Abstract
This study aimed to explore the possible mechanisms of Ling Gui Zhu Gan decoction (LGZGD) in the treatment of nephrotic syndrome (NS) using network pharmacology combined with molecular docking and molecular dynamics simulation. The active ingredients of LGZGD and their targets were retrieved from Traditional Chinese Medicine Systems Pharmacology Database and Swiss Target Prediction database. The NS targets were retrieved from Genecards, OMIM and Drugbank databases. Next, the intersecting targets of drug and disease were imported into the String database for protein-protein interaction network analysis, and the core targets were identified through topological analysis. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed in the Metascape platform. Finally, molecular docking and molecular dynamics simulation were performed for further validation. The network analysis showed that 109 active ingredients of LGZGD were associated with 105 targets in NS. The key active ingredients (quercetin, kaempferol, naringenin, licochalcone A, formononetin, beta-sitosterol) and the core targets (IL6, AKT1, TNF, VEGFA, TP53, JUN, IL1B, CASP3, EGFR, and STAT3) were further identified. Enrichment analysis indicated that multiple biological processes and pathways, including AGE-RAGE, PI3K-Akt, JAK-STAT, and HIF-1 signaling pathways, might be regulated by LGZGD in the treatment of NS. Molecular docking and molecular dynamics simulation results further indicated that the key active ingredients of LGZGD could stably bind to the core targets through hydrogen bonding and hydrophobic interaction. This study demonstrates that the active ingredients of LGZGD may regulate multiple targets, biological processes and signaling pathways in NS. Our findings may provide a theoretical basis for further studies on LGZGD in the treatment of NS.
Keywords: Ling Gui Zhu Gan decoction, molecular docking, molecular dynamics simulation, nephrotic syndrome, network pharmacology
1. Introduction
Nephrotic syndrome (NS) is a common refractory renal disorder with a complex pathology that can be divided into several different pathological types of glomerular disease. The main clinical manifestations of NS include massive proteinuria, hypoproteinemia, hyperlipidemia and edema, among which massive proteinuria and hypoalbuminemia are necessary for the diagnosis. NS progresses rapidly and is prone to recurrence if not treated in time, which may even induce serious infections, thromboembolism and other complications.[1] The conventional treatments for NS include corticosteroids and immunosuppressants, which may lead to immune deficiency, high risk of infection, hormone-related complications, and even the recurrence of the disease.[2,3] Therefore, more alternative therapies are urgently needed for treating NS. Traditional Chinese medicine (TCM) has unique advantages in the treatment of NS. There is growing evidence that the integrated regulation and treatment based on syndrome differentiation of TCM can better alleviate clinical symptoms, modulate immune function, and inhibit the deterioration of renal function.[4]
In TCM theory, NS is generally regarded as the category of “edema,” “water and Qi disease,” “asthenia,” which is closely related to the Spleen and Kidney. The pathogenesis of the disease is attributed to the deficiency of Spleen and Kidney, thereby resulting in the disorder of water homeostasis and edema. Therefore, the treatment strategy of NS by TCM should focus on strengthening the Spleen and Kidney, thereby relieving water retention and swelling. Ling Gui Zhu Gan decoction (LGZGD), a famous herbal formula from Synopsis of Prescriptions of the Golden Chamber written by Zhang Zhongjing in the Eastern Han Dynasty, has the effects of warming Yang, strengthening Spleen and draining dampness. Recent studies revealed that LGZGD could alleviate edema, reduce blood lipids, urine protein, serum creatinine and blood urea nitrogen, thereby protecting renal function and relieving nephrotic syndrome in rats.[5,6] In addition, there is growing evidence that some active ingredients of LGZGD can effectively protect against NS. It has been reported that Poria cocos, the main herb of LGZGD, and its hydroethanolic extract showed therapeutic effect on NS in animal models.[7,8] A recent study also demonstrated that Cinnamomi Ramulus, another main herb of LGZGD, could treat NS by inhibiting MAPK signaling pathway.[9] However, the underlying mechanisms of LGZGD against NS are not yet completely understood due to the complex ingredients of herbal formula.
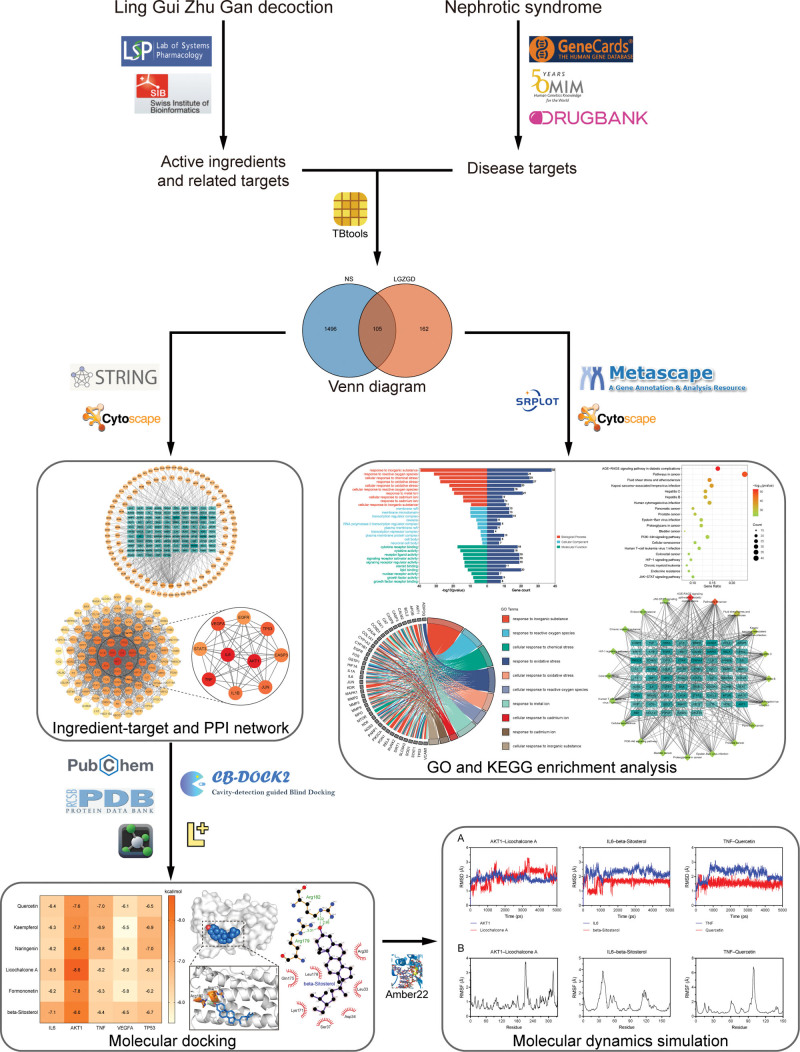
Over the past decade, network pharmacology has been increasingly employed to study the mechanisms of herbal formulae. It is believed that network pharmacology investigates the regulatory effect of drugs on the biomolecular network from a systematic perspective, which is analogous to the holistic view of TCM. This feature of network pharmacology provides a possible way to reveal the complex biological basis of herbal formulae.[10] Moreover, computational techniques, such as molecular docking and dynamics simulation, may help to validate the reliability of network pharmacology. However, to our knowledge, there are no reports using network pharmacology to explore the mechanisms of LGZGD in NS so far. Hence, this study attempted to employ network pharmacology to explore active ingredients, potential targets and corresponding pathways responsible for LGZGD in the treatment of NS. Furthermore, both molecular docking and molecular dynamics simulation were performed to validate the binding ability between ingredients and targets. We hope that our work could provide new ideas for further application and mechanism research of LGZGD in the treatment of NS. The workflow of this study was shown in Figure 1.
