Abstract
An automated fluorescence-based PCR system (a model AG-9600 AmpliSensor analyzer) was investigated to determine whether it could detect Shiga toxin-producing Escherichia coli (STEC). The AmpliSensor PCR assay involves amplification-mediated disruption of a fluorogenic DNA signal duplex (AmpliSensor) that is homologous to conserved target sequences in a 323-bp amplified fragment of Shiga toxin genes stx1, stx2, and stxe. Using the Amplisensor assay, we detected 113 strains of STEC belonging to 50 different serotypes, while 18 strains of non-Shiga-toxin-producing E. coli and 68 strains of other bacteria were not detected. The detection limits of the assay were less than 1 to 5 CFU per PCR mixture when pure cultures of five reference strains were used and 3 CFU per 25 g of food when spiked ground beef samples that were preenriched overnight were used. The performance of the assay was also evaluated by using 53 naturally contaminated meat samples and 48 raw milk samples. Thirty-two STEC-positive samples that were confirmed to be positive by the culture assay were found to be positive when the AmpliSensor assay was used. Nine samples that were found to be positive when the PCR assay was used were culture negative. The system described here is an automated PCR-based system that can be used for detection of all serotypes of STEC in food or clinical samples.
Infection with Shiga toxin-producing Escherichia coli (STEC) (also called verotoxigenic E. coli) in humans has been associated with a spectrum of diseases, including diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (HUS) (14, 21). Foods that have an animal origin, such as beef, have been identified as the main vehicles of these food-borne pathogens. The importance of STEC transmission through the food chain has been illustrated by many outbreaks worldwide. For example, the 1996 outbreak in Japan was initially caused by the consumption of school lunches contaminated with E. coli serotype O157:H7; eventually, there were 9,578 reported cases, 11 people died, and there were more than 90 diagnosed cases of HUS (4). In 1992 and 1993 a multistate outbreak in the United States, which was attributed to consumption of hamburgers contaminated with E. coli O157:H7, involved more than 700 people, and there were four deaths and 51 cases of HUS (7). The 1996 Scottish E. coli O157 outbreak associated with meat products resulted in 490 cases of infection and 18 deaths (5).
More than 100 serotypes of STEC have been associated with human diseases, although serotype O157:H7 is the most commonly reported serotype (1). Other commonly isolated outbreak STEC serotypes include O111:NM, O26:H11, O26:NM, and O103:H2 (18). The strains belonging to these STEC serotypes can produce cytotoxins, which collectively are called Shiga toxin (Stx) (also verotoxin and Shiga-like toxin [23]). Other virulence factors, such as intimin (17, 19), hemolysin (30), and other virulence proteins may also be produced by STEC strains. STEC strains can produce different immunotypes of Stx and other virulence factors which play different roles in the onset of disease (21).
Major social and economic consequences underline the benefit of preventive measures that reduce or eliminate exposure to any type of STEC. The American Gastroenterological Association (2) has recommended that future planning for diagnosis and prevention of hemorrhagic colitis, and HUS in the United States should include testing for non-O157 E. coli strains that produce Stx. In the last 20 years, the Vero cell assay has been the “gold standard” for testing for STEC (9). This method, however, requires 4 to 5 days to complete and also requires tissue culture facilities which are not available in many laboratories. Immunological methods for detection of STEC are more rapid and convenient than the Vero cell assay. However, these methods are designed to detect specific toxin types or specific E. coli serotypes, such as O157:H7. In recent years, molecular methods based on PCR have been developed and successfully used to detect STEC. These PCR methods have been reviewed by Olsen et al. (24) and Scheu et al. (29) in terms of their target genes, detection systems, detection limits, and application to foods. Most of the PCR methods have been developed to detect particular types of Stx producers or specific serotypes of STEC (3, 6, 10, 27, 37, 38). Several of the PCR assays have been designed to detect all serotypes of STEC (12, 20, 25, 28). In previously described PCR methods, however, either ethidium bromide and gel electrophoresis or post-PCR hybridization-capture methods are used to detect PCR products. Gel-based methods are laborious and time-consuming, lack sensitivity and specificity, and are difficult to automate. Post-PCR hybridization-capture methods reportedly are more sensitive and more specific than gel-based methods and are easy to automate, but they require multiple post-PCR signal development steps that are even more time-consuming and sometimes more laborious than gel-based methods. In addition, many of the PCR assays have been evaluated only with pure cultures or spiked samples, and their applicability to naturally contaminated food or clinical samples is unknown.
