Abstract
The occurrence and abundance of microbial fatty acids have been used for the identification of microorganisms in microbial communities. However, these fatty acids can also be used as indicators of substrate usage. For this, a systematic investigation of the discrimination of the stable carbon isotopes by different microorganisms is necessary. We grew 11 strains representing major bacterial and fungal species with four different isotopically defined carbon sources and determined the isotope ratios of fatty acids of different lipid fractions. A comparison of the differences of δ13C values of palmitic acid (C16:0) with the δ13C values of the substrates revealed that the isotope ratio is independent of the growth stage and that most microorganisms showed enrichment of C16:0 with 13C when growing on glycerol. With the exception of Burkholderia gladioli, all microorganism showed depletion of 13C in C16:0 while incorporating the carbons of glucose, and most of them were enriched with 13C from mannose, with the exception of Pseudomonas fluorescens and the Zygomycotina. Usually, the glycolipid fractions are depleted in 13C compared to the phospholipid fractions. The δ13C pattern was not uniform within the different fatty acids of a given microbial species. Generally, tetradecanoic acid (C14:0) was depleted of 13C compared to palmitic acid (C16:0) while octadecanoic acid (C18:0) was enriched. These results are important for the calibration of a new method in which δ13C values of fatty acids from the environment delineate the use of bacterial substrates in an ecosystem.
Certain components of microbial cells are used as biomarkers for microorganisms (35). A variety of such components, ranging from those found in all cells to those specific to groups or species of microorganisms, have been identified (14). Fatty acids in glyco- and phospholipids constitute an important group of biomarkers for microorganisms. Analysis of such fatty acids of microorganisms in biofilms, soils, sediments and water has provided a reproducible and quantitative means of defining the biomass and community structure of microbial assemblages (35). Furthermore, compositional patterns of fatty acids have been used for the classification of bacteria (2, 23).
In principle, fatty acids may also be used in combination with their role as biomarkers for the monitoring the carbon flux in a bacterial community through measurement of the ratios of their carbon isotopes (26, 28, 29, 34). They are documents of nutritional history. There are several reports that the isotope ratio in the bacterial biomass agrees closely with that in bacterial substrate (1, 5, 9, 10). It was also stated in the literature that the lipids are depleted in 13C compared to the biomass, a phenomenon caused by the discrimination of the carbon isotopes during the oxidation of pyruvate to acetyl coenzyme A (13). Monson and Hayes determined the kinetic isotopic fractionation of carbon in the carboxy group of fatty acids in Escherichia coli (24, 26) and the fractionation during dehydrogenation of the fatty acids in Saccharomyces cerevisiae (25). To determine some of the constants in their models, they took the isotopic fractionation constants determined in the desaturation of fatty acids in Corynebacterium diphtheriae and applied them to the yeast Saccharomyces cerevisiae.
However, no systematic investigation was conducted on the degree and strain specificity of isotopic 13C fractionation with regard to the growth substrate. Before fatty acids can be used as chemotaxonomic markers and as indicators of substrate usage in microbial communities, such a calibration study is urgently needed. With the sensitivity nowadays available in gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS) and its advantage of rapid analyses, this method is ideally suited for the analysis of the individual fatty acids. To study the depletion or enrichment of 13C in the fatty acids in relation to the substrate, we grew 17 microorganisms—7 bacteria and 10 fungi—in a minimal medium with different isotopically defined carbon sources. To compare the microorganisms and also to apply the rules of isotopic fractionation derived from changes in δ13C values of fatty acids from microorganisms grown on substrates of the minimal media, all the strains were additionally grown in a complex medium.
MATERIALS AND METHODS
Strains used in this study.
Pseudomonas sp. strain 8001 (a brown-coal degrader isolated from an open-cast mining lake of brown coal), P. fluorescens LMG 1799, P. putida LMG 2171, Burkholderia gladioli LMG 6877 (36), Brevundimonas diminuta LMG 2089T, Sphingomonas parapaucimobilis LMG J7510T, S. trueperi LMG 2142T (20), Rhizopus arrhizus ATCC 11145, Mortierella isabellina DSM 1414, Fusarium solani DSM 62416, Aspergillus niger DSM 2182, Cunninghamella elegans DSM 1908, Mucor circinelloides CBS 394.68, Thamnidium elegans DSM 912, Chaetomium cochlioides DSM 63353, Glomerella cingulata DSM 1166 and Saccharomyces cerevisiae CBS 1505 were used in this study.
These bacteria and fungi were grown in minimal media with four different carbon sources. For comparison, all the strains were also grown in a complex medium (medium E) consisting of 10 g of glucose per liter (δ13C, −10.10‰), 10 g of universal peptone (Merck) per liter (δ13C, −24.00‰), 20 g of malt extract per liter (δ13C, −11.94‰), and 3 g of yeast extract per liter (δ13C, −22.88‰). Minimal medium for bacteria contained 8 g of NH4H2PO4, 0.2 g of yeast extract, 2 g of K2HPO4, 0.5 g of MgSO4 · 7H2O, 0.5 g of Na2SO4, 0.5 g of NaCl, 10 mg of ZnCl2 · 2H2O, 8 mg of MnSO4 · 7H2O, 10 mg of FeSO4 · 7H2O, and 50 mg of CaCl2 in 1 liter of distilled water. Minimal medium for fungi contained 3 g of NaNO3, 1 g of yeast extract, 1.3 g of K2HPO4, 0.5 g of MgSO4 · 7H2O, 50 mg of FeSO4 · 7H2O, and 0.7 g of citric acid monohydrate (δ13C, −24.94‰) in 1 liter of distilled water.
To these minimal media was added 10 g of glycerol (δ13C, −28.57‰), glucose (δ13C, −10.10‰), mannose (δ13C, −23.74‰), or lactose (δ13C, −27.35‰) per liter. To validate the results, Pseudomonas sp. strain 8001 was grown and analyzed twice in all the media and some of the strains were grown twice in some of the media. To rule out the possibility that the microorganisms grew on the yeast extract of the minimal medium, the growth of all strains on minimal medium without substrate was determined. No strain showed detectable growth (within 72 h) on minimal medium without additional carbon source. Also, growth on the minimal medium with each of the four carbon sources and growth on the complex medium was compared. Only strains and substrates showing comparable growth (90% or more biomass produced compared to the complex medium and corrected for the fourfold carbon content of the complex medium) were included in the study, which excluded Brevundimonas diminuta and Sphingomonas parapaucimobilis, which did not grow on the mannose medium. All the strains were grown at 30°C in 2-liter flasks containing 200 ml of medium with rotation at 100 rpm, and the biomass was harvested in the late exponential phase after 72 h.
Polar lipid fatty acid analysis.
Lipids were extracted by a modified Bligh-Dyer procedure (6) as described previously (15). Briefly, wet cells (2 g) were suspended in methanol-dichloromethane-phosphate buffer; treated for 15 min with an ultrasonic probe; and kept overnight at room temperature. The samples were centrifuged to separate the phases, and the dichloromethane phase was dried. This total lipid fraction was fractionated by column chromatography on silica gel and sequential elution with dichloromethane, acetone, and methanol, which resulted in three fractions of different polarity: neutral lipids, glycolipids, and phospholipids. The glycolipid and phospholipid fractions were separately dissolved in 1 ml of dichloromethane-methanol and subjected to a mild-alkali hydrolysis (1 M KOH–methanol). Impurities were separated from the fatty acids by addition of 2 ml of hexane. After removal of the hexane phase, dichloromethane, buffer, and 6 M HCl were added to the aqueous phase. The organic phase was separated and dried, and the free fatty acids were methylated as described previously (15). A 1-ml volume of n-octane containing internal standards (10 ng of n-hexadecane per ml and 12 ng of n-tetracosane per ml) was added to the dried fatty acid methyl esters (FAMEs), which were then analyzed by GC.
