Abstract
Complete metabolism of chlorinated benzenes is not a feature that is generally found in aerobic bacteria but is thought to be due to a novel recombination of two separate gene clusters. Such a recombination could be responsible for adaptation of a natural microbial community in response to contamination with synthetic chemicals. This hypothesis was tested in a chlorobenzene (CB)-contaminated aquifer. CB-degrading bacteria from a contaminated site were characterized for a number of years by examining a combination of growth characteristics and DNA-DNA hybridization, PCR, and DNA sequence data. The genetic information obtained for the CB pathway of the predominant microorganism, Ralstonia sp. strain JS705, revealed a unique combination of (partially duplicated) genes for chlorocatechol degradation and genes for a benzene-toluene type of aromatic ring dioxygenase. The organism was detected in CB-polluted groundwater by hybridizing colonies cultivated on low-strength heterotrophic media with probes for the CB pathway. Southern hybridizations performed to determine the organization of the CB pathway genes and the 16S ribosomal DNA indicated that CB-degrading organisms isolated from different wells at the site were identical to JS705. Physiological characterization by the Biolog test system revealed some differences. The genes for the aromatic ring dioxygenase and dihydrodiol dehydrogenase of JS705 were detected in toluene and benzene degraders from the same site. Our results suggest that recent horizontal gene transfer and genetic recombination of existing genes between indigenous microorganisms were the mechanisms for evolution of the catabolic pathway. Evolution of the CB pathway seems to have created the capacity for natural attenuation of CB at the contaminated site.
The ability of bacteria to adapt in order to exploit novel chemicals as growth substrates has been the subject of intensive study for almost 50 years. Early studies revealed that the enhanced degradation of pesticides applied to agricultural soils (2). Subsequent work revealed that similar phenomena allow bacteria to degrade synthetic contaminants in a variety of ecosystems (5, 6, 16, 23, 36, 38, 39, 46). Some synthetic contaminants, notably chlorinated aromatic compounds like chlorobenzenes (CBs), chlorobiphenyls, and chlorinated herbicides, can be recalcitrant to biodegradation (1, 29), perhaps because they were introduced into the environment only recently. Chloroaromatic compounds are not readily degraded by most microorganisms; however, bacteria that are able to use these compounds as sole sources of carbon and energy have been isolated, mostly from contaminated ecosystems (4, 28, 35, 44).
Three possible mechanisms have been put forward to explain the apparent adaptation of microbial communities. Appropriate strains might (i) have been present in the community in numbers below the detection limit, (ii) have been introduced into the community by dispersal from a distant area, or (iii) have arisen by genetic changes within the indigenous community (35, 44).
Most bacteria that are able to use chlorinated aromatic compounds as sole sources of carbon and energy were isolated from enrichment cultures prepared with inocula from polluted environments (5, 6, 10, 13, 32, 34, 37, 42, 46, 51). Although the strains and the genes for some of the degradative pathways were characterized in detail, whether the strains were representative of the bacteria in the environments from which they were enriched or were artifacts of the enrichment procedure was not known (35, 44). Studies of CB- and 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from contaminated ecosystems suggested that genetic changes were the major cause of the adaptation of the organisms (14, 19, 33, 48, 49). The most obvious changes involved the presence of insertion elements or transposons near or within the genes that encode the metabolic pathways for breakdown of the chloroaromatic compounds. It was postulated that the activity of the mobile elements caused new DNA rearrangements and horizontal transfer of genetic material (19, 43, 50). These observations suggested that “novel” pathways were simply built by adding existing genetic material from different microorganisms to the genome of a single organism. For example, the genes for CB degradation in Pseudomonas sp. strain P51 are organized in two distinct regions (47), one of which is a transposable element containing five genes necessary for the conversion of CB to chlorocatechol (48). These five genes are very similar to genes in bacteria that degrade toluene (49). The other region contains the genes for metabolism of chlorocatechols, which are very similar to genes in bacteria that degrade 3-chlorobenzoate (3-CBA) or 2,4-D (7, 25, 45). The combination of these two regions in strain P51, however, is unique and is not found in bacteria that degrade toluene, 3-CBA, or 2,4-D. Similar observations have been made for other bacteria, including the 2,4-D-degrading organism Ralstonia eutropha JMP134(pJP4) (14, 19). The formation of a pathway for CB degradation in a single microorganism after starting with two different species has also been documented in laboratory experiments (17, 24).
To date, little direct information is available on where and when genetic adaptation has taken place in natural environments. Recent field data suggested that horizontal transfer of naphthalene-degradative genes occurred among members of a soil bacterial community (12). In the present work we investigated the origin of CB-degrading bacteria in contaminated groundwater at Kelly Air Force Base (KAFB) in Texas. CB-degrading bacteria can be isolated only from the CB-contaminated area at KAFB, not from the region outside this area. The contaminated site at KAFB, therefore, provided an opportunity to study the mechanisms responsible for adaptation of the microbial community to degrade CB. The purpose of the study was to determine which of the following hypotheses best explains the adaptation of the microbial community to degrade CB. If the CB-degrading bacteria had been present in the indigenous communities in low numbers prior to contamination, they should have been detectable with sensitive molecular techniques (colony hybridization, which can detect 1 in 104 cells [30], or PCR, which can detect 102 cells per ml [41]) or with selective enrichment and plating techniques. If the CB pathway arose only once and reached KAFB by recent dispersal from elsewhere, all of the CB-degrading bacteria from a variety of sites should contain related genetic material and should be less closely related to indigenous bacteria. If the ability to degrade CB arose once at KAFB, identical genes for the pathway should be distributed throughout the site. The genes should be different from the genes in CB-degrading bacteria elsewhere and should be related to genes in indigenous bacteria. Finally, if the ability to degrade CB arose repeatedly throughout the contaminated site, a variety of gene combinations should be present, and they should be related to genes in indigenous bacteria.
To test the hypotheses described above, a variety of microbial isolation procedures and genetic techniques were employed. DNA probes and primers derived from the CB pathway genes of a predominant CB-degrading organism obtained from polluted groundwater at KAFB were used to screen bacteria isolated from different wells at KAFB under selective and nonselective conditions. The experiments were conducted with groundwater obtained within and outside the contaminated area at intervals over several years.