New and improved PCR systems have been developed for detection of STEC in attempts to perform homogeneous and automated direct detection assays of PCR products without the need for gel electrophoresis. One such system is the TaqMan PCR detection system that was developed for detection of Stx I-producing E. coli (37) and E. coli O157:H7 (13). This system is based on the 5′-nuclease activity of Taq DNA polymerase which hydrolyzes an internal flurogenic probe in order to monitor amplification. Another system is the temperature-dependent fluorescence-PCR system, in which an intercalating dye, SYBR Green, is used to monitor amplification. The temperature-dependent fluorescence-PCR system, in combination with the commercially available BAX system, has been used to detect E. coli O157:H7 (35).
We have previously described an automated PCR method which requires no post-PCR handling steps; this method, which is called the AmpliSensor assay, is used for specific detection of Salmonella spp. in foods (8). The AmpliSensor assay is comprised of the following two steps: (i) an initial asymmetric amplification performed with normal primers, which overproduces one strand of the target; and (ii) subsequent seminested amplification and signal detection with an AmpliSensor primer. The AmpliSensor primer is a double-stranded signal probe labelled with fluorescein isothiocyanate and Texas Red (36). The seminested amplification step results in dissociation of the strands of the AmpliSensor duplex and, consequently, disruption of the fluorescence signal. The extent of signal disruption is proportional to the amount of the AmpliSensor primer incorporated into the amplification product and can be measured cycle by cycle and used for quantification of the initial target. In this paper development of the AmpliSensor assay for detection of all serotypes of STEC is described. The results of studies performed to elucidate characteristics of the assay and the applicability of the assay to food samples are presented.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
A total of 113 STEC strains belonging to 50 different serotypes, 18 non-Stx-producing E. coli strains, and 68 strains of other bacteria were used (Tables 1 through 3). These strains were obtained from the collections of the Laboratory Services Division (University of Guelph), the Health of Animals Laboratory (Health Canada), and the Sunny Brook Health Centre (Toronto, Ontario, Canada) and from the American Type Culture Collection. Cultures were maintained on appropriate agar plates and stored at 4°C.
TABLE 1.
Reference STEC strains used in the AmpliSensor PCR assay
| Strain | Serotype | Toxin typea | Reference(s) |
|---|---|---|---|
| H30 | O26:H11 | Stx1 (SLT-I) | 26 |
| 933 W (C600) | O?(rough) | Stx2 (SLT-II) | 34 |
| E32511 | O157:NM | Stx2 (VT2, SLT-II, SLT-IIc) | 16, 31, 32 |
| 412 (DAB) | O139:K82 | Stx2e (SLT-IIe) | 15 |
| H.I.8 | O128:B12 | Stx2va (SLT-IIva) | 11, 39 |
SLT, Shiga-like toxin; VT, verotoxin.
TABLE 3.
Non-STEC bacteria tested for the presence of stx genes with the AmpliSensor PCR assay
| Species | No. of strains |
|---|---|
| E. coli (non-STEC) | 18 |
| Acinetobacter calcoaceticus | 1 |
| Aeromonas hydrophila | 2 |
| Bacillus sp. | 5 |
| Campylobacter sp. | 6 |
| Candida albicans | 1 |
| Carnobacterium mobile | 1 |
| Citrobacter sp. | 2 |
| Edwardsiella tarda | 1 |
| Enterobacter sp. | 2 |
| Enterococcus fecalis | 2 |
| Klebsiella pneumoniae | 1 |
| Kluyvera ascorbata | 1 |
| Lactobacillus sp. | 3 |
| Listeria sp. | 2 |
| Moraxella osloensis | 1 |
| Morganella morganii | 1 |
| Proteus sp. | 2 |
| Pseudomonas sp. | 2 |
| Salmonella sp. | 16 |
| Sarcina sp. | 1 |
| Serratia marcescens | 1 |
| Shigella sp. | 2 |
| Staphylococcus sp. | 8 |
| Streptococcus sp. | 2 |
| Xanthomonas maltophilia | 1 |
| Yersinia enterocolitica | 1 |
Five strains of STEC (Table 1) were used as reference strains in the detection limit and spiking experiments. The cells were grown at 37°C overnight in brain heart infusion broth (Becton Dickinson). Cells were enumerated by plating dilutions of overnight cultures onto MacConkey agar (Difco) plates and incubating the plates at 37°C overnight. E. coli ATCC 25922 was used as a negative control strain.
Preparation of food samples.