Figure 1.
The workflow of this study.
2. Materials and methods
2.1. Collection of active ingredients and related targets of LGZGD
All herbs of LGZGD, namely Fu Ling (Poria), Gui Zhi (Cinnamomi Ramulus), Bai Zhu (Atractylodis Macrocephalae Rhizoma), and Gan Cao (Glycyrrhizae Radix et Rhizoma), were searched using the Traditional Chinese Medicine Systems Pharmacology Database (old.tcmsp-e.com/tcmsp.php), a systematic pharmacological analysis platform for TCM.[11] Oral bioavailability ≥ 30% and drug-likeness ≥ 0.18 were used as the criteria to screen the active ingredients.[12] Then, the related targets of active ingredients of LGZGD were retrieved from the Traditional Chinese Medicine Systems Pharmacology Database. The Swiss Target Prediction database (www.swisstargetprediction.ch) was used to supplement the ingredients with missing targets, and obtained the targets with the top probability values. Finally, target names were converted into standard gene names using the Uniprot database (www.uniprot.org).[13] The active ingredients and related targets were represented in an Excel file and imported into Cytoscape 3.9.1 software (Cytoscape Consortium, San Diego, CA)[14] to draw a drug-ingredient-target network diagram.
2.2. Collection of NS targets
“Nephrotic syndrome” was used as the keyword in the Genecards database (www.genecards.org),[15] OMIM database (www.omim.org),[16] Drugbank database (go.drugbank.com),[17] to search for the targets related to NS. Among the targets retrieved from the Genecards database, the score value was proportional to the association between target and disease. According to the method previously reported,[18,19] the median score of targets were calculated, and the corresponding targets with scores greater than the median were considered as the targets of NS. Finally, the targets obtained from the 3 databases were merged, and the duplicate values were deleted to obtain the final targets of NS.
2.3. Identification of potential targets and construction of active ingredient-disease target network
The targets obtained from LGZGD and NS were imported into TBtools software[20] to obtain the intersecting targets, which were considered as the potential targets of LGZGD in the treatment of NS. The relationship between active ingredients and disease targets obtained above was represented in an Excel file and imported into Cytoscape 3.9.1 software to draw an active ingredient-disease target network diagram. Network Analyzer plugin in Cytoscape software was used to analyze the topological properties of the network, such as degree, betweenness centrality and closeness centrality. The key active ingredients were identified according to the topological parameters of the nodes in the network.
2.4. Construction of protein-protein interaction (PPI) network
The potential targets obtained above were imported into the STRING 11.5 database (string-db.org). Species was set as “Homo sapiens,” and the “minimum required interaction score” was set to be >0.4, then the PPI network model was constructed.[21] The tsv file was downloaded and then imported into Cytoscape 3.9.1 software to conduct topological analysis and draw the PPI network diagram. The core targets were identified according to the degree of nodes.
2.5. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses
The potential targets were imported into the Metascape platform (www.metascape.org),[22] and the GO and KEGG pathway enrichment analyses were performed respectively. The top 10 GO terms of biological process, cellular component and molecular function along with the top 20 KEGG pathways were retained. The SRplot platform (www.bioinformatics.com.cn) was used to draw bar charts, bubble plots and chord diagrams. The pathway-target network diagram was constructed by Cytoscape 3.9.1 software.
2.6. Molecular docking
Molecular docking is the most widely applied technique that can predict the best matching binding mode of a ligand to a protein.[23] In this present study, molecular docking of the key active ingredients and core targets was performed to validate the mechanisms of LGZGD for the treatment of NS. The structures of the active ingredients of LGZGD were obtained from PubChem database (pubchem.ncbi.nlm.nih.gov), and OpenBabel 3.1.1[24] was used for energy minimization. The crystal structures of the key targets were obtained from RCSB Protein Data Bank (www.rcsb.org). Then, the molecules and proteins were submitted to CB-Dock2 (cadd.labshare.cn/cb-dock2) to employ AutoDock Vina 1.2.0 for template-independent blind docking.[25] The core targets and their best-docked ingredients were selected as the final docking conformations. The interactions between ligand and receptor were analyzed by LigPlot + v.2.25 software[26] and visualized using PyMOL 2.5.0 software (Schrödinger, LLC, New York, NY.
2.7. Molecular dynamics simulation
After molecular docking, the stability of the selected docking models was further verified using molecular dynamics simulations. The Amber22 software (University of California, San Francisco, CA) package was used to perform molecular dynamics simulations. The ff14SB force field was used to calculate system force field parameters. The TIP3P water model was used to solvate the system and corresponding numbers of counterbalance ions were added to neutralize the charges. After the system energy was minimized, the system was heated from 0 K to 300 K within 500 ps. Then, the system was equilibrated using canonical ensembles and isothermal-isobaric simulations at 300 K. Finally, a 50 ns molecular dynamics simulation was performed at constant temperature and pressure with periodic boundary conditions. All covalent bonds involving hydrogen were restricted by the SHAKE method. The root-mean-square deviation and root-mean-square fluctuation (RMSF) were calculated to analyze the molecular dynamics trajectories, and the results were visualized using GraphPad Prism 7 software (GraphPad Software, Boston, MA). The binding free energy was calculated using the MM-PBSA method.
3. Results
3.1. Active ingredients and related targets of LGZGD
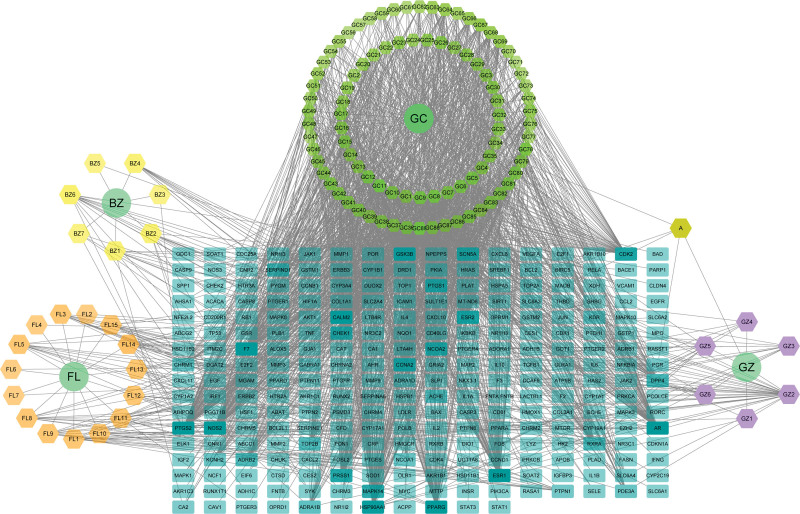
A total of 120 ingredients were identified. Of these ingredients, 7 were from Bai Zhu, 15 from Fu Ling, 7 from Gui Zhi, 92 from Gan Cao, and 1 common ingredient of Gui Zhi and Gan Cao. There were 50 targets of Bai Zhu, 121 targets of Fu Ling, 68 targets of Gui Zhi and 1629 targets of Gan Cao, and the total number of targets was 267 after combining and deleting duplicate values. The drug-ingredient-target network diagram was constructed by Cytoscape software, comprising 389 nodes and 1986 edges (Fig. 2).
