Abstract
Long noncoding RNAs (lncRNAs) play an important role in regulating gene expression. Changes in their expression have been associated with many types of cancer, including thyroid cancer. This study aimed to investigate how changes in the expression of potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1 (KCNQ1OT1) and HAGLR opposite strand lncRNA (HAGLROS) lncRNAs correlate with the development and clinicopathological characteristics of papillary thyroid cancer (PTC). Reverse transcription-quantitative polymerase chain reaction was used to investigate the expression of lncRNAs in both tumor and adjacent normal thyroid tissue samples of the patients. Expressions of KCNQ1OT1 and HAGLROS were upregulated in the patients tumor samples compared to the adjacent normal thyroid samples. KCNQ1OT1 expression was linked to microcarcinoma and gender, while HAGLROS expression was linked to microcarcinoma and tumor size. When only microcarcinoma samples were evaluated, KCNQ1OT1 expression was higher in tumor tissues compared to normal tissues; however, no significant difference was observed in HAGLROS expression. Our data suggests that high expressions of KCNQ1OT1 and HAGLROS might contribute to the development of PTC and disease progression, and both lncRNAs may be potential therapeutic targets in PTC patients.
Keywords: gene expression, HAGLROS, KCNQ1OT1, long noncoding RNA, microcarcinoma, papillary thyroid cancer
1. Introduction
Thyroid cancer is the most common endocrine malignancy. Its incidence is on the rise worldwide.[1,2] According to GLOBOCAN data, the number of new thyroid cancer cases diagnosed in 2020 was 586,000 – ranking at 11th out of all 36 cancer types.[3] Differentiated thyroid cancers are more common among thyroid malignancies. Cases with papillary thyroid cancer (PTC) account for more than 85% of all patients.[4] Today, PTC is a generally curable; however, it can be aggressive and even fatal in some cases. Moreover, the recurrence rate of the disease is approximately thirty percent.[5] Therefore, further studies need to be done on molecular biomarkers associated with PTC development and progression so that early diagnosis can be established for the disease and treatment targets are determined.
Long noncoding RNAs (lncRNA) are an important member of noncoding RNA class. They are larger than 200 nucleotides and not translated into a protein.[6,7] LncRNAs can regulate gene expression at different stages by interacting with mRNA, miRNA, DNA, and proteins. Thus, abnormalities in the expression of lncRNAs have been associated with various forms of cancer, including thyroid.[8] Changes in the expression of some lncRNAs–namely BRAF-activated nonprotein coding RNA, Papillary thyroid carcinoma susceptibility candidate 3, Homeobox transcript antisense intergenic RNA, metastasis associated lung adenocarcinoma transcript 1, Long noncoding RNA H19, Nuclear Enriched Abundant Transcript 1, Maternally expressed gene 3, Growth arrest-specific transcript 5, and Plasmacytoma variant translocation 1–have been associated with thyroid cancer’s development and progression.[9]
The Potassium Voltage-Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1 (KCNQ1OT1) gene is located in the region 11p15.5. The product of this gene is a 91 kb unspliced lncRNA. KCNQ1OT1 is responsible for transcriptionally silencing a gene cluster, including important tumor suppressor genes and KCNQ1, via genomic imprinting.[7] It interacts with epigenetic regulators – for example polycomb repressive complex 2, histone methyltransferase G9a, and DNA methyl transferase 1 in order to silence those genes. The overexpression of KCNQ1OT1 can cause Beckwith-Wiedemann syndrome. In some of children who suffer from this disease, embryonal tumors also point to the role of KCNQ1OT1 in cancer development.[10] As a matter of fact, recent studies have identified increased KCNQ1OT1 expression in retinoblastoma, acute promyelocytic leukemia,[10] bladder,[11] ovarian,[12] and non-small cell lung cancer (NSCLC).[13] However, no study has investigated the expression of KCNQ1OT1 in thyroid cancer.