Red meat samples which were used in the spiking experiments were purchased from a local retail store. Fifty-three naturally contaminated meat samples were obtained from a study of the prevalence of STEC in 327 raw meat products and 744 ready-to-eat meat products from provincially inspected plants in Ontario (40), and 48 naturally contaminated raw milk samples were obtained from a survey of food-borne pathogens in 1,720 Ontario bulk tank milk samples (33). Preenriched food samples were prepared by homogenizing 25 g of meat in 225 ml of nutrient broth (Becton Dickinson) or 25 ml of milk in 225 ml of universal preenrichment broth (Difco) and then incubating the preparations at 37°C overnight. The preenriched samples were subjected to two steps of selective enrichment with MacConkey broth and brain heart infusion broth and then tested for the presence of STEC by using the Vero cell assay, followed by confirmatory tests performed by the method of Clarke et al. (9). Additional confirmatory tests were performed with meat samples by using the hydrophobic grid membrane filter (HGMF) method described by Yee et al. (40). The preenriched samples were also used in the AmpliSensor assays.
In the mock-contamination experiments, only those food samples that were confirmed to be STEC negative by both culture and PCR methods were used. Food samples were mock contaminated in the following two ways: (i) food samples (25 g) were inoculated with 3 to 336 CFU of an STEC strain before homogenization and preenrichment in nutrient broth (these food samples were designated prespiked samples) and (ii) preenriched food samples (250 ml) were inoculated with 1.12 × 102 to 1.12 × 105 CFU of the target cells per ml (these food samples were designated postspiked samples).
Extraction of DNA from pure cultures and from food samples.
DNA were prepared from pure cultures by using an InstaGene matrix (Bio-Rad) as described by Chen et al. (8). Briefly, 10-μl portions of serial dilutions of an overnight culture of STEC were incubated with 200 μl of InstaGene matrix at 56°C for 20 min and then boiled for 10 min. The mixtures were placed on ice for 10 min and then centrifuged at 16,000 × g for 5 min. The supernatants were used for PCR.
DNA were prepared from food samples by using 1-ml pellets of preenriched food samples and a modification of the EnviroAmp Legionella sample preparation kit protocol (Perkin-Elmer) (8). Briefly, a sample pellet was resuspended in 500 μl of the EnviroAmp DNA extraction reagent and boiled for 20 min. The lysate was cooled on ice for 5 min and centrifuged at 16,000 × g for 3 min to remove the cell and food debris. The DNA was then precipitated from 400 μl of the supernatant by using 400 μl of 100% isopropanol and washed once with 500 μl of 75% isopropanol. The DNA pellet was resuspended in 160 μl of sterile distilled water.
Primers and DNA signal duplex for AmpliSensor.
The amplification target was conserved sequences in subunit A of Stx genes stx1, stx2, and stxe, as described by Read et al. (28). The two primers used for asymmetric amplification were the primers described by Read et al. (28), and using these primers resulted in a 323-bp PCR product. The AmpliSensor primer designed in this study was a signal duplex which targeted the sequence 5′-CGTTTTGTCACTGTGACAGCA-3′ in the stx genes and served as the forward primer in seminested amplifications, generating a 59-bp fragment. The oligonucleotide and complement used for the signal duplex reaction were synthesized with amino-modified deoxyribosylthymidine residues at specific positions and then conjugated with fluorescein isothiocyanate and Texas Red, respectively, by using the reaction conditions recommended by the supplier (Molecular Probes). Since the stx-specific sequences were not completely conserved, several base degeneracies were incorporated into all three primers to allow amplification of all types of stx genes.
Asymmetric amplification.
An amplification reaction mixture (20 μl) containing the following reagents was used: amplification buffer (50 mM Tris-HCl [pH 8.9], 40 mM KCl, 4.0 mM MgCl2, 0.1% Triton X-100, 0.05% Tween 20), 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 400 μM dUTP, 0.1 μM forward primer, 0.75 μM reverse primer, 0.625 U of AmpliTaq DNA polymerase (Perkin-Elmer), 0.25 U of AmpErase uracil N-glycosylase (Perkin-Elmer), and 10 μl of template DNA. The reaction mixture was overlaid with 10 μl of mineral oil. The mineral oil and the master mixture (containing all of the PCR components except the template DNA) were automatically dispensed into a 96-well microtiter polycarbonate plate by using a model AG-9600 AmpliSensor analyzer (Biotronics Corporation). DNA from a pure culture of the Stx-producing organism E. coli 933W was used as a positive control, and DNA from E. coli ATCC 25922 and water were used as negative controls. PCR was performed in the 96-well microtiter plates by using a model 9600 Perkin-Elmer GeneAmp PCR system apparatus. The cycling conditions were as follows: incubation at 50°C for 2 min and at 95°C for 5 min; 29 cycles consisting of denaturation at 94°C for 15 s, annealing at 49°C for 1 min, and extension at 72°C for 30 s; incubation at 72°C for 7 min; and incubation at 4°C until seminested amplification was performed.