Bound-lipid extraction.
After the total-lipid fraction was removed at the end of the modified Bligh-Dyer procedure, the aqueous phase containing the cell residue was treated with 6 M HCl to obtain the lipids, which previously had not been extracted. After overnight heating at 40°C and cooling to room temperature, an extraction was performed by adding 25 ml of dichloromethane. The dichloromethane phase was dried by filtration through a sodium sulfate-containing phase separation filter (no. 2200150; Whatman International Ltd., Maidstone, United Kingdom) and reduced in volume by rotary evaporation. These so-called bound lipids were dissolved in 1 ml of dichloromethane-methanol (1:1, vol/vol), and an aliquot of 250 μl was methylated as described previously (15).
Capillary CG analyses were performed on a Hewlett-Packard HP 5890 series II gas chromatograph equipped with a capillary column HP Ultra 2 (5% diphenylpolysiloxane 95% dimethylpolysiloxane; length, 50 m; inside diameter, 0.2 mm; film thickness, 0.11 mm). The oven program was 150°C for 2 min, 150 to 289°C at 4°C/min, followed by an isothermal period of 11 min.
MS.
The GC-MS analyses were performed with a similar gas chromatograph to that described for the analysis of the polar lipid fatty acids (same column and conditions but with helium as the carrier gas) connected to a HP 5989A quadrupole mass spectrometer. The electron impact ion source was maintained at 200°C, and the quadropole temperature was 100°C. The electron energy was 70 eV. The dimethyldisulfide adducts were formed as previously described (27) to determine the position of double bonds in monounsaturated fatty acids, while silylated derivatives (Tri-Sil; Pierce, Rockford, Ill.) indicated the position of OH groups by GC-MS measurements.
Determination of δ13C values of biomass by GC-C-IRMS.
1-0.25 mg of dried biomass was combusted in an element analyzer Fisons EA 1108 with CHN packing and analyzed with the isotope ratio mass spectrometer. The analyses were run five times. The standard deviations for all these analyses were 0.1‰ or better.
GC-C-IRMS.
Measurements were performed on a model 252 isotope ratio mass spectrometer (Finnigan MAT, Bremen, Germany) in triplicate. The apparatus is coupled via a combustion interface with an HP 5890 gas chromatograph. The fatty acid methyl esters were separated on a Restek Rtx-2 column (5% diphenylsiloxane, 95% dimethylsiloxane; length, 60 m; inner diameter, 0.32 mm; film thickness, 0.25 μm). The column effluent was combusted on-line in an oxidation oven (copper-nickel-platinum catalyst at 980°C) and passed through a reactor with elemental copper (600°C) to reducing NOx and remove surplus O2. The combustion gas was dried by a water-permeable membrane (Nafion).
Notation.
The standard notation for expressing high-precision gas isotope ratio MS results in δ being defined as follows:
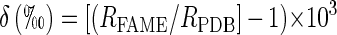 |
1 |
where RFAME and RPDB are the 13C/12C isotope ratios corresponding respectively to the sample and to the international internal standard PeeDee Belemnite, a South Carolinian carbonate rich in 13C (RPDB = 0.0112372 ± 0.0000090). The more 13C a compound contains, the higher δ13C becomes. For the carbon isotopes, a 0.01 (1%) excess in fractional 13C content relative to the natural abundance corresponds approximately to a δPDB of 1,000‰, which is a useful order-of-magnitude approximation.
The carbon isotopic fractionation for the process of conversion of the substrate to the product in question is expressed according to the approach advocated by Hayes (18), using the epsilon (ɛ) notation
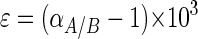 |
2 |
where
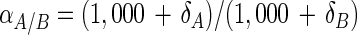 |
3 |
αA/B is the fractionation factor, δA is the δ13C of the substrate, and δB is the δ13C of the fatty acid in question.
Calculation of the isotope ratios of the fatty acids.
The derivatization of the fatty acids introduces one additional carbon which is not present in the parent compound and which alters the original isotope ratio of the fatty acids. However, the derivatization process introduces a distinct reproducible fraction that is constant for each fatty acid (17).
The measured isotope ratios of the FAMEs were corrected for the isotope ratio of the methyl moiety to obtain the isotope ratios of the fatty acids. This was done by using the formula
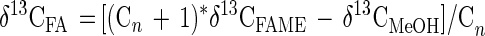 |
4 |
where δ13CFA is the δ13C of the fatty acid, Cn is the number of carbons in the fatty acid, δ13CFAME is the δ13C of the fatty acid methyl ester (FAME), and δ13CMeOH is the δ13C of the methanol used for the methylating reaction (−37.57‰) to calculate the isotope ratios of the fatty acids (3, 17).
Data analysis.
Polar lipid FAMEs were identified by GC-MS analyses with the FAME adducts described above for the determination of the hydroxy and double bond positions. cis-trans isomers were differentiated by comparison with retention times of standards.
RESULTS
Biosynthesis of fatty acids starts in the multienzyme complex fatty acid synthase from acetate and ends with palmitic acid (C16:0). Palmitic acid is then the starting material for dehydro-fatty acids and longer fatty acids, giving this specific fatty acid a central role in the biosynthesis of the vast majority of fatty acids in eubacteria and higher organisms (30). Palmitic acid is present in all eubacteria and higher organisms (22) and we found it in all of our strains during growth in all media and in both the glycolipid and phospholipid fractions (Table 1). It is used here as a standard to determine the depletion or enrichment in 13C between the substrate and the fatty acids.
TABLE 1.
