 (± 4.7 × 102) CFU/g of downstream soil, and 2.2 × 106 (± 8.2 × 105) CFU/g of creek soil. Estimates of acidophilic bacteria ranged from 102 to 103 CFU/g of each sample, but statistical analysis was confounded because the plates yielded few colonies and were contaminated with fungi.
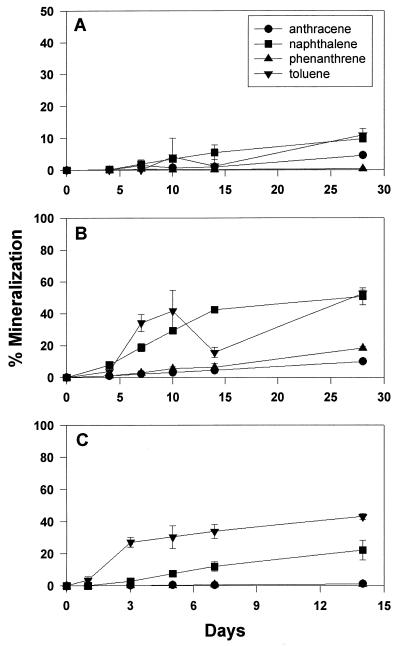
Results from 14C-labeled substrate mineralization experiments indicated that natural samples of acidic pH maintained the capacity for significant biodegradation of aromatic hydrocarbons (Fig. 1). Each of the three soil samples tested possessed the ability to mineralize one or more of the aromatic hydrocarbons tested. Figure 1A shows mineralization of aromatic hydrocarbons by the source soil sample, where carbon dioxide evolution of toluene and naphthalene reached approximately 10% at 28 days. The downstream soil sample showed mineralization of each compound tested (Fig. 1B). Toluene and naphthalene mineralization approached 50% degradation, while phenanthrene and anthracene showed 10 to 20% mineralization. Soil samples from the creek area showed significant toluene mineralization (40%) at 14 days (Fig. 1C). Naphthalene mineralization reached 20% mineralization at 2 weeks, while little phenanthrene and anthracene were degraded.
FIG. 1.
Mineralization of aromatic hydrocarbons in acidic soil samples, showing biodegradation in the source coal pile sample (A), the downstream drainage basin sample (B), and the control creek sample (C). Each soil sample was slurried with its corresponding water sample, which had been filter sterilized.
Microbial enumeration studies revealed representatives of both fungi and yeasts in the soil microbial communities in each sample. A mineralization experiment was designed to test the impact of these microeukaryotes on biodegradation of the test aromatic hydrocarbons. Cycloheximide was used to repress microeukaryotic metabolism during a 14-day mineralization experiment. Cycloheximide treatment of the downstream samples eliminated aromatic hydrocarbon metabolism but did not eliminate mineralization in the creek samples (Table 1). This indicates that the mineralization of aromatic hydrocarbons in the highly acidic downstream sample involves eukaryotic organisms, probably yeasts and/or fungi.
TABLE 1.
Effects of cycloheximide (10 mg/ml) on mineralization of naphthalene and toluene after 14 days
| Sample | % Mineralization (mean ± SD)
|
|
|---|---|---|
| Naphthalene | Toluene | |
| Downstream | 31.74 ± 6.47 | 5.19 ± 1.62 |
| Downstream with cycloheximide | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Creek | 74.77 ± 1.16 | 45.54 ± 15.25 |
| Creek with cycloheximide | 56.57 ± 4.35 | 30.58 ± 19.14 |
Whole-community DNA was extracted from each soil sample and hybridized with molecular probes targeting genes commonly associated with aromatic hydrocarbon degradation under neutrophilic conditions (data not shown). Hybridization analyses with the gene probes nahA (naphthalene dioxygenase), nahH (catechol-2,3-dioxygenase), nahG (salicylate hydroxylase), todC1C2 (toluene dioxygenase), and tomA (toluene ortho-monooxygenase) did not produce positive signals above the calculated detection limit of 0.003 ng of target sequence. The extracted DNA did hybridize positively with a universal 16S rDNA oligonucleotide targeting all forms of life (27, 28), indicating that DNA of sufficient quality for analysis was successfully recovered from the samples. The strongest hybridization signal with the universal 16S rDNA oligonucleotide was seen with DNA recovered from the creek sample, probably due to the presence of a greater biomass. This corresponds with the data from the microbial enumeration analysis, where the creek sample supported 2 orders of magnitude more total heterotrophic organisms than the soils impacted by reduced pH.