Seminested amplification and data acquisition.
After 29 cycles of asymmetric amplification, 4 μl of amplification buffer containing 7.5 ng of the signal duplex was automatically added to each reaction mixture by using the model AG-9600 AmpliSensor analyzer, and cycling was resumed at an annealing temperature of 60°C instead of 49°C. Data were obtained after cycle 1 (in seminested amplification) and every third cycle thereafter for 25 cycles by directly measuring the fluorescence of the amplification mixture with the model AG-9600 AmpliSensor analyzer. The wavelengths used were 485 nm for excitation and 630 nm for emission. A detection index was calculated by using the AG AmpliSensor assay program (Biotronics Corporation) and the following equation: detection index = 1 − (Fs,x/Fn,x), where Fs,x is the fractional decrease in energy transfer of a sample and Fn,x is the fractional decrease in energy transfer of the negative control. An increase in the detection index for each sample was monitored dynamically during seminested amplification. A sample was considered positive if there was a slope as expressed by detection index increase/cycle number increase and if the increase in the detection index was 0.1 or more. The PCR end products were also visualized by using ethidium bromide-stained 2% agarose gels.
RESULTS
Optimization of the assay.
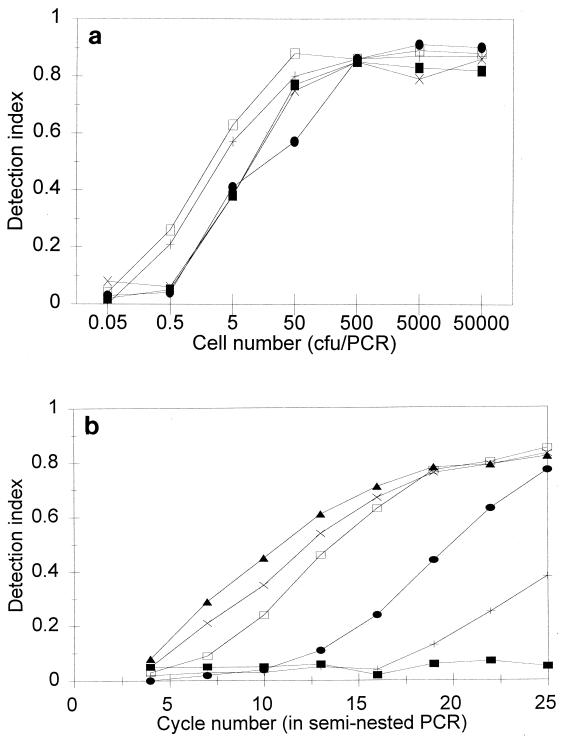
PCR conditions were optimized for amplification of low copy numbers of target DNA sequences. The annealing temperature used for asymmetric amplification, between 49 and 51°C, was optimal for obtaining good yields of PCR products and minimizing nonspecific amplification. An annealing temperature of 60°C was used for seminested amplification; this temperature did not compromise the signal intensity of the PCR products. To detect less than 10 target copies, it was necessary to perform 25 to 30 cycles of asymmetric PCR in the first step for detection of the PCR product in the second step after 15 to 20 cycles.
Specificity, sensitivity, and reproducibility of the AmpliSensor assay.
The specificity of the assay for different STEC serotypes was determined by using 113 STEC strains (Tables 1 and 2) and 86 non-STEC strains (Table 3). All of the reaction mixtures containing STEC resulted in detection index values between 0.5 and 0.9. PCR products of the expected size (323 bp) were visualized by using the agarose gels. All reaction mixtures containing non-STEC had detection index values within 0.0 ± 0.10, and no PCR products of the expected size were observed when gel electrophoresis was performed.
TABLE 2.