Similarly, the HAGLR opposite strand lncRNA (HAGLROS) is located in the region 2q31.1. It has only one transcript: a 699 nucleotide HAGLROS lncRNA.[14] Other recent studies have found that HAGLROS expression increases in various cancers and this lncRNA has oncogenic properties. One study in particular concluded that increased HAGLROS expression contributes to the development of gastric cancer and poor prognosis.[15] Likewise, Zheng et al[16], reported that HAGLROS expression significantly increased in triple negative breast cancer and this increase was associated with poor overall survival. In addition, some studies have reported increased HAGLROS expression in hepatocellular carcinoma (HCC),[17] esophageal squamous cell carcinoma,[18] ovarian cancer,[19] NSCLC,[20] and osteosarcoma.[21] One study based on TCGA dataset reported that HAGLROS was among the 10 top genes that exhibited increased expression in papillary thyroid cancer.[22] However, no other studies on this subject currently exist.
Therefore, this study focused on examining the correlations between the development and clinicopathological features of papillary thyroid cancer and the expression profiles of KCNQ1OT1 and HAGLROS.
2. Methods
2.1. Patients and tissue specimens
Giresun University’s Clinical Research Ethics Committee approved this study (Approval: 05/12/2019/25). Informed consent was obtained from all individual participants included in the study. We used power analysis to estimate the required sample size and found as 54 to achieve 90% power for medium effect size. In our study, this number was exceeded with a sample size of 128. PTC patients to be included in the study were selected from those who had not received any cancer treatment before surgery (such as radiotherapy, chemotherapy or other treatments). A total of 137 PTC patients who had undergone at thyroidectomy at Giresun University’s Faculty of Medicine between 2015 and 2021 were included in the study. Tumor and noncancerous thyroid tissue samples of the each patient included in the study were examined by the pathology department. Pathologists confirmed the diagnosis of PTC and determined the tumor tissue blocks and matched noncancerous thyroid tissue blocks. Tissue samples were obtained from formalin-fixed paraffin-embedded tissue blocks that had been archived in the same pathology department. After RNA extraction, 9 patients were excluded due to poor RNA quality (RNA samples whose A260/A280 ratio was not between 1.8–2.1/ degraded RNA samples). This brought the number down to 128 – expression analysis was conducted on them. The researchers obtained the subjects clinicopathological traits (age, gender, tumor diameter, and TNM stage) from their patient records and summarized them in Table 1. The 6 patients whose lymphovascular invasion findings could not be reached are shown as unknown in table 1.
Table 1.
The relationship of KCNQ1OT1 and HAGLROS expressions with clinicopathological characteristics of PTC patients. Statistical analysis was performed using chi-square test.
| Variables | Number of patients | KCNQ1OT1 expression | Odds ratio (95% CI) | P value | HAGLROS expression | Odds ratio (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||||
| Gender | 3.222 (1.170–8.875) | .019* | 1.620 (0.639–4.109) | .307 | |||||
| Female | 106 | 58 | 48 | 70 | 36 | ||||
| Male | 22 | 6 | 16 | 12 | 10 | ||||
| Age (yr) | 0.796 (0.370–1.712) | .559 | 1.243 (0.553–2.790) | .598 | |||||
| <45 | 37 | 17 | 20 | 25 | 12 | ||||
| ≥45 | 91 | 47 | 44 | 57 | 34 | ||||
| Microcarcinoma | 0.362 (0.173–0.758) | .006* | 0.364 (0.163–0.814) | .012* | |||||
| No | 79 | 32 | 47 | 44 | 35 | ||||
| Yes | 49 | 32 | 17 | 38 | 11 | ||||