Figure 2.
Drug-ingredient-target network. The circular nodes represent drugs, the hexagonal nodes represent active ingredients, and the rectangle nodes represent the action targets. The darker color of the rectangle represents the higher degree value of the target. BZ = Bai Zhu, FL = Fu Ling, GZ = Gui Zhi, GC = Gan Cao.
3.2. Potential targets for LGZGD in the treatment of NS
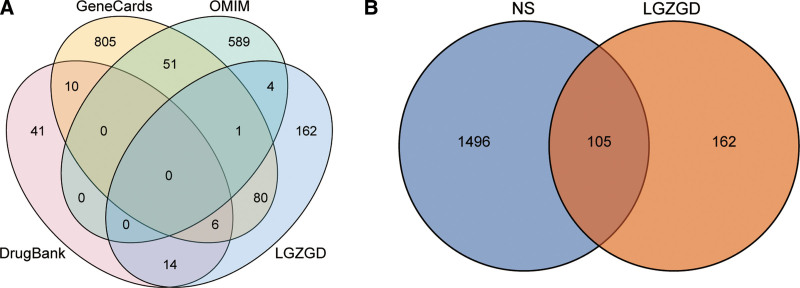
A total of 1905 targets of NS were obtained from Genecards database. The targets with scores greater than the median were set as the targets of NS. The maximum score of NS target obtained by Genecards was 125.21, the minimum score was 0.18, and the median score was 6.42. Therefore, 953 targets with score > 6.42 were set as the targets of NS. After combining the targets obtained from multiple databases and removing duplicate values, 1601 targets related to NS were obtained, with a total of 105 targets intersecting with those of LGZGD (Fig. 3). These 105 intersecting targets were considered as the potential targets for LGZGD in the treatment of NS.
Figure 3.
Venn diagram of intersecting targets. (A) Intersecting targets of LGZGD and different disease databases. (B) Intersecting targets of LGZGD and NS. LGZGD = Ling Gui Zhu Gan decoction, NS = Nephrotic syndrome.
3.3. Active ingredient-disease target network
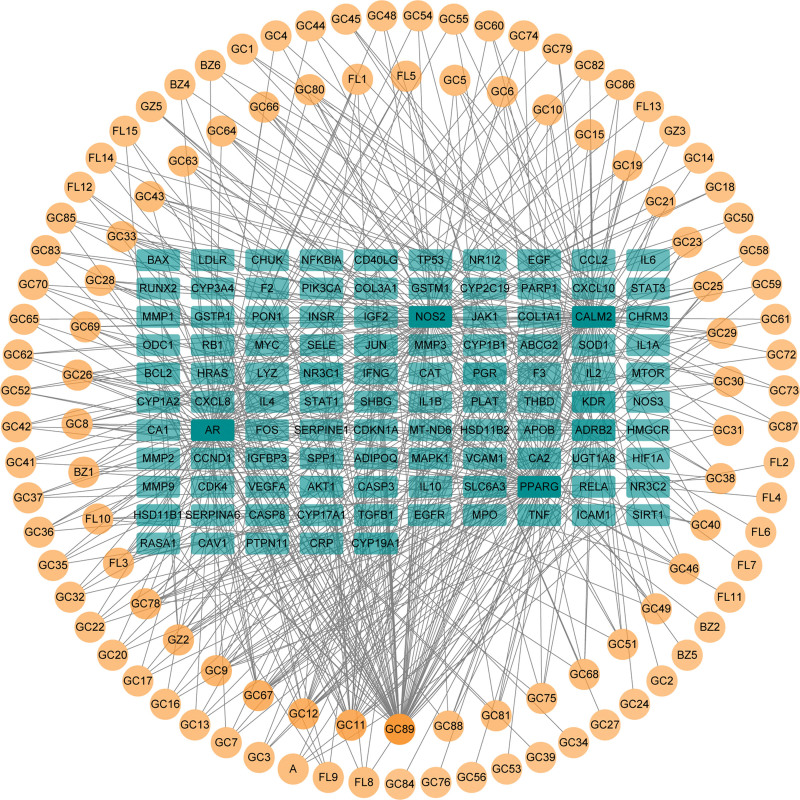
The intersecting targets and active ingredients were imported into Cytoscape software for topological analysis. Then, the active ingredient-disease target network comprising 214 nodes and 530 edges was constructed, where circular nodes represent active ingredients, the rectangle nodes represent the potential targets (Fig. 4). There were 109 active ingredients of LGZGD associated with the NS targets. According to the topological parameters of the nodes, the top-ranked active ingredients included quercetin, kaempferol, naringenin, licochalcone A, formononetin, and beta-sitosterol, which were considered as the key active ingredients of LGZGD (Table 1). Those active ingredients may be the potent active ingredients of LGZGD in the treatment of NS.
Figure 4.
Active ingredient-disease target network. The circular nodes represent active ingredients. The rectangle nodes represent the potential targets. The darker the color, the higher the degree value.
Table 1.
Characteristics of the key active ingredients of Ling Gui Zhu Gan decoction.
| Molecule name | MOL ID | PubChem CID | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|---|---|
| Quercetin | MOL000098 | 5,280,343 | 71 | 0.43210412 | 0.48617512 |
| Kaempferol | MOL000422 | 5,280,863 | 25 | 0.1065087 | 0.42369478 |
| Naringenin | MOL004328 | 439,246 | 15 | 0.06695291 | 0.37147887 |
| Licochalcone A | MOL000497 | 5,318,998 | 14 | 0.04696199 | 0.39962121 |
| Formononetin | MOL000392 | 5,280,378 | 11 | 0.03304877 | 0.38363636 |
| Beta-sitosterol | MOL000358 | 222,284 | 10 | 0.0141624 | 0.31681682 |
3.4. PPI network of potential targets for LGZGD against NS
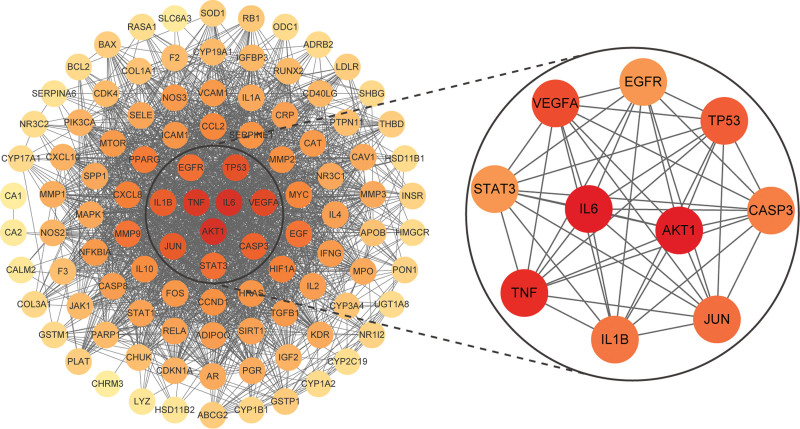
The 105 intersecting targets of LGZGD and NS were imported into the String database, and the PPI network of the potential targets was depicted (Fig. 5), with 105 nodes and 1970 edges. The PPI network diagram was arranged into circles from inside to outside according to the degree value of each node. The higher the degree value, the darker the color. The innermost circle of the PPI network showed the core targets of LGZGD for the treatment of NS, namely IL6, AKT1, TNF, VEGFA, TP53, JUN, IL1B, CASP3, EGFR, and STAT3. The corresponding degree values were 83, 83, 81, 77, 75, 73, 72, 71, 68, and 68, respectively (Table 2). And the top 5 targets were selected for subsequent molecular docking and molecular dynamics simulation. The PPI network diagram indicated that LGZGD may treat NS through multiple targets, and the core targets may play crucial roles in the mechanism of LGZGD for the treatment of NS.