In order to explore the possibility of isolating a pure culture of an obligately acidophilic microorganism capable of growth on PAHs, enrichments were prepared with 10 g of a downstream soil sample and 40 ml of a corresponding filter-sterilized water sample (pH 2.0) saturated with naphthalene. These microcosm enrichments were allowed to shake for 2 weeks at 25°C at 100 rpm, and then 10 μl of the aqueous phase was spread plated on acidophile medium agar plates (9). The acidophile plates were incubated at 25°C for 1 week before individual colonies were restreaked. After two consecutive restreaks, the cultures appeared to be of a single colony type. Each isolate was restreaked twice more to ensure that the culture was axenic. At this point, three distinct isolates were identified based on colony morphologies and growth characteristics. Two isolates, designated SRS 1 and SRS 2, were microscopically identified by size as bacteria, and the third isolate, designated SRS 3, was identified as a yeast. Furthermore, the two bacterial isolates were defined as obligate acidophiles by their ability to grow on acidophile medium plates (pH 3.0) but not on one-fourth strength YEPG medium plates (pH 7.0). The yeast isolate was able to grow on both types of medium. A fourth bacterial isolate, designated SRS 4, was obtained as a contaminant in a fungal culture from the downstream sample.
Partial sequence analysis of small-subunit rDNA provided tentative identification of the bacterial isolates. SRS 1 (1,000 bases sequenced) and SRS 2 (240 bases sequenced) showed 97 and 90% sequence similarity, respectively, with Acidocella sp. (11) and Acidiphilium facilis (14). The heterotrophic yeast was microscopically designated Pichia sp. A partial sequence was also obtained for isolate SRS 4, but it showed little sequence similarity with any organism currently in the database.
Growth assays with simple carbon substrates and mineralization studies were performed on isolates SRS 1, SRS 2, and SRS 3. Growth assays were scored as positive if they showed signs of turbidity (Table 2). The isolate designated SRS 1 grew to turbidity in minimal salts medium (pH 3.0) plus each of the substrates tested except lactose, arginine, and phenylalanine. The isolate designated SRS 2 showed turbidity on only citrate, catechol, and yeast extract. The isolate designated SRS 3 grew on every carbon source except lactose. Biodegradation of toluene, naphthalene, or phenanthrene was at background levels, with no more than 3% mineralization by any of the isolates under any of the conditions tested (Table 3). However, salicylic acid was mineralized to various degrees by all three isolates. SRS 1 showed a marked increase in mineralization of salicylic acid when incubated with filter-sterilized water from the downstream area. SRS 2 also showed an increase in salicylic acid mineralization when incubated with the natural water sample. SRS 3 mineralized salicylic acid under all conditions tested, with the largest amount of 14C-labeled carbon dioxide occurring in minimal salts medium (pH 3.0).
TABLE 2.
Carbon source utilization spectrum for soil isolates
| Carbon source | Isolate
|
||
|---|---|---|---|
| SRS 1 | SRS 2 | SRS 3 | |
| Maltose | + | − | + |
| Galactose | + | − | + |
| Glucose | + | − | + |
| Lactose | − | − | − |
| Citrate | + | + | + |
| Acetate | + | − | + |
| Succinate | + | − | + |
| Salicylate | + | + | + |
| Catechol | + | + | + |
| Yeast extract | + | + | + |
| Arginine | − | − | + |
| Phenylalanine | − | − | + |
| Acidophile medium | + | + | + |
| 1/4 YEPG | − | − | + |
TABLE 3.