| Tumor diameter (cm) | 1.772 (0.839-3.745) | .132 | 2.842 (1.319–6.121) | .007* | |||||
| <2 | 86 | 47 | 39 | 62 | 24 | ||||
| ≥2 | 42 | 17 | 25 | 20 | 22 | ||||
| Lymphovascular invasion | 0.739 (0.305–1.788) | .501 | 1.456 (0.596–3.556) | ||||||
| No | 97 | 47 | 50 | 63 | 34 | .408 | |||
| Yes | 25 | 14 | 11 | 14 | 11 | ||||
| Unknown | 6 | 5 | 1 | ||||||
| Primary tumor | .353 | .499 | |||||||
| T1 | 103 | 55 | 48 | - | 68 | 35 | - | ||
| T2 | 19 | 7 | 12 | 10 | 9 | ||||
| T3 | 6 | 2 | 4 | 4 | 2 | ||||
| Lymph node metastasis | 0.238 (0.026–2.191) | .365 | 0.433 (0.047–3.997) | .654 | |||||
| No | 123 | 60 | 63 | 78 | 45 | ||||
| Yes | 5 | 4 | 1 | 4 | 1 | ||||
| TNM staging | 1.000 (0.137–7.325) | 1.000 | 0.585 (0.059–5.794) | 1.000 | |||||
| I | 124 | 62 | 62 | 79 | 45 | ||||
| II | 4 | 2 | 2 | 3 | 1 | ||||
| Extrathyroidal extension | 0.360 (0.107–1.215) | .089 | 0.450 (0.119–1.705) | .231 | |||||
| No | 114 | 54 | 60 | 71 | 43 | ||||
| Yes | 14 | 10 | 4 | 11 | 3 | ||||
| Multicentricity | 0.664 (0.321–1.374) | .269 | 0.924 (0.434–1.967) | .838 | |||||
| No | 82 | 38 | 44 | 52 | 30 | ||||
| Yes | 46 | 26 | 20 | 30 | 16 | ||||
| Multifocality | 1.296 (0.639–2.629) | .472 | 1.589 (0.764–3.305) | .214 | |||||
| No | 76 | 40 | 36 | 52 | 24 | ||||
| Yes | 52 | 24 | 28 | 30 | 22 | ||||
HAGLROS = HAGLR opposite strand lncRNA, KCNQ1OT1 = potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1, PTC = papillary thyroid cancer,
* P < .05 was considered statistically significant.
2.2. RNA isolation and reverse transcription - quantitative polymerase chain reaction (RT-qPCR)
First, 5-µm thick samples were taken from the paraffin blocks using a microtome. Next, they were treated with 1 ml xylene to remove the paraffin, and then washed with 100%, 70% and 50% ethanol. Then, total RNA extraction was carried out using a RNeasy formalin-fixed paraffin-embedded kit (Qiagen GmbH) according to the manufacturer’s instructions. RNA concentration and purity were assessed using a NanoDrop 1/1C Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, Inc.). Next, the RNA samples were subjected to agarose gel electrophoresis to evaluate their integrity. The samples that proved to be of poor quality were excluded from the study. Afterwards, complementary DNA synthesis was conducted according to the manufacturer’s instructions using a Revert Aid RT Reverse Transcription kit (Thermo Fisher Scientific, Inc.). Next, all of samples were stored at − 80 °C until expression analysis. To avoid detection bias, the samples were randomly numbered without specifying whether they were tumors or controls, so that researchers who would do laboratory analyzes did not know whether the sample was a tumor or a control sample. Expression analyses were conducted using a Light Cycler 480 SYBR-Green I Master mix (Roche Diagnostics GmbH)–final volume: 20 µL–on a Light Cycler 480 Real-Time PCR system (Roche Diagnostics GmbH) according to the manufacturer’s instructions. The following cycling protocol was performed for the qRT amplification: initial denaturation at 95◦C for 1 minute, followed by 45 cycles at 95°C for 10 seconds, 57°C for 30 seconds, and 72°C for 30 seconds. The β actin gene was used as an endogenous control to normalize the expression of lncRNAs. All of the samples were tested in triplicates. Relative gene expression levels of the target lncRNAs were examined using the 2−ΔΔCT method.[23] All of the primer sequences for the qRT PCR were as follows: 5’-GGGAGCTGTTGTCCCTTACC-3’ (forward) and 5’-TTCGGAGTGGTAACTGTGCC-3’ (reverse) for KCNQ1OT1; 5’-CTGATCCACCTGGAACACTTT-3’ (forward) and 5’-CTCCTCCGCTTGCGATTT-3’ (reverse) for HAGLROS; 5’-TCTACAATGAGCTGCGTGTG -3’ (forward) and 5’-GGTCTCAAACATGATCTGGGT-3’ (reverse) for β actin.