Figure 5.
Protein-protein interaction network of the potential targets. The innermost circle indicates the top 10 targets identified according to degree values. The darker the color, the higher the degree value.
Table 2.
Characteristics of the top 10 targets of Ling Gui Zhu Gan decoction.
| Target name | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|
| IL6 | 83 | 0.05699103 | 0.83064516 |
| AKT1 | 83 | 0.04112476 | 0.83064516 |
| TNF | 81 | 0.03657549 | 0.81746032 |
| VEGFA | 77 | 0.01862779 | 0.79230769 |
| TP53 | 75 | 0.02211144 | 0.78030303 |
| JUN | 73 | 0.037832 | 0.77443609 |
| IL1B | 72 | 0.01722133 | 0.76296296 |
| CASP3 | 71 | 0.01564577 | 0.75735294 |
| EGFR | 68 | 0.01990443 | 0.74100719 |
| STAT3 | 68 | 0.01006593 | 0.74100719 |
3.5. GO enrichment analysis
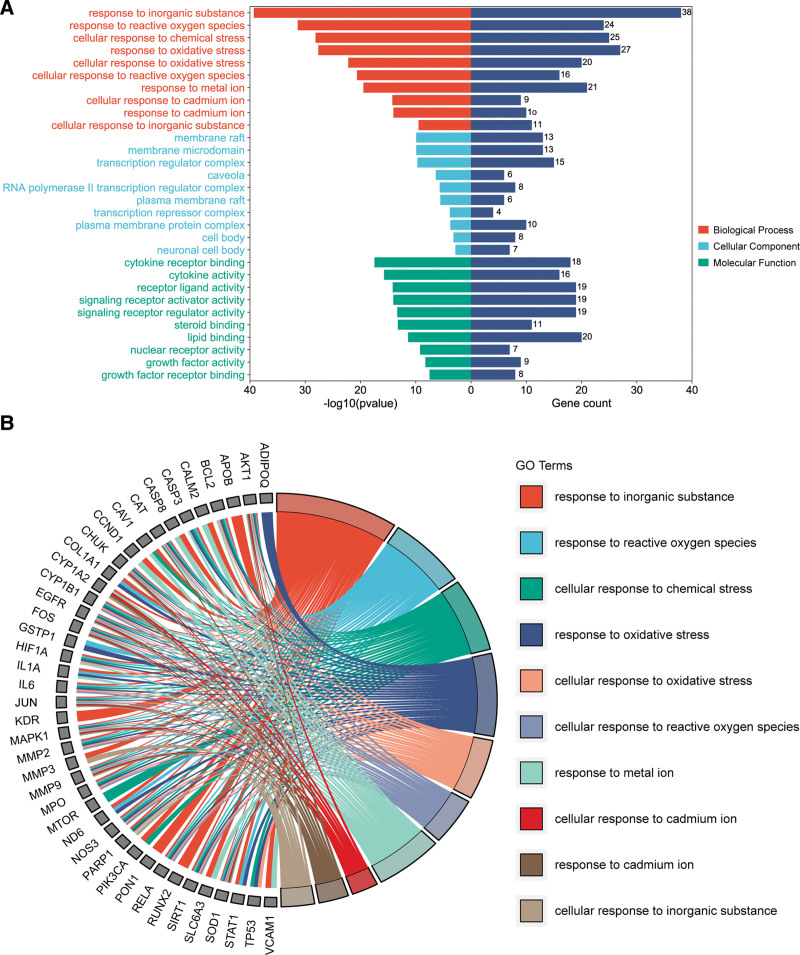
GO enrichment analysis was performed on the potential targets for LGZGD in the treatment of NS, and the top 10 terms were selected to draw the bar chart and chord diagram (Fig. 6). Among them, the GO terms of biological process enrichment analysis were 1595, mainly involving response to inorganic substance, response to reactive oxygen species, cellular response to chemical stress, and cellular response to oxidative stress. The GO terms of cellular component enrichment analysis were 60, mainly involving membrane raft, membrane microdomain, transcription regulator complex and caveola. The GO terms of molecular function enrichment analysis were 130, mainly involving cytokine receptor binding, cytokine activity, receptor ligand activity and signaling receptor activator activity. Taken together, GO analysis results indicated that the mechanism of LGZGD for the treatment of NS might involve multiple biological processes, which reflected the multi-level action characteristics of LGZGD.
Figure 6.
GO enrichment analysis. (A) Top 10 terms of biological process, cell component and molecular function. (B) Chord diagram of top 10 terms of biological process. GO = Gene Ontology.
