Abstract
Epitopes recognized by T Cells are a collection of short peptide fragments derived from specific antigens or proteins. Immunological research to study T cell responses is hindered by the extreme degree of heterogeneity of epitope targets, which are usually derived from multiple antigens; within a given antigen, hundreds of different T cell epitopes can be recognized, differing from one individual to the next because T cell epitope recognition is restricted by their ability to bind to major histocompatibility complex (MHC) molecules, which are extremely polymorphic in different individuals. Testing large pools encompassing hundreds of peptides is technically challenging because of logistic consideration ad solvent-induced toxicity. To address this issue, we developed the MegaPool (MP) approach based on sequential lyophilization of large numbers of peptides that can be used in a variety of assays to measure T cell responses, including ELISPOT, Intracellular Cytokine Staining (ICS), and Activation Induced Marker (AIM) assays and have been validated in the study of infectious diseases, allergies, and autoimmunity. Here we describe the procedures for generating and testing “MegaPools”, starting with peptide synthesis and lyophilization, as well as guidelines and recommendations for their handling and experimental usage. Overall, the MP approach is a powerful strategy for studying T cell responses and understanding the immune system’s role in health and disease.
Basic Protocol 1: A method for the generation of peptide pools (“MegaPools”)
Basic Protocol 2: MegaPool testing and quantitation of antigen-specific T Cell responses
Keywords: AIM, Epitopes, Infectious diseases, Lyophilization, T cell, SARS-CoV-2
Introduction
T cell responses are key elements of the body’s reaction to vaccination and infection and have likely evolved to provide a synergistic line of defense alongside antibody responses. T cells recognize epitopes, which are peptide fragments derived from the cellular processing of protein antigens, through their T cell receptors (TCRs), which are highly specialized proteins on the surface of T cells (Punt, Stranford, Jones, & Owen 2018). T cells can be grouped into a series of subsets based on their function or protein/gene expression. There are several types of T cells, with the most prevalent being CD4+ T cells, also known as helper T cells, and CD8+ T cells, which are referred to as cytotoxic T cells or killer T cells.
Because of their key role in adaptive immunity, measuring T cell responses is an important component of vaccine and diagnostic evaluations, in immunotherapy applications and in the general study on immunopathology. The most common and well-known methodologies to measure antigen-specific T cell responses are cytokine-based assays (e.g. IGRA, ELISPOT and Intracellular Cytokine Staining (ICS)) (Tian et al., 2018), Tetramer/multimer staining assays (Klenerman, Cerundolo, & Dunbar, 2002), and Activation Induced Marker (AIM) assays, which are agnostic towards particular cytokine functionality (Dan et al., 2016; Poloni et al., 2023) (see: Basic Protocol 2 section). The different approaches represent the fundamental tensions that exist between ease of performance, robustness, cost and throughput, on the one hand, and physiological relevance,, complexity, depth of analysis and granularity of information on the other. A comprehensive discussion of the available techniques is beyond the scope of the present protocol and some recent reviews have discussed the topic (Gondre-Lewis et al., 2023; Peters, Nielsen, & Sette, 2020; Sidney, Peters, & Sette, 2020).
In the following protocol, we will describe the generation and use of specialized reagents for probing and characterizing both CD4+ and CD8+ T cell responses. These reagents allow for the efficient elucidation of adaptive T cell immunity against any target of known sequence, and in the context of infectious disease, cancer, allergy and autoimmunity. With the capacity to probe large sequences, the regents described below are of value for use in epitope discovery, crucial for vaccine design, and diagnostic purposes.
Human T cell responses generally recognize multiple epitopes restricted by a diverse set of HLA molecules
Measurement of human T cell responses has to contend with the large diversity of epitope targets recognized (Livingstone & Fathman, 1987; Peters et al., 2020). This diversity originates from two main interdependent sources: the diversity of the antigens recognized, and the diversity of HLA molecules, which bind and present epitopes recognized by T cells. Importantly, this diversity is consequential because responses to different antigens and epitopes can be directly related to the preservation of T cell responses, the counteracting of immune escape, or associated with differential disease outcomes (Dillon et al., 2015; Schulten, Westernberg, et al., 2018) (Grifoni & Sette, 2022).
The genes encoding HLA molecules are polygenic and highly polymorphic. In humans, there are three main class I loci (A, B and C) and several different class II loci (DRA, DRB1, DRB3/4/5, DPA, DPB and DQA and DQB), resulting in the expression of four different types of HLA class II heterodimers. Because of heterozygosity and the high degree of HLA polymorphism, each individual can express up to 14 different HLA molecules (8 HLA class II and 6 HLA class I). Given that each HLA molecule binds a different set of peptide specificities, HLA polygeny and polymorphism is a powerful force, amplifying the diversity of epitope repertoires recognized in humans (for review, see e.g., (Little & Parham, 1999; Madden, 1995; Parham, 1988; Sidney, Peters, Frahm, Brander, & Sette, 2008; Stevanovic, 2002).
At the same time, this large diversity poses unique challenges for approaches to measure T cell responses (McKinney et al., 2013; Nilsson, Grifoni, Tarke, Sette, & Nielsen, 2021; Sidney et al., 2020). Utilizing just a handful of epitopes and HLA molecules will most likely result in biased and incomplete coverage of the T cell responses, and provide a skewed and incomplete assessment of responses. This situation is made more complex by the fact that different HLA variants are expressed at different frequencies in different ethnic groups and geographic regions (Gonzalez-Galarza et al., 2020). Perhaps unsurprisingly, epitopes presented by HLA molecules that are the most frequent in Caucasians populations, are the ones that have been most highly characterized to date in the literature. This ethnic bias in coverage, a result of approaches utilizing few epitope specificities, has been repeatedly noted, and is a serious limitation that needs to be addressed in developing approaches to study T cell responses in human populations (Sette, Chesnut, Livingston, Wilson, & Newman, 2000; Sette et al., 2001; Sidney et al., 2020).
The MegaPool (MP) approach
To address several of the concerns discussed above, including the need to probe for responses to a wide breadth of epitopes, and in the context of diverse populations, our group developed the MegaPool (MP) approach (Carrasco Pro et al., 2015; Sidney et al., 2020). This approach, based on testing large pools of peptides and/or epitopes, provides an efficient means for comprehensive analysis of T cell responses in virtually any donor and has many advantages compared to other competing technologies (Table 1). To provide a comprehensive evaluation of diverse targets in a diverse population, it may be necessary to test peptides pools that can encompass several hundred different peptides. However, pooling such large numbers of peptides can result in a stock solution that is relatively diluted when each separate peptide is considered. Also, requiring the addition of a relatively large amount of solvent (such as DMSO) into test cell cultures will contribute to cell toxicity, if a sufficiently high enough amount of each peptide is required (de Abreu Costa et al., 2017; Verheijen et al., 2019). To overcome this limitation, we developed a sequential lyophilization approach that achieves much higher concentrations of each peptide in viable amounts of solvent, thus eliminating or reducing DMSO (or other solvent) toxicity (see: Basic Protocol 1 section).
Table 1.
MegaPool approach comparison with competing technologies
| Other technologies | Advantages | Limitations |
|---|---|---|
|
| ||
| MegaPool approach | • Exhaustive HLA coverage • Comprehensive coverage of diverse and broad epitope repertoire • Enhanced specificity • Ease and speed of production • Allows ex vivo assessment bypassing proteolysis • Broad applicability (i.e. AIM, ICS, ELISPOT assays using either PBMCs or whole blood) |
• Epitope Masking • Limited Information on fine specificity • Requires specialized equipment |
| Multimers/tetramers | • Allows ex vivo assessment bypassing proteolysis • Enhanced specificity • Enhanced sensitivity |
• Need for predefinition of exact epitope and HLA restriction • Increased complexity/need for specialized reagents • Higher cost • Limited applicability, throughput and repertoire |
| Use of whole organism (Live pathogens, cell lysates, supernatant or extracts) | • Broad range of antigens • Native conformation of antigens • Allow measuring post-translationally modified epitopes |
• Potential toxicity/biosafety issues • Cell lysates do not contain non-structural proteins or secreted antigens • Requires longer time for antigen-processing • Complexity and heterogeneity • Limited applicability |
|
| ||
| Antigens (Recombinant or purified) | • Allow measuring post-translationally modified epitopes • Reduced complexity and heterogeneity |
• Potential toxicity/biosafety issues if protein is a toxin or has immunomodulatory effects • Requires longer time for antigen-processing • Limited applicability |
There are three main approaches to generate the MPs that we routinely utilize in our research. The first corresponds to the use of sets of overlapping peptides spanning the entire sequence of an antigen(s) of interest (Maecker et al., 2001). The second is based on assembling sets of predicted HLA binding epitopes (Peters et al., 2020). Finally, sets can be assembled to consider previously identified and experimentally defined epitopes (Dhanda et al., 2019). Each of these approaches is associated with distinctive advantages and disadvantages.
The use of overlapping peptides is the most comprehensive and unbiased, as it does not rely on any predictive algorithm, or a priori knowledge of which epitopes are recognized in a given human population and is irrespective of known HLA phenotype. By selecting a 15-mers size overlapping by 10 amino acids, it further allows to simultaneously assess CD4+ and CD8+ T cell responses in flow cytometry approaches by setting appropriate gating on the responding T cell populations. By definition, however, this approach requires the highest number of peptides, flow-cytometry based techniques, which are less high-throughput and cost-effective, and consequently, the highest number of cells required for testing if individual epitope identification is further desired.
MPs based on predictive approaches allow to reduce the number of peptides tested. However, this approach requires careful consideration of HLA polymorphism to ensure good and appropriate population coverage. Furthermore, this approach requires separate pools for CD4+ and CD8+ T cell detection, and will also detect a fraction of the total response, depending on the comprehensiveness of the HLA predictions and allele coverage. Additionally, there is the intrinsic limitation that predictive algorithms, while highly effective and accurate in most cases, are not 100% sensitive and specific, meaning that some epitopes may be missed.
Finally, MPs based on curated epitopes are effective, since they include epitopes experimentally shown to be recognized by T cells. Epitope sequences can be retrieved by using web databases such as IEDB (https://www.iedb.org). The main limitation of this approach is that, by definition, it relies on the assumption that the epitope repertoire associated with a given indication has already been thoroughly defined in multiple HLA alleles and accounting for diverse ethnic backgrounds representative of the general worldwide population.
In general, MPs are envisioned for determining antigen-specific T cell responses for any particular indication, such as but not limited to allergies, autoimmunity, and bacterial or viral infectious diseases. However, MPs designed with overlapping peptides spanning an individual antigen or a combination of several could be initially used to screen for donor responsiveness, and further introduced into an epitope screening pipeline, and sequentially deconvoluted to map individual peptide CD4+ T cell reactivity. This multi-step approach has been recently employed for genome-wide screening and epitope identification of SARS-CoV-2, common cold coronaviruses, and Bordetella pertussis (da Silva Antunes et al., 2023; Tarke, Sidney, Kidd, et al., 2021; Tarke et al., 2023).
In the following sections, we will give a brief account of the main MPs we have defined to probing and characterizing both CD4+ and CD8+ T cell responses for several different indications (Table 2). These pools exemplify each of the considerations above, and demonstrate how they can be efficient tools for characterizing immune responses in a wide range of different immunological contexts.
Table 2.