Isotope ratios of common fatty acids from phospholipids, glycolipids, and bound lipids of the strains studied in minimal media with different substrates and in a complex medium (medium E)
| Species | Substratea | Fractionb | Isotope ratio (‰)c in:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| C16:0 | C18:1ω7 | C16:1ω7 | C18:0 | C14:0 | C24:0 | C18:1ω6 | |||
| Brevundimonas diminuta | Glycerol (−28.6‰) | PL | −27.8 ± 0.0 | −26.6 ± 0.0 | −27.2 ± 0.4 | ||||
| GL | −26.1 ± 0.2 | −26.6 ± 0.1 | −24.8 ± 0.9 | −26.7 ± 0.0 | |||||
| BL | −26.9 ± 0.1 | −27.7 ± 0.1 | |||||||
| Glucose (−10.1‰) | PL | −16.7 ± 0.8 | −14.2 ± 0.2 | −12.9 ± 0.0 | −16.8 ± 0.0 | ||||
| GL | −19.8 ± 0.3 | −20.3 ± 0.0 | −17.7 ± 0.7 | ||||||
| BL | −18.2 ± 0.0 | −14.2 ± 0.0 | |||||||
| Lactose (−27.4‰) | PL | −26.0 ± 0.0 | −24.8 ± 0.1 | −24.0 ± 0.2 | −23.6 ± 0.9 | −27.6 ± 0.3 | |||
| GL | −26.3 ± 1.7 | −26.9 ± 1.8 | |||||||
| BL | −23.1 ± 0.0 | −23.7 ± 0.3 | −24.9 ± 1.3 | ||||||
| Medium E | PL | −31.1 ± 0.1 | −28.8 ± 0.2 | −21.6 ± 0.1 | −19.6 ± 0.3 | −26.6 ± 0.3 | |||
| GL | −31.1 ± 0.3 | −26.6 ± 0.9 | −26.3 ± 2.0 | ||||||
| BL | −29.8 ± 0.0 | −27.7 ± 0.0 | |||||||
| Burkholderia gladioli | Glycerol (−28.6‰) | PL | −26.0 ± 0.4 | −26.1 ± 0.0 | −26.9 ± 0.7 | −26.9 ± 0.7 | −26.1 ± 0.2 | ||
| GL | −24.4 ± 0.1 | −22.7 ± 0.2 | −24.9 ± 1.2 | −23.1 ± 0.2 | |||||
| BL | −24.2 ± 0.1 | −26.8 ± 0.1 | −26.6 ± 0.1 | −261 | −26.6 ± 0.2 | ||||
| Glucose (−10.1‰) | PL | −8.9 ± 0.1 | −18.1 ± 0.6 | −12.6 ± 0.1 | |||||
| GL | −14.0 ± 0.6 | −17.4 ± 0.4 | −14.9 ± 3.0 | −19.9 ± 0.8 | −14.2 ± 0.2 | ||||
| BL | −8.6 ± 0.1 | −19.6 ± 0.2 | −12.0 ± 0.6 | ||||||
| Mannose (−23.7‰) | PL | −22.4 ± 0.2 | −26.4 ± 0.6 | ||||||
| GL | −22.1 ± 0.0 | −24.6 ± 0.0 | |||||||
| BL | −20.7 ± 0.1 | −22.3 ± 0.0 | |||||||
| Lactose (−27.4‰) | PL | −24.8 ± 0.0 | −26.0 ± 0.0 | −23.9 ± 0.4 | −23.6 ± 0.3 | −27.6 ± 0.0 | |||
| GL | −28.7 ± 1.3 | −24.6 ± 0.6 | −26.4 ± 0.6 | ||||||
| BL | −21.1 ± 0.1 | −24.3 ± 0.0 | −22.9 ± 0.6 | −23.4 ± 0.3 | |||||
| Medium E | PL | −19.0 ± 0.0 | −20.9 ± 0.6 | −23.9 ± 0.2 | |||||
| GL | −19.8 ± 0.0 | −26.3 ± 0.4 | −23.6 ± 2.6 | −23.4 ± 0.6 | |||||
| BL | −17.3 ± 0.1 | −27.2 ± 0.4 | |||||||
| Pseudomonas fluorescens | Glycerol (−28.6‰) | PL | −27.6 ± 0.1 | −26.9 ± 0.1 | −26.2 ± 0.0 | −26.2 ± 0.1 | −29.9 ± 0.1 | −28.4 ± 0.3 | |
| GL | −27.3 ± 0.2 | −26.8 ± 0.3 | −26.7 ± 0.2 | −27.0 ± 3.3 | −29.6 ± 0.9 | ||||
| BL | −26.6 ± 0.0 | −24.6 ± 2.0 | −26.2 ± 0.1 | −29.0 ± 1.6 | |||||
| Glucose (−10.1‰) | PL | −12.8 ± 0.0 | −12.6 ± 0.1 | −12.6 ± 0.1 | −11.6 ± 0.0 | −13.0 ± 0.3 | |||
| GL | −16.2 ± 0.1 | −13.4 ± 0.1 | −13.6 ± 0.1 | −14.9 ± 0.6 | −17.8 ± 0.6 | ||||
| BL | −13.2 ± 0.2 | −13.0 ± 1.6 | −13.2 ± 0.1 | ||||||
| Mannose (−23.7‰) | PL | −26.8 ± 0.0 | −24.3 ± 0.2 | −24.3 ± 0.3 | −26.9 ± 2.9 | ||||
| GL | −26.6 ± 0.1 | −24.3 ± 0.2 | −24.0 ± 0.1 | −24.9 ± 0.2 | |||||
| BL | −24.3 ± 0.1 | −23.0 ± 0.2 | −24.0 ± 0.3 | −24.1 ± 1.0 | |||||
| Lactose (−27.4‰) | PL | −24.6 ± 0.2 | −23.9 ± 0.7 | −23.9 ± 0.2 | −23.2 ± 0.4 | −26.0 ± 0.0 | |||
| GL | −26.8 ± 1.0 | −26.3 ± 0.1 | −23.9 ± 0.6 | ||||||
| BL | −23.2 ± 0.1 | −23.1 ± 0.1 | −26.3 ± 0.6 | −23.2 ± 0.7 | |||||
| Medium E | PL | −26.6 ± 0.3 | −24.0 ± 0.2 | ||||||
| GL | −24.9 ± 0.2 | −23.6 ± 0.1 | −23.8 ± 0.0 | ||||||
| BL | −23.8 ± 0.1 | −24.7 ± 0.1 | −33.1 ± 0.1 | −42.0 ± 0.2 | |||||
| Pseudomonas putida | Glycerol (−28.6‰) | PL | −24.0 ± 0.2 | −26.9 ± 0.1 | −22.6 ± 0.2 | −23.0 ± 0.0 | −26.6 ± 0.1 | −28.8 ± 0.1 | |
| GL | −23.8 ± 0.1 | −24.9 ± 0.4 | −22.3 ± 0.1 | −26.2 ± 0.1 | −28.3 ± 0.3 | ||||
| BL | −22.1 ± 0.1 | −26.2 ± 0.1 | −26.2 ± 0.0 | −28.2 ± 0.1 | |||||
| Glucose (−10.1‰) | PL | −11.7 ± 0.1 | −13.3 ± 0.0 | −11.8 ± 0.0 | −11.6 ± 0.3 | −16.4 ± 2.3 | −16.3 ± 0.3 | ||
| GL | −12.7 ± 0.8 | −13.2 ± 0.1 | −12.6 ± 0.4 | −13.6 ± 0.3 | −16.0 ± 1.6 | ||||
| BL | −11.6 ± 0.0 | −11.8 ± 0.1 | −13.7 ± 1.0 | ||||||
| Mannose (−23.7‰) | PL | −18.2 ± 0.3 | −22.8 ± 0.2 | −26.3 ± 2.7 | −20.6 ± 0.2 | ||||