Serotypes of STEC strains tested for the presence of stx genes with the AmpliSensor PCR assay
| Serotype | No. of strains | Serotype | No. of strains | |
|---|---|---|---|---|
| O?(Rough) | 1 | O91:NM | 2 | |
| O?:H2 | 1 | O98:NM | 1 | |
| O?:H7 | 1 | O103:H2 | 3 | |
| O?:H8 | 1 | O111:H8 | 1 | |
| O?:H19 | 1 | O111:H11 | 2 | |
| O?:H21 | 1 | O111:NM | 5 | |
| O?:NM | 2 | O113:H21 | 1 | |
| O1:H20 | 1 | O116:H21 | 3 | |
| O2:H29 | 1 | O121:H7 | 3 | |
| O5:NM | 6 | O121:H19 | 1 | |
| O7:H43 | 3 | O126:H8 | 1 | |
| O8:H? | 1 | O128:B12 | 1 | |
| O8:H9 | 1 | O132:NM | 3 | |
| O8:H19 | 3 | O136:H12 | 1 | |
| O15:H27 | 1 | O136:H16 | 1 | |
| O15:NM | 1 | O139:K82 | 1 | |
| O22:H8 | 9 | O142:H38 | 1 | |
| O26:H11 | 7 | O145:NM | 1 | |
| O26:NM | 2 | O153:H25 | 2 | |
| O38:H21 | 1 | O153:NM | 1 | |
| O45:H2 | 3 | O157:H7 | 10 | |
| O80:NM | 1 | O157:NM | 6 | |
| O84:H2 | 3 | O163:H19 | 1 | |
| O91:H14 | 3 | O163:NM | 2 | |
| O91:H21 | 5 | O165:NM | 1 |
The limit of detection of the assay was determined by using dilutions of overnight pure cultures of five reference STEC strains (Table 1). The detection limit was 1 to 5 CFU per PCR, as shown in Fig. 1.
FIG. 1.
(a) Detection of STEC in pure cultures of five reference STEC strains by the AmpliSensor PCR assay. Detection index values were obtained after 25 cycles of the seminested PCR. Symbols: ■, STEC strain H30; +, STEC strain 933W; •, STEC strain 32511; □, STEC strain 412; ×, STEC strain HI8. (b) Detection of STEC in a pure culture of strain H30 by the AmpliSensor PCR assay: extent of amplification versus cycle number and initial copy number of target DNA. Symbols: ■, 0.46 CFU/PCR; +, 4.6 CFU/PCR; •, 46 CFU/PCR; □, 460 CFU/PCR; ×, 4,600 CFU/PCR ▴, 46,000 CFU/PCR.
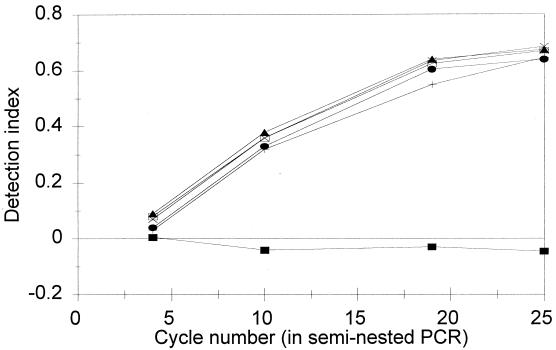
The reproducibility of the assay was studied on different days by using three replicates of an overnight culture of strain 933W. Figure 2 shows the relationship between the log number of cells and the detection index values when strain 933W was used. The interassays were reproducible. All of the subsequent spiking experiments were performed in duplicate.
FIG. 2.
Detection of STEC in a pure culture of strain 933W by the AmpliSensor PCR assay. Detection index values were obtained after 22 cycles of the seminested PCR. Symbols: ■, run 1; +, run 2; •, run 3.
Detection of STEC in spiked ground beef.
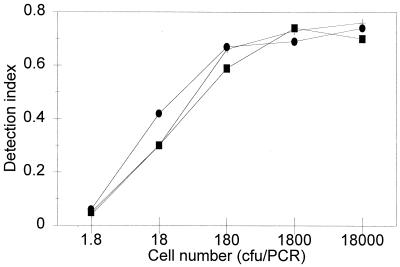
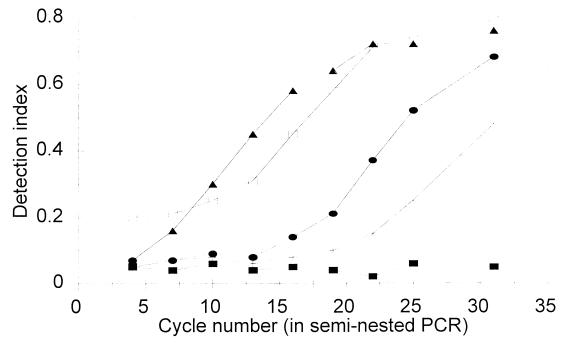
To test the limit of detection of the assay for STEC in foods, samples of ground beef were prespiked or postspiked with target cells. Figure 3 shows the cycle-dependent accumulation of the PCR products for the prespiked samples tested. The detection limit was 3 CFU/25 g. In samples that exhibited cycle-dependent accumulation of the PCR product, a DNA band at 323 bp was detected by agarose gel electrophoresis (results not shown). Figure 4 shows the relationships between the log numbers of cells and the detection index values in a cycle-dependent manner for postspiked ground beef samples. The detection limit was 112 CFU/ml (equivalent to 8.6 CFU/PCR).