List of MPs developed and targets of responses
| Indication | Pathogen/Organism | Pool Name | Source | Peptide/Epitope type | Number of peptides | Reference |
|---|---|---|---|---|---|---|
|
| ||||||
| Infectious diseases (Viruses) | Cytomegalovirus | CD4 CMV | Whole proteome | Experimentally defined | 313 | Carrasco Pro et al., 2015 (Carrasco Pro et al., 2015) |
| Cytomegalovirus | CD4 CMV235 | Whole proteome | Experimentally defined | 235 | Dhanwani et al., 2021 (Dhanwani et al., 2021) | |
| Cytomegalovirus | CD4 CMV | Whole proteome | Experimentally defined | 198 | Carrasco Pro et al., 2015 (Carrasco Pro et al., 2015) | |
| Dengue Virus | DENV_CD8 | Whole proteome | Predicted and experimentally defined | 268 | Grifoni et al., 2017 (Grifoni, Angelo, Lopez, et al., 2017) | |
| Dengue virus | DENV_CD4 | Whole proteome | Experimentally defined | 180 | Grifoni et al., 2017 (Grifoni, Angelo, Lopez, et al., 2017) | |
| Dengue/Zika virus cross reactive | DENV/ZIKV_CR | Whole proteome | Experimentally defined | 94 | Schoeust et al, 2021 (Schouest et al., 2021) | |
| Ectromelia, Vaccinia and Variola virus | OP-CD4-E MP | Whole proteome | Experimentally defined | 300 | Grifoni et al., 2022 (Grifoni et al., 2022) | |
| Epstein-Barr virus | CD8 EBV | Whole proteome | Experimentally defined | 218 | Carrasco Pro et al., 2015 (Carrasco Pro et al., 2015) | |
| Epstein-Barr virus | CD4 EBV | Whole proteome | Experimentally defined | 83 | Dan et al., 2016 (Dan et al., 2016) | |
| HCoV-229E | 229E | Whole proteome | Spike (S) Overlapping/ Rest of proteome (R) Predicted | 225 | da Silva Antunes et al., 2021 (da Silva Antunes, Pallikkuth, et al., 2021) | |
| HCoV-HKU1 | HKU1 | Whole proteome | Spike (S) Overlapping/ Rest of proteome (R) Predicted | 320 | da Silva Antunes et al., 2021 (da Silva Antunes, Pallikkuth, et al., 2021) | |
| HCoV-NL63 | NL63 | Whole proteome | Spike (S) Overlapping/ Rest of proteome (R) Predicted | 280 | da Silva Antunes et al., 2021 (da Silva Antunes, Pallikkuth, et al., 2021) | |
| HCoV-OC43 | OC43 | Whole proteome | Spike (S) Overlapping/ Rest of proteome (R) Predicted | 294 | da Silva Antunes et al., 2021 (da Silva Antunes, Pallikkuth, et al., 2021) | |
| Human immunodeficiency virus | HIV CD4 | Whole proteome | Experimentally defined | 164 | Al-Kolla et al., 2022 (Al-Kolla et al., 2022) | |
| Human immunodeficiency virus | HIV CD8 | Whole proteome | Experimentally defined | 187 | Al-Kolla et al., 2022 (Al-Kolla et al., 2022) | |
| Influenza A | HA-Influenza MP | Hemagglutinin (HA) | Experimentally defined/Predicted | 161 | Meckiff et al., 2020 (Meckiff et al., 2020) | |
| Influenza A | Flu-Other | Non-Hemagglutinin proteome | Experimentally defined | 169 | Yu et al. 2021 (Yu, Grifoni, et al., 2021) | |
| Influenza A | flu-CD8 | Whole proteome | Experimentally defined | 400 | Poon et al., 2021 (Poon, Byington, et al., 2021) | |
| Japanese encephalitis virus | JEV CD4 | Whole proteome | Predicted | 239 | Grifoni et al., 2020 (Grifoni, Voic, et al., 2020) | |
| Japanese encephalitis virus | JEV CD8 | Whole proteome | Predicted | 310 | Grifoni et al., 2020 (Grifoni, Voic, et al., 2020) | |
| Metapneumovirus | HMPV | Whole proteome | Predicted | 107 | Meckiff et al., 2020 (Meckiff et al., 2020) | |
| Monkeypox virus | MPx-CD4-P MP | Whole proteome | Predicted | 276 | Grifoni et al., 2022 (Grifoni et al., 2022) | |
| Monkeypox virus | MPx-CD8-P P1-P5 MP | Whole proteome | Predicted | 1647 | Grifoni et al., 2022 (Grifoni et al., 2022) | |
| Parainfluenza virus | HPIV | Whole proteome | Predicted | 256 | Meckiff et al., 2020 (Meckiff et al., 2020) | |
| Respiratory syncytial virus | RSV | Whole proteome | Experimentally defined | 216 | Yu et al. 2022 (Yu, Narowski, et al., 2022) | |
| Rhinovirus | Rhinovirus | Whole proteome | Experimentally defined | 136 | Grifoni et al., 2019 (Grifoni, Mahajan, et al., 2019) | |
| SARS-CoV-2 | CD4RE | Non-spike proteome | Experimentally defined | 284 | Grifoni et al., 2021 (Grifoni et al., 2021) | |
| SARS-CoV-2 | CD8RE | Non-spike proteome | Experimentally defined | 621 | Grifoni et al., 2021 (Grifoni et al., 2021) | |
| SARS-CoV-2 | CD8A | Whole proteome | Predicted | 314 | Grifoni et al., 2021 (Grifoni et al., 2021) | |
| SARS-CoV-2 | CD8B | Whole proteome | Predicted | 314 | Grifoni et al., 2021 (Grifoni et al., 2021) | |
| SARS-CoV-2 | CD4R | Non-spike proteome | Predicted | 221 | Grifoni et al., 2020 (Grifoni, Sidney, et al., 2020) | |
| SARS-CoV-2 | SARS2 | Whole proteome | Spike (S) Overlapping/ Rest of proteome (R) Predicted | 474 | da Silva Antunes et al., 2021 (da Silva Antunes, Pallikkuth, et al., 2021) | |
| SARS-CoV-2 | CD4E | Whole proteome | Experimentally defined | 280 | Tarke et al., 2021 (Tarke, Sidney, Kidd, et al., 2021) | |
| SARS-CoV-2 | CD8E | Whole proteome | Experimentally defined | 454 | Tarke et al., 2021 (Tarke, Sidney, Kidd, et al., 2021) | |
| Vaccinia and Variola virus | OP-CD8-E MP | Whole proteome | Experimentally defined | 238 | Grifoni et al., 2022 (Grifoni et al., 2022) | |
| Varicella zoster virus | VZV | Whole proteome | Glycoprotein E (gE) Experimemntaly definied / Rest of proteome (R) Predicted | 335 | Voic et al., 2020 (Voic et al., 2020) | |
| West Nile | WNV CD4 | Whole proteome | Predicted | 244 | Grifoni et al., 2020 (Grifoni, Voic, et al., 2020) | |
| West Nile | WNV CD8 | Whole proteome | Predicted | 324 | Grifoni et al., 2020 (Grifoni, Voic, et al., 2020) | |
| Yellow Fever | CD4_YF; CD4_YF_rev | Whole proteome | Predicted and experimentally defined | 215;275 | Mateus et al, 2020; Grifoni et al., 2020 (Grifoni, Voic, et al., 2020; Mateus, Grifoni, Voic, et al., 2020) | |
| Yellow Fever | YF_CD8 | Whole proteome | Predicted | 368 | Grifoni et al., 2020 (Grifoni, Voic, et al., 2020) | |
| Zika virus | ZIKV CD4 | Whole proteome | Predicted | 209 | Grifoni et al., 2017; Grifoni et al., 2020 (Grifoni, Pham, et al., 2017; Grifoni, Voic, et al., 2020) | |
| Zika virus | ZIKV CD8 | whole proteome | Predicted | 309 | Grifoni et al., 2017; Grifoni et al., 2020 (Grifoni, Pham, et al., 2017; Grifoni, Voic, et al., 2020) | |
|
| ||||||
| Infectious diseases (Bacteria) | Bordetella pertussis | BP(E)VAC | Acelullar vaccine antigens (FHA, Fim2/3, PRN and PtTox) | Experimentally defined | 132 | Dan et al., 2016 (Dan et al., 2016) |
| Bordetella pertussis | BP(E)R | Non-vaccine antigens proteome | Experimentally defined | 170 | da Silva Antunes et al., 2023 (da Silva Antunes et al., 2023) | |
| Clostridium tetani | TT | Tetanus toxoid | Experimentally defined | 125 | da Silva Antunes et al., 2017 (da Silva Antunes et al., 2017) | |
| Mycobacterium avium | MAC-specific pool 1–3 | Whole proteome | Predicted | 628 | Lindestam Arlehamn et al., 2022 (Lindestam Arlehamn et al., 2022) | |
| Mycobacterium avium and Mycobacterium tuberculosis | MAC/Mtb-specific pool 1–3 | Whole proteome | Predicted | 440 | Lindestam Arlehamn et al., 2022 (Lindestam Arlehamn et al., 2022) | |
| Mycobacterium tuberculosis | MTB300 | Whole proteome | Experimentally defined | 300 | Lindestam Arlehamn et al., 2016 (Lindestam Arlehamn et al., 2016) | |
| Mycobacterium tuberculosis | Type 1 | Whole proteome | Experimentally defined | 113 | Scriba et al., 2017 (Scriba et al., 2017) | |
| Mycobacterium tuberculosis | Type 2 | Whole proteome | Experimentally defined | 122 | Scriba et al., 2017 (Scriba et al., 2017) | |
| Mycobacterium tuberculosis | MTBCD8 | Whole proteome | Experimentally defined | 113 | Pomaznoy et al., 2020 (Pomaznoy et al., 2020) | |
| Mycobacterium tuberculosis | ATB116 | Whole proteome | Experimentally defined | 116 | Panda et al., 2023 (pre-print) | |
| Non-tuberculous mycobacteria | NTM-specific pool 1–3 | Whole proteome | Predicted | 516 | Lindestam Arlehamn et al., 2022 (Lindestam Arlehamn et al., 2022) | |
|
| ||||||
| Allergy | Cockroach | CR | Major cockroach allergens | Experimentally defined | 228 | Schulten et al., 2019 (Schulten et al., 2019) |
| House Dust Mite | HDM | Major house dust mite allergens | Experimentally defined | 75 | Hinz et al., 2015 (Hinz et al., 2015) | |
| Mouse | LoMo | Mouse urinary oligopeptides (Low molecular weight fraction) | Experimentally defined | 225 | da Silva Antunes et al., 2018 (da Silva Antunes, Pham, et al., 2018) | |
| Mouse | HiMo | High molecular weight fraction of mouse allergen extracts | Experimentally defined | 106 | Schulten et al., 2018 (Schulten, Westernberg, et al., 2018) | |
| Timothy Grass | TG P20 | Major TG allergens | Experimentally defined | 20 | Schulten et al., 2013 (Schulten et al., 2013) | |
| Timothy Grass | PUTGA P19 | Non-dominant TG allergens | Experimentally defined | 19 | Schulten et al., 2013 (Schulten et al., 2013) | |
|
| ||||||
| Cow Milk | MT111 | Cow milk extract | Experimentally defined | 111 | Lewis et al., 2023 (Lewis et al., 2023) | |
|
| ||||||
| Autoimmuniy | Self | 𝛼-syn | Alpha-Synuclein | Experimentally defined | 13 | Lindestam Arlehamn et al., 2020 (Lindestam Arlehamn et al., 2020) |
| Self | APOB | Apolipoprotein B | Experimentally defined | 20 | Roy et al., 2022 (Roy et al., 2022) | |
Allergens
MPs have been used extensively to characterize allergen specific T cell responses. The first use of MPs was associated with the characterization of timothy grass responses, addressing both known and novel allergens (Hinz et al., 2016; Oseroff et al., 2010; Schulten et al., 2013), and was later expanded to a broad collection of allergens (Oseroff, Sidney, Vita, et al., 2012). Further studies investigated Japanese Cedar (Oseroff et al., 2016), Cockroach (Birrueta et al., 2019; da Silva Antunes, Sutherland, et al., 2021; Dillon et al., 2015; Oseroff, Sidney, Tripple, et al., 2012), Dust Mite (Hinz et al., 2015; Seumois et al., 2020), murine (da Silva Antunes, Pham, et al., 2018; Grifoni, da Silva Antunes, et al., 2019; Schulten, Westernberg, et al., 2018) and cow milk (Lewis et al., 2023) allergens.
Some of the salient and original applications of MP include the characterization of T cell responses to antigens identified by immunoproteomic approaches (Oseroff et al., 2017; Schulten et al., 2013), epitopes differentially recognized in different clinical manifestations (asthma versus rhinitis) (Dillon et al., 2015) or different levels of sensitization (Schulten et al., 2019), and detection of T cell responses in non-allergic subjects. Interestingly, by probing these reagents allergic and non-allergic subjects were both found to mount T cell responses, but were associated with different phenotypes (da Silva Antunes, Pham, et al., 2018; Grifoni, da Silva Antunes, et al., 2019; Hinz et al., 2016; Schulten, Westernberg, et al., 2018; Yu, Westernberg, et al., 2021). Moreover, specific mouse-allergen derived epitopes were identified by coupling mass-spectrometry and bioinformatic approaches (da Silva Antunes, Pham, et al., 2018). Dust mite peptide pools were used to define the transcriptional profiles associated with allergic asthma (Seumois et al., 2020), and timothy grass and cockroach specific MPs were used to follow T cell reactivity in the context of allergen specific immunotherapy (Rudman Spergel et al., 2021; Schulten et al., 2016; Schulten, Tripple, et al., 2018; Schulten et al., 2014). Lastly, timothy grass and mouse allergen MPs were used to follow the modulation of T cell responses associated with allergen exposure in non-allergic subjects (Hinz et al., 2016; Yu, Westernberg, et al., 2021) and most recently to reveal recognition of allergen-specific responses by Gamma Delta T cells (Yu, Wang, Garrigan, Sutherland, et al., 2022).
Autoimmunity
MPs have also been utilized in the study of autoimmunity, specifically in the study of neurodegenerative and cardiovascular diseases. In the case of Parkinson’s Disease (PD), MPs derived from alpha-synuclein (Lindestam Arlehamn et al., 2020; Singhania, Pham, et al., 2021; Sulzer et al., 2017), tau (Lindestam Arlehamn et al., 2019), and other neuronal-associated antigens (Dhanwani et al., 2020) have been studied. Similar studies have been conducted in the context of Alzheimer’s Disease (AD). The results thus far indicate an autoimmune component in PD, possibly associated with early disease stages (Lindestam Arlehamn et al., 2020), with enhanced reactivity against alpha-synuclein (Sulzer et al., 2017), and possibly other antigens. No increased reactivity against neuronal antigens has thus far been associated with AD (Dhanwani et al., 2020).
In the case of cardiovascular disease, pools of predicted class II binding peptides derived from apolipoprotein B (APOB) were utilized to identify dominant epitopes, and responses to these APOB epitopes correlated with coronary artery disease severity (Roy et al., 2022). In both neurodegenerative and cardiovascular disease applications, an initial restimulation step was necessary to detect antigen-specific responses, potentially reflective of lower frequency of autoimmune T cells compared to other indications.
Bacterial antigens
Several studies have described MPs encompassing different bacterial targets. The first target was MTB, where a genome-wide screen revealed an unprecedented breadth of responses targeting many known and novel antigens (Lindestam Arlehamn et al., 2013). This resulted in the development of the MTB300 MP (Lindestam Arlehamn et al., 2016), which was utilized in over 14 different studies since 2020 (Chihab et al., 2023; Day et al., 2021; Du Bruyn et al., 2022; Du Bruyn et al., 2021; Foreman et al., 2022; Hoft et al., 2023; Kauffman et al., 2021; Ogongo et al., 2021; Patankar et al., 2020; Pomaznoy et al., 2020; Riou et al., 2020; Riou et al., 2021; Robison et al., 2021; Sakai et al., 2021; White et al., 2021; Wood et al., 2020; Woodworth et al., 2021). In addition to being used primarily for the characterization of human CD4+ T cell reactivity, this pool has also been used, based on the similarity of peptide binding repertoires, to probe reactivity in non-human primates (Foreman et al., 2022; Kauffman et al., 2021; Mothe et al., 2015; Sakai et al., 2021; White et al., 2021; Wood et al., 2020), and mice (Patankar et al., 2020). Additional MPs were designed to explore the relationship with conservation in other Mycobacteria (Lindestam Arlehamn et al., 2022), to perform a quantitative analysis of TCR and epitope repertoire composition (Glanville et al., 2017; Scriba et al., 2017), to study specific gene deficiencies in humans (Kong et al., 2018; Martinez-Barricarte et al., 2018), to study the role of CD8+ T cell responses (Pomaznoy et al., 2020), and to derive immune signatures associated with transcriptional profiles and different disease outcomes (Singhania, Dubelko, et al., 2021).
Bordetella pertussis is another target where MPs have been described and validated (da Silva Antunes, Babor, et al., 2018; da Silva Antunes et al., 2023; da Silva Antunes, Quiambao, et al., 2021; da Silva Antunes et al., 2020). Briefly, a series of studies defined epitopes encoded in the antigens included in the acellular Pertussis (aP) vaccine currently in use (Bancroft et al., 2016). These epitopes were used to generate MPs that revealed a long-lasting polarization of responses as a function of the original priming, and unveiled transcriptomic profiles associated with human CD4+ T cell responses to vaccine antigens (da Silva Antunes, Babor, et al., 2018), in addition to clonality assessment of TCR repertoires (Singhania, Pham, et al., 2021). Recently, a whole-genome screening of Bordetella pertussis revealed a highly diverse T cell repertoire and identified epitopes derived from antigens not included in the aP vaccine (da Silva Antunes et al., 2023), which resulted in the development of several MPs that aid the characterization of CD4+ T cell responses to these antigens. Additional studies described and validated CD4+ T cell human epitopes from the Tetanus toxoid protein, which is also in the multivalent Tetanus Diphtheria acellular Pertussis (TDaP) vaccine (da Silva Antunes et al., 2017).