3.6. KEGG pathway enrichment analysis
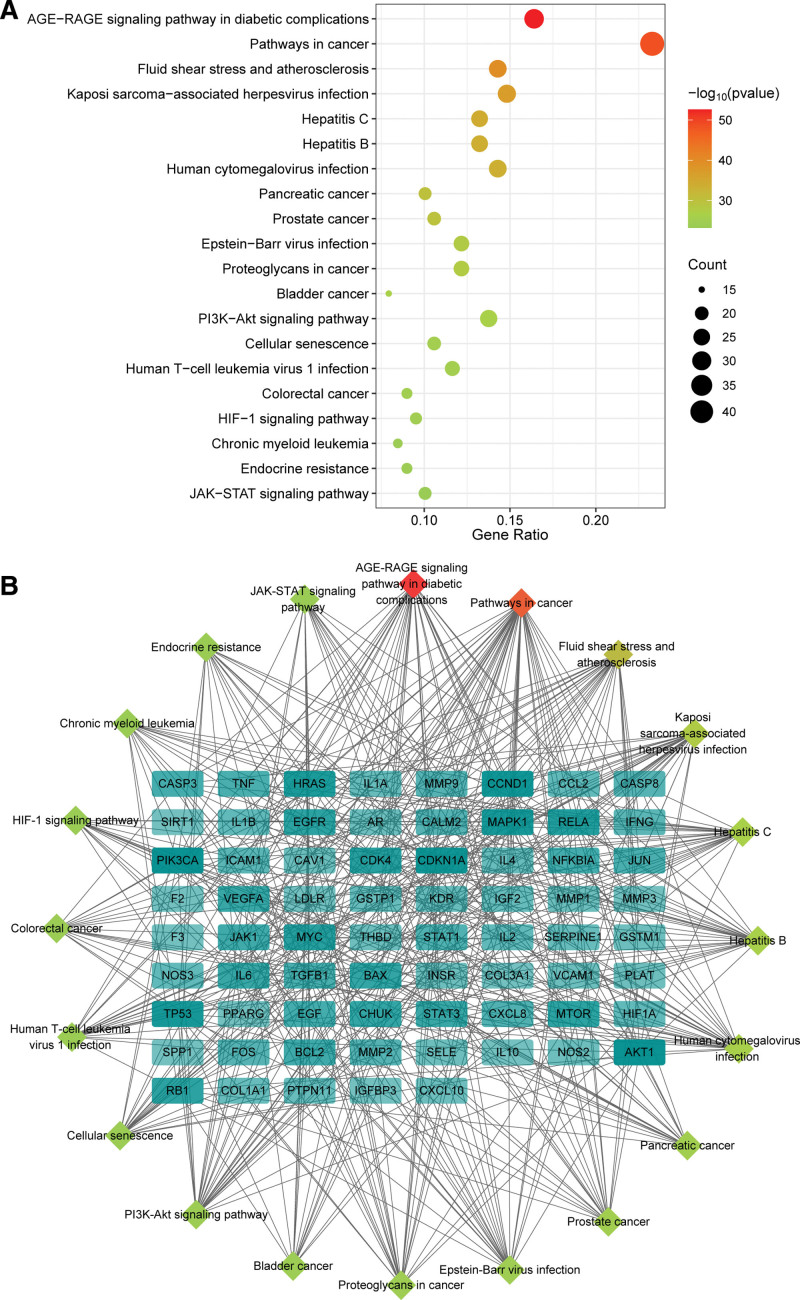
KEGG pathway enrichment analysis of the potential targets yielded 189 pathways, and the top 20 KEGG terms ranked by the P value were presented in Figure 7A. The results showed that the potential targets were mainly involved in AGE-RAGE signaling pathway in diabetic complications, Pathways in cancer, Fluid shear stress and atherosclerosis, Kaposi sarcoma-associated herpesvirus infection, and Hepatitis C. In addition, KEGG-enriched pathways and corresponding targets were imported into Cytoscape 3.9.1 to construct the pathway-target network, comprising 89 nodes and 464 edges (Fig. 7B).
Figure 7.
KEGG enrichment analysis. (A) Top 20 terms of KEGG enrichment analysis. (B) The pathway-target network. The rhombic nodes represent KEGG pathways. The rectangle nodes represent the gene, and the darker color represents the higher degree value of the gene. KEGG = Kyoto Encyclopedia of Genes and Genomes.
3.7. Molecular docking
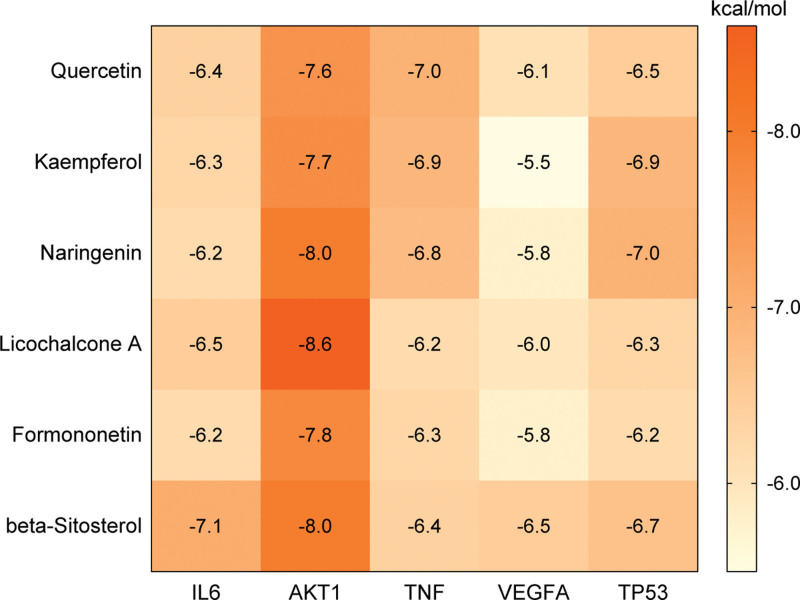
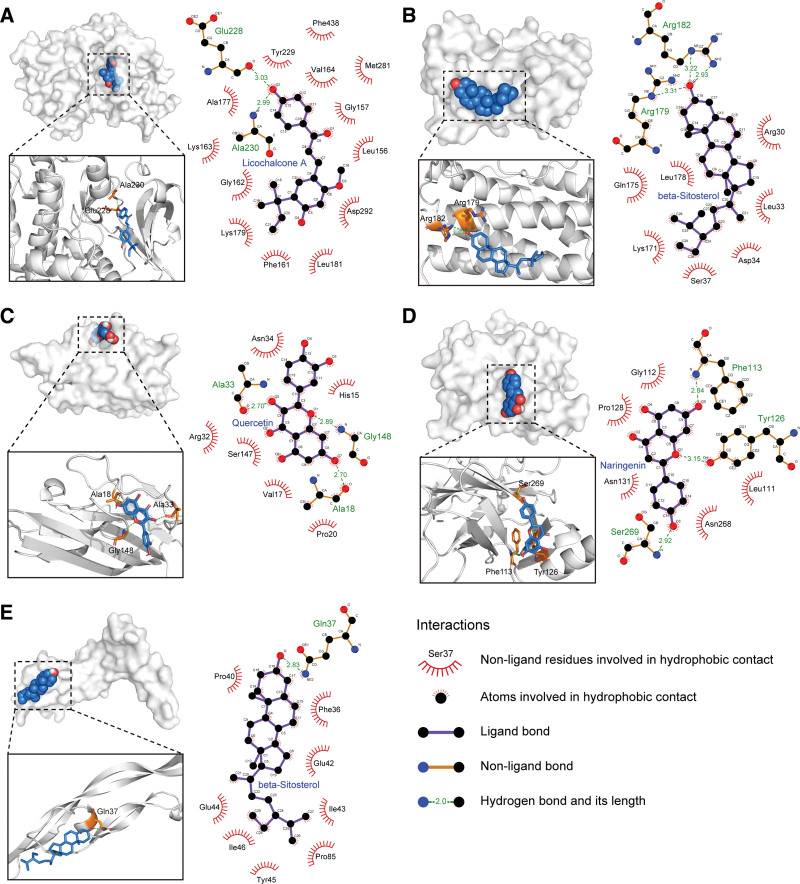
In the present study, the key active ingredients along with the top 5 targets, namely AKT1 (4gv1), IL6 (1alu), TNF (5uui), VEGFA (4kzn) and TP53 (3d06), were selected for molecular docking. The docking details were shown in Figure 8. The top 5 targets with their best-docked ingredients, namely AKT1–Licochalcone A, IL6–beta-Sitosterol, TNF–Quercertin, TP53–Naringenin, VEGFA–beta-Sitosterol, were selected for further analysis. The 5 pairs of docking models were imported into LigPlot software (EMBL-EBI, Hinxton, Cambridgeshire, UK) to analyze the interactions between ligand and receptor, which were visualized using PyMOL software to show the exact binding sites. As illustrated in Figure 9, licochalcone A bound to AKT1 via 2 hydrogen bonds and 13 hydrophobic contacts; beta-sitosterol bound to IL6 via 3 hydrogen bonds and 7 hydrophobic contacts; quercertin bound to TNF via 3 hydrogen bonds and 6 hydrophobic contacts; naringenin bound to TP53 via 3 hydrogen bonds and 5 hydrophobic contacts; beta-sitosterol bound to VEGFA via 1 hydrogen bonds and 8 hydrophobic contacts. According to molecular docking theory, docking energy is negatively correlated to binding activity, and a lower docking energy indicates stronger interactions between ligand and receptor. The binding energy values of the 5 docking models were all < −6 kcal/mol, and the active ingredients firmly bound to the active sites of corresponding targets through hydrogen bonding and hydrophobic interaction. These results indicated that the key active ingredients had good binding affinity with the core targets of NS.