MATERIALS AND METHODS
Site description.
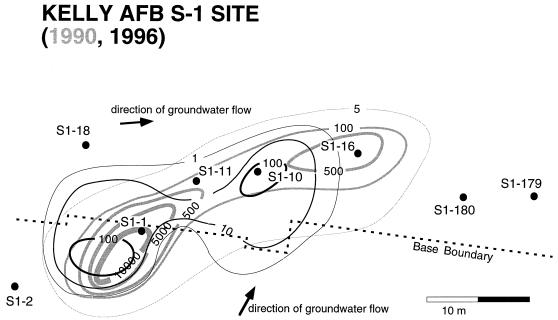
Groundwater samples were collected from monitoring wells located in a CB plume emanating from a former waste solvent storage area at KAFB near San Antonio, Tex. (Fig. 1). The storage facility, known as site S1, was used from 1960 until 1973 (26). The contaminated aquifer consists of silty sand and clay 7.6 to 10.4 m below the surface. During 1993 and 1994 a program was begun to halt the flow of contaminants from the source area by pumping water from a series of wells just downstream of the source area. The wells that were sampled were selected based on the 1990 plume map (Fig. 1) and included a transect along the length of the CB plume, as well as uncontaminated wells outside the plume. Possibly due in part to a 2-year drought in Texas during 1995 and 1996, water was present only sporadically in one of the wells (well S1-1) located close to the source.
FIG. 1.
Schematic overview of the CB plume and sampling wells at KAFB site S1. The interpolated isoconcentrations (in micrograms per liter) were derived from measurements obtained at observation wells in 1990 (light lines) and 1996 (dark lines). The direction of the groundwater flow is indicated by the arrow.
Sample collection and isolation of bacteria with specific degradative abilities.
The sampling strategy used for the last 6 years is shown in Table 1; 2 to 3 well volumes was removed from each sampling well, and samples (1 liter) were collected aseptically. The numbers of heterotrophic CFU were estimated by spreading serial dilutions onto one-quarter-strength tryptic soy agar plates and incubating the plates for 3 days at 30°C. Specific capacities to degrade aromatic substances were determined by spreading appropriate dilutions of groundwater on one-quarter-strength minimal medium (MSB) (40) solidified with 1.8% (wt/vol) agar. Aromatic substrates were provided individually; CB, toluene, and benzene were each provided as a vapor (10). 3-CBA and 2,4-D were dissolved in the media at a concentration of 2.5 mM. Plates were incubated for 5 days at 30°C and examined for the presence of colonies. When no visible colonies appeared, the plates were incubated for an additional 20 days and reexamined twice weekly. Large colonies that grew on each substrate were retested along with appropriate controls to confirm that the substrate was degraded (10). When the plating method was used, the detection limit for CB-degrading bacteria in groundwater was 20 CFU/ml.
TABLE 1.
Sampling dates and summary of studies performed at KAFB
| Well | Results for the following sampling datesa:
|
|||||
|---|---|---|---|---|---|---|
| August 1992 | January 1993 | August 1994 | September 1996 | December 1996 | January 1998 | |
| S1-1 | I+, JS705 isolated | I+ | E+, CH+, SH+, PCR+ | —b | I− | I+ |
| S1-2 | I− | I− | E−, CH+/−, SH−, PCR+/− | I+, PCR+ | I−, E− | |
| S1-10 | I+, CH+, SH+ | |||||
| S1-11 | I+, CH+, SH+ | |||||
| S1-16 | I−, E− | |||||
| S1-18 | I−, E− | |||||
| S1-179 | I−, E− | |||||
| S1-180 | I−, SH−, PCR+ | I−, E− | ||||
I+, CB, benzene, and toluene degraders were isolated; I−, benzene and toluene degraders were isolated but CB degraders were not isolated; E+, positive for enrichment on CB, benzene, toluene, and 1,4-dichlorobenzene but not for enrichment on 3-CBA; E−, no enrichment on CB, 1,4-dichlorobenzene, or 3-CBA but positive for toluene or benzene; CH+, colony hybridization occurred with clc and mcb probes of JS705; CH+/−, colony hybridization occurred with either the clc probe or the mcb probe; SH+, hybridization to clc, mcb, or 16S rDNA probes of isolated strains in Southern analysis identical to hybridization of strain JS705; SH−, no hybridization or different hybridization patterns; PCR+, PCR-amplified fragment from total DNA isolated from well water positive with all primers specific for JS705; PCR+/−, fragment positive with only one primer set.
—, well was dry.
Groundwater samples from wells outside the CB plume were used as inocula for enrichment cultures to which CB was supplied in the vapor phase (50 ml of groundwater was mixed with 50 ml of MSB). The cultures were incubated at 30°C with shaking (250 rpm). After 2 months of incubation, the cultures were sampled to determine whether CB-degrading bacteria were present, as described above. All confirmed CB-degrading isolates and 300 benzene- and toluene-degrading isolates were grouped on the basis of their Gram reactions and cell and colony morphology and were characterized by using Biolog GN or GP (Biolog, Hayward, Calif.) microplates as appropriate. Strains were also compared after PCR amplification of parts of their 16S ribosomal DNA (rDNA) with primers V1.1 and V3.2, restriction enzyme digestion of the amplification products with HaeIII and Sau3AI, and separation of the restriction fragments by agarose gel electrophoresis.
Bacteria and plasmids.
Escherichia coli DH5α, the host bacterium used for all cloning experiments and plasmid isolations, was cultivated on Luria-Bertani medium (31) supplemented with ampicillin (50 μg/ml) when appropriate. Ralstonia sp. strain JS705 was isolated from well S1-1 by growing it on CB. It was cultivated on MSB medium supplemented with CB vapor in closed flasks at 30°C. Other bacterial strains isolated during this study are listed in Table 2.
TABLE 2.