MPs for Flaviviruses
In terms of application of the MP approach to viral targets, early efforts focused on flaviviruses, and in particular on dengue virus (DENV). Initial studies using predicted and experimentally confirmed epitopes spanning the entire proteome of the four DENV serotypes defined a wealth of CD4+ and CD8+ epitopes recognized by T cells derived from individuals previously infected with DENV from different endemic areas, such as Sri Lanka and Nicaragua (Grifoni, Angelo, Lopez, et al., 2017; Grifoni, Moore, et al., 2019; Tian, Grifoni, Sette, & Weiskopf, 2019; Weiskopf et al., 2015; Weiskopf et al., 2016; Weiskopf et al., 2014) and participants in vaccine studies utilizing a live attenuated vaccine (Angelo et al., 2017), a challenge model (Grifoni, Angelo, Sidney, et al., 2017), as well as HLA transgenic mice (Weiskopf et al., 2011). This led to the generation of MPs of experimentally defined epitopes covering both CD4+ and CD8+ T cell responses, which were validated in endemic different populations such as Sri Lanka, Nicaragua, India and Brazil (Grifoni, Angelo, Lopez, et al., 2017; Weiskopf et al., 2015).
The analysis of T cell responses associated with infection and vaccination with different flaviviruses was further expanded to other members of this viral family, such as Yellow Fever (YF), Japanese encephalomyelitis (JEV), West Nile virus (WNV) and Zika virus (ZIKV) (Grifoni, Pham, et al., 2017; Grifoni, Voic, et al., 2020; Mateus, Grifoni, Voic, et al., 2020). These pools were utilized to broadly describe patterns of reactivity and cross-reactivity (Grifoni, Pham, et al., 2017; Schouest et al., 2021). Overall, the results illustrated how the MP approach is broadly applicable to viral infection and vaccination targets.
SARS-CoV-2 and coronaviruses
MPs have played a key role in the study and elucidation of T cell responses to SARS-CoV-2 in the context of infection and vaccination. The first description of CD4+ and CD8+ T cell responses in the context of convalescent individuals defined the key characteristics of a successful immune response utilizing MPs of overlapping peptides spanning the entire proteome, but also MPs based on the use of bioinformatically predicted epitopes (Grifoni, Sidney, et al., 2020; Grifoni, Weiskopf, et al., 2020). These studies highlighted, as previously described for other systems, that MPs based on predicted HLA binders are remarkably effective, and recapitulate at least 50% of the total response, while requiring substantially less peptides and cells for the analysis (Grifoni, Weiskopf, et al., 2020).
These SARS-CoV-2 pools were made broadly available to the scientific community to over 110 laboratories in four different continents, and were utilized in a large number of studies, resulting thus far in over 80 publications (Ansari et al., 2021; Ansari et al., 2022; Apostolidis et al., 2021; Arunachalam et al., 2022; Banki et al., 2022; Bhuiyan et al., 2022; Blixt et al., 2022; Boland et al., 2022; Bosteels et al., 2022; Bowen, Addetia, et al., 2022; Bowen, Park, et al., 2022; Brasu et al., 2022; Bueno et al., 2022; Cheon et al., 2021; Chiuppesi et al., 2022; Costa et al., 2022; da Silva Antunes, Pallikkuth, et al., 2021; Dan et al., 2021; Dentone et al., 2022; Galvez et al., 2022; Gao, Cai, Grifoni, et al., 2022; Gao, Cai, Wullimann, et al., 2022; Garcia-Valtanen et al., 2022; Gazzinelli-Guimaraes et al., 2022; Geers et al., 2023; GeurtsvanKessel et al., 2022; Goel et al., 2021; Grifoni et al., 2021; Grifoni, Sidney, et al., 2020; Grifoni, Weiskopf, et al., 2020; Hassan et al., 2020; He et al., 2022; Hsieh et al., 2022; Jin et al., 2021; Keeton et al., 2021; Keeton et al., 2022; Keeton et al., 2023; Lederer et al., 2022; Madelon, Heikkila, et al., 2022; Madelon, Lauper, et al., 2022; Mateus et al., 2021; Mateus, Grifoni, Tarke, et al., 2020; Meckiff et al., 2020; Mele et al., 2021; Melo-Gonzalez et al., 2021; Murugesan et al., 2021; Nelson et al., 2022; Ogbe et al., 2021; Painter et al., 2021; Paul et al., 2022; Peluso et al., 2022; Perez-Gomez et al., 2022; Petrone et al., 2021; Petrone, Picchianti-Diamanti, et al., 2022; Petrone, Tortorella, et al., 2022; Pino et al., 2021; Poon, Byington, et al., 2021; Poon, Rybkina, et al., 2021; Premkumar et al., 2020; Ramirez et al., 2022; Rydyznski Moderbacher et al., 2022; Rydyznski Moderbacher et al., 2020; Schultz et al., 2022; Shaan Lakshmanappa et al., 2021; Singh et al., 2022; Soto et al., 2022; Sun et al., 2022; Tarke, Coelho, et al., 2022; Tarke, Potesta, et al., 2022; Tarke, Sidney, Kidd, et al., 2021; Tarke, Sidney, Methot, et al., 2021; Ukey et al., 2022; Valencia et al., 2022; Van Damme et al., 2020; Vikkurthi et al., 2022; Weiskopf et al., 2020; Williams et al., 2023; Yu, Narowski, et al., 2022; Yu, Wang, Garrigan, Goodwin, et al., 2022; Zhang et al., 2022). While an in-depth review is beyond the scope of this report, the MPs helped clarify a diverse set of topics and concerns related to SARS-CoV-2, including elucidation of responses in the acute phase of infection, responses to vaccination, breakthrough infections, kinetics and features of responses in the memory phase, responses in vulnerable and immunocompromised individuals, health care workers, and responses in children.
Several additional insights of SARS-CoV-2 infection were also addressed with specific MPs. These include the demonstration of cross-reactive pre-existing memory T cell responses (Mateus, Grifoni, Tarke, et al., 2020), and the demonstration that both CD4+ and CD8+ T cell responses are remarkably preserved in the context of the different variants that originate throughout the pandemic (Tarke, Coelho, et al., 2022; Tarke, Sidney, Methot, et al., 2021). Additional MPs were subsequently generated, based on experimentally defined epitopes, and used to derived immunodiagnostic strategies to address vaccination and infection history (Grifoni et al., 2021; Tarke, Sidney, Kidd, et al., 2021; Yu, Wang, Garrigan, Goodwin, et al., 2022). In parallel, specific MPs were also derived to follow responses to other coronavirus species, such as the main common cold coronaviruses (da Silva Antunes, Pallikkuth, et al., 2021; Tarke et al., 2023; Yu, Narowski, et al., 2022).
Broad application to study responses to human viral pathogens
While perhaps flaviviruses and SARS-CoV-2 represent “poster child” applications of MPs in the study of viral disease, several additional viral indications have been addressed by the MP approach. These include influenza, where pools addressing different subtypes and antigens have been validated (Meckiff et al., 2020; Poon, Byington, et al., 2021; Yu, Grifoni, et al., 2021), and other common respiratory viruses such as Rhinovirus (Grifoni, Mahajan, et al., 2019), Metapneumovirus (HMPV) (Meckiff et al., 2020), Parainfluenza virus (HPIV) (Meckiff et al., 2020), and Respiratory syncytial virus (RSV) (Yu, Narowski, et al., 2022), as well as herpesviruses such as Epstein-Barr Virus (EBV) (Carrasco Pro et al., 2015; Dan et al., 2016), Cytomegalovirus (CMV) (Carrasco Pro et al., 2015; Dhanwani et al., 2021), and Varicella Zoster Virus (VZV) (Voic et al., 2020). Recently, MPs were developed and validated for detection of HIV-specific and CD4+ and CD8+ T cells (Al-Kolla et al., 2022). Murine poxvirus epitopes were first described in the mid 2000s providing rational for the characterization of smallpox vaccines (Pasquetto et al., 2005; Tscharke et al., 2005), followed more recently by the design and validation of specific MPs broadly encompassing orthopox and MPOX-derived epitopes for monitoring of infection and vaccination (Grifoni et al., 2022).
BASIC PROTOCOL 1: A method for the generation of peptide pools (“MegaPools”)
This section describes guidelines on how to produce MPs. The initial step encompasses synthesis of peptide sets that are produced and lyophilized, as common practice in the field. They can be purchased from any supplier. However, all MPs depicted in this manuscript were generated from peptides synthesized by TC peptide lab or Mimotopes. Each peptide is then solubilized, and a pool generated by combining equivalent amounts of each solubilized peptide. The resulting pool is then re-lyophilized. If necessary, the procedure is repeated until a white, reasonably fluffy, powder (lyocake) is obtained. Any synthesized peptides can be pooled for use into a MP. Generally, given the high cost of large sets of peptides, purified preparations are not necessary and the of use crude peptides is acceptable. In the vast majority of cases these will achieve >70% purity. Further, large scale synthesis of crude peptide can usually be obtained in microtube racks, which allow efficient dilution of peptide stocks using, for example, multi-channel pipettors. Purified peptides are excellent if resources are available, or use necessitates. The final MP amount required should be calculated based on the number of samples or assays that need to be tested, but we recommend starting this protocol with 1–2mg per individual peptide.
Materials
Peptides (TC peptide lab (San Diego, CA) or Mimotopes (Victoria, Australia).
Falcon Conical Tubes, 15 and 50mL
Fast - Freeze™ Flask Adaptors ¾” to ¾” (Labconco® Cat# 7457200)
Fast - Freeze™ Flask 900 mL (Labconco® Cat# 7540901)
Fast - Freeze™ Tube Holder 50mL (Labconco® Cat# 7379500)
Fast - Freeze™ Filter Paper 50mL (Labconco®A-75448)
FreeZone® Benchtop Freeze Dryer, 4.5L Capacity (Labconco® Cat#720401000)
Peptide preparation and pooling
-
1Resuspend peptides intended for use in MPs at a high concentration (≥20 mgs/ml) in 100% DMSO.
- For some peptides, it may be necessary to reduce concentration and/or use water. Similarly, in a few cases, precipitates might from in the solubilized pools. This can be addressed by further diluting the pools in water.
-
2Pool a uniform and desirable amount of each peptide in a suitable vessel, such as a 15 or 50 ml conical tube.
- To facilitate efficient lyophilization, the final total volume of pooled peptides should not exceed 20% of the capacity of the vessel.
-
3
Dilute pooled sample 2:1 with H2O (i.e., max 15 ml total volume in a 50 ml conical tube).
-
4Place diluted sample in a −80°C freezer for 24 hours.
- Ideally, samples should be frozen at an angle (~45 degrees) to maximize surface area of the liquid sample in the tube. The rack used to hold the pools in the lyophilized will ideally be frozen at the same time, to facilitate maintenance of sample temperature (and prevent thawing) when subsequently moving to the lyophilizer.
Lyophilization
-
5
Operate the lyophilization unit as per the manufacturer’s specifications. If using the 4.5L Capacity Benchtop Freeze Dryer unit, set conditions at −105°C at 0.1–0.01 mBar. Verify that all parameters have been set as prescribed by the manufacturer.
-
6
Remove the accessory drying chamber (manifold) from the collector chamber lid, and using a soft, lint-free cloth or paper towel, wipe the port gasket(s) and sealing surfaces of the drying chamber/manifold and collector chamber lid to remove any dirt or contaminants that could cause a vacuum leak or contaminate your sample.
-
7Check that each sample valve is closed or in the “vent” position, then start the instrument collector and allow the refrigeration system to reach its specified operating temperature (−105°C for our model). Start the vacuum when collector temperature is at −40°C or colder.
- Once the system pressure is at 0.1–0.01 mBar, and the temperature is −105°C, the system is ready for use.
-
8Moving quickly to avoid thawing, retrieve samples from the freezer, and using a 50ml tube holder, place the samples into a pre – chilled Fast - Freeze™ Flask.
- Do not overload the beaker; utilize additional flasks if necessary.
-
9Slowly open the port valves to commence lyophilization.
- Abrupt opening may result in burping the sample against jar filters, or into the instrument, potentially causing cross contamination. Vacuum pressure will temporarily increase, but if the vessel is properly sealed and mounted it will go back down to operating range (e.g., 0.1–0.01 mBar).
-
10
Check sample and system periodically to ensure proper temperature and pressure is maintained, and that the sample has not thawed.
-
11
When the sample has completely dried, close the pressure valve and remove the beaker from the lyophilizer. It may take up to 24 hours to dry a sample, depending on the volume and sample mixture, but most samples are typically dried between 16–18 hours.
-
12Resuspend the sample in water using the same initial volume of water as was used to dilute the original sample (i.e. 2 -parts water to 1-part sample in DMSO), and then refreeze the sample.
- This will facilitate further removal of any left over DMSO.
-
13After the sample is frozen, return it to the freeze dryer, repeating steps 1–8 above, as necessary. When the sample is completely dried, close the valve and carefully remove the flask from the machine.
- A successfully dried product will have a consistent white almost fluffy powder appearance.
MPs resuspension and storage
-
14Resuspend the lyophilized MP in 100% DMSO (SIGMA D2650) down to 1mg/mL/peptide.
- The final concentration of each individual peptide in the MP is uniform as long as the same amount of peptide is pooled, irrespective of the number of peptides contained in each MP. In order to use the MP in cell stimulation assays, the peptides should be resuspended in enough volume to go into solution, but not too concentrated (viscous) for accurate pipetting
-
15Perform additional dilutions with H2O, if concentrations lower than 1mg/mL are necessary.
- Avoid dilution with DMSO to minimize cytotoxicity in cell culture. In this case, it is advised to maintain a 1 mg/ml DMSO mother stock, but dilute smaller aliquots.
-
16
Aliquot diluted pools in small aliquots (default volume aliquots of 25 or 50μL are optimal) to prevent freeze-thaw cycles.
-
17
Store aliquoted MPs at −20°C and inspect overtime for appearance. If routinely precipitation is noted, re- lyophilize the MPs.
BASIC PROTOCOL 2: MegaPool testing and quantitation of antigen-specific T Cell responses
MPs are a complex mixture of multiple peptides and therefore unfeasible to perform mass spectrometry analysis for quality purposes. The assessment of quality of MPs is generally assessed by testing the bioactivity on a T cell assay. This section describes guidelines on how to to assess the quality of MPs and their use in an immunological assay to quantify antigen-specific responses. Specifically, the immunological assay described herein is an Activation Induced Marker (AIM) assay using PBMCs as biological source material. This protocol also describes the steps of thawing and washing cryopreserved PBMCs prior to cell culture and stimulation, and how to perform a flow cytometry staining for membrane markers. This will allow identifying co-stimulatory receptors that are expressed on activated T cells after antigen-specific stimulation with MPs. Although this protocol is focused on an AIM assay, MPs can be used in other assay modalities to assess antigen-specific responses and/or validate MPs quality such as ICS or ELISPOT assays, and by using whole blood instead of PBMCs.