2.3. Statistical analyses
Statistical analysis was conducted on SPSS 15.0 (SPSS Inc., Chicago, IL). Expression levels of lncRNAs between tumor and normal tissues were compared using the paired Student t test. The correlation between the clinicopathological features and the expressions of lncRNAs was evaluated using the chi-square test. For analyses purposes, the patient group was divided into 2 smaller groups – low and high expression – according to their median lnc expression values. Continuous variables were expressed in mean ± standard deviation. P values <.05 were considered statistically significant. Figures were created using GraphPad Prism version 8.0.
3. Results
3.1. KCNQ1OT1 and HAGLROS are significantly upregulated in PTC tissues
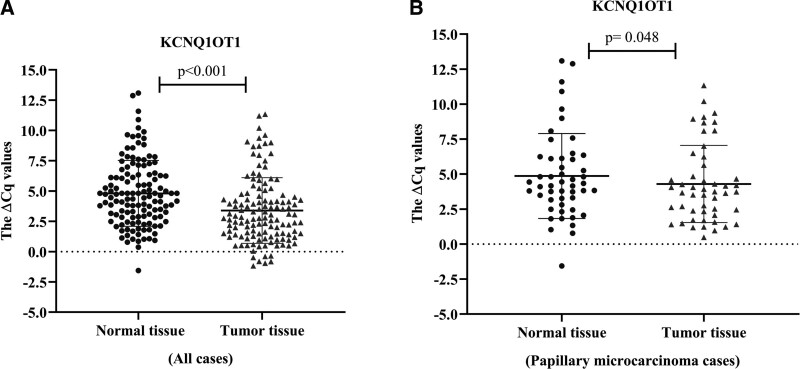
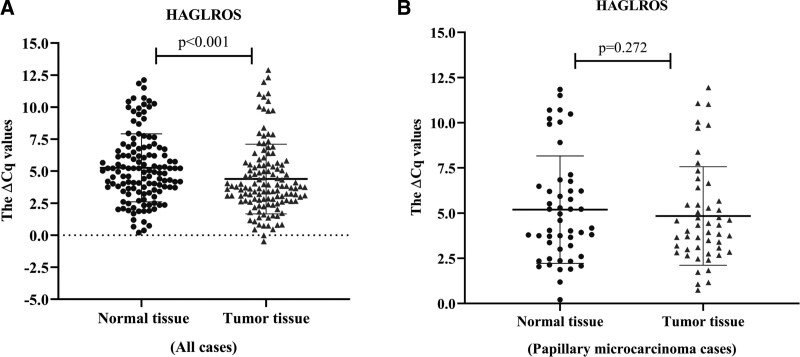
The relative expression levels of KCNQ1OT1 and HAGLROS in the tumor and normal thyroid tissues (n = 128 for each group) were identified using qRT PCR. Both KCNQ1OT1 (fold change: 2.67, P < .001) and HAGLROS expressions (fold change: 1.83, P < .001) were significantly higher in tumor tissues compared to adjacent normal thyroid tissues (Figs. 1A and 2A).
Figure 1.
(A) Relative expression of KCNQ1OT1 in tumor and normal tissues of all cases (n = 128) b Relative expression of KCNQ1OT1 in tumor and normal tissues of papillary microcarcinoma cases (n = 48). All of the data are presented as the mean ± standard deviation. Paired Student t test was used for statistical analysis. P < .05 was statistically significant.
Figure 2.
(A) Relative expression of HAGLROS in tumor and normal tissues of all cases (n = 128) b Relative expression of HAGLROS in tumor and normal tissues of papillary microcarcinoma cases (n = 48). All of the data are presented as the mean ± standard deviation. Paired Student t test was used for statistical analysis. P < .05 was statistically significant.