FIG. 3.
Detection of STEC in prespiked ground beef by the AmpliSensor PCR assay: extent of amplification versus cycle number and initial copy number of target DNA. Symbols: ■, no STEC; +, 3 CFU/25 g; •, 6 CFU/25 g; □, 33 CFU/25 g; ×, 66 CFU/25 g; ▴, 336 CFU/25 g.
FIG. 4.
Detection of STEC in postspiked ground beef by the AmpliSensor PCR assay: extent of amplification versus cycle number and initial copy number of target DNA. Symbols: ■, no STEC; +, 112 CFU/ml; •, 1,120 CFU/ml; □, 11,200 CFU/ml; ▴, 112,000 CFU/ml.
Detection of STEC in naturally contaminated foods.
To evaluate the applicability of the assay to naturally contaminated foods, 53 meat samples and 48 raw milk samples were tested by using the AmpliSensor assay. The results are summarized in Tables 4 through 6. All of the detection index values fell into the following two groups: higher than 0.4 and within 0.0 ± 0.10. Of the 101 samples, 32 (17 meat samples and 15 raw milk samples) were positive by both the culture and AmpliSensor PCR methods, 59 (28 meat samples and 31 raw milk samples) were negative by both methods, 9 (7 meat samples and 2 raw milk samples) were positive by the AmpliSensor PCR method alone, and 1 was positive by the culture method alone. One sample which was positive by the culture method alone was examined further by using the HGMF method and the PCR method with less inhibitory selective enrichment broth instead of preenrichment broth. The sample remained negative, and culture contamination was suspected.
TABLE 4.
Detection of STEC in naturally contaminated raw milk samples by the AmpliSensor PCR assay and the culture method
| Sample | Detection indexa | STEC isolated |
|---|---|---|
| VE2-58 | 0.59 | Yes |
| VE4-11 | 0.68 | Yes |
| VE4-31 | 0.71 | Yes |
| VE5-7 | 0.72 | Yes |
| VE5-18 | 0.66 | Yes |
| VE7-34 | 0.64 | Yes |
| VE7-46 | 0.55 | No |
| VE11-38 | 0.73 | Yes |
| VE12-59 | 0.77 | Yes |
| VE14-67 | 0.68 | Yes |
| VE15-56 | 0.53 | Yes |
| VE16-37 | 0.69 | Yes |
| VE16-64 | 0.64 | Yes |
| VE16-75 | 0.73 | Yes |
| VE20-28 | 0.66 | Yes |
| VE21-45 | 0.68 | No |
| VE22-120 | 0.7 | Yes |
| STEC strain 933W | 0.7 | Yes |
| E. coli ATCC 25922 | 0 | No |
Detection index values were determined at the end of 25 cycles of seminested amplification. All remaining 31 samples had detection index values within 0.0 ± 0.10, and no STEC were isolated by the culture method.
TABLE 6.
Detection of STEC in naturally contaminated meat samples by the AmpliSensor PCR assay and the culture method: samples negative for STEC by the PCR assay or the culture method
| Sample origin | No. |
|---|---|
| Beefjerky | 1 |
| Beerwurst | 1 |
| Bologna | 2 |
| Boneless beef | 3 |
| Boneless pork | 1 |
| Ground beef | 7 |
| Ground beef and pork | 1 |
| Ham | 2 |
| Hot liver sausage | 1 |
| Kielbasa | 1 |
| Kobassa | 1 |
| Meatloaf | 1 |
| Pepperoni | 2 |
| Roast beef | 1 |
| Salami | 1 |
| Smoked sausage | 1 |
| Specwurst | 1 |
| Total | 28 |
DISCUSSION
Specificity and sensitivity of the AmpliSensor assay.
The results obtained in our study demonstrate the specificity of the primers based on the tests performed with 113 strains of STEC belonging to 50 common serotypes and 86 non-STEC strains. This indicates that the degenerate primers did not compromise the specificity of the assay at a measurable level. The degeneracy of the primers may be compensated for because three primers are used in the system. The specificity of the two primers used for asymmetric amplification was demonstrated previously in the study of Read et al. (28), in which all 223 STEC strains tested and 2 of the 148 non-STEC strains tested were identified as STEC. The only two positive non-STEC strains in the study of Read et al. were Shigella dysenteriae type 1 strains. However, detection of S. dysenteriae type 1 in foods was considered advantageous (28).