Properties of isolates
| Strain | Wella | Growth on:
|
Taxon as determined by:
|
|||
|---|---|---|---|---|---|---|
| CB | Benzene | Toluene | Biolog tests | 16S rDNA analysis | ||
| JS702 | S1-1 | NIb | ||||
| JS705 | S1-1 | + | Comamonas | Ralstonia | ||
| JS707 | S1-1 | − | Alcaligenes faecalis | |||
| JS711 | S1-1 | + | Comamonas | |||
| JS727 | RI3-1W | + | NI | |||
| JS743 | S1-11 | + | + | Alcaligenes eutrophus | ||
| JS744 | S1-11 | + | + | Comamonas acidovorans | ||
| JS745 | S1-11 | + | + | NI | ||
| JS749 | S1-180 | + | + | Alcaligenes eutrophus | ||
| JS751 | S1-11 | + | + | Comamonas acidovorans | ||
| JS755 | S1-11 | + | CDC group IVc-2 | |||
| JS756 | S1-11 | + | Comamonas acidovorans | |||
| JS757 | S1-11 | + | + | + | Alcaligenes eutrophus | |
| JS758 | S1-11 | + | NI | |||
| JS759 | S1-11 | + | CDC group IVc-2 | |||
Wells S1-1, S1-11, and S1-180 were wells at KAFB site 1 (Fig. 1); well RI3-1W was at Robins Air Force Base.
NI, not identified.
The plasmids used for heterologous hybridization with strain JS705 were pTCB111 (containing a tcbAaAb gene fragment) (49), pDTG351 (containing todC1C2BADE) (52), and pDC1001 (containing a 1.8-kb EcoRI-HindIII fragment of pDC100 with clcA) (7). Vectors pUC18 (31), pT7Blue T (Novagen, Madison, Wis.), and pGEM-T Easy (Promega Corp., Madison, Wis.) were used to clone JS705 DNA fragments. We constructed pCBA24, which contains a 4.8-kb PstI fragment in pUC18, pCBA21, which contains a 4.5-kb PstI fragment, and pCBA31, which contains a 3.5-kb BamHI fragment in pUC18 (see below). pCBA32 contains a randomly derived 3.8-kb BamHI fragment of the genomic DNA of JS705. Plasmid pCBA117 contains a 1,504-bp region of the 16S rDNA of strain JS705 that was amplified by PCR with two eubacterial primers, primers 6F and 1510R (Table 3). The fragment was cloned in plasmid pT7Blue T, and the sequences of both strands were determined.
TABLE 3.
Primers used
| Primer set | Primer | Sequence (5′-3′) | Product size | Locationa |
|---|---|---|---|---|
| clc | CGAGTTGCCGAGGTCGCAGGC | 763 bp | +9-29 | |
| TCATGCCACTGTCTCCGTAGCCG | −772-750 | |||
| mcb | GTCAGGAAGGCGGGCGACTACTTTAC | 617 bp | +183-209 | |
| GATATAGAAGCCGCTGCCATGTC | −800-777 | |||
| clcA-mcbF | GGGCAAATGGGTGGACGATGACTG | 839 bp | +626-649 | |
| GCGGCGCCGGGACAACTC | −170-153 | |||
| 16S rDNA | V1.1 | GCGGCGTGCCTAATACATGC | 659 bp | +41-60 |
| V3.2 | ATCTACGCATTTCACCGCTAC | −705-685 | ||
| 16S rDNA | 6F | GGAGAGTTAGATCTTGGCTCAG | 1.5 kb | +6-28 |
| 1510R | GTGCTGCAGGGTTACCTTGTTACGACT | −1510-1483 |
Location relative to the start site of the gene.
Colony and Southern hybridizations.
The water from wells S1-1 and S1-2 used for colony hybridization was serially diluted with filter-sterilized groundwater from the site. Samples (100 μl) were spread onto PTYG agar containing (per liter) 0.5 g of peptone, 0.5 g of tryptone, 0.25 g of yeast extract, 0.5 g of glucose, 700 mg of MgCl2, and 100 mg of CaCl2. Plates were incubated at 20°C for 4 weeks. Plates containing 103 to 104 colonies were used for colony blotting and hybridization. Colony blotting was performed on Qiabrane nylon membranes (Qiagen, Basel, Switzerland) by using previously described procedures (31). Spots that reacted positively in the DNA-DNA hybridization experiments were matched with the colonies on the original agar plates and were restreaked onto PTYG agar to obtain pure cultures.
DNA-DNA hybridizations were performed with different gene probes at 62°C under stringent conditions by using previously described procedures (31). The probes used were the 4.5-kb PstI fragment of plasmid pCBA21 and the 3.5-kb BamHI fragment of plasmid pCBA31 (Fig. 2).
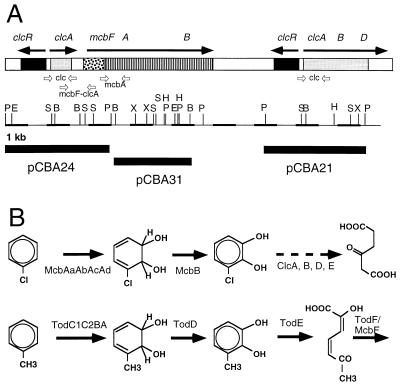
FIG. 2.
Genetic and physical organization of the CB pathway genes of strain JS705. (A) The upper line shows the relative positions of the aromatic ring dioxygenase gene cluster (mcb) and the genes for chlorocatechol degradation (clc) and their orientations (as determined by DNA sequencing). Below this are the approximate locations of the primer sets used (Table 3). Restriction site abbreviations: P, PstI; E, EcoRI; S, SalI; B, BamHI; X, XhoI; H, HindIII. At the bottom the cloned fragments of the region and the plasmid designations are indicated. (B) Initial enzymatic steps in the degradation of CB and toluene. The initial dioxygenation and rearomatization steps in toluene degradation are carried out by enzymes which can also mediate CB conversion.
Isolating DNA from groundwater and PCR amplification.