| GL | −17.6 ± 0.2 | −22.1 ± 0.3 | −14.7 ± 0.6 | −19.2 ± 1.6 | −19.0 ± 0.6 | −22.4 ± 0.3 | |||
| BL | −16.8 ± 0.0 | −21.3 ± 0.0 | −17.3 ± 1.2 | −19.6 ± 0.4 | |||||
| Lactose (−27.4‰) | PL | −24.6 ± 0.3 | −24.7 ± 0.2 | −23.0 ± 0.6 | −23.8 ± 0.3 | −26.7 ± 0.0 | |||
| GL | −26.6 ± 0.2 | −26.4 ± 0.2 | −21.6 ± 1.2 | ||||||
| BL | −22.6 ± 0.1 | −23.8 ± 0.2 | −23.3 ± 0.1 | ||||||
| Medium E | PL | −18.4 ± 0.2 | −20.6 ± 0.2 | −17.7 ± 0.3 | −18.0 ± 0.2 | −22.8 ± 2.8 | −22.8 ± 0.4 | ||
| GL | −19.2 ± 0.1 | −21.6 ± 0.8 | −18.6 ± 0.0 | −22.4 ± 4.2 | −23.9 ± 0.6 | ||||
| BL | −16.8 ± 0.0 | −20.0 ± 0.2 | −22.8 ± 0.0 | −26.0 ± 0.4 | |||||
| Pseudomonas sp. strain 8001 | Glycerol (−28.6‰) | PL | −27.8 ± 0.1 | −27.4 ± 0.1 | −26.7 ± 0.1 | −26.6 ± 0.3 | −30.6 ± 0.4 | −29.8 ± 0.1 | |
| GL | −27.6 ± 0.0 | −26.4 ± 0.9 | −26.2 ± 0.7 | −31.7 ± 0.0 | −30.0 ± 0.8 | −28.8 ± 0.3 | |||
| BL | −26.3 ± 0.1 | −26.3 ± 0.1 | −27.2 ± 0.1 | −30.6 ± 0.1 | |||||
| Glucose (−10.1‰) | PL | −13.1 ± 0.2 | −13.0 ± 0.1 | −13.2 ± 0.4 | −12.3 ± 2.8 | −14.6 ± 0.6 | |||
| GL | −13.6 ± 0.2 | −13.2 ± 0.1 | −12.8 ± 0.1 | −12.3 ± 0.3 | −18.7 ± 0.3 | ||||
| BL | −12.6 ± 0.1 | −16.6 ± 0.1 | −19.0 ± 2.9 | ||||||
| Mannose (−23.7‰) | PL | −20.0 ± 0.2 | −20.3 ± 0.0 | −18.6 ± 0.1 | −18.8 ± 0.1 | −20.6 ± 1.3 | −22.7 ± 0.6 | ||
| GL | −21.1 ± 0.2 | −21.3 ± 0.9 | −20.6 ± 1.6 | −22.4 ± 1.1 | −21.6 ± 0.8 | ||||
| BL | −18.8 ± 0.1 | −20.7 ± 0.1 | −22.6 ± 0.3 | −22.3 ± 0.6 | |||||
| Lactose (−27.4‰) | PL | −26.1 ± 0.0 | −24.9 ± 0.1 | −24.3 ± 0.0 | −23.6 ± 0.1 | −28.1 ± 0.6 | |||
| GL | −26.9 ± 0.7 | −26.6 ± 0.1 | −24.9 ± 0.6 | ||||||
| BL | −23.3 ± 0.1 | −23.2 ± 0.0 | −26.2 ± 0.3 | −22.9 ± 0.3 | |||||
| Medium E | PL | −21.6 ± 0.1 | −19.9 ± 0.1 | −20.2 ± 0.1 | −17.8 ± 0.2 | −26.6 ± 0.0 | −23.4 ± 0.3 | ||
| GL | −23.0 ± 0.1 | −22.0 ± 0.2 | −21.9 ± 0.1 | −19.9 ± 0.3 | |||||
| BL | −19.6 ± 0.1 | −19.0 ± 0.1 | −26.1 ± 0.1 | −18.1 ± 0.1 | −22.2 ± 0.3 | ||||
| Sphingomonas trueperi | Glycerol (−28.6‰) | PL | −29.8 ± 0.0 | −26.1 ± 0.1 | −30.8 ± 0.8 | ||||
| GL | −28.6 ± 0.1 | −26.7 ± 0.1 | −27.1 ± 0.4 | ||||||
| BL | −27.3 ± 0.1 | −26.2 ± 0.1 | |||||||
| Glucose (−10.1‰) | PL | −13.3 ± 0.1 | −12.7 ± 0.2 | ||||||
| GL | −12.1 ± 0.1 | −12.7 ± 1.0 | −11.0 ± 0.0 | ||||||
| BL | −12.3 ± 0.2 | −9.2 ± 0.0 | |||||||
| Mannose (−23.7‰) | PL | ||||||||
| GL | |||||||||
| BL | −26.9 ± 0.1 | −26.7 ± 0.7 | −26.4 ± 0.6 | ||||||
| Lactose (−27.4‰) | PL | −26.9 ± 0.2 | −28.7 ± 0.1 | −24.8 ± 0.1 | −28.8 ± 0.3 | −28.3 ± 1.2 | |||
| GL | −31.2 ± 0.9 | −33.1 ± 1.4 | −26.4 ± 0.1 | ||||||
| BL | −27.4 ± 0.1 | −26.4 ± 0.6 | |||||||
| Medium E | PL | −20.7 ± 0.0 | −19.9 ± 0.4 | −21.7 ± 1.4 | |||||
| GL | −22.0 ± 0.3 | −20.9 ± 0.1 | −20.1 ± 1.1 | ||||||
| BL | −20.4 ± 0.4 | −19.4 ± 0.1 | |||||||
| Sphingomonas parapauco-mobilis | Glycerol (−28.6‰) | PL | −26.6 ± 0.0 | −26.4 ± 0.2 | −26.0 ± 0.4 | −26.1 ± 0.0 | −24.1 ± 0.1 | ||
| GL | −29.2 ± 0.1 | −27.2 ± 0.7 | −28.0 ± 0.4 | ||||||
| BL | −29.7 ± 0.0 | −26.9 ± 0.0 | −26.8 ± 0.2 | ||||||
| Glucose (−10.1‰) | PL | −10.9 ± 0.6 | −11.3 ± 0.3 | −10.1 ± 0.4 | −12.6 ± 0.4 | ||||
| GL | −14.1 ± 0.6 | −13.4 ± 1.3 | −13.0 ± 0.8 | −16.2 ± 6.2 | |||||
| BL | −10.6 ± 0.2 | ||||||||
| Lactose (−27.4‰) | PL | −23.2 ± 0.3 | −23.6 ± 0.0 | −22.6 ± 0.6 | −21.1 ± 0.3 | −24.9 ± 0.2 | |||
| GL | −27.4 ± 0.2 | −29.6 ± 0.2 | |||||||
| BL | −23.6 ± 0.1 | −27.6 ± 0.0 | −28.2 ± 0.1 | −30.2 ± 0.2 | −22.6 ± 0.6 | ||||
| Medium E | PL | −22.3 ± 1.7 | −26.3 ± 0.9 | −28.4 ± 1.6 | |||||
| GL | −26.3 ± 0.2 | −22.6 ± 0.0 | −22.9 ± 0.1 | −26.9 ± 0.6 | |||||
| BL | −22.9 ± 0.0 | −20.3 ± 0.1 | −26.0 ± 1.3 | ||||||
| Aspergillus niger | Glycerol (−28.6‰) | PL | −22.6 ± 0.1 | −23.6 ± 0.6 | −21.1 ± 1.1 | ||||
| GL | −24.7 ± 2.0 | −23.3 ± 0.6 | |||||||
| BL | −21.6 ± 0.2 | −22.8 ± 0.4 | −22.6 ± 0.4 | ||||||
| Glucose (−10.1‰) | PL | −19.4 ± 0.1 | |||||||
| GL | −19.4 ± 0.2 | −18.0 ± 0.6 | |||||||
| BL | −18.6 ± 0.1 | −14.6 ± 0.1 | |||||||
| Mannose (−23.7‰) | PL | −18.0 ± 0.3 | −18.8 ± 0.6 | ||||||
| GL | −18.9 ± 0.2 | −17.9 ± 0.7 | |||||||