Mineralization of aromatic hydrocarbons by soil isolates
| Isolate | Compound | % Mineralization in:
|
||
|---|---|---|---|---|
| Acidophile medium | Minimal salts medium | Natural water sample | ||
| SRS 1 | Salicylic acid | 2.29 | 5.12 | 51.02 |
| Toluene | 0.00 | 0.01 | 0.50 | |
| Naphthalene | 0.01 | 1.76 | 1.73 | |
| Phenanthrene | 1.64 | 0.51 | 0.83 | |
| SRS 2 | Salicylic acid | 4.67 | 13.0 | 35.10 |
| Toluene | 0.03 | 1.30 | 0.73 | |
| Naphthalene | 0.02 | 1.45 | 2.36 | |
| Phenanthrene | 2.27 | 0.95 | 0.68 | |
| SRS 3 | Salicylic acid | 26.40 | 67.30 | 36.87 |
| Toluene | 0.00 | 0.88 | 0.00 | |
| Naphthalene | 1.40 | 1.50 | 0.88 | |
| Phenanthrene | 1.50 | 1.20 | 0.98 | |
The ability of model mixed microbial cultures to biodegrade aromatic hydrocarbons at pH 2.0 was evaluated with the previously mentioned isolates, along with five fungal isolates, slurried in filter-sterilized water from the downstream drainage basin area (Table 4). Microorganisms isolated from the downstream sample were used for the mixed-community mineralizations. The organisms included two obligate acidophilic bacteria (SRS 1 and SRS 2), a heterotrophic yeast (SRS 3), and a fungus (fungus 3). Initial work with SRS 1, SRS 2, SRS 3, and fungus 3 revealed the ability of this mixed culture to mineralize naphthalene. However, microscopic analysis of the fungus 3 enrichment culture revealed that it was contaminated with bacteria. The neutrophilic bacterium designated SRS 4 was recovered from the contaminated fungal culture but proved unimportant in the mineralization of naphthalene either individually or in mixed culture. SRS 4 grew rapidly on one-fourth YEPG at pH 7.0 but very slowly on acidophile medium at pH 3.0. No other organisms were isolated from the contaminated fungal culture. Only the microbial consortium including the contaminated fungus 3 and unidentified, uncultured organisms were able to mineralize naphthalene to carbon dioxide. Fungus 3 was isolated in pure culture by successive transfers in liquid acidophile medium containing the antibiotics streptomycin, kanamycin, and rifampin. The fungal culture was determined to be axenic by microscopic analysis. When SRS 1, SRS 2, SRS 3, SRS 4, and uncontaminated fungus 3 were mixed together, naphthalene mineralization did not occur. This supports the hypothesis that other uncultured organisms capable of mineralizing naphthalene exist in the undefined microbial consortium. While the model microbial mixed cultures constructed with pure cultures were unsuccessful in the mineralization of naphthalene, salicylic acid was mineralized to a greater extent (Table 4). Mineralization approached 100% after 1 week in mixed cultures but remained under 52% by any individual isolate.
TABLE 4.
Mineralization of aromatic hydrocarbons by soil microorganisms
| Microorganism(s)a | % Mineralization
|
|
|---|---|---|
| Salicylate | Naphthalene | |
| SRS 1 | 51.02 ± 25.99 | 1.73 ± 1.22 |
| SRS 2 | 35.10 ± 6.84 | 2.36 ± 0.90 |
| SRS 3 | 36.87 ± 4.82 | 0.88 ± 0.33 |
| SRS 4 | 41.31 ± 6.02 | 0.00 ± 0.00 |
| Fungus 3 | 38.67 ± 0.37 | 0.00 ± 0.00 |
| SRS 1, SRS 2 | 82.60 ± 4.49 | 0.00 ± 0.00 |
| SRS 1, SRS 3 | 79.78 ± 1.48 | 0.84 ± 0.02 |
| SRS 2, SRS 3 | 64.89 ± 3.40 | 0.19 ± 0.10 |
| SRS 1, SRS 2, SRS 3 | 97.55 ± 3.87 | 0.43 ± 0.01 |
| SRS 1, SRS 2, SRS 3, fungus 1 | 93.88 ± 11.33 | 1.45 ± 0.66 |
| SRS 1, SRS 2, SRS 3, fungus 2 | 90.04 ± 0.89 | 0.35 ± 0.07 |
| SRS 1, SRS 2, SRS 3, fungus 3 | 32.08 ± 1.72 | 0.00 ± 0.00 |
| SRS 1, SRS 2, SRS 3, fungus 4 | 73.57 ± 4.21 | 0.47 ± 0.52 |
| SRS 1, SRS 2, SRS 3, fungus 5 | 86.59 ± 6.75 | 0.45 ± 0.44 |
| Undefined mixed cultureb | 93.34 ± 4.44 | 27.13 ± 15.73 |
a Microorganisms were suspended in a filter-sterilized water sample from the downstream drainage basin area (pH 2.0).
b The undefined mixed culture consisted of SRS 1, SRS 2, SRS 3, fungus 3, and unidentified bacterial contaminants of the original fungus 3 culture.