3.2. Associations of KCNQ1OT1 and HAGLROS expressions with clinicopathological features of the PTC patients
The patients (n = 128) were divided into 2 groups – low and high expression – to examine the relationship between the clinicopathological features and the expression levels of lncRNAs. Accordingly, the expression level of KCNQ1OT1 was associated with the gender (P = .019) and microcarcinoma (P = .006), while HAGLROS expression was associated with microcarcinoma (P = .012) and tumor diameter (P = .007). However, no significant correlation was determined between the expressions of lncRNAs and other clinical parameters – namely age, lymphovascular invasion, lymph node metastasis, primary tumor, TNM stage, extra thyroidal extension, multicentricity, and multifocality (Table 1). Hence, the expression of lncRNAs was subsequently examined only in papillary microcarcinoma samples (tumor size of ≤ 1). The expression of KCNQ1OT1 was higher in papillary microcarcinoma tumor samples than it was in the normal thyroid tissues (P = .048); however, no significant difference was found in HAGLROS expression (P = .272), (Figs. 1B and 2B). On the other hand, upon examining the papillary carcinoma samples only, both KCNQ1OT1 and HAGLROS expressions were also significantly higher in tumor tissues than they were in the normal ones.
4. Discussion
Recent studies have revealed that lncRNAs can regulate the expression of genes involved in various cellular processes at different levels, thus playing a key role in the regulation of biological processes. Indeed, this data supports the notion that lncRNAs are key to tumorigenesis and disease progression.[24] In the literature, changes in the expression of lncRNAs have been associated with the development and progression of many types of cancer.[25] On the other hand, KCNQ1OT1 has been associated with the development and prognosis of colorectal cancer, sarcoma, osteosarcoma, tongue squamous cell carcinoma, and breast cancer.[7] However, to the best of our knowledge, no one has yet studied the relationship between these lncRNAs and PTC development (including its clinical features)–beyond one study investigating the relationship between HAGLROS and PTC.[22]
One study reported that the expression of KCNQ1OT1 was upregulated in tumor samples of NSCLC patients and NSCLC cell lines. The findings of the present study revealed that KCNQ1OT1 regulated JAG1 expression by sponge miR-129-5p, and thereby increased proliferation, migration and invasion in NSCLC cells. Therefore, it seems as though KCNQ1OT1 might be an important therapeutic target for NSCLC.[13] Another study reported that KCNQ1OT1 upregulated CAPN10 expression through sponging miR-142-5p, thereby promoting proliferation and migration of ovarian cancer cells.[12] Gong et al[26], found that expression of KCNQ1OT1 was upregulated in tumor samples of glioma patients and glioma cell lines. They have also showed that KCNQ1OT1 enhances Cyclin E2 expression by acting as miR-370 sponge, and thus contributes to glioma tumorigenesis. Zhang et al[27], argued that increased expression of KCNQ1OT1 contributed to the development of tongue squamous cell carcinoma, and is associated with poor prognosis. The authors also reported that KCNQ1OT1 expression was linked to Gleason score, T stage, and lymph node status. The findings of the present study revealed that the expression of KCNQ1OT1 was upregulated in PTC tumor samples compared to the adjacent noncancerous thyroid tissues. Moreover, its expression was associated with microcarcinoma and gender. In addition, there was also elevated KCNQ1OT1 expression in the tumor samples of papillary microcarcinoma patient group. The results of the present study were compatible with the literature and indicated that increased KCNQ1OT1 expression might contribute to the development of PTC. Furthermore, this increased expression of KCNQ1OT1 in the papillary microcarcinoma tumor samples suggested that this lncRNA might play a role in the early stages of PTC tumorigenesis. Therefore, future studies ought to evaluate its usability as an early diagnosis marker with larger numbers of samples. The incidence of PTC is 3 times greater in women than it is in men.[4] Likewise, it was found in the present study that the ratio of female patients to male patients was approximately 5:1 and 16 of the 22 male patients fell in the high expression group (Table 1). In other words, the increase in KCNQ1OT1 expression in male PTC patients might be more significant and is more likely to cause disease. However, since there is no other study in the literature that found a relationship between KCNQ1OT1 expression and gender, these data should be confirmed by other studies with more PTC patients.