| BL | −18.4 ± 0.6 | −19.7 ± 0.1 | −18.3 ± 0.2 | ||||||
| Lactose (−27.4‰) | PL | −27.6 ± 0.9 | −27.7 ± 0.0 | ||||||
| GL | −27.6 ± 1.6 | −26.6 ± 0.4 | |||||||
| Medium E | PL | −14.1 ± 0.2 | −14.2 ± 0.9 | −14.3 ± 0.4 | |||||
| GL | −16.1 ± 0.2 | −17.0 ± 1.6 | −16.4 ± 0.6 | ||||||
| BL | −14.1 ± 0.2 | −17.7 ± 0.0 | −13.9 ± 0.1 | −13.7 ± 0.0 | |||||
| Fusarium solani | Glycerol (−28.6‰) | PL | −26.6 ± 0.1 | −26.0 ± 1.0 | −26.6 ± 0.6 | ||||
| GL | −26.6 ± 0.6 | −29.6 ± 2.9 | |||||||
| BL | −26.6 ± 0.0 | −27.9 ± 0.1 | −26.1 ± 0.0 | −22.6 ± 0.0 | |||||
| Glucose (−10.1‰) | PL | −20.4 ± 0.3 | |||||||
| GL | −16.6 ± 0.3 | −17.3 ± 0.2 | −16.6 ± 0.0 | −16.4 ± 0.6 | |||||
| BL | −18.3 ± 0.2 | −19.6 ± 0.0 | −14.9 ± 0.0 | ||||||
| Mannose (−23.7‰) | PL | −19.9 ± 2.3 | −20.8 ± 0.8 | ||||||
| GL | −18.4 ± 0.0 | ||||||||
| BL | −22.4 ± 0.0 | −26.8 ± 0.1 | −24.3 ± 0.2 | −26.8 ± 0.2 | −23.4 ± 0.1 | ||||
| Lactose (−27.4‰) | PL | −23.9 ± 0.2 | −26.4 ± 0.1 | −24.2 ± 0.1 | |||||
| GL | −28.1 ± 0.4 | −30.3 ± 0.3 | −28.2 ± 3.0 | ||||||
| Medium E | PL | −19.4 ± 0.6 | −20.3 ± 0.4 | ||||||
| GL | −20.1 ± 0.6 | −20.4 ± 2.2 | −19.9 ± 0.6 | ||||||
| Mortierella isabellina | Glycerol (−28.6‰) | PL | −26.6 ± 0.1 | −26.1 ± 2.4 | −26.7 ± 0.0 | ||||
| GL | −26.1 ± 0.2 | −28.3 ± 0.6 | −26.4 ± 0.0 | −26.4 ± 0.7 | |||||
| BL | −26.2 ± 0.2 | ||||||||
| Glucose (−10.1‰) | PL | −17.8 ± 0.6 | −18.4 ± 3.6 | ||||||
| GL | −18.3 ± 0.0 | −18.7 ± 0.8 | |||||||
| BL | −16.7 ± 0.2 | −16.9 ± 0.1 | −16.8 ± 0.8 | −12.1 ± 0.3 | |||||
| Mannose (−23.7‰) | PL | −26.1 ± 0.2 | −26.9 ± 0.4 | −28.0 ± 1.1 | −27.7 ± 3.9 | ||||
| GL | −27.1 ± 0.0 | −27.2 ± 0.9 | −28.0 ± 0.2 | −27.3 ± 0.3 | |||||
| BL | −26.8 ± 0.2 | −26.0 ± 0.0 | −27.4 ± 0.1 | −26.7 ± 0.2 | |||||
| Lactose (−27.4‰) | PL | −28.9 ± 0.2 | −29.8 ± 0.7 | −30.2 ± 0.4 | |||||
| GL | −29.2 ± 0.3 | −30.4 ± 0.2 | −29.8 ± 0.6 | −28.1 ± 1.9 | |||||
| Medium E | PL | −10.4 ± 0.1 | −13.6 ± 0.2 | −11.6 ± 0.7 | −9.0 ± 0.2 | ||||
| GL | −16.7 ± 0.1 | −17.3 ± 0.2 | −16.4 ± 0.8 | −16.0 ± 0.0 | −20.6 ± 0.1 | ||||
| BL | −13.4 ± 0.0 | −14.9 ± 0.0 | −16.3 ± 0.3 | −16.8 ± 0.3 | −12.8 ± 0.0 | −12.4 ± 0.2 | |||
| Rhizopus arrhizus | Glycerol (−28.6‰) | PL | −28.0 ± 0.2 | −27.4 ± 0.3 | −32.4 ± 2.3 | ||||
| GL | −27.6 ± 0.0 | −28.1 ± 0.2 | −30.2 ± 0.7 | ||||||
| BL | −26.8 ± 0.1 | −26.9 ± 0.1 | −27.9 ± 0.9 | −26.4 ± 1.8 | −29.3 ± 0.9 | ||||
| Glucose (−10.1‰) | PL | −16.4 ± 0.0 | −16.6 ± 0.6 | ||||||
| GL | −13.8 ± 0.0 | −16.2 ± 0.2 | −18.6 ± 4.3 | ||||||
| BL | −12.2 ± 0.0 | −14.6 ± 0.1 | −14.6 ± 0.0 | −9.6 ± 0.1 | |||||
| Mannose (−23.7‰) | PL | −26.8 ± 0.0 | −28.0 ± 0.1 | −31.6 ± 0.3 | |||||
| GL | −28.1 ± 0.2 | −29.0 ± 0.1 | −27.8 ± 1.0 | ||||||
| Lactose (−27.4‰) | PL | −27.7 ± 0.1 | −28.1 ± 0.1 | ||||||
| GL | −26.0 ± 0.0 | −26.6 ± 0.2 | −29.7 ± 0.0 | ||||||
| BL | −26.9 ± 0.0 | −28.3 ± 0.1 | −26.1 ± 0.1 | −23.7 ± 0.1 | |||||
| Medium E | PL | −14.8 ± 0.1 | −16.6 ± 0.4 | −16.0 ± 0.1 | −16.0 ± 0.4 | −19.6 ± 0.3 | |||
| GL | −16.3 ± 0.6 | −13.3 ± 0.3 | −16.2 ± 1.7 | ||||||
| Cunninghamella elegans | Mannose (−23.7‰) | PL | −28.6 ± 0.3 | −29.7 ± 1.0 | |||||
| GL | −29.3 ± 0.0 | −28.8 ± 0.3 | −29.7 ± 0.0 | −32.8 ± 0.2 | |||||
| Mucor circinel-loides | Mannose (−23.7‰) | PL | −22.6 ± 0.2 | −22.8 ± 0.1 | −22.0 ± 0.6 | −26.9 ± 0.6 | |||
| GL | −23.9 ± 0.2 | −23.0 ± 0.2 | −23.6 ± 0.3 | ||||||
| Thamnidium elegans | Mannose (−23.7‰) | PL | −22.1 ± 0.2 | −23.6 ± 1.3 | |||||
| GL | −26.6 ± 0.0 | −26.6 ± 0.2 | −28.1 ± 0.3 | −26.2 ± 1.3 | |||||
| Chaetomium cochlioides | Mannose (−23.7‰) | PL | −29.2 ± 0.2 | −29.9 ± 2.1 | |||||
| GL | −29.9 ± 0.0 | −29.1 ± 1.3 | −28.6 ± 0.1 | ||||||
| Glomerella cingulata | Mannose (−23.7‰) | PL | −26.4 ± 0.1 | −26.9 ± 0.2 | |||||
| GL | −26.4 ± 0.2 | −27.1 ± 0.1 | −29.6 ± 1.7 | ||||||
| Saccharomyces cerevisiae | Mannose (−23.7‰) | PL | −24.6 ± 0.0 | ||||||
| GL | −24.8 ± 0.2 | −36.1 ± 0.1 | −26.3 ± 0.1 | ||||||
For better comparison, the isotope ratio of the substrate is given in parentheses.