Samples of groundwater from wells S1-1, S1-2, and S1-180 (0.5 liter) were centrifuged for 10 min at 10,000 × g in sterile 250 ml centrifuge tubes. Sterilized double-distilled H2O was processed as a control. DNA was isolated from the cell pellet essentially as described by Hallier-Soulier et al. (11).
PCR was used to detect DNA identical to the CB pathway genes of strain JS705. The following three sets of primers were developed and synthesized: (i) primers for specific detection of a region in the clcA gene, (ii) primers for detection of the gene for the large subunit of the CB dioxygenase of strain JS705 (mcbAa), and (iii) primers for detection of the region between the clcA and mcbF genes of strain JS705 (Fig. 2 and Table 3). The composition of the PCR mixture was the composition specified by the manufacturer (Life Technologies, Gaithersburg, Md.). Amplification products either were analyzed on an agarose gel, blotted, and hybridized for verification or were cloned and sequenced.
DNA techniques.
Analysis of DNA fragments on agarose gels, cloning of DNA fragments, plasmid isolation, restriction digestion, purification of DNA fragments, and radioactive labeling were all performed by using previously described procedures (31) or the specifications of the supplier. DNA sequencing was performed with double-stranded DNA templates by cycle sequencing by using fluorescently labeled primers (IRD800; MWG, Ebersberg, Germany) and the protocol of a Thermo Sequenase kit (Amersham Life Sciences, Little Chalfont, United Kingdom). Sequence reaction products were separated with an automated DNA sequence analyzer (model 400L; LiCOR, Lincoln, Nebr.).
Nucleotide sequence accession numbers.
The nucleotide sequence of JS705 rDNA has been deposited in the GenBank database under accession no. AF027407. The nucleotide sequences of the clcR and clcA genes and of the mcbFAaAbAcAd genes have been deposited in the GenBank database under accession no. AJ006307.
RESULTS
Detection and isolation of DNA encoding the CB degradation pathway of strain JS705.
Strain JS705 was isolated from CB-contaminated groundwater at KAFB in 1992 (Table 1). Biolog GN microplate analysis resulted in identification of the strain as a member of the genus Comamonas. However, a 16S rDNA sequence analysis indicated that the highest identity (97.4% in a 1,497-bp overlap region) was with R. eutropha (GenBank accession no. M32021). The 16S rDNA sequence of strain JS705 also clustered nearest to R. eutropha 16S rDNA sequence, as determined by the Ribosomal Database Project program SuggestTree (20), when a subsequence alignment was examined. Therefore, we named strain JS705 Ralstonia sp. strain JS705.
Total genomic DNA of JS705 was analyzed by Southern hybridization to determine whether genetic material similar to the genes for the CB degradation pathway of Pseudomonas sp. strain P51 (45, 49), for the toluene degradation pathway of Pseudomonas putida F1 (52), and for the chlorocatechol degradation pathway of P. putida(pAC27) (7) was present. Very strong hybridization signals were obtained with DNA probes containing the clcRABD genes (Fig. 3) and DNA probes containing the tcbAaAb genes or the todC1C2AB genes (data not shown). Three restriction fragments were cloned from strain JS705 into pUC18. One 4.5-kb PstI fragment (pCBA21) contained a clcRABD gene cluster that exhibited 99% DNA sequence identity to the gene cluster of P. putida(pAC27) (7) (Fig. 2). A 4.8-kb PstI fragment (pCBA24) contained a partial duplication of the clcRABD gene cluster. The duplicated genes, clcR and clcA, were upstream of a gene similar to todF (21), which was tentatively designated mcbF. The third cloned fragment, a 3.5-kb BamHI fragment (pCBA31), contained the genes for an aromatic ring dioxygenase (tentatively designated mcbAaAbAcAd) similar to the dioxygenases encoded by the tcbAaAbAcAd (49) and todC1C2BA genes (52). The identities of all of the genes were confirmed by DNA sequencing (GenBank accession no. AJ006307). We hybridized these three cloned fragments with total DNA of strain JS705 that was digested with different combinations of restriction enzymes, and we constructed a physical map of the region where the CB pathway genes are located (Fig. 2). Our results revealed a unique and distinctive arrangement of genes for CB degradation in strain JS705. Although not all of the genetic and biochemical details were analyzed, we presumed, based on the strong sequence similarities between the CB genes of JS705 and the CB genes of other strains, that the CB pathway of strain JS705 is encoded by the single region analyzed here (Fig. 2).
FIG. 3.
Southern hybridization of Ralstonia sp. strain JS705 total DNA with the 1.8-kb EcoRI-HindIII fragment of plasmid pDC1001 which carries clcA (7). Multiple bands indicate the duplicated nature of the clc gene fragments in strain JS705. Lanes X, B, P, S, E, and H, DNA digested with XhoI, BamHI, PstI, SalI, EcoRI, and HindIII, respectively.
Direct screening of colonies grown from groundwater samples by DNA-DNA hybridization.
To determine the abundance of strain JS705 genetic material in groundwater, in 1994 we screened (using the colony hybridization technique) 104 colonies from well S1-1 (grown on PTYG agar) and 105 colonies from well S1-2. Using the CB pathway probes (the inserts of plasmids pCBA21 and pCBA31), we detected 5 to 10 signals on each membrane blotted from plates containing 103 colonies from contaminated well S1-1. A total of 10 membranes, each containing 104 colonies from uncontaminated well S1-2, were examined, and only one signal was found for each probe. All positive spots on well S1-1-derived membranes reacted with both of the CB pathway probes. The two positive spots from well S1-2 samples did not cross-react.
To verify that the hybridizing colonies contained genetic material related to the genetic material of strain JS705, we isolated DNA from nine positive colonies grown on the original agar plates from well S1-1 samples that had been used for hybridization. In 6 of the 9 colonies we detected a proper-size DNA fragment when PCR were performed with the clc primers (data not shown). Nonspecific amplification products were obtained with DNA from the other three cultures. Total DNA isolated from the six positive cultures, digested with XhoI, BamHI, or HindIII, and hybridized with probes for the clc genes from JS705(pCBA21) and for the mcb genes (pCBA31) produced patterns similar to the JS705 pattern (data not shown). The 16S rDNA that were amplified with primers V1.1 and V3.2 (Table 3) and digested with Sau3AI and HaeIII produced the same restriction patterns as JS705 16S rDNA produced, which indicates that the organisms were taxonomically very similar (Fig. 4). Neither of the two positively reacting spots from the well S1-2 groundwater samples could be recovered because of the high colony densities on the original plates.