Various studies on different cancer types have revealed that KCNQ1OT1 contains a binding site for various miRNAs and contributes to tumorigenesis with competing endogenous RNA (ceRNA) function.[7] Of these, miRNAs, miR-296-5p, miR-211-5p, miR-124-3p, miR-204, miR-145-5p, miR-506, 129-5p, miR-153, miR-15a and miR-9-5p also act as tumor suppressor miRNAs in PTC.[7,28] Although the interaction between KCNQ1OT1 and these miRNAs in PTC needs to be confirmed by functional studies, it nevertheless suggests that KCNQ1OT1 may contribute to PTC tumorigenesis, thereby supporting the data of the current study.
A study by Tang et al[29], reported that HAGLROS was overexpressed in tumor samples from HCC patients and HCC cell lines. They moreover suggested that HAGLROS upregulated the expression of karyopherin α2 by sponging for miR-26b-5p and inactivated the p53 signaling pathway, thus contributing to malignant progression of HCC. A study in head and neck squamous cell carcinoma reported that HAGLROS was 1 of 13 optimal diagnostic biomarkers.[30] Another study suggested that the expression of HAGLROS increased in triple negative breast cancer and high HAGLROS expression in particular was associated with poor overall survival.[16] Shu et al[31], also reported that high HAGLROS expression contributed to the development and poor prognosis of diffuse large B-cell lymphoma through interaction with miR-100. Guo et al[22],–in their study on papillary thyroid cancer TCGA data – demonstrated that HAGLROS was among the top 10 aberrantly expressed lncRNAs. In the present study, it was determined that HAGLROS expression was significantly higher in PTC tumor samples compared to the adjacent normal thyroid tissues. Moreover, HAGLROS expression was associated with microcarcinoma and tumor size. On the other hand, HAGLROS expression did not change significantly in the papillary microcarcinoma samples alone. The data of the present study are similar to that of Guo et al, as well as studies conducted on different cancer type and pointing out that HAGLROS plays a role in the development and progression of PTC. On the other hand, the absence of any significant change in HAGLROS expression in the papillary microcarcinoma patient group suggests that the possible role of HAGLROS in PTC tumorigenesis begins to become evident after the disease reaches a certain stage.
The contribution of HAGLROS to PTC tumorigenesis may be possible by playing the role of ceRNA. Studies in different cancers have reported that HAGLROS harbors a binding site for miR-206, miR-152 and miR-26b-5p and functions as ceRNA.[18,21,29,32] These miRNAs were also tumor suppressor miRNAs whose expression was downregulated in PTC.[28,33–35] Hence, HAGLROS might contribute to PTC tumorigenesis via these miRNAs.
The present study has two limitations: The lack of functional studies to the support expression data, and; The partially small number of patients. Nevertheless, we have managed to demonstrate that KCNQ1OT1 and HAGLROS lncRNAs are highly expressed in PTC tissues, KCNQ1OT1expression is high in microcarcinoma samples, and HAGLROS expression is associated with tumor size. The results of the present study indicated that KCNQ1OT1 and HAGLROS may contribute to PTC development and disease progression and they have the potential to be used as therapeutic targets for PTC. Large-scale studies involving patients from different populations would be beneficial for the generalizability of our results.
Acknowledgments
The authors thank Dr Ceren Varer Akpinar for the medical statistics.
Author contributions
Conceptualization: Fadime Mutlu Icduygu, Egemen Akgun, Asuman Ozgoz, Demet Sengul, Ebru Alp.
Data curation: Fadime Mutlu Icduygu, Egemen Akgun, Asuman Ozgoz, Kuyas Hekimler Ozturk, Demet Sengul, Ebru Alp.
Formal analysis: Egemen Akgun, Asuman Ozgoz, Kuyas Hekimler Ozturk.
Investigation: Fadime Mutlu Icduygu, Kuyas Hekimler Ozturk.
Methodology: Fadime Mutlu Icduygu, Egemen Akgun, Demet Sengul, Ebru Alp.
Validation: Fadime Mutlu Icduygu, Ebru Alp.
Visualization: Fadime Mutlu Icduygu.
Writing – original draft: Fadime Mutlu Icduygu.
Writing – review & editing: Asuman Ozgoz, Demet Sengul, Ebru Alp, Egemen Akgun.