The detection limits of the AmpliSensor assay were 1 to 5 CFU/PCR when pure cultures of five reference STEC strains that produce different types of toxins were used and 8.6 CFU/PCR when postspiked ground beef samples were used. No noticeable difference in amplification efficiency was observed when different stx genes were amplified. These detection limits are similar to the detection limit of the 5′ nuclease assay for stx1 (10 ± 5 CFU) (37) and 1 to 2 logs lower than the detection limit of the gel-based PCR assays (102 CFU) (20, 28). In this study, a 2-log decrease in the detection limit was also observed when the same PCR products were analyzed by the AmpliSensor assay compared-to agarose gels (results not shown).
Other features of the assay.
The model AG-9600 AmpliSensor analyzer is an automated system for dispensing PCR reagents and for detecting PCR products. For both the PCR process and PCR product detection, only one 96-well microplate and one pipetting step (for the addition of template DNA) are required. The simplicity of the procedure increases the reproducibility of the assay and reduces the chance of laboratory DNA contamination. The AmpliSensor assay also employs uracil N-glucosylase and dUTP (22) to minimize potential problems caused by PCR product carryover contamination.
Applicability of the AmpliSensor assay to food samples.
One important criterion that is necessary for a rapid STEC testing method to be applicable to food samples is that the rapid method must be as sensitive as or more sensitive than conventional methods (which have a theoretical level of detection of 1 CFU/25 g or 1 CFU/25 ml of food). In this study we were able to detect 3 CFU of STEC in 25 g of food following overnight preenrichment. In the prespiked samples the concentration of the target cells after preenrichment was not determined. However, the detection limit of 112 CFU/mL for the postspiked samples suggests that overnight preenrichment is sufficient to ensure the sensitivity of the assay because the concentration of STEC in food samples usually is 104 CFU/ml or more after overnight preenrichment.
The AmpliSensor assay was also successfully used with 101 naturally contaminated food samples. When the AmpliSensor assay was used, nine additional STEC-positive samples (seven meat samples and two raw milk samples) were detected compared to the number of positive samples detected by the traditional culture method, suggesting that the AmpliSensor assay is more sensitive than the culture method. In a previous study, the same seven meat samples that were positive only by the AmpliSensor PCR method were analyzed by using the HGMF method, a method that was found to be more efficient than the traditional method for recovering STEC (40). STEC belonging to different serotypes were isolated from three of the seven samples, suggesting that the culture method was not sensitive enough to detect of all of the STEC-positive samples. In addition to the greater sensitivity of the PCR method, the following factors may also result in PCR-positive and culture-negative detection: (i) the target cells may be injured or dead and therefore nonculturable but detectable by the PCR method and (ii) the presence of S. dysenteriae type 1 could result in positive reactions in the PCR assay (28) but no recovery of STEC.
It should be noted that the prevalence of STEC in the food samples tested in this study (32 of 101 samples [31.7%]) is not the true prevalence because the naturally contaminated samples used in this study were selected from two previous prevalence studies (33, 40). The samples used included all of the culture-positive samples identified in the prevalence studies plus randomly selected STEC-negative samples.
TABLE 5.
Detection of STEC in naturally contaminated meat samples by the AmpliSensor PCR assay and the culture method: samples positive for STEC by the PCR assay or the culture method
| Sample origin or taxon | Sample or strain | Detection indexa | STEC isolated |
|---|---|---|---|
| Boneless beef | 344 | −0.02 | Yes |
| 449 | 0.67 | No | |
| 470 | 0.67 | No | |
| 476 | 0.58 | Yes | |
| 543 | 0.5 | Yes | |
| 599 | 0.68 | Yes | |
| 1024 | 0.71 | No | |
| 1029 | 0.49 | Yes | |
| Boneless pork | 311 | 0.41 | No |
| 409 | 0.7 | Yes | |
| 685 | 0.67 | Yes | |
| 756 | 0.64 | No | |
| 911 | 0.56 | No | |
| 918 | 0.68 | No | |
| Ground beef | 82 | 0.77 | Yes |
| 132 | 0.61 | Yes | |
| 373 | 0.49 | Yes | |
| 548 | 0.66 | Yes | |
| 842 HAL | 0.63 | Yes | |
| 850 HAL | 0.69 | Yes | |
| Ground pork | 194 | 0.52 | Yes |
| 244 | 0.62 | Yes | |
| Kielbasa | 280 | 0.68 | Yes |
| Pepperoni | 428 | 0.65 | Yes |
| Tyrolee salami | 879 | 0.7 | Yes |
| STEC | 933W | 0.7 | Yes |
| E. coli | ATCC 25922 | 0 | No |
Detection index values were determined at the end of 25 cycles of seminested amplification.