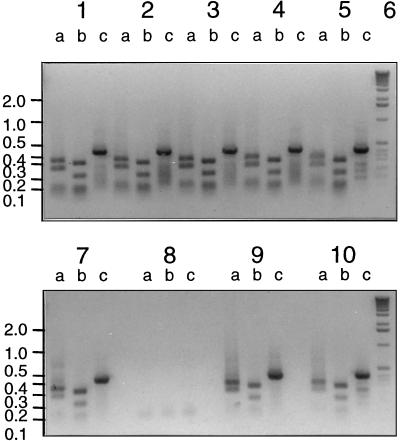
FIG. 4.
Restriction digests of a PCR product amplified with the 16S rDNA primers from colonies that were positive in colony hybridization experiments. The positions of size markers (in kilobases) are indicated on the left. Lanes a, Sau3AI digests; lanes b, HaeIII digests; lanes c, undigested products. Lanes 1 through 5 and 7 contained six independently derived colonies; lane 6 contained the 1-kb marker; lanes 8 contained no DNA; and lanes 9 and 10 contained JS705.
Screening of DNA purified from the groundwater samples by PCR.
Because no CB-degrading bacteria were detected in wells outside the CB-contaminated area in 1992 and 1993, we used PCR to determine whether genetic material similar or identical to the genetic material encoding the CB pathway in strain JS705 could be detected in uncontaminated groundwater. Purified DNA obtained from groundwater from wells S1-1 and S1-2 in 1994 and 1996 and from well S1-180 in 1996 were used as targets in the PCR (Table 1). Proper-size amplification products were detected in the DNA from wells S1-1 and S1-180 with mcbA primers. The identities of the products were confirmed by hybridization with the insert of pCBA31 and by DNA sequencing (data not shown). No amplification product for mcbA was obtained with DNA from well S1-2 in 1994. When the clcA gene primers were used, amplification products were detected in DNA from groundwater from wells S1-1, S1-2, and S1-180. The identities of these products were confirmed by hybridization with the clcA probe. When the primer set for the combination clcA-mcbF was used, we detected amplifiable targets in samples from well S1-1 in 1994 and from well S1-180 in 1996. Using DNA isolated from well S1-2 in 1996 resulted in amplification products with all three primer sets used for the CB pathway genes of strain JS705 (data not shown).
Screening of CB degraders isolated from different wells on KAFB.
The results of the 1994 screening study of DNA isolated from groundwater with probes derived from JS705 led us to examine additional wells at site S1 in 1996 and 1998 (Table 1). Samples were also inoculated onto plates containing toluene, benzene, 3-CBA, or 2,4-D to determine whether the parental genes for the CB pathway of JS705 could be recovered in other bacteria from site S1.
The total numbers of bacteria recovered from the various wells varied by 2 orders of magnitude (Table 4). CB-degrading bacteria were isolated without enrichment from wells S1-10, S1-11, and S1-2 in 1996. Growth of the CB-degrading strains was immediate and rapid, indicating that the bacteria were active in the groundwater (22, 23). The uncontaminated wells (wells S1-18, S1-179, and S1-180) yielded no bacteria that were able to grow on CB either by direct isolation or in enrichment cultures in 1996. Well S1-2 was negative when it was sampled in 1993 and 1994, but in 1996 it yielded a small number of CB-degrading isolates. However, well S1-2 is only slightly upgradient of the plume source, and it is possible that contaminated water reached the well in 1996. In contrast to samples obtained in 1993 and 1994, in December 1996 well S1-1 yielded no CB-degrading bacteria. However, this well was dry in September 1996. Samples were obtained from wells S1-1, S1-2, and S1-180 in 1998, and CB-degrading bacteria were found only in well S1-1. These results were consistent with observations made in 1993 and 1994 but conflicted with results obtained in December 1996.
TABLE 4.
Numbers of culturable bacteria in groundwater in September and December 1996
| Well | No. of bacteria (CFU/ml) with the following substrates:
|
|||
|---|---|---|---|---|
| Tryptic soy agar | CB | Benzene | Toluene | |
| S1-1 | 3.8 × 105 ± 0.2 × 105 | NDa | NEb | NE |
| S1-2 | 8.6 × 103 ± 2 × 103 | 8.1 × 102 ± 1.3 × 102 | NE | NE |
| S1-10 | 1.7 × 105 ± 0.1 × 105 | 1.4 × 104 ± 0.4 × 104 | 9.2 × 104 ± 0.5 × 104 | 3.9 × 104 ± 0.3 × 104 |
| S1-11 | 4.1 × 105 ± 0.3 × 105 | 3.0 × 104 ± 0.8 × 104 | 8.4 × 104 ± 0.5 × 104 | 4.9 × 104 ± 0.3 × 104 |
| S1-16 | 2.6 × 103 ± 0.6 × 103 | ND | NE | NE |
| S1-18 | 2.6 × 104 ± 0.5 × 104 | ND | NE | NE |
| S1-179 | 4.5 × 103 ± 1.3 × 103 | ND | NE | NE |
| S1-180 | 2.7 × 105 ± 0.5 × 105 | ND | 3.2 × 103 ± 0.3 × 103 | 2.5 × 103 ± 0.6 × 103 |
ND, not detectable.
NE, no enumeration.
The 55 CB-degrading isolates obtained in 1996 accounted for about 10% of the total CFU grown on tryptic soy agar. Two CB-degrading isolates also degraded benzene, and one degraded benzene and toluene. A total of 220 benzene-degrading isolates and 110 toluene-degrading isolates were recovered from the three wells sampled in September 1996. None of the isolates selected on benzene or toluene could degrade CB.