PL, phospholipid; GL, glycolipid; BL, bound lipid.
Results are means and standard deviations (n = 4; P < 0.05).
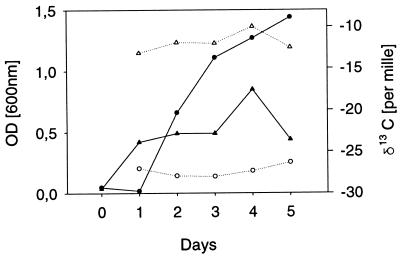
For the interpretation of the results, it is important to know whether the isotope ratios in fatty acids depend on the growth stage of the microorganisms. Previous authors reported that apart from methane oxidizers, the isotope ratios in fatty acids are independent of the growth stage of the microorganisms. Because this observation is crucial for our study, we determined the isotope ratio in C16:0 at different growth stages but found no significant differences (Fig. 1). The result corroborated the findings of Summons et al. (32).
FIG. 1.
Growth curves and isotope ratios of the palmitic acid (C16:0) of Pseudomonas putida in defined medium with glucose (triangles) and glycerol (circles). Biomass was determined as the optical density at 600 nm (OD600) and is shown as solid symbols, whereas the isotope ratios of C16:0 are given as open symbols.
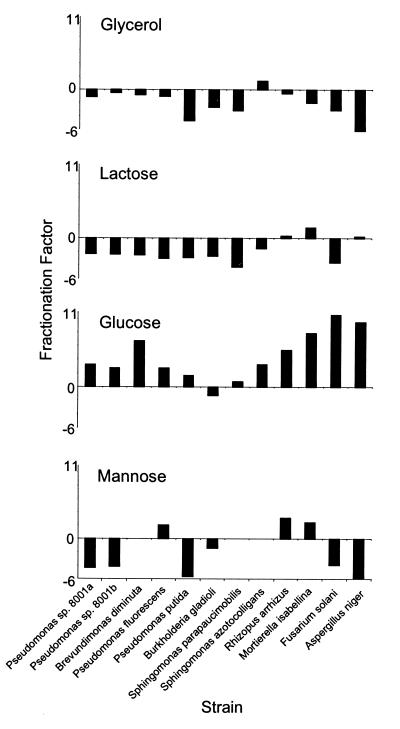
The δ13C values determined for the individual fatty acids revealed that fatty acids were depleted in 13C in relation to glucose in all the microorganisms examined (with the single exception of two lipid fractions of Burkholderia gladioli) (Table 1). The depletion was stronger within the fungi than within the bacteria. The extent of the depletion ranged from a δ13C of 0.45‰ in C16:0 of the phospholipids of Burkholderia gladioli to a δ13C of 11.34‰ in C16:0 of the phospholipids of Fusarium solani. Most bacteria and the Ascomycotina possessed C16:0 enriched in 13C when growing in glycerol. However, C16:0 was depleted in 13C in Sphingomonas trueperi, while the Zygomycotina had only a slightly changed isotope ratio during the incorporation of the carbons of glycerol into C16:0. The situation was also not uniform with lactose, since Brevundimonas, Burkholderia, and Pseudomonas formed C16:0 slightly enriched in 13C while Sphingomonas and the fungi formed C16:0 slightly depleted in 13C (Fig. 2).
FIG. 2.
Carbon isotopic fractionation, ɛ, of C16:0 from four different carbon sources (for definitions, see Materials and Methods). The four diagrams have the same scale. Pseudomonas sp. strain 8001 was grown twice with all substrates and is shown as 8001a and 8001b to show biological reproducibility.
With mannose as the substrate, a broad range of isotope differentiation was observed. C16:0 was enriched in 13C by Burkholderia gladioli and all Pseudomonas spp. except P. fluorescens. The two fungi belonging to the Zygomycotina showed a distinctly different isotopic fractionation from those of the Ascomycotina. To explore this outstanding effect further, three more fungi of the Zygomycotina (Cunninghamella elegans DSM 1908, Mucor circinelloides CBS 394.68, and Thamnidium elegans DSM 912) and Ascomycotina (Chaetomium cochlioides DSM 63353, Glomerella cingulata DSM 1166, and Saccharomyces cerevisiae CBS 1505) were tested in the minimal medium with mannose. The 10 fungi displayed a very broad range of isotope ratios in C16:0, ranging from a δ13C of 17.96‰ to 29.22‰ in the phospholipids. No clear difference could be found between members of the Ascomycotina and those of the Zygomycotina, although the latter usually formed C16:0 more depleted in 13C than the former did. With one exception, C16:0 from glycolipid fractions was depleted in 13C compared to that from phospholipid fractions corroborating our earlier findings.
Knowing the extent of the different uptake of 13C from the individual substrates by the individual strains, the use of the different carbon sources in a complex medium (medium E in Table 1) with its many different types of substrates can be better understood. The data for fatty acids from the four fungal strains grown on complex medium are all very similar to those found for fatty acids from the fungi grown on glucose as the sole carbon source. These results show that the fungi all used glucose from the medium, indicated by the δ13C values of C16:0 in the two fractions of polar lipids. In contrast to the fungi, the pattern of substrate utilization in bacteria was less obvious, since a much broader range of δ13C values was observed for their fatty acids. However, the range is very different from that observed with the same strains with glucose; therefore, glucose can be excluded as the main source of carbon for bacteria in this complex medium.
DISCUSSION
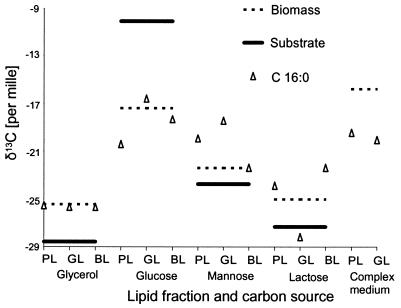
The findings in previous studies that carbon of whole bacterial cells reflect within 3‰ the ratio of substrate carbon isotopes (1, 5, 10) proved to be a rough estimate for fatty acids (Fig. 3). Using minimal media with isotopically defined substrates, we could show that each substrate is individually processed, leading to a range of isotope ratios in the biomass and C16:0. Glucose showed the largest difference in carbon isotopes in C16:0 in relation to the substrate (13). A comparison of the variations in δ13C values of C16:0 to the δ13C values of the substrates revealed that most microorganisms, except Sphingomonas spp., formed C16:0 enriched in 13C (ɛ < 0) when growing on glycerol. With the exception of Burkholderia gladioli, all the strains formed C16:0 (ɛ > 0) depleted in 13C while incorporating the carbons of glucose, most of them formed C16:0 enriched in 13C from mannose with the clear exception of P. fluorescens and the Zygomycotina, and lactose was incorporated with a carbon isotopic fractionation between ɛ = −9 and ɛ = +4 (Fig. 2).
FIG. 3.
Isotope ratios of biomass and C16:0 from the different fractions of polar and bound lipids of Fusarium solani compared with the isotope ratio of the corresponding substrate. Abbreviations: PL, phospholipids; GL, glycolipids; BL, bound lipids. For clarity, the isotope ratio of the substrate and the biomass is shown three times for direct comparison with those of the fatty acid.