Abbreviations:
- ceRNA
- competing endogenous RNA
- HAGLROS =
- HAGLR opposite strand lncRNA
- HCC
- hepatocellular carcinoma
- KCNQ1OT1 =
- potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1
- lncRNA
- long noncoding RNA
- NSCLC
- non-small cell lung cancer
- PTC
- papillary thyroid cancer
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This study was funded by Giresun University’s Scientific Research Projects Committee (Grant Number: SAĞ-BAP-A-270220-54).
The authors have no funding and conflicts of interest to disclose.
How to cite this article: Mutlu Icduygu F, Akgun E, Ozgoz A, Hekimler Ozturk K, Sengul D, Alp E. Upregulation and the clinical significance of KCNQ1OT1 and HAGLROS lncRNAs in papillary thyroid cancer: An observational study. Medicine 2023;102:29(e34379).
Contributor Information
Egemen Akgun, Email: egemen.akgun@gmail.com.
Asuman Ozgoz, Email: biologistasu@gmail.com.
Kuyas Hekimler Ozturk, Email: kuyash@gmail.com.
Demet Sengul, Email: demet.sengul@giresun.edu.tr.
Ebru Alp, Email: ebrualmaz@hotmail.com.
References
- [1].Zaridze D, Maximovitch D, Smans M, et al. Thyroid cancer overdiagnosis revisited. Cancer Epidemiol. 2021;74:102014. [DOI] [PubMed] [Google Scholar]
- [2].Lee AW, Mendoza RA, Aman S, et al. Thyroid cancer incidence disparities among ethnic Asian American populations, 1990–2014. Ann Epidemiol. 2022;66:28–36. [DOI] [PubMed] [Google Scholar]
- [3].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- [4].Remer LF, Lee CI, Picado O, et al. Sex differences in papillary thyroid cancer. J Surg Res. 2022;271:163–70. [DOI] [PubMed] [Google Scholar]
- [5].Santiago K, Chen Wongworawat Y, Khan S. Differential microRNA-signatures in thyroid cancer subtypes. J Oncol. 2020;2020:2052396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang H, Li L, Wen K. Interactions between long non-coding RNAs and RNA-binding proteins in cancer. Oncol Rep. 2021;46:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cagle P, Qi Q, Niture S, et al. KCNQ1OT1: an oncogenic long noncoding RNA. Biomolecules. 2021;11:1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zamaraev AV, Volik PI, Sukhikh GT, et al. Long non-coding RNAs: a view to kill ovarian cancer. Biochim Biophys Acta Rev Cancer. 2021;1876:188584. [DOI] [PubMed] [Google Scholar]
- [9].Peng X, Zhang K, Ma L, et al. The role of long non-coding RNAs in thyroid cancer. Front Oncol. 2020;10:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tang D, Luo Y, Jiang Y, et al. LncRNA KCNQ1OT1 activated by c-Myc promotes cell proliferation via interacting with FUS to stabilize MAP3K1 in acute promyelocytic leukemia. Cell Death Dis. 2021;12:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li Y, Shi B, Dong F, et al. LncRNA KCNQ1OT1 facilitates the progression of bladder cancer by targeting MiR-218-5p/HS3ST3B1. Cancer Gene Ther. 2020;28:212–20. [DOI] [PubMed] [Google Scholar]
- [12].Liu H, Chen R, Kang F, et al. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 axis. Mol Genet Genomic Med. 2020;8:e1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Y, Zhang L, Yang J, et al. LncRNA KCNQ1OT1 promotes cell proliferation, migration and invasion via regulating miR-129-5p/JAG1 axis in non-small cell lung cancer. Cancer Cell Int. 2020;20:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Available at: https://www.ncbi.nlm.nih.gov/gene/102800310. [access date April 12, 2023]
- [15].Chen JF, Wu P, Xia R, et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zheng L, He J, Li M, et al. Clinical significance and mechanism of long noncoding RNA HAGLROS in triple negative breast cancer. Pathol Res Pract. 2022;231:153810. [DOI] [PubMed] [Google Scholar]
- [17].Wei H, Hu J, Pu J, et al. Long noncoding RNA HAGLROS promotes cell proliferation, inhibits apoptosis and enhances autophagy via regulating miR-5095/ATG12 axis in hepatocellular carcinoma cells. Int Immunopharmacol. 2019;73:72–80. [DOI] [PubMed] [Google Scholar]