CB degraders isolated from well S1-1 in 1993 and before and from wells S1-10 and S1-11 in 1996 were characterized by PCR and Southern hybridization to determine whether they were similar to strain JS705. All of these organisms hybridized to the clc probe (insert from pCBA21), to the mcb probe (insert from pCBA31), and to a 16S rDNA probe (insert from pCBA117) with the same pattern as JS705 (Fig. 5). CB degraders isolated from other CB-contaminated sites (one example of which is shown in Fig. 5, lane 9) produced distinctly different patterns. DNA amplified by PCR from the 55 CB-degrading strains isolated from site S1 with the three primer sets used for the CB pathway genes from JS705 and with primers V1.1 and V3.2 and digested with the appropriate restriction enzymes produced the same restriction patterns as JS705 (data not shown). These results indicate that the genes for the CB pathway were the same in all of the CB-degrading isolates from site S1. The different CB-degrading strains could not be distinguished by genetic screening, although differences were detected in Biolog test results (Table 2).
FIG. 5.
Southern hybridization of total DNA of selected CB degraders isolated from wells at KAFB with the pCBA21 insert for the clc genes (A) and with 16S rDNA derived from JS705 (B). DNA were digested with BamHI. Lane 1, strain JS702; lane 2, strain JS707 (a non-CB degrader); lane 3, strain JS711; lane 4, strain JS755; lane 5, strain JS756; lane 6, strain JS757; lane 7, strain JS758; lane 8, strain JS759; lane 9, strain JS727; lane 10, strain JS705. Most strains were isolated from KAFB; the only exception was strain JS727, which was isolated from Robins Air Force Base in Georgia in 1993.
Detection of genes identical to the mcb genes of strain JS705 in toluene- and benzene-degrading bacteria.
Twelve representative toluene- and/or benzene-degrading bacteria isolated from site S1 were analyzed to determine the presence of gene fragments similar to the CB pathway genes of strain JS705. Four isolates from well S1-11 and one isolate from S1-180 yielded a product after PCR amplification with the mcb primer set. No amplification products were obtained with the clc or mcbF-clcA primer set. DNA sequencing of the mcb products revealed that three of the five strains (JS743, JS745, and JS751) had a 617-bp sequence that was identical to the sequence of the JS705 fragment (data not shown).
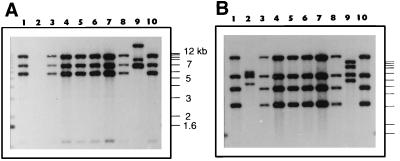
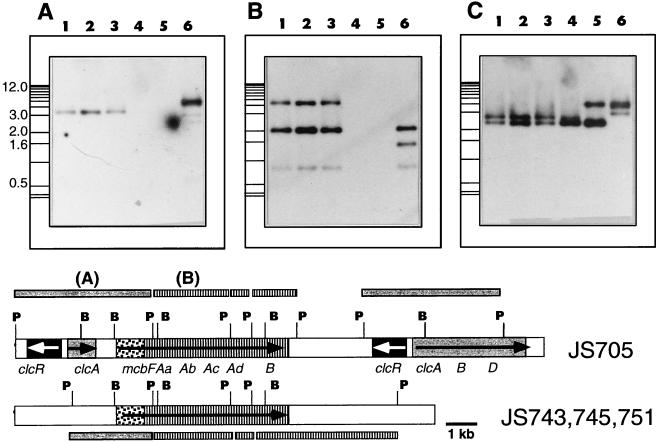
Hybridization of the PstI fragment of pCBA24 with PstI-digested total DNA resulted in one positively reacting band in strains JS743, JS745, and JS751 and two different bands in JS705 (Fig. 6A). When BamHI-digested DNA was used, only one corresponding BamHI fragment (1.3 kb) was found (data not shown). The 1.3-kb BamHI fragment encompassed the mcbF gene in JS705 (Fig. 2 and 6), and the results suggested that mcbF was present in the three toluene- and benzene-degrading organisms examined. The other two strains (JS744 and JS749) hybridized weakly with the PstI insert of pCBA24, suggesting that there was sequence similarity with parts of the probe (probably the mcbF part) but no identity (since the restriction patterns were different). No evidence of the clcRA genes (Fig. 6A) or of the clcBD genes (i.e., the pCBA21 insert (data not shown) was found in the benzene- and toluene-degrading bacteria. Hybridization of the pCBA31 insert with BamHI-digested DNA samples yielded a single band having the same size in JS705, JS743, JS745, and JS751 (data not shown). When the same probe and PstI-digested samples were used, two of three bands were the same in JS705, JS743, JS745, and JS751 (Fig. 6B). The 1.4-kb PstI fragment that hybridized in JS705 DNA was replaced by a 4.5-kb band in JS743, JS745, and JS751. Hybridizations of the same blots with a probe for the 16S rDNA of strain JS705 revealed clear differences (Fig. 6C). Strains JS743, JS745, and JS751 produced identical band patterns, whereas the other two toluene and benzene degraders (JS744 and JS749) and JS705 were clearly different. These results indicated that the genes for the initial steps in CB degradation in JS705 were identical to the genes in the indigenous toluene and benzene degraders (JS743, JS745, and JS751).
FIG. 6.
Southern hybridization of total DNA of selected toluene and benzene degraders from wells S1-11 and S1-180 and of JS705 with probes for the clc-mcbF genes (insert of pCBA24) (A), for the mcbAB genes (pCBA31) (B), and for 16S rDNA (C). All DNAs were digested with PstI. Lane 1, strain JS743; lane 2, strain JS751; lane 3, strain JS745; lane 4, strain JS744; lane 5, strain JS749; lane 6, strain JS705. The positions of size markers (1-kb ladder) are indicated on the left. An interpretation of the hybridization results is shown at the bottom.