A comparison of the isotope ratios of C16:0 in the different lipid fractions of the same strain, shown for Pseudomonas putida in Fig. 3, reveals differences of up to 7.5‰ (B. gladioli). Such surprisingly large differences need further analysis because stable isotope ratios of less than 5‰ have been used in studies of the carbon flux in ecosystems (8, 11, 16). The correlation of the isotope ratios of the fatty acids of a given lipid fraction with the ones of another lipid fraction of the same strain and carbon source revealed that all fatty acids of a lipid fraction have an offset of their isotope ratios against the other lipid fractions. This points to the biosynthesis of the polar lipids itself as the cause of the observed fractionation, and not the biosynthesis of the fatty acids. Although there are exceptions, the glycolipid fractions are usually depleted in 13C compared to the phospholipid fractions. The bound lipids, although originating from a variety of different compounds, are more similar to the phospholipids than to the glycolipids in their isotope ratio. They usually present a lipid fraction most highly enriched in 13C of the three types investigated in this study. Phospholipids, especially phosphatidate, play a central role in the biosynthesis of glycolipids (7, 31, 33) and bound lipids (11a, 21, 35a). Phosphatidic acid is cleaved by phosphatidate phosphatase to diacylglycerol, which is the substrate for hexosyltransferases, opening the field to glycolipids (4). Most enzymes investigated in this respect show, to a greater or lesser extent, carbon isotopic fractionations; therefore, the isotope ratio of an end product of a biosynthesis is the sum of all these fractionations (13, 24). This probably explains why the isotope ratio of C16:0 obtained from the phospholipid fraction is not identical to the one from the glycolipid or bound-lipid fraction.
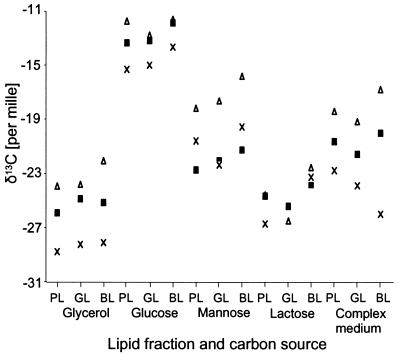
The discrimination of carbon isotopes was not uniform within the different fatty acids of a strain. Each strain showed its own pattern of variance within the δ13C data for the fatty acids, which also depended on the substrate. However, some empirical rules were found. For the saturated fatty acids, a pronounced shift in δ13C values was observed. Generally, myristic acid (C14:0) was depleted in 13C compared to C16:0 while stearic acid (C18:0) was enriched. The results with Rhizopus arrhizus indicated that the simple rule that the longer the fatty acid the heavier, cannot be applied in general. Here, tetracosanoic acid (C24:0) is more depleted of 13C than is C16:0 or C18:0. Growing on glycerol, Pseudomonas strain 8001, P. fluorescens, and P. putida showed enriched 13C in 9-hexadecenoic acid (C16:1ω7) and 11-octadecenoic acid (C18:1ω7) compared to C16:0. The fatty acids 12-octadecenoic acid (C18:1ω6) and C18:1ω7, differing only in the position of the double bond, displayed different isotope ratios, pointing to the reaction of the desaturase as the source of these differences (Fig. 4). Within the fungi, such a common pattern could not be observed, and no clear tendency for the individual fatty acids could be found. The range of δ13C values for the individual fatty acids was somewhat smaller for the fungi than for the bacteria.
FIG. 4.
Isotope ratios of different fatty acids of Pseudomonas putida in four different minimal media and one complex medium. Open triangles, C16:0; solid squares, C18:1ω7; crosses, C18:1ω6. GL, glycolipids; PL, phospholipids; BL, bound lipids.
Isotopic fractionation of C16:0 of polar lipids occurs (i) during transport of the substrate into the cell and its degradation to acetate, (ii) during the synthesis of palmitic acid, and (iii) during its transportation to the membrane and its esterification to the polar lipids. From our data, it is possible to estimate the contributions of isotopic fractionation resulting from transport and degradation of the substrate, the fractionation caused by the synthesis of palmitic acid, and the fractionation resulting from the synthesis of the polar lipids. The transportation of the substrate and its breakdown to acetate is different for each substrate, and so different isotopic fractionations can be expected in a given strain. The synthesis of C16:0 from acetate, however, is the same in a given microorganism; therefore, the isotopic fractionation should also be the same. However, the comparison of isotopic fractionation of C16:0 from microorganisms, grown on different substrates, indicates that for most strains growing in glucose, the first part of this process may be the more important one because of the large 13C depletion in C16:0, while for all the other substrates tested the synthesis of the fatty acid may be the main contribution to the isotopic fractionation in the fatty acids. One important exception to this observation is mannose; for most microorganisms, the transport and the degradation of the substrate again seem to provide the main contribution to the observed differences in isotope ratios of the fatty acids.
The same fatty acid, analyzed from different lipid fractions, shows different isotope ratios. Usually the isotope ratio in a given fatty acid obtained from the phospholipid fraction is more similar to that in the same fatty acid from the bound lipid than to that in the fatty acid from the glycolipid fraction. The glycolipid fatty acids are very often depleted in 13C compared to those from the phospholipid or bound-lipid fraction, although some exceptions were observed. These differences cannot be explained by isotopic fractionation during the synthesis of the fatty acids but must be attributed to the biosynthesis of the polar lipid, i.e., the transportation of the fatty acid to the membrane and its esterification to the polar lipids. Therefore, the enzyme(s) responsible for the differing isotopic fractionation of the glycolipids must act after the formation of phosphatidic acid, the last common intermediate of both glycolipids and phospholipids.
This study demonstrated that the fatty acids of polar lipids document the nutritional history of the strains and can serve as powerful tools in combination with GC-C-IRMS for the study of carbon flux in microbial communities. The application of the rules derived above can help to improve the interpretation of the results obtained from complex communities utilizing a wide range of substrates. Fatty acids from polar lipids can be used to identify substrates in extracting microbial lipids from the environment and determining their stable carbon isotope ratios (19). In this manner, it should be possible to identify bacterial substrates, especially in ecosystems where substrates have a wide range of isotope ratios. The flux of carbon in microbial communities can be studied by analyzing lipids specific to different bacterial groups (2, 28). Potential applications of this method include the flux of nutrients into different cell types in medical research (12) and the study of the degradation of pollutants by microorganisms.
ACKNOWLEDGMENTS
Michaela Blank, Silke Hardtke, Dagmar Duttmann, and Andrea Brinkop are thanked for their technical assistance. We acknowledge the suggestions and recommendations of two unknown reviewers, which helped us to greatly improve the manuscript. The International Atomic Energy Agency, Vienna, Austria), is acknowledged for providing free reference materials for the calibration of the IRMS.
This work was supported by grants from the German Federal Ministry for Science, Education and Research (projects 0319433B and 0319433C).