DISCUSSION
The data presented show conclusively that biodegradation of aromatic hydrocarbons can occur in extremely acidic environments. While involvement of individual heterotrophic, prokaryotic acidophiles has not been eliminated, biodegradation in the downstream area of this particular drainage basin likely requires microeukaryotic organisms, such as filamentous fungi and yeasts. Since 14CO2 evolution from fungi has been shown to be rare (6), complete mineralization of the aromatic contaminants may involve a complex interaction between several distinct groups of microorganisms.
It is well understood that the biotransformation of PAH compounds by fungi primarily exists as a detoxification mechanism, with the secondary metabolites formed having lower toxicity than the parent compound (30). Moreover, In der Wiesch et al. (13) showed that the addition of soil microorganisms to a fungal culture increased mineralization of the PAH pyrene. This study suggested that initial fungal, extracellular enzymatic attacks on the PAHs produced intermediates that were available for further degradation by soil microorganisms. We hypothesize that a similar situation may explain the aromatic hydrocarbon mineralization activity detected under extremely acidic conditions. This hypothesis is supported by the demonstration that cycloheximide treatment eliminates mineralization of both naphthalene and toluene in the sediment sample recovered from the downstream drainage basin area. The organisms in the soil samples able to degrade these compounds did not hybridize to gene probes targeting enzymes known to be involved in the biochemical degradation of aromatic hydrocarbons. This finding further supports the involvement of microeukaryotic organisms in the degradation of aromatic hydrocarbons in this environment. Attempts to isolate pure cultures of aromatic-hydrocarbon-degrading acidophilic microorganisms are ongoing. However, 16S rRNA sequence analyses suggest the presence of acidophilic bacteria in these samples.
Metabolism of salicylic acid was shown to be greatly affected by addition of the water sample from the downstream area. Increased mineralization of salicylic acid by a consortium of organisms, compared with individual organisms, was seen. These data suggest that an unknown cofactor or nutrient may enhance the levels of salicylic acid metabolism by acidophilic bacteria.
Additional support for the biodegradation of aromatic contaminants by acidophilic bacteria comes from the work of Quentmeier and Friedrich (23). Their experiments showed that plasmids encoding either phenol degradation or antibiotic resistance from neutrophilic bacteria could be acquired by the acidophilic bacterium Acidiphilium cryptum by conjugation. Once the genes were successfully transferred into the acidophile, the encoded proteins were shown to be functional. This directly supports the hypothesis that genes encoding enzymes involved in degrading aromatic compounds can be acquired and expressed in heterotrophic acidophiles.
To date, the great majority of studies focusing on aromatic hydrocarbon biodegradation have been performed with organisms isolated from nonextreme environments. Considering that aromatic hydrocarbon contamination has been documented within extreme environments, clearly research is needed to ascertain the ability of these environments to biodegrade such compounds. This study lends credence to the hypothesis that microbially based degradation mechanisms are at work in extreme acidic ecosystems. Furthermore, this study suggests that instead of a single organism being responsible for complete mineralization of aromatic contaminants to carbon dioxide and water, biodegradation in these environments may be the result of complex interactions within the microbial community. Such consortium-based systems have recently been reported for compounds generally considered to be recalcitrant in nature (2, 3, 7, 17).
ACKNOWLEDGMENTS
Funding for this project was provided by The Center for Environmental Biotechnology and The Waste Management Research and Education Institute of The University of Tennessee—Knoxville.