Materials
Peripheral blood mononuclear cells (PBMCs)
Benzonase (Sigma, E8263)
Spike MP, CD4RE MP, CD8RE MP and EBV MP
Phosphate-buffered saline (PBS, GIBCO BRL 10010–023)
HR5 media (see recipe)
MACS Buffer (see recipe)
CD40 antibody (Miltenyi Biotech, 130–094-133)
Conjugated antibodies (see Table 3)
Table 3 –
List of antibodies used in Basic Protocol 2
| Membrane Antibody | Fluorochrome | Clone/Source/catalog |
|---|---|---|
| CD8 | BUV496 | RPA-T8/BD/612942 |
| CD3 | AF532 | UCHT1/Life Tech/58-0038-42 |
| LIVE/DEAD | aqua | eBioscience/65-0866-18 |
| CD14 | V500 | M5E2/BD/561391 |
| CD19 | V500 | HIB19/BD/561121 |
| CD4 | BV605 | RPA-T4/BD/562658 |
| CD40L | PerCP-ef710 | 24-31/eBioscience/46-1548-42 |
| CD69 | PE | FN50/BD/555531 |
| OX40 | PE-Cy7 | Ber-ACT35/Biolegend/350012 |
| CD137 | APC | 4B4-1/Biolegend/309810 |
Laminar flow hood (Labconco Purifier BSC Class II, or equivalent)
Tissue culture microscope (Nikon or equivalent)
Falcon Polypropylene conical tubes and cell culture flasks
96-well round bottom cell culture–treated plate (Grenier Bio-One, cat. no. 655180)
Multichannel pipettes (1000, 200, 20 and 0.5 μl)
ZE5 Flow cytometer (Biorad)
Thawing of PBMCs
-
1
Submerge cryovials with frozen PBMCs into a 37°C water bath. Hold the vials in the bath until all but a tiny bit of ice remains in the vial. Do not allow the cells to warm completely to 37°C.
-
2
Transfer the cells into the cold conical propylene tubes with cold culture medium (10 mL HR5+20 μL benzonase per vial used) and centrifuge the cells at 1200 rpm at 4°C for 7 min.
-
3
Count the cells and determine cell viability.
-
4Add HR5 media to adjust the cell concentration to 10×106 cells/ml.
- If using fresh PBMCs advance to step 5.
Cell Stimulation
-
5Prepare MP stimulation solution at the desirable concentrations. Adjust the volume depending on the number of samples used per each experiment.
- When a new MP is used for the first time, it is advisable to perform a titration with cells known to respond to the stimulus. If it’s not feasible to do a titration, the default recommended final concentrations (for each peptide) are 1 μg/mL for CD4+ and CD8+ MPs, unless indicated differently for each individual MP.
-
6
Plate in 96 well U-bottom, 100μL of stimuli at a 2X concentration.
-
7Plate 100μL of cells at a concentration of 10×106 cells/ml to plate 1×106 cells per each well.
- The assay can be performed by plating a range of 0.5–2×106 cells/well. If lower cells are available, consider removing one of the conditions from the assay. If more than 2×106 cells per condition are available, consider to carry out the experiment in duplicate or triplicate.
-
8
Optional: If the membrane staining will include the activation marker CD40L, then add CD40 antibody in the solution per each donor before plating for stimulation.
-
9
Incubate plate for a total of 24 hours at 37°C/5% CO2.
Flow Cytometry antibody surface staining
-
10To perform the membrane staining after 24 hours stimulation, prepare a cocktail of antibodies as shown in the Table 3.
- Volumes and fluorochromes combinations are optimized for the Biorad ZE5, please change the fluorochrome combinations and perform an additional antibody titration if using a different instrument.
-
11
After stimulation, spin 96 U-bottom plate at 1400 rpm at 4°C for 2 min.
-
12
Wash plate using 200μL PBS at 1400 rpm at 4°C for 2 min.
-
13
Add in each well 100μL of antibody mix.
-
14
Incubate for 30 min at 4°C. Protect from light.
-
15
After incubation, add 100μL MACS buffer and spin plate at 1400 rpm at 4°C for 2 min.
-
16
Wash 2X plate using 100μL MACs buffer at 1400 rpm at 4°C for 2 min.
-
17
Resuspend in 100μL of PBS per 1×106 cells plated (if you plate 0.5×106 cells resuspend in 50μL; if you plate 2×106 cells resuspend in 200μL).
-
18
Acquire samples in a Flow Cytometry machine.
REAGENTS AND SOLUTIONS
HR5 media
1000mL RPMI (Omega RP-21)
50mL Heat Inactivated Human Serum AB (GeminiBio 100–512)
10mL Pen-Strep (GeminiBio 400–109)
10mL L-Glutamine (Glutamax) (Gibco 35050061)
MACS Buffer
500ml PBS pH 7.4 In Vitrogen (#10010–0230)
2.5g BSA Sigma (A-3294) stored at 4°C (Deli cabinet)
2ml 0.5 M EDTA
Filter through .2μ filter and degas for 30 minutes
Store at 4°C
COMMENTARY
Background information
To evaluate the targets of T cell responses to diverse pathogens or allergens, testing peptide pools with hundreds of peptides may be necessary. However, challenges associated with pooling and testing a large number of peptides can hinder the identification and quantification of T cell responses. Here, we developed a sequential lyophilization method to increase peptide concentrations and reduce solvent toxicity for further use in biological assays and assessment of antigen-specific responses such as, but not restricted to, AIM assays.
AIM assays are based on TCR-dependent upregulation of co-stimulatory immune molecules, independent of cytokine response, and have been successfully used in various studies to detect T cells specific to viruses, bacteria or allergens. In this particular protocol, we are using as example SARS-CoV-2 MPs (see section below: Understanding results), which have been applied in more than 80 studies thus far, and OX40, 4–1BB and CD69 as activation markers. In this assay, CD4+ T cell responses are measured utilizing OX40+CD137+ dual marker combination and CD8+ T cell responses measured utilizing 4–1BB+CD137+ dual marker combination. The use of other activation markers and/or marker combination can be used as discussed previously (da Silva Antunes et al., 2017; Dan et al., 2016; Reiss et al., 2017).
Critical Parameters
To avoid contamination or vacuum leaks prior to start up the MP lyophilization, remove any baffling (if equipped) and wipe with a soft cloth or paper towel to remove any remaining moisture within the drying chamber, and ensure that the collector chamber and drain line are free of any residual moisture. It is recommended that all the samples be covered using a filter paper, parafilm or Kim wipe as an extra precaution to avoid contamination of samples during the freeze dry process.
Because of the intrinsic solubility and numbers of epitopes, there can be great variability in solubility and viscosity between different MPs, and even different batches of the same MP. Additionally, storage conditions and multiple freeze-thaw cycles can affect the stability of the peptides in the MP. To maximize cellular responses and lengthen the shelf life of the pool, once the MP is removed from the freezer, it can be stored at 4°C for 2 weeks or refrozen. Avoid more than 5 freezing-thawing cycles.
When assessing MP reactivity, the most important and mandatory control in the AIM assay is a control of specificity or negative control. For that purpose, all assays should be performed with a DMSO stimulation control in parallel, at a concentration matching the same exact concentration of MPs used. It is highly recommended to perform this control in triplicate if enough PBMCs are available. The DMSO signal should then be subtracted to the MP-specific signal.
The second most important control in this assay is a positive control. These control is used to ensure that a detection of signal is observed by validating the expression and/or upregulation of the different activation markers. Typical positive controls used in this assay are polyclonal stimulation agents such as PHA, PMA/Ion, α-CD3/CD28 or SEB.
Another control, but optional, is the use of a MP that is not associated with the biological target that the user is interested, typically named as irrelevant MP control. This allows for an internal assay control of the donor response or sample being interrogated. Typical MPs used as irrelevant controls are MPs associated with generalized responses across a human population like those elicited after vaccination (e.g. Pertussis, Tetanus or SARS-CoV-2) or after infection with ubiquitous pathogens (e.g. CMV or EBV).
Another optional control is the inclusion of PBMC samples from donors that are not expected to respond to a specific MP (negative donors), such as for example using HIV negative donors to test reactivity to an HIV MP. This control will identify if any undesired reactivity of a MP or issues with the MP synthesis occurred (e.g. lyophilization contamination). Alternatively, PBMC samples from donors that are expected to respond to a specific MP can be included (positive or control donors), such as donors with a particular indication, donors that have experienced a previous clinical diagnosed disease or undergone a particular vaccination schedule (e.g. Pertussis, Tetanus or SARS-CoV-2). This control can be particularly valuable in the monitoring and assessment of longitudinal response stability (da Silva Antunes, Sutherland, et al., 2021).
Troubleshooting
Fluctuations in the vacuum readings as well as a loud suctioning grunt during the machine start up is often an indicator of a vacuum leak. This can be resolved by making sure all the knobs on the valves are in the “Vented” position which closes the valves, allowing the drying chamber pressure to drop to working condition. It is important to also inspect the lid as well as the gaskets on the chamber/manifold for proper placement and/or any damage or degradation that may cause any unwanted vacuum leaks.
If samples begin to thaw before being placed into the freeze dry unit, it is necessary to have them refrozen to facilitate freeze drying efficiency and avoid sample loss. Liquid Nitrogen can be employed to help with this after a short period in the −80°C freezer. Flasks can be further insulated using a neoprene cozy and, or aluminum foil, to minimize sample thawing. The cozy can also function to protect the vessel from breakage in case it falls.
While the Labconco® Benchtop Freeze Dryer is our Lyophilizer of choice, solvent removal from peptide pools could be performed using a standard Speed Vacuum Concentrator, which will remove a large amount of solvent relatively quickly. However, this type of device does not do as complete a job as a Freeze Dryer, and thus may pose some issues with DMSO toxicity in the final sample.
Regarding the AIM assay If using frozen PBMCs that are not thawed properly, viability and cell recovery can be compromised, and the cells may not perform optimally in the AIM assay. In general, cells should be thawed quickly and kept cold while still in the presence of DMSO. Cells with DMSO intercalated into their membranes are very fragile, and must be pelleted and handled gently.
When performing the surface staining steps, it is important to minimize the exposure to light as the antibodies used for this assay are conjugated with fluorochromes that are sensitive to light. The volume for each antibody depends on the Flow cytometer used to acquire the sample and on the manufacturers’ recommendation. As such it is advisable to perform an antibody titration to assess the optimal configuration based on the instrument. Similarly, fluorochromes combinations are machine-dependent and should be chosen according to the Flow cytometer used for the specific experiment.
When interpreting results some MPs pools can be used to simultaneously detect CD4+ and CD8+ T cell responses, while others are optimized to detect exclusively CD4+ or CD8+ T cell responses. If unsure, please refer to the details about the specificity of the MP you are using.
The number of peptides that can be pooled into a single MP depends on several factors that affect solubility, such as the number of peptides with a high proportion of hydrophobic amino acids, the size of peptides, or the presence of salts in the peptide preparations, to name a few. As a general rule MPs should not exceed 300–400 peptides. However, a strategy to test higher number of peptides could be to use multiple pools for simultaneously stimulation directly in the culture assay (Yu et al., 2022).
Understanding results
Within this segment, we describe and visually outline the detection and characterization of antigen-specific responses to SARS-CoV-2 after employing the AIM assay, as mentioned above. To detect SARS-CoV-2 T cell reactivity, a MP of overlapping peptides spanning the entire Spike sequence and two MPs optimized for detection of non-Spike (remainder of SARS-CoV-2 proteome) reactivity in CD4+ and CD8+ T cells were employed (CD4RE and CD8RE, respectively). The combination of these pools allowed to discriminate 4 groups of subjects with different SARS-CoV-2 infection and COVID-19 vaccine status as described below. An EBV MP was used as control (Figure 1). CD4+ and CD8+ T cells responses were calculated as percent of total CD4+ (OX40+CD137+) or CD8+ (CD69+CD137+) T cells. The background was removed from each stimulation by subtracting the signal from wells stimulated with DMSO. The gating strategy utilized is shown in Figure 2, as well as an example of reactive CD4+ and CD8+ T cell responses to SARS-CoV-2 Spike, CD4RE and CD8RE MPs and EBV MP from a representative donor.
Figure 1. SARS-CoV-2-specific CD4+ and CD8+ T cell responses in 4 study groups of subjects with different SARS-CoV-2 infection and COVID-19 vaccine status.
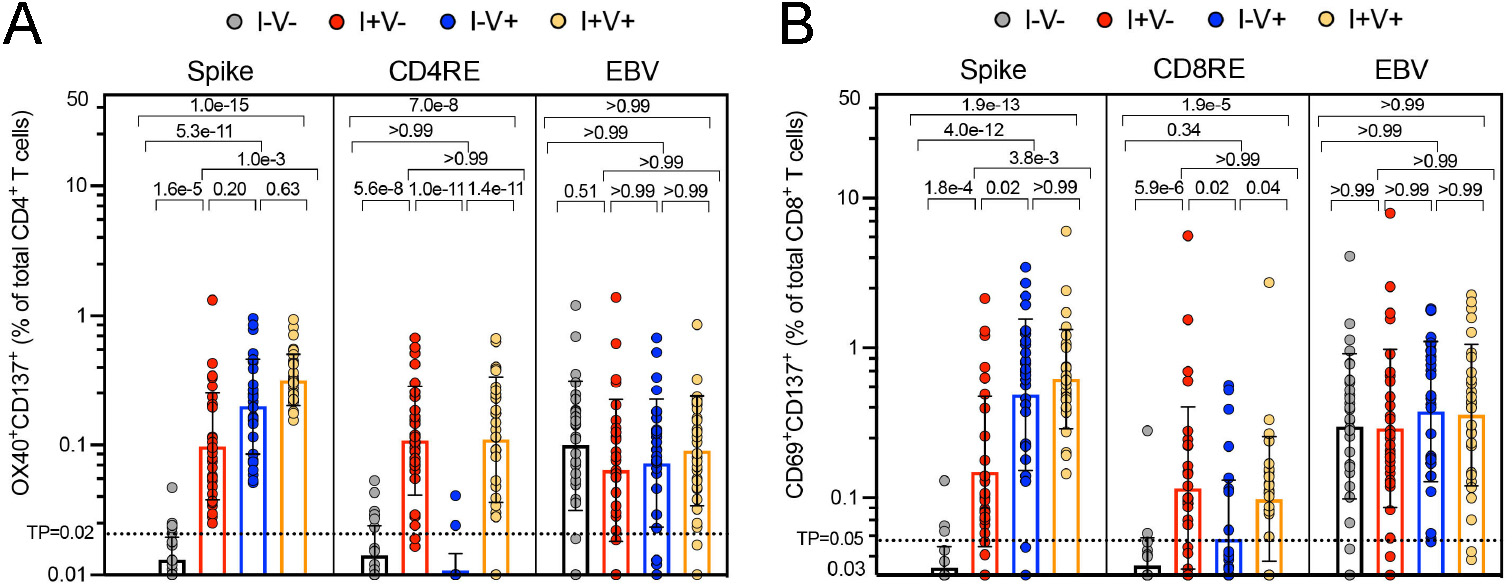
A T cell-based classification scheme has been previously developed with the aid of MPs that can discriminate responses exclusively from COVID-19 vaccination (i.e. using a MP targeting Spike) and from SARS-CoV-2 infections (i.e. using MPs targeting the remainder of SARS-CoV-2 proteome). T cell reactivity was assessed by AIM assay and SARS-CoV-2-and EBV specific CD4+ and CD8+ T cell responses were measured as percentages of AIM+ (OX40+CD137+) CD4+ T cells (A) or AIM+ (CD69+CD137+) CD8+ T cells (B) after stimulation of PBMCs with peptides pools encompassing spike-only (Spike) MPs or the experimentally defined CD4RE and CD8RE MPs representing all the proteome without spike. EVB MP was used as a control. Graphs show individual response to each MP plotted as background subtracted against DMSO negative control. Geometric mean with standard deviation (SD) for the 4 different groups is shown. Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons was performed, and p values < 0.05 considered statistically significant. I−V−, unexposed and unvaccinated (n = 30); I+V−, infected and non-vaccinated (n = 30); I+V+, infected and then vaccinated (n = 30); I−V+, non-infected and vaccinated (n = 30). Threshold of positivity (TP) is indicated. Median response each group is shown. This figure was adapted from (Yu, Wang, Garrigan, Goodwin, et al., 2022).