Figure 8.
Heatmap of molecular docking results. The darker the color, the greater the binding activity between the active ingredient and the target.
Figure 9.
The molecular docking models of the key active ingredients and the targets. (A) AKT1–Licochalcone A, −8.6 kcal/mol; (B) IL6–beta-Sitosterol, −7.1 kcal/mol; (C) TNF–Quercertin, −7.0 kcal/mol; (D) TP53–Naringenin, −7.0 kcal/mol; (E) VEGFA–beta-Sitosterol, −6.5 kcal/mol.
3.8. Molecular dynamics simulation
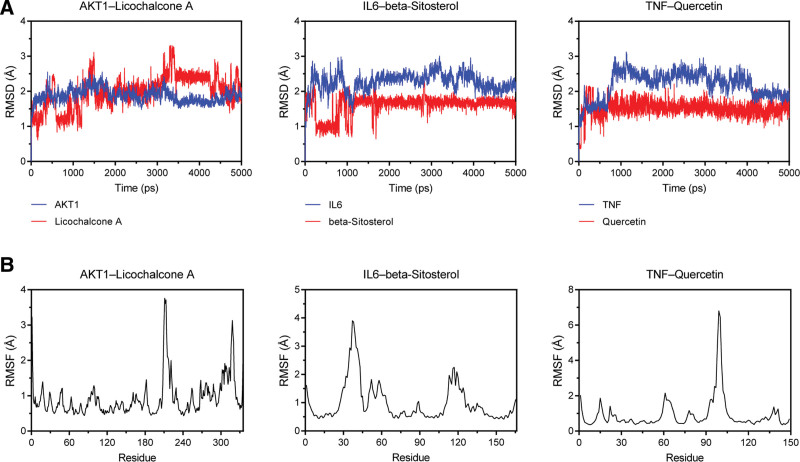
To better investigate the stability and dynamic interactions of protein-ligand complexes, the 3 complexes with best binding activity, namely AKT1–Licochalcone A, IL6–beta-Sitosterol and TNF–Quercertin, were selected for molecular dynamics simulations. The root-mean-square deviation analysis is generally used to evaluate the deviation extent for a group of atoms in the complex. As shown in Figure 10A, the RMSF curves of all the 3 systems showed a certain degree of fluctuations in the early stages and tended to stabilize after around 10 ns, which indicated that the binding of the ligands to the corresponding receptors was relatively stable, especially in the IL6–beta-Sitosterol complex. The RMSF analysis is generally used to depict the fluctuation of the amino acid residues in the complex. As shown in Figure 10B, AKT1–Licochalcone A complex fluctuated more considerably near the residues 211 and 318; IL6–beta-Sitosterol complex fluctuated more considerably near the residues 37 and 117; TNF–Quercertin fluctuated more considerably near the residue 99. These results indicated that the complexes had greater flexibility in these residue regions. In addition, the binding free energies of the above 3 complexes were calculated, the details of which are listed in Table 3. Of the 3 complexes, the IL6–beta-Sitosterol complex had the lowest total binding free energy (−21.85 kcal/mol), while van der Waals energy and nonpolar solvation energy were conducive to the binding of IL6 to beta-sitosterol. Altogether, these results suggested that the protein-ligand complexes exhibited good stability during molecular dynamics simulations.
Figure 10.
Molecular dynamics simulations of the protein-ligand complexes. (A) Root-mean-square deviation analysis, (B) Root-mean-square fluctuation analysis.
Table 3.
Binding free energies of the protein-ligand complexes calculated by MM-PBSA method.
| Energy | AKT1–Licochalcone A | IL6–beta-Sitosterol | TNF–quercetin |
|---|---|---|---|
| ΔEvdw | −39.14 ± 2.22 | −29.20 ± 2.55 | −29.81 ± 2.38 |
| ΔEele | −6.63 ± 5.28 | 0.67 ± 1.32 | −10.98 ± 6.07 |
| ΔGPB | 33.95 ± 7.99 | 9.96 ± 3.17 | 31.20 ± 6.20 |
| ΔGSA | −3.95 ± 0.12 | −3.28 ± 0.22 | −2.91 ± 0.12 |
| ΔGbind | −15.78 ± 4.20 | −21.85 ± 2.54 | −12.50 ± 3.63 |
All values are in kcal/mol.
ΔGbind = binding free energy, ΔEele = electrostatic energy, ΔGPB = polar solvation energy calculated by Poisson Boltzmann (PB) equation, ΔGSA = nonpolar solvation energy, ΔEvdw = van der Waals energy.
4. Discussion
LGZGD is composed of 4 herbs, namely Fu Ling (Poria), Gui Zhi (Cinnamomi Ramulus), Bai Zhu (Atractylodis Macrocephalae Rhizoma), and Gan Cao (Glycyrrhizae Radix et Rhizoma). In TCM theory, it is mainly used for “warming Yang, strengthening spleen and draining dampness.” It is widely used in the treatment of many systemic diseases in addition to NS. Modern pharmacology reveals that LGZGD has anti-inflammatory, diuretic, anti-swelling, immune improving, anti-allergic and antitumor effects. For instance, Fu Ling has antibacterial, antitumor effects, and has been found to regulate immune system. Gui Zhi has antibacterial, anti-inflammatory, anti-allergy effects. Bai Zhu has a significant diuretic effect. Gan Cao has diuretic and anti-inflammatory effects.[27] However, the mechanisms of LGZGD in the treatment of NS are not fully understood. In the present study, we investigated the underlying mechanisms of LGZGD against NS using the network pharmacology combined with molecular docking and dynamics simulation.