DISCUSSION
Natural attenuation can be an important process by which pollutants disappear from the environment without high treatment costs. This process is suitable for natural chemicals, such as petroleum hydrocarbons, which are readily metabolizable by ubiquitous microorganisms in most ecosystems (3, 18). CBs, however, are among the synthetic chemicals which are not usually metabolized by natural communities (10, 27, 44, 46). Our observations at KAFB site S1 demonstrated that spontaneous natural attenuation of CBs has occurred since 1990 and that the size of the contaminant plume has decreased. Since active CB-degrading bacteria were detected almost exclusively in groundwater from wells with the highest CB concentrations, our results provide evidence that the population of CB degraders was maintained by mineralization of CB. When the CB was depleted, the population of CB degraders fell to undetectable levels in the groundwater. Clearly, our samples reflected only what was present in the groundwater itself and, therefore, probably underestimated the biomass in the system.
The results of our investigations support the hypothesis that the ability to degrade CB was not present initially but arose once, relatively recently, in the microorganisms at site S1. The CB degradation pathway in the form that is present in strains at KAFB probably arose by relatively simple horizontal gene transfer and gene recombination between different ancestral strains. The host that finally obtained the CB pathway genes gained a selective advantage and colonized most of the CB-contaminated area.
Our arguments that support this hypothesis and not the alternative hypotheses described in the introduction are as follows. First, the CB-degrading bacteria predominant in the groundwater at the site, as exemplified by Ralstonia sp. strain JS705, all had the same genetic organization of the genes for CB degradation. This organization was unique and clearly different from the genetic organizations observed in CB-degrading bacteria from other sites, such as Robins Air Force Base (Fig. 5), or in Pseudomonas sp. strain P51 (47). Second, some of the CB degradation genes in strain JS705 were identical to genes found in indigenous bacteria at the site. Thus, the possibility that the bacterium degrading CBs at site S1 arose elsewhere and was transported to KAFB seems highly unlikely.
Third, our results indicate that CB-degrading bacteria were not present in low numbers at the site before contamination. Despite several attempts, we could not consistently isolate CB-degrading bacteria from wells outside the contaminated zone by direct plating and enrichment techniques or by colony hybridization with heterotrophically grown bacterial colonies; CB-degrading bacteria were isolated only from wells within the contaminated zone. Clearly, it is impossible to prove unequivocally that there were no CB-degrading bacteria outside the contaminated zone or at site S1 before CB contamination was present. Furthermore, we did not sample the subsurface solids, which may harbor a larger population of bacteria, including the ancestors of the CB pathway bacteria or other CB-degrading bacteria. However, within the limits of our analyses (the detection limits were 0.02 CFU/ml for the enrichment technique and 1 in 105 culturable microorganisms for colony hybridization), no CB degraders were detected in uncontaminated subsurface material.
The temporal and spatial variability in the water levels at the site during the study period probably resulted in some anomalous findings. For example, one of the wells sampled in this study (well S1-16) could be considered formerly contaminated. This well did not yield CB-degrading bacteria, which was consistent with the hypothesis that the presence of CBs is required to support a detectable population of CB-degrading bacteria in the water phase. We examined DNA isolated from well S1-180 in 1996 and detected the typical CB pathway genes of strain JS705 by PCR; this finding was not confirmed by isolation of CB degraders. In 1996 well S1-2 also contained CB degraders and was positive for the CB pathway genes of strain JS705 as determined by PCR. However, samples collected at other times yielded no culturable CB degraders from wells S1-2 and S1-180, both of which were outside the CB-contaminated zone. The 1996 results, therefore, can be interpreted only in the context of the observations made in other years.
The genes for CB degradation in strain JS705 were composed of three distinct regions (Fig. 2), two of which (the genes for chlorocatechol degradation) were partial duplicates of each other. The duplicated regions contained genes for chlorocatechol degradation in strain JS705 that were almost identical to genes described previously in bacteria that degrade 3-CBA (7). However, the clc genes could not be traced directly in indigenous non-CB-degrading microorganisms by classical plating and enrichment on 3-CBA or 2,4-D; they could be traced only by PCR performed with DNA samples from uncontaminated groundwater obtained at site S1 (Table 1). This result suggests that the clc genes are present in indigenous bacteria at the site; however, they may be in a cryptic form or may encode parts of an unknown metabolic pathway. Alternatively, more sensitive radiolabeling and isolation techniques (9) might have to be used to detect the metabolic activities of indigenous bacteria that are able to use 3-CBA, 2,4-D, or a related compound. Such techniques have been used to detect oligotrophic microorganisms carrying genes for 2,4-D and 3-CBA metabolism in pristine environments (8, 9, 15).
The third region of CB pathway genes in strain JS705, which encodes the CB ring dioxygenase and dihydrodiol dehydrogenase, was found in a benzene and toluene degrader at site S1. This DNA region, which contained the mcbFAB genes, was identical in JS705 and the benzene and toluene degrader. Since the genetic background of the benzene and toluene degraders was different from the genetic background of JS705, this is the best evidence which we have at present that the genes for the CB pathway originated from existing genes in two different ancestral strains, were recombined to form one large gene region, and were horizontally transferred at least once to the present host. The dramatic reduction in the size of the contaminant plume over the last 6 years and the distribution of CB degraders in groundwater of the contaminated wells provide compelling evidence for the activity of the CB-degrading bacteria. This is the first strong evidence for adaptation due to genetic recombination among bacteria in a groundwater aquifer, resulting in the formation of a novel pathway for chlorobenzene degradation, and the subsequent disappearance of chlorobenzenes from that environment.
ACKNOWLEDGMENTS
We thank Chuck Somerville and Gene Madsen for helpful technical discussions and Glenn Johnson and Rik Eggen for reviewing the manuscript.
We thank the Air Force Office of Scientific Research for financial support.