Figure 2. Gating strategy and representative CD4+ and CD8+ T cell responses plots of the activation induced marker (AIM) assay.
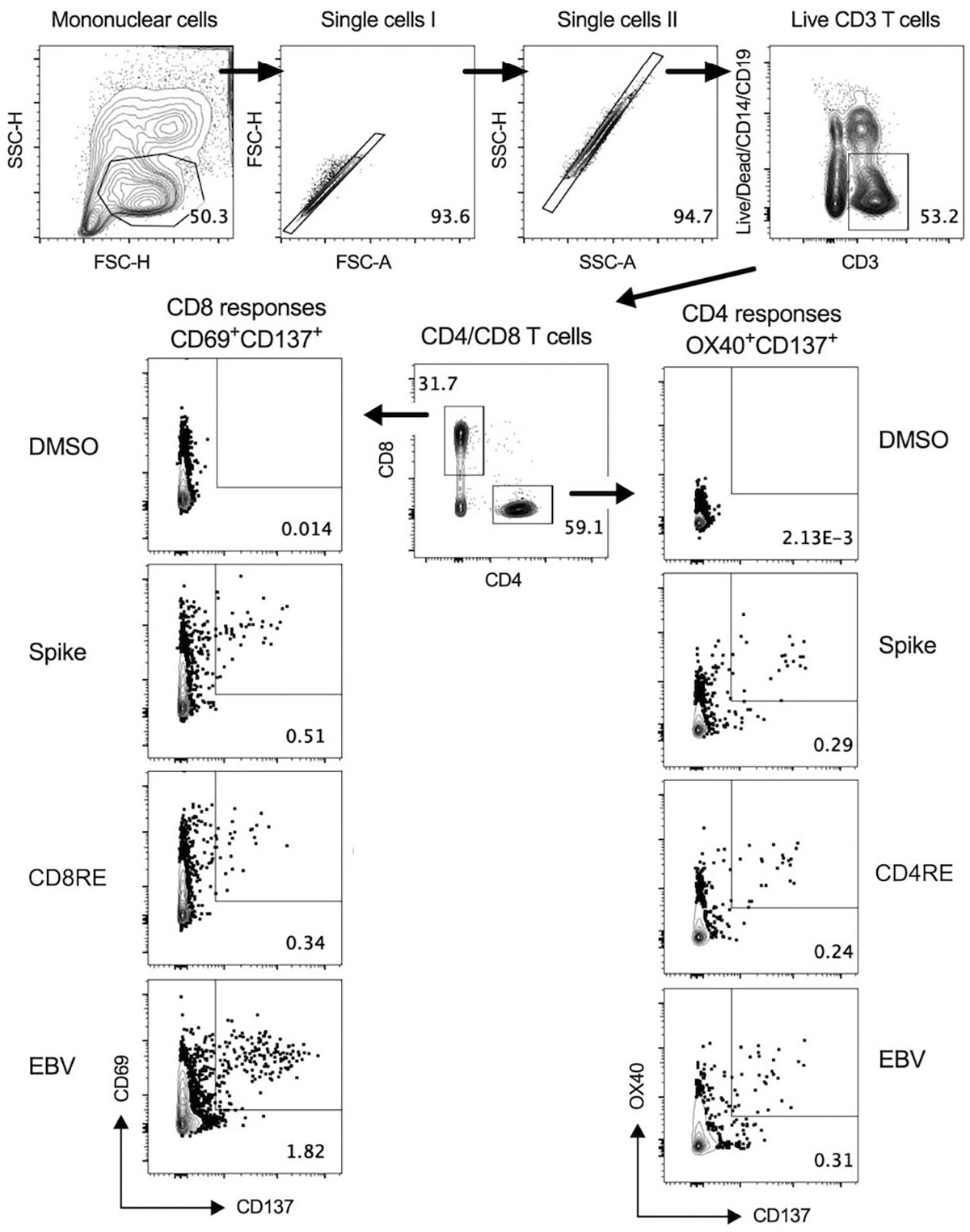
Representative gating of live CD3+ T cells, CD4+ T cells, CD8+ T cells and reactive OX40+CD137+ CD4 or CD69+CD137+ CD8 T cells from donor PBMCs is shown. Briefly, mononuclear cells were gated out of all events followed by subsequent singlet gating. Live CD3+ cells were gated as Live/Dead-CD14-CD19-CD3+. Cells were then gated as CD4+CD8− or CD4−CD8+ T cells, and reactive OX40+CD137+ CD4+ or CD69+CD137+ CD8+ T cells were gated and calculated as percent of total CD4+ or CD8+ T cells. Representative CD4+ and CD8+ antigen-specific responses plots after stimulating with DMSO (negative control) and SARS-CoV-2 or EBV specific MPs are shown. This figure was adapted from (Yu, Wang, Garrigan, Goodwin, et al., 2022)
As expected Spike SARS-CoV-2 specific-T cells can be detected in individuals that have been vaccinated with COVID-19 vaccines (non-infected and vaccinated; I−V+) and infected with SARS-CoV-2 (infected and non-vaccinated; I+V−) or both (Infected and then vaccinated; I+V+). Conversely no responses are observed in individuals neither infected nor vaccinated (non-infected, non-vaccinated; I−V−). Importantly, CD4RE and CD8RE responses are only detected in convalescent subjects (I+V− or I+V+) and not in unexposed (I−V−) or vaccinated (I−V+) subjects, as expected since all vaccinated donors included in this study received exclusively Spike-based mRNA vaccines. Also, EBV reactivity was detected and observed at equal levels across all the groups. Overall this data demonstrates the attributes of the MP approach and versatility of MP design to detect antigen-specific responses to SARS-CoV-2 and EBV, complex pathogens composed of multiple antigens. Importantly, due to the intrinsic nature and diversity of the epitopes selected, the MPs employed in this section allowed to discriminate responses specific to COVID-19 vaccination or SARS-CoV-2 infection, highlighting the potential use of MPs as immunodiagnostic tools. For complete details on the use and execution of this protocol for this particular experimental setting and/or interpretation of the data, please refer to (Yu, Wang, Garrigan, Goodwin, et al., 2022).
Time Considerations
On average, it takes about 16–18 hours per run to completely lyophilize a sample, and two runs in total with an additional dilution step to extract all the DMSO from a peptide sample. For the lyophilization of any MP, if time is of the essence, the drying process can be accelerated first with the speedvac, then the lyophilizer utilized to further remove residual DMSO, though this does incur the potential risks involved with additional sample handling. Use of a speedvac must also proceed with caution, as doing the process too quickly risks loss of sample along with the solvent.
The AIM assay requires 2 days to be completed. The thawing of PBMCs should take less then 1 hour, although more time may be needed if more than 10 samples are used at the same time. If using fresh PBMCs plan ahead the time allocated for blood separation before PBMCs are processed. The T cell culturing period with MPs should be exactly 24h. The surface staining should be performed in the following day and immediately after stimulation. If not possible the plate should be removed from the incubator and placed at 4°C until the surface staining is performed. Samples should be then immediately acquired in a Flow Cytometer machine. However, fixation of the PBMCs samples can be performed after the last step of the protocol to extend the period before sample acquisition.
ACKNOWLEDGMENTS
Data reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award number U19 AI142742 and contract number 75N93019C00065. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST STATEMENT
A. Sette is a consultant for Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress, and Repertoire. S.C. is a consultant for Avalia. La Jolla Institute for Immunology (LJI) has filed for patent protection for various aspects of MPs design. All other authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
Materials availability: MPs described in this protocol will be made available to the scientific community upon request, depending on material availability, and following execution of a material transfer agreement (MTA), by contacting A.S. (alex@lji.org).
Literature Cited
- Al-Kolla R, Grifoni A, Crotty S, Sette A, Gianella S, & Dan J (2022). Design and validation of HIV peptide pools for detection of HIV-specific CD4+ and CD8+ T cells. PLoS One, 17(8), e0268370. doi: 10.1371/journal.pone.0268370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, Peters B, … Weiskopf D (2017). Human CD4(+) T Cell Responses to an Attenuated Tetravalent Dengue Vaccine Parallel Those Induced by Natural Infection in Magnitude, HLA Restriction, and Antigen Specificity. J Virol, 91(5). doi: 10.1128/JVI.02147-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Arya R, Sachan S, Jha SN, Kalia A, Lall A, … Gupta N (2021). Immune Memory in Mild COVID-19 Patients and Unexposed Donors Reveals Persistent T Cell Responses After SARS-CoV-2 Infection. Front Immunol, 12, 636768. doi: 10.3389/fimmu.2021.636768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Sachan S, Jit BP, Sharma A, Coshic P, Sette A, … Gupta N (2022). An efficient immunoassay for the B cell help function of SARS-CoV-2-specific memory CD4(+) T cells. Cell Rep Methods, 2(6), 100224. doi: 10.1016/j.crmeth.2022.100224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolidis SA, Kakara M, Painter MM, Goel RR, Mathew D, Lenzi K, … Bar-Or A (2021). Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat Med, 27(11), 1990–2001. doi: 10.1038/s41591-021-01507-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam PS, Feng Y, Ashraf U, Hu M, Walls AC, Edara VV, … Pulendran B (2022). Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci Transl Med, 14(658), eabq4130. doi: 10.1126/scitranslmed.abq4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft T, Dillon MB, da Silva Antunes R, Paul S, Peters B, Crotty S, … Sette A (2016). Th1 versus Th2 T cell polarization by whole-cell and acellular childhood pertussis vaccines persists upon re-immunization in adolescence and adulthood. Cell Immunol, 304-305, 35–43. doi: 10.1016/j.cellimm.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banki Z, Mateus J, Rossler A, Schafer H, Bante D, Riepler L, … Kimpel J (2022). Heterologous ChAdOx1/BNT162b2 vaccination induces stronger immune response than homologous ChAdOx1 vaccination: The pragmatic, multi-center, three-arm, partially randomized HEVACC trial. EBioMedicine, 80, 104073. doi: 10.1016/j.ebiom.2022.104073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan TR, Al Banna H, Kaisar MH, Karmakar PC, Hakim A, Akter A, … Qadri F (2022). Correlation of antigen-specific immune response with disease severity among COVID-19 patients in Bangladesh. Front Immunol, 13, 929849. doi: 10.3389/fimmu.2022.929849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrueta G, Frazier A, Pomes A, Glesner J, Filep S, Schal C, … Schulten V (2019). Variability in German Cockroach Extract Composition Greatly Impacts T Cell Potency in Cockroach-Allergic Donors. Front Immunol, 10, 313. doi: 10.3389/fimmu.2019.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt L, Gao Y, Wullimann D, Muren Ingelman-Sundberg H, Muschiol S, Healy K, … Osterborg A (2022). Hybrid immunity in immunocompromised patients with CLL after SARS-CoV-2 infection followed by booster mRNA vaccination. Blood, 140(22), 2403–2407. doi: 10.1182/blood.2022016815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland BS, Goodwin B, Zhang Z, Bloom N, Kato Y, Neill J, … Dan JM (2022). Preserved SARS-CoV-2 Vaccine Cell-Mediated Immunogenicity in Patients With Inflammatory Bowel Disease on Immune-Modulating Therapies. Clin Transl Gastroenterol, 13(4), e00484. doi: 10.14309/ctg.0000000000000484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosteels C, Van Damme KFA, De Leeuw E, Declercq J, Maes B, Bosteels V, … Lambrecht BN (2022). Loss of GM-CSF-dependent instruction of alveolar macrophages in COVID-19 provides a rationale for inhaled GM-CSF treatment. Cell Rep Med, 3(12), 100833. doi: 10.1016/j.xcrm.2022.100833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen JE, Addetia A, Dang HV, Stewart C, Brown JT, Sharkey WK, … Veesler D (2022). Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science, 377(6608), 890–894. doi: 10.1126/science.abq0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen JE, Park YJ, Stewart C, Brown JT, Sharkey WK, Walls AC, … Veesler D (2022). SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci Immunol, 7(78), eadf1421. doi: 10.1126/sciimmunol.adf1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasu N, Elia I, Russo V, Montacchiesi G, Stabile SA, De Intinis C, … Pace L (2022). Memory CD8(+) T cell diversity and B cell responses correlate with protection against SARS-CoV-2 following mRNA vaccination. Nat Immunol, 23(10), 1445–1456. doi: 10.1038/s41590-022-01313-z [DOI] [PubMed] [Google Scholar]
- Bueno SM, Abarca K, Gonzalez PA, Galvez NMS, Soto JA, Duarte LF, … Kalergis AM (2022). Safety and Immunogenicity of an Inactivated Severe Acute Respiratory Syndrome Coronavirus 2 Vaccine in a Subgroup of Healthy Adults in Chile. Clin Infect Dis, 75(1), e792–e804. doi: 10.1093/cid/ciab823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, & Sette A (2015). Automatic Generation of Validated Specific Epitope Sets. J Immunol Res, 2015, 763461. doi: 10.1155/2015/763461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon IS, Li C, Son YM, Goplen NP, Wu Y, Cassmann T, … Sun J (2021). Immune signatures underlying post-acute COVID-19 lung sequelae. Sci Immunol, 6(65), eabk1741. doi: 10.1126/sciimmunol.abk1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihab LY, Kuan R, Phillips EJ, Mallal SA, Rozot V, Davis MM, … Group SS (2023). Expression of specific HLA class II alleles is associated with an increased risk for active tuberculosis and a distinct gene expression profile. HLA, 101(2), 124–137. doi: 10.1111/tan.14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F, Zaia JA, Frankel PH, Stan R, Drake J, Williams B, … Diamond DJ (2022). Safety and immunogenicity of a synthetic multiantigen modified vaccinia virus Ankara-based COVID-19 vaccine (COH04S1): an open-label and randomised, phase 1 trial. Lancet Microbe, 3(4), e252–e264. doi: 10.1016/S2666-5247(22)00027-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PR, Correia CA, Marmorato MP, Dias JZC, Thomazella MV, Cabral da Silva A, … Silveira CGT (2022). Humoral and cellular immune responses to CoronaVac up to one year after vaccination. Front Immunol, 13, 1032411. doi: 10.3389/fimmu.2022.1032411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Babor M, Carpenter C, Khalil N, Cortese M, Mentzer AJ, … Sette A (2018). Th1/Th17 polarization persists following whole-cell pertussis vaccination despite repeated acellular boosters. J Clin Invest, 128(9), 3853–3865. doi: 10.1172/JCI121309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Garrigan E, Quiambao LG, Dhanda SK, Marrama D, Westernberg L, … Sette A (2023). T cell reactivity to Bordetella pertussis is highly diverse regardless of childhood vaccination. Cell Host Microbe, 31(8), 1404–1416 e1404. doi: 10.1016/j.chom.2023.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Pallikkuth S, Williams E, Dawen Yu E, Mateus J, Quiambao L, … Sette A (2021). Differential T-Cell Reactivity to Endemic Coronaviruses and SARS-CoV-2 in Community and Health Care Workers. J Infect Dis, 224(1), 70–80. doi: 10.1093/infdis/jiab176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Paul S, Sidney J, Weiskopf D, Dan JM, Phillips E, … Lindestam Arlehamn CS (2017). Definition of Human Epitopes Recognized in Tetanus Toxoid and Development of an Assay Strategy to Detect Ex Vivo Tetanus CD4+ T Cell Responses. PLoS One, 12(1), e0169086. doi: 10.1371/journal.pone.0169086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Pham J, McMurtrey C, Hildebrand WH, Phillips E, Mallal S, … Sette A (2018). Urinary Peptides As a Novel Source of T Cell Allergen Epitopes. Front Immunol, 9, 886. doi: 10.3389/fimmu.2018.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Quiambao LG, Soldevila F, Sutherland A, Peters B, & Sette A (2021). Lack of evidence supporting a role of IFN-beta and TGF-beta in differential polarization of Bordetella pertussis specific-T cell responses. Cytokine, 137, 155313. doi: 10.1016/j.cyto.2020.155313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Quiambao LG, Sutherland A, Soldevila F, Dhanda SK, Armstrong SK, … Sette A (2020). Development and Validation of a Bordetella pertussis Whole-Genome Screening Strategy. J Immunol Res, 2020, 8202067. doi: 10.1155/2020/8202067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R, Sutherland A, Frazier A, Schulten V, Pomes A, Glesner J, … Sette A (2021). Heterogeneity of magnitude, allergen immunodominance, and cytokine polarization of cockroach allergen-specific T cell responses in allergic sensitized children. Clin Transl Allergy, 11(8), e12073. doi: 10.1002/clt2.12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, … Crotty S (2016). A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J Immunol, 197(3), 983–993. doi: 10.4049/jimmunol.1600318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, … Crotty S (2021). Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science, 371(6529). doi: 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TA, Penn-Nicholson A, Luabeya AKK, Fiore-Gartland A, Du Plessis N, Loxton AG, … team T. s. (2021). Safety and immunogenicity of the adjunct therapeutic vaccine ID93 + GLA-SE in adults who have completed treatment for tuberculosis: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Respir Med, 9(4), 373–386. doi: 10.1016/S2213-2600(20)30319-2 [DOI] [PubMed] [Google Scholar]