We thank T. Hazen and M. Franck of the Westinghouse Savannah River Laboratory for providing the soil and water samples used in this study. DNA sequencing was performed at the Molecular Biology Resource Facility of the University of Tennessee—Knoxville.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Alvey S, Crowley D E. Survival and activity of an atrazine-mineralizing bacterial consortium in rhizosphere soil. Environ Sci Technol. 1996;30:1595–1603. [Google Scholar]
- 3.Assaf N A, Turco R F. Accelerated biodegradation of atrazine by a microbial consortium is possible in culture and soil. Biodegradation. 1994;5:29–35. doi: 10.1007/BF00695211. [DOI] [PubMed] [Google Scholar]
- 4.Balkwill D L, Rucinsky T E, Casida L E. Release of microorganisms from soil with respect to transmission electron microscopy viewing and plate counts. Antonie Leeuwenhoek. 1977;43:73–87. doi: 10.1007/BF02316212. [DOI] [PubMed] [Google Scholar]
- 5.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 6.Cerniglia C E, Sutherland J B, Crow S A. Fungal metabolism of aromatic hydrocarbons. In: Winkelmann G, editor. Microbial degradation of natural products. Weinheim, Germany: VCH Press; 1992. pp. 193–217. [Google Scholar]
- 7.De Souza M L, Newcombe D, Alvey S, Crowley D E, Hay A, Sadowski M J, Wackett L P. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerson D, Moyer C. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol. 1997;63:4784–4792. doi: 10.1128/aem.63.12.4784-4792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison A P., Jr Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int J Syst Bacteriol. 1981;31:327–332. [Google Scholar]
- 10.Harrison A P. The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu Rev Microbiol. 1984;38:265–292. doi: 10.1146/annurev.mi.38.100184.001405. [DOI] [PubMed] [Google Scholar]
- 11.Hiraishi A. Phylogeny of acidophilic chemoorganotrophic bacteria. 1996. GenBank accession no. D86510. [Google Scholar]
- 12.Holmes D S, Quigley C M. A rapid boiling method for preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 13.In der Wiesch C, Martens R, Zadrazil F. Two-step degradation of pyrene by white-rot fungi and soil microorganisms. Appl Microbiol Biotechnol. 1996;46:653–659. doi: 10.1007/s002530050876. [DOI] [PubMed] [Google Scholar]
- 14.Johnston W H, Stapleton R D, Sayler G S. Direct extraction of microbial DNA from soils and sediments. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial manual. Dordrecht, The Netherlands: Kluwer; 1996. pp. 1.3.2:1–9. [Google Scholar]
- 15.Kishimoto N, Kosako Y, Wakao N, Tano T, Hiraishi A. Transfer of Acidiphilium facilis and Acidiphilium aminolytica to the genus Acidocella gen. nov., and emendation of the genus Acidiphilium. Syst Appl Microbiol. 1995;18:85–91. [Google Scholar]
- 16.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley and Sons; 1991. pp. 115–148. [Google Scholar]
- 17.Mandelbaum R T, Wackett L P, Allan D L. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl Environ Microbiol. 1993;59:1695–1701. doi: 10.1128/aem.59.6.1695-1701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 19.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 20.Pizarro J, Jedlicki E, Orellano O, Espejo R T. Bacterial populations in samples of bioleached copper ore as revealed by analysis of DNA obtained before and after cultivation. Appl Environ Microbiol. 1996;62:1323–1328. doi: 10.1128/aem.62.4.1323-1328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pothuluri J V, Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons. In: Chaundry G R, editor. Biological degradation and bioremediation of toxic chemicals. Portland, Oreg: Dioscorides Press; 1994. pp. 92–124. [Google Scholar]
- 22.Promega Corporation. Promega technical bulletin 009. Madison, Wis: Promega Corporation; 1992. [Google Scholar]
- 23.Quentmeier A, Friedrich C G. Transfer and expression of degradative and antibiotic resistance plasmids in acidophilic bacteria. Appl Environ Microbiol. 1994;60:973–978. doi: 10.1128/aem.60.3.973-978.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanseverino J, Werner C, Flemming J, Applegate B, King J M H, Sayler G S. Molecular diagnostics of polycyclic aromatic hydrocarbon biodegradation in manufactured gas plant soils. Biodegradation. 1993;4:303–321. doi: 10.1007/BF00695976. [DOI] [PubMed] [Google Scholar]
- 25.Sayler G S, Shields M S, Tedford E, Breen A, Hooper S, Sirotkin K, Davis J. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985;49:1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley and Sons; 1991. pp. 205–248. [Google Scholar]
- 27.Stahl D A, Flescher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stapleton, R. D., and G. S. Sayler. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer. Microb. Ecol., in press. [DOI] [PubMed]
- 29.Straub K L, Benz M, Schink B, Widdel F. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol. 1996;62:1458–1460. doi: 10.1128/aem.62.4.1458-1460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutherland J B. Detoxification of polycyclic aromatic hydrocarbons by fungi. J Ind Microbiol. 1992;9:53–62. doi: 10.1007/BF01576368. [DOI] [PubMed] [Google Scholar]
- 31.Sutherland J B, Rafii F, Kahn A A, Cerniglia C E. Mechanisms of polycyclic aromatic hydrocarbon degradation. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: Wiley-Liss; 1995. pp. 269–306. [Google Scholar]
- 32.Wichalacz P L, Unz R F. Acidophilic, heterotrophic bacteria of acid mine waters. Appl Environ Microbiol. 1981;41:1254–1261. doi: 10.1128/aem.41.5.1254-1261.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]