- [18].Gai L, Huang Y, Zhao L, et al. Long non-coding RNA HAGLROS regulates the proliferation, migration, and apoptosis of esophageal cancer cells via the HAGLROS-miR-206- NOTCH3 axis. J Gastrointest Oncol. 2021;12:2093–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang M, Zhai Z, Zhang Y, et al. Gynecologic oncology clinical significance and oncogene function of long noncoding RNA HAGLROS overexpression in ovarian cancer. Arch Gynecol Obstet. 2019;300:703–10. [DOI] [PubMed] [Google Scholar]
- [20].Li L, Zhu H, Li X, et al. Long non-coding RNA HAGLROS facilitates the malignant phenotypes of NSCLC cells via repressing miR-100 and up-regulating SMARCA5. Biomed J. 2021;44:S305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou K, Xu J, Yin X, et al. Long noncoding RNA HAGLROS promotes cell invasion and metastasis by sponging miR-152 and upregulating ROCK1 expression in osteosarcoma. Comput Math Methods Med. 2020;2020:7236245. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [22].Guo K, Chen L, Wang Y, et al. Long noncoding RNA RP11-547D24.1 regulates proliferation and migration in papillary thyroid carcinoma: identification and validation of a novel long noncoding RNA through integrated analysis of TCGA database. Cancer Med. 2019;8:3105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [24].Liu SJ, Dang HX, Lim DA, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21:446–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bhan A, Soleimani M, Mandal SS. Long non-coding RNA (lncRNA) and cancer: a new paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gong W, Zheng J, Liu X, et al. Knockdown of long non-coding RNA KCNQ1OT1 restrained glioma cells’ malignancy by activating miR-370/CCNE2 axis. Front Cell Neurosci. 2017;11:84. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [27].Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hitu L, Gabora K, Bonci EA, et al. MicroRNA in papillary thyroid carcinoma: a systematic review from 2018 to June 2020. Cancers (Basel). 2020;12:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tang G, Zhao H, Xie Z, et al. Long non-coding RNA HAGLROS facilitates tumorigenesis and progression in hepatocellular carcinoma by sponging miR-26b-5p to up-regulate karyopherin α2 (KPNA2) and inactivate p53 signaling. Bioengineered. 2022;13:7829–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu Y, Guo G, Li J, et al. Screening key lncRNAs with diagnostic and prognostic value for head and neck squamous cell carcinoma based on machine learning and mRNA-lncRNA co-expression network analysis. Cancer Biomark. 2020;27:195–206. [DOI] [PubMed] [Google Scholar]
- [31].Shu L, Guo K, Lin ZH, et al. Long non-coding RNA HAGLROS promotes the development of diffuse large B-cell lymphoma via suppressing miR-100. J Clin Lab Anal. 2022;36:e24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang WL, Yu DJ, Zhong M. LncRNA HAGLROS accelerates the progression of lung carcinoma via sponging microRNA-152. Eur Rev Med Pharmacol Sci. 2019;12:6531–8. [DOI] [PubMed] [Google Scholar]
- [33].Stokowy T, Gawel D, Wojtas B. Differences in miRNA and mRNA profile of papillary thyroid cancer variants. Int J Endocrinol. 2016;2016:1427042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zheng H, Fu Q, Ma K, et al. Circ_0079558 promotes papillary thyroid cancer progression by binding to miR-26b-5p to activate MET/AKT signaling. Endocr J. 2021;68:1247–66. [DOI] [PubMed] [Google Scholar]
- [35].Zhou A, Pan H, Sun D, et al. miR-26b-5p inhibits the proliferation, migration and invasion of human papillary thyroid cancer in a β-catenin-dependent manner. Onco Targets Ther. 2020;13:1593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]