- de Abreu Costa L, Henrique Fernandes Ottoni M, Dos Santos MG, Meireles AB, Gomes de Almeida V, de Fatima Pereira W, … Eustaquio Alvim Brito-Melo G (2017). Dimethyl Sulfoxide (DMSO) Decreases Cell Proliferation and TNF-alpha, IFN-gamma, and IL-2 Cytokines Production in Cultures of Peripheral Blood Lymphocytes. Molecules, 22(11). doi: 10.3390/molecules22111789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentone C, Fenoglio D, Ponzano M, Cerchiaro M, Altosole T, Franciotta D, … Bassetti M (2022). Healthcare Worker Study Cohort to Determine the Level and Durability of Cellular and Humoral Immune Responses after Two Doses of SARS-CoV-2 Vaccination. Vaccines (Basel), 10(11). doi: 10.3390/vaccines10111784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC, … Peters B (2019). IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res, 47(W1), W502–W506. doi: 10.1093/nar/gkz452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanwani R, Dhanda SK, Pham J, Williams GP, Sidney J, Grifoni A, … Benedict CA (2021). Profiling Human Cytomegalovirus-Specific T Cell Responses Reveals Novel Immunogenic Open Reading Frames. J Virol, 95(21), e0094021. doi: 10.1128/JVI.00940-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanwani R, Pham J, Premlal ALR, Frazier A, Kumar A, Pero ME, … Lindestam Arlehamn CS (2020). T Cell Responses to Neural Autoantigens Are Similar in Alzheimer’s Disease Patients and Age-Matched Healthy Controls. Front Neurosci, 14, 874. doi: 10.3389/fnins.2020.00874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon MB, Schulten V, Oseroff C, Paul S, Dullanty LM, Frazier A, … Sette A (2015). Different Bla-g T cell antigens dominate responses in asthma versus rhinitis subjects. Clin Exp Allergy, 45(12), 1856–1867. doi: 10.1111/cea.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Bruyn E, Ruzive S, Howlett P, Jacobs AJ, Arlehamn CSL, Sette A, … Riou C (2022). Comparison of the frequency and phenotypic profile of Mycobacterium tuberculosis-specific CD4 T cells between the site of disease and blood in pericardial tuberculosis. Front Immunol, 13, 1009016. doi: 10.3389/fimmu.2022.1009016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Bruyn E, Ruzive S, Lindestam Arlehamn CS, Sette A, Sher A, Barber DL, … Riou C (2021). Mycobacterium tuberculosis-specific CD4 T cells expressing CD153 inversely associate with bacterial load and disease severity in human tuberculosis. Mucosal Immunol, 14(2), 491–499. doi: 10.1038/s41385-020-0322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman TW, Nelson CE, Kauffman KD, Lora NE, Vinhaes CL, Dorosky DE, … Barber DL (2022). CD4 T cells are rapidly depleted from tuberculosis granulomas following acute SIV co-infection. Cell Rep, 39(9), 110896. doi: 10.1016/j.celrep.2022.110896 [DOI] [PubMed] [Google Scholar]
- Galvez NMS, Pacheco GA, Schultz BM, Melo-Gonzalez F, Soto JA, Duarte LF, … Kalergis AM (2022). Differences in the immune response elicited by two immunization schedules with an inactivated SARS-CoV-2 vaccine in a randomized phase 3 clinical trial. Elife, 11. doi: 10.7554/eLife.81477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cai C, Grifoni A, Muller TR, Niessl J, Olofsson A, … Buggert M (2022). Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med, 28(3), 472–476. doi: 10.1038/s41591-022-01700-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cai C, Wullimann D, Niessl J, Rivera-Ballesteros O, Chen P, … Buggert M (2022). Immunodeficiency syndromes differentially impact the functional profile of SARS-CoV-2-specific T cells elicited by mRNA vaccination. Immunity, 55(9), 1732–1746 e1735. doi: 10.1016/j.immuni.2022.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Valtanen P, Hope CM, Masavuli MG, Yeow AEL, Balachandran H, Mekonnen ZA, … Grubor-Bauk B (2022). SARS-CoV-2 Omicron variant escapes neutralizing antibodies and T cell responses more efficiently than other variants in mild COVID-19 convalescents. Cell Rep Med, 3(6), 100651. doi: 10.1016/j.xcrm.2022.100651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli-Guimaraes PH, Sanku G, Sette A, Weiskopf D, Schaughency P, Lack J, & Nutman TB (2022). Antigenic Determinants of SARS-CoV-2-Specific CD4(+) T Cell Lines Reveals M Protein-Driven Dysregulation of Interferon Signaling. Front Immunol, 13, 883159. doi: 10.3389/fimmu.2022.883159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geers D, Sablerolles RSG, van Baarle D, Kootstra NA, Rietdijk WJR, Schmitz KS, … group S. r. (2023). Ad26.COV2.S priming provided a solid immunological base for mRNA-based COVID-19 booster vaccination. iScience, 26(1), 105753. doi: 10.1016/j.isci.2022.105753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel CH, Geers D, Schmitz KS, Mykytyn AZ, Lamers MM, Bogers S, … de Vries RD (2022). Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol, 7(69), eabo2202. doi: 10.1126/sciimmunol.abo2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, … Davis MM (2017). Identifying specificity groups in the T cell receptor repertoire. Nature, 547(7661), 94–98. doi: 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, … Wherry EJ (2021). mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science, 374(6572), abm0829. doi: 10.1126/science.abm0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondre-Lewis TA, Jiang C, Ford ML, Koelle DM, Sette A, Shalek AK, & Thomas PG (2023). NIAID workshop on T cell technologies. Nat Immunol, 24(1), 14–18. doi: 10.1038/s41590-022-01377-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, … Jones AR (2020). Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res, 48(D1), D783–D788. doi: 10.1093/nar/gkz1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Angelo M, Sidney J, Paul S, Peters B, de Silva AD, … Weiskopf D (2017). Patterns of Cellular Immunity Associated with Experimental Infection with rDEN2Delta30 (Tonga/74) Support Its Suitability as a Human Dengue Virus Challenge Strain. J Virol, 91(8). doi: 10.1128/JVI.02133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Angelo MA, Lopez B, O’Rourke PH, Sidney J, Cerpas C, … Weiskopf D (2017). Global Assessment of Dengue Virus-Specific CD4(+) T Cell Responses in Dengue-Endemic Areas. Front Immunol, 8, 1309. doi: 10.3389/fimmu.2017.01309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, da Silva Antunes R, Westernberg L, Pham J, Birrueta G, Peters B, … Schulten V (2019). Characterization and epitope identification of the T cell response in non-allergic individuals exposed to mouse allergen. World Allergy Organ J, 12(4), 100026. doi: 10.1016/j.waojou.2019.100026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Mahajan S, Sidney J, Martini S, Scheuermann RH, Peters B, & Sette A (2019). A survey of known immune epitopes in the enteroviruses strains associated with acute flaccid myelitis. Hum Immunol, 80(11), 923–929. doi: 10.1016/j.humimm.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Moore E, Voic H, Sidney J, Phillips E, Jadi R, … Sette A (2019). Characterization of Magnitude and Antigen Specificity of HLA-DP, DQ, and DRB3/4/5 Restricted DENV-Specific CD4+ T Cell Responses. Front Immunol, 10, 1568. doi: 10.3389/fimmu.2019.01568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Pham J, Sidney J, O’Rourke PH, Paul S, Peters B, … Sette A (2017). Prior Dengue Virus Exposure Shapes T Cell Immunity to Zika Virus in Humans. J Virol, 91(24). doi: 10.1128/JVI.01469-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, & Sette A (2022). From Alpha to omicron: The response of T cells. Curr Res Immunol, 3, 146–150. doi: 10.1016/j.crimmu.2022.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, & Sette A (2021). SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe, 29(7), 1076–1092. doi: 10.1016/j.chom.2021.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, & Sette A (2020). A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe, 27(4), 671–680 e672. doi: 10.1016/j.chom.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Voic H, Dhanda SK, Kidd CK, Brien JD, Buus S, … Sette A (2020). T Cell Responses Induced by Attenuated Flavivirus Vaccination Are Specific and Show Limited Cross-Reactivity with Other Flavivirus Species. J Virol, 94(10). doi: 10.1128/JVI.00089-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, … Sette A (2020). Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell, 181(7), 1489–1501 e1415. doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Zhang Y, Tarke A, Sidney J, Rubiro P, Reina-Campos M, … Sette A (2022). Defining antigen targets to dissect vaccinia virus and monkeypox virus-specific T cell responses in humans. Cell Host Microbe, 30(12), 1662–1670 e1664. doi: 10.1016/j.chom.2022.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, … Diamond MS (2020). A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell, 183(1), 169–184 e113. doi: 10.1016/j.cell.2020.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WT, Yuan M, Callaghan S, Musharrafieh R, Song G, Silva M, … Andrabi R (2022). Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques. Sci Transl Med, 14(657), eabl9605. doi: 10.1126/scitranslmed.abl9605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz D, Oseroff C, Pham J, Sidney J, Peters B, & Sette A (2015). Definition of a pool of epitopes that recapitulates the T cell reactivity against major house dust mite allergens. Clin Exp Allergy, 45(10), 1601–1612. doi: 10.1111/cea.12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz D, Seumois G, Gholami AM, Greenbaum JA, Lane J, White B, … Sette A (2016). Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity. Clin Exp Allergy, 46(5), 705–719. doi: 10.1111/cea.12692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft SG, Kauffman KD, Sakai S, Lindestam Arlehamn CS, Sette A, Hoft DF, … Barber DL (2023). Imprinting of Gut-Homing Receptors on Mtb-Specific Th1* Cells Is Associated with Reduced Lung Homing after Gavage BCG Vaccination of Rhesus Macaques. mBio, e0022023. doi: 10.1128/mbio.00220-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LE, Song J, Grifoni A, Shimizu C, Tremoulet AH, Dummer KB, … Franco A (2022). T Cells in Multisystem Inflammatory Syndrome in Children (MIS-C) Have a Predominant CD4+ T Helper Response to SARS-CoV-2 Peptides and Numerous Virus-Specific CD4− CD8− Double-Negative T Cells. Int J Mol Sci, 23(13). doi: 10.3390/ijms23137219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Kim C, Xia Q, Gould TM, Cao W, Zhang H, … Goronzy JJ (2021). Activation of mTORC1 at late endosomes misdirects T cell fate decision in older individuals. Sci Immunol, 6(60). doi: 10.1126/sciimmunol.abg0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman KD, Sakai S, Lora NE, Namasivayam S, Baker PJ, Kamenyeva O, … Barber DL (2021). PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci Immunol, 6(55). doi: 10.1126/sciimmunol.abf3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R, Richardson SI, Moyo-Gwete T, Hermanus T, Tincho MB, Benede N, … Burgers WA (2021). Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe, 29(11), 1611–1619 e1615. doi: 10.1016/j.chom.2021.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, … Riou C (2022). T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature, 603(7901), 488–492. doi: 10.1038/s41586-022-04460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R, Tincho MB, Suzuki A, Benede N, Ngomti A, Baguma R, … Riou C (2023). Impact of SARS-CoV-2 exposure history on the T cell and IgG response. Cell Rep Med, 4(1), 100898. doi: 10.1016/j.xcrm.2022.100898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenerman P, Cerundolo V, & Dunbar PR (2002). Tracking T cells with tetramers: new tales from new tools. Nat Rev Immunol, 2(4), 263–272. doi: 10.1038/nri777 [DOI] [PubMed] [Google Scholar]
- Kong XF, Martinez-Barricarte R, Kennedy J, Mele F, Lazarov T, Deenick EK, … Casanova JL (2018). Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol, 19(9), 973–985. doi: 10.1038/s41590-018-0178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K, Bettini E, Parvathaneni K, Painter MM, Agarwal D, Lundgreen KA, … Locci M (2022). Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell, 185(6), 1008–1024 e1015. doi: 10.1016/j.cell.2022.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SA, Sutherland A, Soldevila F, Westernberg L, Aoki M, Frazier A, … Peters B (2023). Identification of cow milk epitopes to characterize and quantify disease-specific T cells in allergic children. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2023.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, Benson B, Kuan R, Dill-McFarland KA, Peterson GJ, Paul S, … Hawn TR (2022). T-cell deficiency and hyperinflammatory monocyte responses associate with Mycobacterium avium complex lung disease. Front Immunol, 13, 1016038. doi: 10.3389/fimmu.2022.1016038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, Dhanwani R, Pham J, Kuan R, Frazier A, Rezende Dutra J, … Sette A (2020). alpha-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat Commun, 11(1), 1875. doi: 10.1038/s41467-020-15626-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, … Sette A (2013). Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog, 9(1), e1003130. doi: 10.1371/journal.ppat.1003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, McKinney DM, Carpenter C, Paul S, Rozot V, Makgotlho E, … Sette A (2016). A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans. PLoS Pathog, 12(7), e1005760. doi: 10.1371/journal.ppat.1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, Pham J, Alcalay RN, Frazier A, Shorr E, Carpenter C, … Sette A (2019). Widespread Tau-Specific CD4 T Cell Reactivity in the General Population. J Immunol, 203(1), 84–92. doi: 10.4049/jimmunol.1801506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AM, & Parham P (1999). Polymorphism and evolution of HLA class I and II genes and molecules. Rev Immunogenet, 1(1), 105–123. [PubMed] [Google Scholar]