In this study, we found that there were 109 active ingredients of LGZGD that may modulate 105 targets related to NS. Then, we identified 6 key active ingredients of LGZGD, including quercetin, kaempferol, naringenin, licochalcone A, formononetin and beta-sitosterol. These active ingredients may play important roles in the mechanism of LGZGD for treatment of NS. Several studies have shown that quercetin, isorhamnetin and kaempferol have anti-inflammatory and antioxidant effects.[28–31] Kaempferol has been demonstrate to reduce mesangial matrix expansion, glomerular basement membrane and podocyte loss by inhibiting apoptosis and promoting autophagy through AMPK/mTOR pathways, thereby protecting the kidney.[32] It was found that Astragalus membranaceus could inhibit renal fibrosis and inflammation, as well as reduce blood creatinine and cystatin, thus playing a protective role in the kidney.[33,34] Moreover, studies have shown that naringenin may inhibit the expression of inflammatory cells, attenuate oxidative stress and the accumulation of extracellular matrix in the glomerulus, exhibiting renal protective effect.[35,36]
Furthermore, the PPI network analysis of the potential targets for LGZGD against NS identified the core targets, including IL6, AKT1, TNF, VEGFA, and TP53. It is widely believed that the development of NS is closely associated with the dysfunctions of organismal immune, inflammatory response, as well as renal filtration membrane permeability,[3,37] which may cause a variety of metabolic disorders and complications, including hypercoagulable state, bacterial infection and renal failure.[38] It has been revealed that increasing renal tubular mitochondrial AKT1 in mitochondria-targeted constitutively active AKT1 mice reduced azotemia, tubular injury, renal fibrosis and glomerulosclerosis with significantly improved survival after ischemia-reperfusion injury.[39] IL6 is a multifunctional cytokine with diverse biological activities for its unique receptor system, which may be involved in the regulation of immune response, acute phase response and inflammation, etc. Clinical studies demonstrated that IL6 plays a critical role in the pathogenesis of autoimmune diseases.[40]
The KEGG pathway analysis showed that the regulatory pathways of LGZGD against NS involved multiple signaling pathways, including AGE-RAGE, PI3K-Akt, JAK-STAT, and HIF-1 signaling pathways. It has been revealed that the toxic effects of AGEs could directly damage renal cells and tissues, while AGEs-RAGE signaling pathway could activate inflammatory signals, resulting in renal inflammatory response and endothelial cell injury, and exacerbating kidney injury.[41,42] Renal fibrosis is the common pathological basis for the progression of many chronic kidney diseases to an advanced stage. It is believed that inhibiting renal fibrosis plays a crucial role in the treatment of kidney diseases. There is evidence that activation of PI3K-Akt signaling pathway exacerbates extracellular matrix (ECM) deposition in the kidney, leading to the progress of renal fibrosis.[43] Moreover, several studies have demonstrated that JAK-STAT signaling pathway is involved in the pathogenesis of inflammatory and autoimmune diseases, inhibiting pro-fibrotic factors in the kidney, and playing a important role in inflammatory infiltration and renal tubulointerstitial fibrosis.[44,45] The HIF-1 signaling pathway is associated with the progression of chronic kidney injury, while HIF-1α can induce renal fibrosis, mainly mediating epithelial-mesenchymal trans differentiation of renal tubular epithelial cells. It has been proposed that kidney injury may be improved by inhibiting the HIF-1 signaling pathway.[46,47] In addition, combined with previous reports, the development of NS may involve the combined actions of multiple biological processes and signals. Collectively, our findings suggest that LGZGD may exert therapeutic effects in NS by regulating multiple targets and pathways, which is consistent with the multi-channel overall regulation of TCM theory.
To further probe the exact mechanism of LGZGD on NS, molecular docking was performed to validate the interactions between the key active ingredients and core targets. Our results suggest that the key active ingredients of LGZGD have good affinity with the core targets of NS. The active ingredients could bind to the core targets through different interactions, including hydrogen bonding and hydrophobic effect. Moreover, molecular dynamics simulation was performed to assess the stability of the protein-ligand complex in 50 ns. As expected, the results of molecular dynamics simulation were consistent with those of molecular docking, indicating that the complexes formed by the active ingredients and the receptors have good stability, especially the IL6–beta-Sitosterol complex. These findings suggest that the active ingredients from LGZGD may regulate the therapeutic targets of NS, and IL6 is possibly the pivotal target. However, we have to note that, due to the limitations of bioinformatics approaches, our work is merely a preliminary exploration of the possible mechanisms of LGZGD in NS. Specifically, the findings of network pharmacology still need to be further verified on cell or animal models, even combined with clinical researches, to achieve more convincing results.
5. Conclusion
The present study systematically explored the active ingredients, potential targets, and signaling pathways of LGZGD for the treatment of NS using network pharmacology combined with molecular docking and molecular dynamics simulation. Our findings provide evidence that the active ingredients of LGZGD, including quercetin, kaempferol, naringenin, licochalcone A, formononetin, beta-sitosterol, may modulate the core targets (IL6, AKT1, TNF, VEGFA, TP53, JUN, IL1B, CASP3, EGFR, and STAT3) to regulate multiple biological processes and pathways. Collectively, LGZGD may exert beneficial effects in the treatment of NS through multi-ingredient, multi-target, and multi-pathway actions. Moreover, our study may provide a clinical reference for the treatment of NS with LGZGD and the clinical expansion of the classical prescription. However, the specific mechanism of LGZGD is very complicated and still needs to be further verified by in vitro and in vivo experiments.
Acknowledgments
This work was supported by the Project of Guangdong Provincial Bureau of Traditional Chinese Medicine (20201305).
Author contributions
Conceptualization: Li Shi, Bo Fu.
Formal analysis: Lei Li, Xuyi Kuang.
Funding acquisition: Bo Fu.
Methodology: Li Shi, Denggui Luo.
Project administration: Airong Qi, Bo Fu.
Visualization: Li Shi, Yuanjun Deng.
Writing – original draft: Li Shi, Yuanjun Deng.
Writing – review & editing: Airong Qi, Bo Fu.
Abbreviations:
- GO
- gene ontology
- KEGG
- Kyoto encyclopedia of genes and genomes
- LGZGD
- Ling Gui Zhu Gan decoction
- NS
- nephrotic syndrome
- PPI
- protein-protein interaction
- RMSF
- root-mean-square fluctuation
- TCM
- Traditional Chinese medicine
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
How to cite this article: Shi L, Deng Y, Luo D, Li L, Kuang X, Qi A, Fu B. Exploration of the possible mechanisms of Ling Gui Zhu Gan decoction in nephrotic syndrome based on network pharmacology, molecular docking and molecular dynamics simulation. Medicine 2023;102:29(e34446).
Contributor Information
Li Shi, Email: sl220324@163.com.
Yuanjun Deng, Email: yuanjun.deng@qq.com.
Denggui Luo, Email: 405985835@qq.com.
Lei Li, Email: llmwm11@163.com.
Xuyi Kuang, Email: q5673464@163.com.
Airong Qi, Email: 81863418@163.com.