REFERENCES
- 1.Abelson P, Hoering T. Carbon isotope fractionation in formation of amino acids by photosynthetic organisms. Proc Natl Acad Sci USA. 1961;47:623–632. doi: 10.1073/pnas.47.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham W-R, Meyer H, Lindholst S, Vancanneyt M, Smit J. Phospho- and sulfolipids as biomarkers of Caulobacter, Brevundimonas and Hyphomonas. Syst Appl Microbiol. 1997;20:522–539. [Google Scholar]
- 3.Abrajano T A, Jr, Murphy D E, Fang J, Comet P, Brooks J M. 13C/12C ratios in individual fatty acids of marine mytilids with and without bacterial symbionts. Org Geochem. 1994;12:611–617. [Google Scholar]
- 4.Alban C, Joyard J, Douce R. Comparison of glycerolipid biosynthesis in non-green plastids from sycamore (Acer pseudoplatanus) cells and cauliflower (Brassica oleracea) buds. Biochem J. 1989;259:775–783. doi: 10.1042/bj2590775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair N, Leu A, Muñoz E, Olsen J, Kwong E, DesMarais D. Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl Environ Microbiol. 1985;50:996–1001. doi: 10.1128/aem.50.4.996-1001.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh E G, Dyer W J. A rapid method for total lipid extraction and purification. Can J Biochem Physiol. 1969;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 7.Cho S H, Thompson G A., Jr On the metabolic relationships between monogalactosyldiacylglycerol and digalactosyldiacylglycerol molecular species in Dunaliella salina. J Biol Chem. 1987;262:7686–7693. [PubMed] [Google Scholar]
- 8.Cifuentes L A, Coffin R B, Solorzano L, Cardenas W, Espinoza J, Twilley R. Isotopic and elemental variations of carbon and nitrogen in a mangrove estuary. Coastal Shelf Sci. 1996;43:781–800. [Google Scholar]
- 9.Coffin R, Fry B, Peterson B, Wright R. Identification of bacterial carbon sources with stable isotope analysis. Limnol Oceanogr. 1989;34:1306–1310. [Google Scholar]
- 10.Coffin R, Devereux R, Price W, Cifuentes L. Analyses of stable isotopes of nucleic acids to trace sources of dissolved substrates used by estumarine bacteria. Appl Environ Microbiol. 1990;66:2012–2020. doi: 10.1128/aem.56.7.2012-2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin R B, Cifuentes L A, Pritchard P H. Effect of remedial nitrogen applications on algae and heterotrophic organisms on oil contaminated beaches in Prince William Sound, AK. Mar Environ Res. 1997;1:27–39. [Google Scholar]
- 11a.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 12.Demmelmair, H., T. Sauerwald, B. Koletzko, and T. Richter. 1997. New insights into lipid and fatty acid metabolism via stable isotopes. Eur. J. Pediatr. 166(Suppl. 1):70–74. [DOI] [PubMed]
- 13.DeNiro M, Epstein S. Mechanism of carbon isotope fractionation associated with lipid synthesis. Science. 1977;197:261–263. doi: 10.1126/science.327543. [DOI] [PubMed] [Google Scholar]
- 14.De Rosa M, Gambacorta A, Gliozzi A. Structure, biosynthesis, and physiochemical properties of archaebacterial lipids. Microbiol Rev. 1986;60:70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrickson H L, Cappenberg T E, de Leeuw J. Polar lipid fatty acid compositions of Lake Vechten seston—an ecological application of lipid analysis. FEMS Microb Ecol. 1986;38:381–396. [Google Scholar]
- 16.Fry B, Sherr E B. δ13C measurements as indicators of carbon flow in marine and freshwater ecosystems. Contrib Mar Sci. 1984;27:13–47. [Google Scholar]
- 17.Goodman K J, Brenna J T. High sensitivity tracer detection using high-precision gas chromatography-combustion isotope ratio mass spectrometry and highly enriched [U-13C]-labeled precursors. Anal Chem. 1992;64:1088–1096. doi: 10.1021/ac00034a004. [DOI] [PubMed] [Google Scholar]
- 18.Hayes J M. Factors controlling 13C contents of sedimentary organic compounds: principles and evidence. Mar Geol. 1993;113:111–126. [Google Scholar]
- 19.Jahnke L L, Summons R E, Dowling L M, Zahiralis K D. Identification of methanotrophic lipid biomarkers in cold-seep Mussel gills: chemical and isotopic analysis. Appl Environ Microbiol. 1996;61:676–682. doi: 10.1128/aem.61.2.576-582.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kämpfer P, Denner E B M, Meyer S, Moore E R B, Busse H-J. Classification of “Pseudomonas azotocolligans” Anderson 1966, 132, in the genus Sphingomonas as Sphingomonas trueperi sp. nov. Int J Syst Bacteriol. 1997;47:677–683. doi: 10.1099/00207713-47-2-577. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy E P. Membrane-derived oligosaccharides (periplasmic β-d-glucans) of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1064–1074. [Google Scholar]
- 22.Lechevalier H, Lechevalier M P. Chemotaxonomic use of lipids—an overview. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, United Kingdom: Academic Press, Ltd.; 1988. pp. 869–902. [Google Scholar]
- 23.Lechevalier M P. Lipids in bacterial taxonomy: a taxonomist’s view. Crit Rev Microbiol. 1977;7:109–210. doi: 10.3109/10408417709102311. [DOI] [PubMed] [Google Scholar]
- 24.Monson K D, Hayes J M. Biosynthetic control of the natural abundance of carbon-13 at specific positions within fatty acids in Escherichia coli. Evidence regarding the coupling of fatty acid and phospholipid synthesis. J Biol Chem. 1980;266:11436–11441. [PubMed] [Google Scholar]
- 25.Monson K D, Hayes J M. Biosynthetic control of the natural abundance of carbon-13 at specific positions within fatty acids in Saccharomyces cerevisiae. Isotopic fractionations in lipid synthesis as evidence for peroxisomal regulation. J Biol Chem. 1982;267:6668–6676. [PubMed] [Google Scholar]
- 26.Monson K D, Hayes J M. Carbon isotopic fractionation in the biosynthesis of bacterial fatty acids. Ozonolysis of unsaturated fatty acids as a means of determining the intramolecular distribution of carbon isotopes. Geochim Cosmochim Acta. 1982;46:139–149. [Google Scholar]
- 27.Nichols P D, Guckert J B, White D C. Determination of monounsaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by GC-MS of their dimethyl disulfide adducts. J Microbiol Methods. 1986;6:49–66. [Google Scholar]
- 28.Pel R, Oldenhuis R, Brand W, Vos A, Gottschal J, Zwart K B. Stable-isotope analysis of a combined nitrification-denitrification sustained by thermophilic methanotrophs under low-oxygen conditions. Appl Environ Microbiol. 1997;63:474–481. doi: 10.1128/aem.63.2.474-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelz O, Hesse C, Tesar M, Coffin R B, Abraham W-R. Development of methods to measure carbon isotope ratios of bacterial biomarkers in the environment. Isot Environ Health Stud. 1997;33:131–144. [Google Scholar]
- 30.Rawn J D. Biochemistry. Burlington, N.C: Neil Patterson Publishers; 1989. pp. 427–456. [Google Scholar]
- 31.Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya T. Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc Natl Acad Sci USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summons R E, Jahnke L L, Roksandic Z. Carbon isotopic fractionation in lipids from methanotrophic bacteria: Relevance for interpretation of the geochemical record of biomarkers. Geochim Cosmochim Acta. 1994;68:2863–2863. doi: 10.1016/0016-7037(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 33.Taranto P A, Keenan T W, Potts M. Rehydration induces rapid onset of lipid biosynthesis in desiccated Nostoc commune (Cyanobacteria) Biochim Biophys Acta. 1993;1168:228–237. doi: 10.1016/0005-2760(93)90129-w. [DOI] [PubMed] [Google Scholar]
- 34.Tunlid A, Ek H, Westerdahl G, Odham G. Determination of carbon-13-enrichment in bacterial fatty acids using chemical ionization mass spectrometry with negative ion detection. J Microbiol Methods. 1987;7:77–89. [Google Scholar]
- 35.White D C. Analysis of microorganisms in terms of quantity and activity in natural environments. Symp Soc Gen Microbiol. 1983;34:37–66. [Google Scholar]
- 35a.Wu H C. Biosynthesis of lipoproteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1005–1014. [Google Scholar]
- 36.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1261–1276. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]