- Livingstone AM, & Fathman CG (1987). The structure of T-cell epitopes. Annu Rev Immunol, 5, 477–501. doi: 10.1146/annurev.iy.05.040187.002401 [DOI] [PubMed] [Google Scholar]
- Madden DR (1995). The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol, 13, 587–622. doi: 10.1146/annurev.iy.13.040195.003103 [DOI] [PubMed] [Google Scholar]
- Madelon N, Heikkila N, Sabater Royo I, Fontannaz P, Breville G, Lauper K, … Eberhardt CS (2022). Omicron-Specific Cytotoxic T-Cell Responses After a Third Dose of mRNA COVID-19 Vaccine Among Patients With Multiple Sclerosis Treated With Ocrelizumab. JAMA Neurol, 79(4), 399–404. doi: 10.1001/jamaneurol.2022.0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madelon N, Lauper K, Breville G, Sabater Royo I, Goldstein R, Andrey DO, … Eberhardt CS (2022). Robust T-Cell Responses in Anti-CD20-Treated Patients Following COVID-19 Vaccination: A Prospective Cohort Study. Clin Infect Dis, 75(1), e1037–e1045. doi: 10.1093/cid/ciab954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, … Picker LJ (2001). Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods, 255(1–2), 27–40. doi: 10.1016/s0022-1759(01)00416-1 [DOI] [PubMed] [Google Scholar]
- Martinez-Barricarte R, Markle JG, Ma CS, Deenick EK, Ramirez-Alejo N, Mele F, … Casanova JL (2018). Human IFN-gamma immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol, 3(30). doi: 10.1126/sciimmunol.aau6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Dan JM, Zhang Z, Rydyznski Moderbacher C, Lammers M, Goodwin B, … Weiskopf D (2021). Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science, 374(6566), eabj9853. doi: 10.1126/science.abj9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, … Weiskopf D (2020). Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science, 370(6512), 89–94. doi: 10.1126/science.abd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Voic H, Angelo MA, Phillips E, Mallal S, … Weiskopf D (2020). Identification of Novel Yellow Fever Class II Epitopes in YF-17D Vaccinees. Viruses, 12(11). doi: 10.3390/v12111300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, … Sette A (2013). A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics, 65(5), 357–370. doi: 10.1007/s00251-013-0684-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff BJ, Ramirez-Suastegui C, Fajardo V, Chee SJ, Kusnadi A, Simon H, … Vijayanand P (2020). Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell, 183(5), 1340–1353 e1316. doi: 10.1016/j.cell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele D, Calastri A, Maiorano E, Cerino A, Sachs M, Oliviero B, … Varchetta S (2021). High Frequencies of Functional Virus-Specific CD4(+) T Cells in SARS-CoV-2 Subjects With Olfactory and Taste Disorders. Front Immunol, 12, 748881. doi: 10.3389/fimmu.2021.748881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Gonzalez F, Soto JA, Gonzalez LA, Fernandez J, Duarte LF, Schultz BM, … Bueno SM (2021). Recognition of Variants of Concern by Antibodies and T Cells Induced by a SARS-CoV-2 Inactivated Vaccine. Front Immunol, 12, 747830. doi: 10.3389/fimmu.2021.747830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Lindestam Arlehamn CS, Dow C, Dillon MBC, Wiseman RW, Bohn P, … Sette A (2015). The TB-specific CD4(+) T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis (Edinb), 95(6), 722–735. doi: 10.1016/j.tube.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan K, Jagannathan P, Pham TD, Pandey S, Bonilla HF, Jacobson K, … Banaei N (2021). Interferon-gamma Release Assay for Accurate Detection of Severe Acute Respiratory Syndrome Coronavirus 2 T-Cell Response. Clin Infect Dis, 73(9), e3130–e3132. doi: 10.1093/cid/ciaa1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Namasivayam S, Foreman TW, Kauffman KD, Sakai S, Dorosky DE, … Barber DL (2022). Mild SARS-CoV-2 infection in rhesus macaques is associated with viral control prior to antigen-specific T cell responses in tissues. Sci Immunol, eabo0535. doi: 10.1126/sciimmunol.abo0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JB, Grifoni A, Tarke A, Sette A, & Nielsen M (2021). PopCover-2.0. Improved Selection of Peptide Sets With Optimal HLA and Pathogen Diversity Coverage. Front Immunol, 12, 728936. doi: 10.3389/fimmu.2021.728936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbe A, Kronsteiner B, Skelly DT, Pace M, Brown A, Adland E, … Dunachie S (2021). T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat Commun, 12(1), 2055. doi: 10.1038/s41467-021-21856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogongo P, Tezera LB, Ardain A, Nhamoyebonde S, Ramsuran D, Singh A, … Leslie A (2021). Tissue-resident-like CD4+ T cells secreting IL-17 control Mycobacterium tuberculosis in the human lung. J Clin Invest, 131(10). doi: 10.1172/JCI142014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Christensen LH, Westernberg L, Pham J, Lane J, Paul S, … Sette A (2017). Immunoproteomic analysis of house dust mite antigens reveals distinct classes of dominant T cell antigens according to function and serological reactivity. Clin Exp Allergy, 47(4), 577–592. doi: 10.1111/cea.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Pham J, Frazier A, Hinz D, Sidney J, Paul S, … Sette A (2016). Immunodominance in allergic T-cell reactivity to Japanese cedar in different geographic cohorts. Ann Allergy Asthma Immunol, 117(6), 680–689 e681. doi: 10.1016/j.anai.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, … Sette A (2010). Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol, 185(2), 943–955. doi: 10.4049/jimmunol.1000405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, … Sette A (2012). Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production. J Immunol, 189(2), 679–688. doi: 10.4049/jimmunol.1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, … Sette A (2012). T cell responses to known allergen proteins are differently polarized and account for a variable fraction of total response to allergen extracts. J Immunol, 189(4), 1800–1811. doi: 10.4049/jimmunol.1200850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter MM, Mathew D, Goel RR, Apostolidis SA, Pattekar A, Kuthuru O, … Wherry EJ (2021). Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity, 54(9), 2133–2142 e2133. doi: 10.1016/j.immuni.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P (1988). Function and polymorphism of human leukocyte antigen-A,B,C molecules. Am J Med, 85(6A), 2–5. doi: 10.1016/0002-9343(88)90369-5 [DOI] [PubMed] [Google Scholar]
- Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, … Sette A (2005). HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol, 175(8), 5504–5515. doi: 10.4049/jimmunol.175.8.5504 [DOI] [PubMed] [Google Scholar]
- Patankar YR, Sutiwisesak R, Boyce S, Lai R, Lindestam Arlehamn CS, Sette A, & Behar SM (2020). Limited recognition of Mycobacterium tuberculosis-infected macrophages by polyclonal CD4 and CD8 T cells from the lungs of infected mice. Mucosal Immunol, 13(1), 140–148. doi: 10.1038/s41385-019-0217-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Sibbertsen F, Weiskopf D, Lutgehetmann M, Barroso M, Danecka MK, … Dunay GA (2022). Specific CD4+ T Cell Responses to Ancestral SARS-CoV-2 in Children Increase With Age and Show Cross-Reactivity to Beta Variant. Front Immunol, 13, 867577. doi: 10.3389/fimmu.2022.867577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso MJ, Spinelli MA, Deveau TM, Forman CA, Munter SE, Mathur S, … Henrich TJ (2022). Postacute sequelae and adaptive immune responses in people with HIV recovering from SARS-COV-2 infection. AIDS, 36(12), F7–F16. doi: 10.1097/QAD.0000000000003338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gomez A, Gasca-Capote C, Vitalle J, Ostos FJ, Serna-Gallego A, Trujillo-Rodriguez M, … Teams C-GW (2022). Deciphering the quality of SARS-CoV-2 specific T-cell response associated with disease severity, immune memory and heterologous response. Clin Transl Med, 12(4), e802. doi: 10.1002/ctm2.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Nielsen M, & Sette A (2020). T Cell Epitope Predictions. Annu Rev Immunol, 38, 123–145. doi: 10.1146/annurev-immunol-082119-124838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L, Petruccioli E, Vanini V, Cuzzi G, Gualano G, Vittozzi P, … Goletti D (2021). Coinfection of tuberculosis and COVID-19 limits the ability to in vitro respond to SARS-CoV-2. Int J Infect Dis, 113 Suppl 1, S82–S87. doi: 10.1016/j.ijid.2021.02.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L, Picchianti-Diamanti A, Sebastiani GD, Aiello A, Lagana B, Cuzzi G, … Goletti D (2022). Humoral and cellular responses to spike of delta SARS-CoV-2 variant in vaccinated patients with immune-mediated inflammatory diseases. Int J Infect Dis, 121, 24–30. doi: 10.1016/j.ijid.2022.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L, Tortorella C, Aiello A, Farroni C, Ruggieri S, Castilletti C, … Goletti D (2022). Humoral and Cellular Response to Spike of Delta SARS-CoV-2 Variant in Vaccinated Patients With Multiple Sclerosis. Front Neurol, 13, 881988. doi: 10.3389/fneur.2022.881988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M, Abid T, Pereira Ribeiro S, Edara VV, Floyd K, Smith JC, … Kasturi SP (2021). A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci Immunol, 6(61). doi: 10.1126/sciimmunol.abh3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloni C, Schonhofer C, Ivison S, Levings MK, Steiner TS, & Cook L (2023). T-cell activation-induced marker assays in health and disease. Immunol Cell Biol. doi: 10.1111/imcb.12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomaznoy M, Kuan R, Lindvall M, Burel JG, Seumois G, Vijayanand P, … Lindestam Arlehamn CS (2020). Quantitative and Qualitative Perturbations of CD8(+) MAITs in Healthy Mycobacterium tuberculosis-Infected Individuals. Immunohorizons, 4(6), 292–307. doi: 10.4049/immunohorizons.2000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MML, Byington E, Meng W, Kubota M, Matsumoto R, Grifoni A, … Farber DL (2021). Heterogeneity of human anti-viral immunity shaped by virus, tissue, age, and sex. Cell Rep, 37(9), 110071. doi: 10.1016/j.celrep.2021.110071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MML, Rybkina K, Kato Y, Kubota M, Matsumoto R, Bloom NI, … Farber DL (2021). SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci Immunol, 6(65), eabl9105. doi: 10.1126/sciimmunol.abl9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, … de Silva AM (2020). The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol, 5(48). doi: 10.1126/sciimmunol.abc8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt J, Stranford S, Jones P, & Owen J. (2018). Kuby Immunology (8th ed.): W. H. Freeman. [Google Scholar]
- Ramirez SI, Grifoni A, Weiskopf D, Parikh UM, Heaps A, Faraji F, … Vaccines AST (2022). Bamlanivimab therapy for acute COVID-19 does not blunt SARS-CoV-2-specific memory T cell responses. JCI Insight, 7(24). doi: 10.1172/jci.insight.163471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Baxter AE, Cirelli KM, Dan JM, Morou A, Daigneault A, … Kaufmann DE (2017). Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS One, 12(10), e0186998. doi: 10.1371/journal.pone.0186998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Du Bruyn E, Ruzive S, Goliath RT, Lindestam Arlehamn CS, Sette A, … Wilkinson RJ (2020). Disease extent and anti-tubercular treatment response correlates with Mycobacterium tuberculosis-specific CD4 T-cell phenotype regardless of HIV-1 status. Clin Transl Immunology, 9(9), e1176. doi: 10.1002/cti2.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, du Bruyn E, Stek C, Daroowala R, Goliath RT, Abrahams F, … consortium H (2021). Relationship of SARS-CoV-2-specific CD4 response to COVID-19 severity and impact of HIV-1 and tuberculosis coinfection. J Clin Invest, 131(12). doi: 10.1172/JCI149125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison HM, Chapman CA, Zhou H, Erskine CL, Theel E, Peikert T, … Escalante P (2021). Risk assessment of latent tuberculosis infection through a multiplexed cytokine biosensor assay and machine learning feature selection. Sci Rep, 11(1), 20544. doi: 10.1038/s41598-021-99754-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Sidney J, Lindestam Arlehamn CS, Phillips E, Mallal S, Armstrong Suthahar SS, … Ley K (2022). Immunodominant MHC-II (Major Histocompatibility Complex II) Restricted Epitopes in Human Apolipoprotein B. Circ Res, 131(3), 258–276. doi: 10.1161/CIRCRESAHA.122.321116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman Spergel AK, Sever ML, Johnson J, Gill MA, Schulten V, … Frazier A, Infectious Diseases Inner City Asthma, C. (2021). Development of nasal allergen challenge with cockroach in children with asthma. Pediatr Allergy Immunol, 32(5), 971–979. doi: 10.1111/pai.13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C, Kim C, Mateus J, Plested J, Zhu M, Cloney-Clark S, … Crotty S (2022). NVX-CoV2373 vaccination induces functional SARS-CoV-2-specific CD4+ and CD8+ T cell responses. J Clin Invest, 132(19). doi: 10.1172/JCI160898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, … Crotty S (2020). Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell, 183(4), 996–1012 e1019. doi: 10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Lora NE, Kauffman KD, Dorosky DE, Oh S, Namasivayam S, … Barber DL (2021). Functional inactivation of pulmonary MAIT cells following 5-OP-RU treatment of non-human primates. Mucosal Immunol, 14(5), 1055–1066. doi: 10.1038/s41385-021-00425-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouest B, Grifoni A, Pham J, Mateus J, Sydney J, Brien JD, … Weiskopf D (2021). Pre-existing T Cell Memory against Zika Virus. J Virol, 95(12). doi: 10.1128/JVI.00132-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Frazier A, Calatroni A, Kattan M, Bacharier LB, O’Connor GT, … Sette A (2019). The association of allergic sensitization patterns in early childhood with disease manifestations and immunological reactivity at 10 years of age. Clin Exp Allergy, 49(8), 1087–1094. doi: 10.1111/cea.13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Greenbaum JA, Hauser M, McKinney DM, Sidney J, Kolla R, … Peters B (2013). Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci U S A, 110(9), 3459–3464. doi: 10.1073/pnas.1300512110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Tripple V, Aasbjerg K, Backer V, Lund G, Wurtzen PA, … Peters B (2016). Distinct modulation of allergic T cell responses by subcutaneous vs. sublingual allergen-specific immunotherapy. Clin Exp Allergy, 46(3), 439–448. doi: 10.1111/cea.12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Tripple V, Seumois G, Qian Y, Scheuermann RH, Fu Z, … Peters B (2018). Allergen-specific immunotherapy modulates the balance of circulating Tfh and Tfr cells. J Allergy Clin Immunol, 141(2), 775–777 e776. doi: 10.1016/j.jaci.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Tripple V, Sidney J, Greenbaum J, Frazier A, Alam R, … Sette A (2014). Association between specific timothy grass antigens and changes in TH1- and TH2-cell responses following specific immunotherapy. J Allergy Clin Immunol, 134(5), 1076–1083. doi: 10.1016/j.jaci.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten V, Westernberg L, Birrueta G, Sidney J, Paul S, Busse P, … Sette A (2018). Allergen and Epitope Targets of Mouse-Specific T Cell Responses in Allergy and Asthma. Front Immunol, 9, 235. doi: 10.3389/fimmu.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz BM, Melo-Gonzalez F, Duarte LF, Galvez NMS, Pacheco GA, Soto JA, … Bueno SM (2022). A Booster Dose of CoronaVac Increases Neutralizing Antibodies and T Cells that Recognize Delta and Omicron Variants of Concern. mBio, 13(4), e0142322. doi: 10.1128/mbio.01423-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriba TJ, Carpenter C, Pro SC, Sidney J, Musvosvi M, Rozot V, … Arlehamn CSL (2017). Differential Recognition of Mycobacterium tuberculosis-Specific Epitopes as a Function of Tuberculosis Disease History. Am J Respir Crit Care Med, 196(6), 772–781. doi: 10.1164/rccm.201706-1208OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Chesnut R, Livingston B, Wilson C, & Newman M (2000). HLA-binding peptides as a therapeutic approach for chronic HIV infection. IDrugs, 3(6), 643–648. [PubMed] [Google Scholar]
- Sette A, Livingston B, McKinney D, Appella E, Fikes J, Sidney J, … Chesnut R (2001). The development of multi-epitope vaccines: epitope identification, vaccine design and clinical evaluation. Biologicals, 29(3–4), 271–276. doi: 10.1006/biol.2001.0297 [DOI] [PubMed] [Google Scholar]
- Seumois G, Ramirez-Suastegui C, Schmiedel BJ, Liang S, Peters B, Sette A, & Vijayanand P (2020). Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci Immunol, 5(48). doi: 10.1126/sciimmunol.aba6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaan Lakshmanappa Y, Elizaldi SR, Roh JW, Schmidt BA, Carroll TD, Weaver KD, … Iyer SS (2021). SARS-CoV-2 induces robust germinal center CD4 T follicular helper cell responses in rhesus macaques. Nat Commun, 12(1), 541. doi: 10.1038/s41467-020-20642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, & Sette A (2008). HLA class I supertypes: a revised and updated classification. BMC Immunol, 9, 1. doi: 10.1186/1471-2172-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, & Sette A (2020). Epitope prediction and identification- adaptive T cell responses in humans. Semin Immunol, 50, 101418. doi: 10.1016/j.smim.2020.101418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Obregon-Perko V, Lapp SA, Horner AM, Brooks A, Macoy L, … Chahroudi A (2022). Limited induction of SARS-CoV-2-specific T cell responses in children with multisystem inflammatory syndrome compared with COVID-19. JCI Insight, 7(4). doi: 10.1172/jci.insight.155145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania A, Dubelko P, Kuan R, Chronister WD, Muskat K, Das J, … Lindestam Arlehamn C (2021). CD4+CCR6+ T cells dominate the BCG-induced transcriptional signature. EBioMedicine, 74, 103746. doi: 10.1016/j.ebiom.2021.103746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania A, Pham J, Dhanwani R, Frazier A, Rezende Dutra J, Marder KS, … Lindestam Arlehamn CS (2021). The TCR repertoire of alpha-synuclein-specific T cells in Parkinson’s disease is surprisingly diverse. Sci Rep, 11(1), 302. doi: 10.1038/s41598-020-79726-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto JA, Melo-Gonzalez F, Gutierrez-Vera C, Schultz BM, Berrios-Rojas RV, Rivera-Perez D, … Kalergis AM (2022). Inactivated Vaccine-Induced SARS-CoV-2 Variant-Specific Immunity in Children. mBio, 13(6), e0131122. doi: 10.1128/mbio.01311-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic S (2002). Structural basis of immunogenicity. Transpl Immunol, 10(2–3), 133–136. doi: 10.1016/s0966-3274(02)00059-x [DOI] [PubMed] [Google Scholar]
- Sulzer D, Alcalay RN, Garretti F, Cote L, Kanter E, Agin-Liebes J, … Sette A (2017). T cells from patients with Parkinson’s disease recognize alpha-synuclein peptides. Nature, 546(7660), 656–661. doi: 10.1038/nature22815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Ramos I, Coelho CH, Grifoni A, Balinsky CA, Vangeti S, … Letizia AG (2022). Asymptomatic or symptomatic SARS-CoV-2 infection plus vaccination confers increased adaptive immunity to variants of concern. iScience, 25(10), 105202. doi: 10.1016/j.isci.2022.105202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, … Sette A (2022). SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell, 185(5), 847–859 e811. doi: 10.1016/j.cell.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Potesta M, Varchetta S, Fenoglio D, Iannetta M, Sarmati L, … Sette A (2022). Early and Polyantigenic CD4 T Cell Responses Correlate with Mild Disease in Acute COVID-19 Donors. Int J Mol Sci, 23(13). doi: 10.3390/ijms23137155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, … Sette A (2021). Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med, 2(2), 100204. doi: 10.1016/j.xcrm.2021.100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, … Sette A (2021). Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med, 2(7), 100355. doi: 10.1016/j.xcrm.2021.100355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A, Zhang Y, Methot N, Narowski TM, Phillips E, Mallal S, … Grifoni A (2023). Targets and cross-reactivity of human T cell recognition of Common Cold Coronaviruses. bioRxiv. doi: 10.1101/2023.01.04.522794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, da Silva Antunes R, Sidney J, Lindestam Arlehamn CS, Grifoni A, Dhanda SK, … Sette A (2018). A Review on T Cell Epitopes Identified Using Prediction and Cell-Mediated Immune Models for Mycobacterium tuberculosis and Bordetella pertussis. Front Immunol, 9, 2778. doi: 10.3389/fimmu.2018.02778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Grifoni A, Sette A, & Weiskopf D (2019). Human T Cell Response to Dengue Virus Infection. Front Immunol, 10, 2125. doi: 10.3389/fimmu.2019.02125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, … Yewdell JW (2005). Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med, 201(1), 95–104. doi: 10.1084/jem.20041912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukey R, Bruiners N, Mishra H, Mishra PK, McCloskey D, Onyuka A, … Gennaro ML (2022). Dichotomy between the humoral and cellular responses elicited by mRNA and adenoviral vector vaccines against SARS-CoV-2. BMC Med, 20(1), 32. doi: 10.1186/s12916-022-02252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A, Vergara C, Thio CL, Vince N, Douillard V, Grifoni A, … Duggal P (2022). Trans-ancestral fine-mapping of MHC reveals key amino acids associated with spontaneous clearance of hepatitis C in HLA-DQbeta1. Am J Hum Genet, 109(2), 299–310. doi: 10.1016/j.ajhg.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme KFA, Tavernier S, Van Roy N, De Leeuw E, Declercq J, Bosteels C, … Lambrecht BN (2020). Case Report: Convalescent Plasma, a Targeted Therapy for Patients with CVID and Severe COVID-19. Front Immunol, 11, 596761. doi: 10.3389/fimmu.2020.596761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen M, Lienhard M, Schrooders Y, Clayton O, Nudischer R, Boerno S, … Caiment F (2019). DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci Rep, 9(1), 4641. doi: 10.1038/s41598-019-40660-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikkurthi R, Ansari A, Pai AR, Jha SN, Sachan S, Pandit S, … Gupta N (2022). Inactivated whole-virion vaccine BBV152/Covaxin elicits robust cellular immune memory to SARS-CoV-2 and variants of concern. Nat Microbiol, 7(7), 974–985. doi: 10.1038/s41564-022-01161-5 [DOI] [PubMed] [Google Scholar]
- Voic H, de Vries RD, Sidney J, Rubiro P, Moore E, Phillips E, … Grifoni A (2020). Identification and Characterization of CD4(+) T Cell Epitopes after Shingrix Vaccination. J Virol, 94(24). doi: 10.1128/JVI.01641-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, … Sette A (2015). The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol, 89(1), 120–128. doi: 10.1128/JVI.02129-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, … Sette A (2016). HLA-DRB1 Alleles Are Associated With Different Magnitudes of Dengue Virus-Specific CD4+ T-Cell Responses. J Infect Dis, 214(7), 1117–1124. doi: 10.1093/infdis/jiw309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, & Sette A (2014). Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J Virol, 88(19), 11383–11394. doi: 10.1128/JVI.01108-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, … de Vries RD (2020). Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol, 5(48). doi: 10.1126/sciimmunol.abd2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, … Sette A (2011). Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol, 187(8), 4268–4279. doi: 10.4049/jimmunol.1101970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AD, Sibley L, Sarfas C, Morrison A, Gullick J, Clark S, … Sharpe S (2021). MTBVAC vaccination protects rhesus macaques against aerosol challenge with M. tuberculosis and induces immune signatures analogous to those observed in clinical studies. NPJ Vaccines, 6(1), 4. doi: 10.1038/s41541-020-00262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GP, Muskat K, Frazier A, Xu Y, Mateus J, Grifoni A, … Sette A (2023). Unaltered T cell responses to common antigens in individuals with Parkinson’s disease. J Neurol Sci, 444, 120510. doi: 10.1016/j.jns.2022.120510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MP, Wood LF, Templeton M, Fisher B, Lippy A, Jones CI, … Sodora DL (2020). Transient Immune Activation in BCG-Vaccinated Infant Rhesus Macaques Is Not Sufficient to Influence Oral Simian Immunodeficiency Virus Infection. J Infect Dis, 222(1), 44–53. doi: 10.1093/infdis/jiz382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth JS, Clemmensen HS, Battey H, Dijkman K, Lindenstrom T, Laureano RS, … Mortensen R (2021). A Mycobacterium tuberculosis-specific subunit vaccine that provides synergistic immunity upon co-administration with Bacillus Calmette-Guerin. Nat Commun, 12(1), 6658. doi: 10.1038/s41467-021-26934-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Grifoni A, Sutherland A, Voic H, Wang E, Frazier A, … da Silva Antunes R (2021). Balanced Cellular and Humoral Immune Responses Targeting Multiple Antigens in Adults Receiving a Quadrivalent Inactivated Influenza Vaccine. Vaccines (Basel), 9(5). doi: 10.3390/vaccines9050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Narowski TM, Wang E, Garrigan E, Mateus J, Frazier A, … Sette A (2022). Immunological memory to common cold coronaviruses assessed longitudinally over a three-year period pre-COVID19 pandemic. Cell Host Microbe, 30(9), 1269–1278 e1264. doi: 10.1016/j.chom.2022.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Wang E, Garrigan E, Goodwin B, Sutherland A, Tarke A, … da Silva Antunes R (2022). Development of a T cell-based immunodiagnostic system to effectively distinguish SARS-CoV-2 infection and COVID-19 vaccination status. Cell Host Microbe, 30(3), 388–399 e383. doi: 10.1016/j.chom.2022.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Wang E, Garrigan E, Sutherland A, Khalil N, Kearns K, … da Silva Antunes R (2022). Ex vivo assays show human gamma-delta T cells specific for common allergens are Th1-polarized in allergic donors. Cell Rep Methods, 2(12), 100350. doi: 10.1016/j.crmeth.2022.100350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Westernberg L, Grifoni A, Frazier A, Sutherland A, Wang E, … Sette A (2021). B cells modulate mouse allergen-specific T cells in nonallergic laboratory animal-care workers. JCI Insight, 6(4). doi: 10.1172/jci.insight.145199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Mateus J, Coelho CH, Dan JM, Moderbacher CR, Galvez RI, … Crotty S (2022). Humoral and cellular immune memory to four COVID-19 vaccines. Cell, 185(14), 2434–2451 e2417. doi: 10.1016/j.cell.2022.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials availability: MPs described in this protocol will be made available to the scientific community upon request, depending on material availability, and following execution of a material transfer agreement (MTA), by contacting A.S. (alex@lji.org).