References
- [1].Kodner C. Diagnosis and management of nephrotic syndrome in adults. Am Fam Physician. 2016;93:479–85. [PubMed] [Google Scholar]
- [2].Tullus K, Webb H, Bagga A. Management of steroid-resistant nephrotic syndrome in children and adolescents. Lancet Child Adolesc Health. 2018;2:880–90. [DOI] [PubMed] [Google Scholar]
- [3].Fujisawa H, Nakayama Y, Nakao S, et al. Effectiveness of immunosuppressive therapy for nephrotic syndrome in a patient with late-onset Fabry disease: a case report and literature review. BMC Nephrol. 2019;20:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang XQ, Wang L, Tu YC, et al. Traditional Chinese medicine for refractory nephrotic syndrome: strategies and promising treatments. Evid Based Complement Alternat Med. 2018;2018:8746349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jiang Y. Study on the protective effect of Ling Gui Zhu Gan decoction on rats with nephrotic syndrome model. Chin Natl Health Med. 2016;28:60–2. [Google Scholar]
- [6].Yuan C. Effects of self-prepared Ling Gui Zhu Gan decoction combined with Western medicine on renal function in patients with nephrotic syndrome. Chin Foreign Med Sci. 2017;36:177–9. [Google Scholar]
- [7].Zan JF, Shen CJ, Zhang LP, et al. Effect of Poria Cocos hydroethanolic extract on treating Adriamycin-induced rat model of nephrotic syndrome. Chin J Integr Med. 2017;23:916–22. [DOI] [PubMed] [Google Scholar]
- [8].Lee SM, Lee YJ, Yoon JJ, et al. Effect of Poria Cocos on puromycin aminonucleoside-induced nephrotic syndrome in rats. Evid Based Complement Alternat Med. 2014;2014:570420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He D, Li Q, Du G, et al. An integration of network pharmacology and experimental verification to investigate the mechanism of Guizhi to treat nephrotic syndrome. Front Pharmacol. 2021;12:755421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang X, Wang ZY, Zheng JH, et al. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19:1–11. [DOI] [PubMed] [Google Scholar]
- [11].Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ding S, Wang W, Song X, et al. Based on network pharmacology and molecular docking to explore the underlying mechanism of huangqi gegen decoction for treating diabetic nephropathy. Evid Based Complement Alternat Med. 2021;2021:9928282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1–30. [DOI] [PubMed] [Google Scholar]
- [16].Hamosh A, Amberger JS, Bocchini C, et al. Online Mendelian Inheritance in Man (OMIM): Victor McKusick’s magnum opus. Am J Med Genet A. 2021;185:3259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dan WC, He QY, Qu Y, et al. A study on the molecular mechanism of Hovenia pill in regulating dyslipidemia based on network pharmacology. World Sci Tech Mod Chin Med. 2019;21:2396–405. [Google Scholar]
- [19].Cao ZH, Liu Y, Sun L. Analysis of the potential mechanism of action of Ling Gui Zhu Gan decoction in the treatment of vertigo based on network pharmacology. J Chin Med. 2022;37:1962–70. [Google Scholar]
- [20].Chen C, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202. [DOI] [PubMed] [Google Scholar]
- [21].Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci . 2019;20:4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].O’Boyle NM, Banck M, James CA, et al. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu Y, Yang X, Gan J, et al. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50:W159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–86. [DOI] [PubMed] [Google Scholar]
- [27].Pan YL, Mi J. Professor Mi Jie’s experience in Treating renal edema with Ling Gui Zhu Gan decoction. Asia Pac Tradit Med. 2019;15:99–101. [Google Scholar]
- [28].Li Y, Yao J, Han C, et al. Quercetin, inflammation and immunity. Nutrients. 2016;8:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gong G, Guan YY, Zhang ZL, et al. Isorhamnetin: a review of pharmacological effects. Biomed Pharmacother. 2020;128:11030. [DOI] [PubMed] [Google Scholar]
- [30].Nakamura N, Hayasaka S, Zhang XY, et al. Effects of baicalin, baicalein, and wogonin on interleukin-6 and interleukin-8 expression, and nuclear factor-kappab binding activities induced by interleukin-1beta in human retinal pigment epithelial cell line. Exp Eye Res. 2003;77:195–202. [DOI] [PubMed] [Google Scholar]
- [31].Yang YZ, Tang YZ, Liu YH. Wogonoside displays anti-inflammatory effects through modulating inflammatory mediator expression using RAW264.7 cells. J Ethnopharmacol. 2013;148:271–6. [DOI] [PubMed] [Google Scholar]
- [32].Sheng H, Zhang D, Zhang J, et al. Kaempferol attenuated diabetic nephropathy by reducing apoptosis and promoting autophagy through AMPK/mTOR pathways. Front Med (Lausanne). 2022;9:986825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yao CA, Lin CH. Treatment with the herbal formulation Eefooton slows the progression of chronic kidney disease: a case report. Medicine (Baltim). 2019;98:e17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou X, Sun X, Gong X, et al. Astragaloside IV from Astragalus membranaceus ameliorates renal interstitial fibrosis by inhibiting inflammation via TLR4/NF-кB in vivo and in vitro. Int Immunopharmacol. 2017;42:18–24. [DOI] [PubMed] [Google Scholar]
- [35].Jia R, Li WJ, Li YM, et al. Exploring the protective effect and mechanism of naringenin on renal injury in rats with diabetic nephropathy. J Mod Integr Med. 2019;28:2072–7. [Google Scholar]
- [36].Wen L, Chen L, Sun Y, et al. Naringin alleviates renal injury in rats with diabetic nephropathy through inhibition of TGF-β1/smad signaling pathway. Basic Med Clin. 2016;36:896–901. [Google Scholar]
- [37].Han W, Ma QQ, Feng WX, et al. Changes in renal function indices, lipid levels and safety observations in patients with primary nephrotic syndrome treated with tacrolimus combined with prednisone. PLA J Med. 2022;34:64–6. [Google Scholar]
- [38].Tsuji K, Kitamura S, Wada J. MicroRNAs as biomarkers for nephrotic syndrome. Int J Mol Sci . 2020;22:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lin HY, Chen Y, Chen YH, et al. Tubular mitochondrial AKT1 is activated during ischemia reperfusion injury and has a critical role in predisposition to chronic kidney disease. Kidney Int. 2021;99:870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mihara M, Hashizume M, Yoshida H, et al. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond). 2012;122:143–59. [DOI] [PubMed] [Google Scholar]
- [41].Wu XQ, Zhang DD, Wang YN, et al. AGE/RAGE in diabetic kidney disease and ageing kidney. Free Radic Biol Med. 2021;171:260–71. [DOI] [PubMed] [Google Scholar]
- [42].Dozio E, Vettoretti S, Caldiroli L, et al. Advanced glycation end products (AGE) and soluble forms of AGE receptor: emerging role as mortality risk factors in CKD. Biomedicines. 2020;8:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu S, Zhao H, Yang W, et al. The alcohol extract of coreopsis tinctoria nutt ameliorates diabetes and diabetic nephropathy in db/db mice through miR-192/miR-200b and PTEN/AKT and ZEB2/ECM pathways. Biomed Res Int. 2019;2019:5280514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Banerjee S, Biehl A, Gadina M, et al. JAK-STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs. 2017;77:521–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Matsui F, Meldrum KK. The role of the Janus kinase family/signal transducer and activator of transcription signaling pathway in fibrotic renal disease. J Surg Res. 2012;178:339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wei X, Zhu X, Jiang L, et al. Recent advances in understanding the role of hypoxia-inducible factor 1α in renal fibrosis. Int Urol Nephrol. 2020;52:1287–95. [DOI] [PubMed] [Google Scholar]
- [47].Lu L, Li J, Le Y, et al. Inhibitor of growth 4 (ING4) inhibits hypoxia-induced EMT by decreasing HIF-1α and snail in HK2 cells. Acta Histochem. 2019;121:695–703. [DOI] [PubMed] [Google Scholar]