REFERENCES
- 1.Alexander M. Biodegradation of chemicals of environmental concern. Science. 1981;211:132–138. doi: 10.1126/science.7444456. [DOI] [PubMed] [Google Scholar]
- 2.Audus L J. The decomposition of 2:4-dichlorophenoxyacetic acid and 2-methyl-4-chlorophenoxyacetic acid in soil. J Sci Food Agric. 1952;3:268–275. [Google Scholar]
- 3.Bouwer E J. Bioremediation of organic contaminants in the subsurface. In: Mitchell R, editor. Environmental microbiology. New York, N.Y: Wiley-Liss, Inc.; 1992. pp. 287–318. [Google Scholar]
- 4.Chaudhry G R, Chapalamadugu S. Biodegradation of halogenated organic compounds. Microbiol Rev. 1991;55:59–79. doi: 10.1128/mr.55.1.59-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 7.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulthorpe R R, Rhodes A N, Tiedje J M. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl Environ Microbiol. 1996;62:1159–1166. doi: 10.1128/aem.62.4.1159-1166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigler B E, Nishino S F, Spain J C. Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1988;54:294–301. doi: 10.1128/aem.54.2.294-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallier-Soulier S, Ducrocq V, Mazure N, Truffaut N. Detection and quantification of degradative genes in soils contaminated by toluene. FEMS Microbiol Ecol. 1996;20:121–133. [Google Scholar]
- 12.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickey W J, Focht D D. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl Environ Microbiol. 1990;56:3842–3850. doi: 10.1128/aem.56.12.3842-3850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamagata Y, Fulthorpe R R, Tamura K, Takami H, Forney L J, Tiedje J M. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1997;63:2266–2272. doi: 10.1128/aem.63.6.2266-2272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karns J S, Kilbane J J, Duttagupta S, Chakrabarty A M. Metabolism of halophenols by 2,4,5-trichlorophenoxyacetic acid-degrading Pseudomonas cepacia. Appl Environ Microbiol. 1983;46:1176–1181. doi: 10.1128/aem.46.5.1176-1181.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröckel L, Focht D D. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl Environ Microbiol. 1987;53:2470–2475. doi: 10.1128/aem.53.10.2470-2475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leahy J G, Colwell R R. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leveau J H J, van der Meer J R. Genetic characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutropha JMP134. Gene. 1997;202:103–114. doi: 10.1016/s0378-1119(97)00460-5. [DOI] [PubMed] [Google Scholar]
- 20.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menn F M, Zylstra G J, Gibson D T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase. Gene. 1991;104:91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 22.Nishino S F, Spain J C, Belcher L A, Litchfield C D. Chlorobenzene degradation by bacteria isolated from contaminated groundwater. Appl Environ Microbiol. 1992;58:1719–1726. doi: 10.1128/aem.58.5.1719-1726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishino S F, Spain J C, Pettigrew C A. Biodegradation of chlorobenzene by indigenous bacteria. Environ Toxicol Chem. 1994;13:871–877. [Google Scholar]
- 24.Oltmanns R H, Rast H G, Reineke W. Degradation of 1,4-dichlorobenzene by enriched and constructed bacteria. Appl Microbiol Biotechnol. 1988;28:609–616. [Google Scholar]
- 25.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettigrew C A, Haigler B E, Spain J C. Simultaneous biodegradation of chlorobenzene and toluene by a Pseudomonas strain. Appl Environ Microbiol. 1991;57:157–162. doi: 10.1128/aem.57.1.157-162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reineke W. Microbial degradation of halogenated aromatic compounds. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 319–360. [Google Scholar]
- 28.Reineke W, Knackmuss H-J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 29.Reineke W, Knackmuss H-J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984;47:395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rochelle P A, Wetherbee M K, Olson B H. Distribution of DNA sequences encoding narrow- and broad-spectrum mercury resistance. Appl Environ Microbiol. 1991;57:1581–1589. doi: 10.1128/aem.57.6.1581-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sander P, Wittich R-M, Fortnagel P, Wilkes H, Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl Environ Microbiol. 1991;57:1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlömann M. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 34.Schraa G, Boone M L, Jetten M S M, van Neerven A R W, Colberg P J, Zehnder A J B. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986;52:1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spain J. Synthetic chemicals with potential for natural attenuation. Bioremed J. 1997;1:1–9. [Google Scholar]
- 36.Spain J C. Microbial adaptation in aquatic ecosystems. In: Racke K D, Coats J R, editors. Enhanced biodegradation of pesticides in the environment. Miami Beach, Fla: American Chemical Society; 1989. pp. 183–190. [Google Scholar]
- 37.Spain J C, Nishino S F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987;53:1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spain J C, van Veld P A. Adaptation of natural microbial communities to degradation of xenobiotic compounds: effects of concentration, exposure, time, inoculum, and chemical structure. Appl Environ Microbiol. 1983;45:428–435. doi: 10.1128/aem.45.2.428-435.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spain J C, van Veld P A, Monti C A, Pritchard P H, Cripe C R. Comparison of p-nitrophenol biodegradation in field and laboratory test systems. Appl Environ Microbiol. 1984;48:944–950. doi: 10.1128/aem.48.5.944-950.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 41.Steffan R J, Atlas R M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- 42.Thiele J, Müller R, Lingens F. Enzymatic dehalogenation of chlorinated nitroaromatic compounds. Appl Environ Microbiol. 1988;54:1199–1202. doi: 10.1128/aem.54.5.1199-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meer J R. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek Int J Microbiol. 1997;71:159–178. doi: 10.1023/a:1000166400935. [DOI] [PubMed] [Google Scholar]
- 44.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Meer J R, Eggen R I L, Zehnder A J B, de Vos W M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991;173:2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer J R, Roelofsen W, Schraa G, Zehnder A J B. Degradation of low concentrations of dichlorobenzenes and 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 in nonsterile soil columns. FEMS Microbiol Ecol. 1987;45:333–341. [Google Scholar]
- 47.van der Meer J R, van Neerven A R W, de Vries E J, de Vos W M, Zehnder A J B. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Meer J R, Zehnder A J B, de Vos W M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werlen C, Kohler H-P E, van der Meer J R. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J Biol Chem. 1996;271:4009–4016. doi: 10.1074/jbc.271.8.4009. [DOI] [PubMed] [Google Scholar]
- 50.Wyndham R C, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 51.Wyndham R C, Straus N A. Chlorobenzoate catabolism and interactions between Alcaligenes and Pseudomonas species from Bloody Run Creek. Arch Microbiol. 1988;150:230–236. doi: 10.1007/BF00407785. [DOI] [PubMed] [Google Scholar]
- 52.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todClC2BADE genes and their expression in Echerichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]