ACKNOWLEDGMENTS
This research was supported by the Ontario Food Quality and Safety Research Fund.
We are grateful to provincial food inspectors for collecting the food samples and to M. Rozwadowski and M. Steele for performing the culture assays.
REFERENCES
- 1.Acheson D W K, Keusch G T. Which Shiga toxin-producing types of E. coli are important? ASM News. 1996;62:302–306. [Google Scholar]
- 2.American Gastroenterological Association. Consensus conference statement: Escherichia coli O157:H7 infections—an emerging national health crisis. Gastroenterology. 1995;108:1923–1934. [PubMed] [Google Scholar]
- 3.Begum D, Jackson M P. Direct detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Mol Cell Probes. 1995;9:259–264. doi: 10.1016/s0890-8508(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim K. Escherichia coli O157 outbreak in Japan: lessons for Australia. Aust Vet J. 1997;75:108. doi: 10.1111/j.1751-0813.1997.tb14168.x. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury J. Report on Scottish E. coli O157 outbreak released. Lancet. 1997;349:1073. [Google Scholar]
- 6.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. Morbid Mortal Weekly Rep. 1993;42:258–263. [PubMed] [Google Scholar]
- 8.Chen S, Yee A, Griffiths M, Wu K Y, Wang C-N, Rahn K, De Grandis S A. A rapid, sensitive and automated method for detection of Salmonella species in foods using AG-9600 AmpliSensor analyzer. J Appl Microbiol. 1997;83:314–321. doi: 10.1046/j.1365-2672.1997.00226.x. [DOI] [PubMed] [Google Scholar]
- 9.Clarke R C, McEwen S A, Gannon V P, Lior H, Gyles C L. Isolation of verocytotoxin-producing Escherichia coli from milk filters in south-western Ontario. Epidemiol Infect. 1989;102:253–260. doi: 10.1017/s0950268800029927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannon V P J, Teerling C, Masrei S A, Gyles C L. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J Gen Microbiol. 1990;136:1125–1135. doi: 10.1099/00221287-136-6-1125. [DOI] [PubMed] [Google Scholar]
- 12.Gannon V P J, King R K, Kim J Y, Golsteyn Thomas E J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green R L, Matsuura M, Schreiber E, Ho M S Y, Yagi L A, Lai L T Y, Flood S J A. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Detection of Escherichia coli O157:H7 in foods using a rapid fluorogenic PCR-based assay system, abstr. P-81; p. 417. [Google Scholar]
- 14.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 15.Gyles C L, De Grandis S A, MacKenzie C, Brunton J L. Cloning and nucleotide sequence analysis of the genes determining verocytotoxin production in a porcine edema disease isolate of Escherichia coli. Microb Pathog. 1988;5:419–426. doi: 10.1016/0882-4010(88)90003-4. [DOI] [PubMed] [Google Scholar]
- 16.Head S C, Petric M, Richardson S, Roscoe M, Karmali M A. Purification and characterization of verocytotoxin 2. FEMS Microbiol Lett. 1988;51:211–216. [Google Scholar]
- 17.Jerse A E, Kaper J B. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R, Clarke R C, Wilson J B, Read S, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmal M A, Lior H, Mcewen S A, Spika J S, Gyles C L. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J Food Prot. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 19.Kaper J B. Molecular genetics of attaching and effacing E. coli. In: Karmali M A, Goglio A G, editors. Recent advances in verocytotoxin-producing Escherichia coli infections. Proceedings of the 2nd International Symposium and Workshop, Bergamo, Italy, 1994. Amsterdam, The Netherlands: Elsevier Science/North-Holland Publishing Co.; 1994. pp. 223–231. [Google Scholar]
- 20.Karch H, Meyer T. Single primer pair for amplifying segments of distinct Shiga-like-toxin genes by polymerase chain reaction. J Clin Microbiol. 1989;27:2751–2757. doi: 10.1128/jcm.27.12.2751-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewy Z G, Mecca J, Diaco R. Enhancement of Borrelia burgdorferi PCR by uracil N-glycosylase. J Clin Microbiol. 1994;32:135–138. doi: 10.1128/jcm.32.1.135-138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien A D, Holmes R K. Shiga and Shiga-like toxins. Microbiol Rev. 1987;51:206–220. doi: 10.1128/mr.51.2.206-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen J E, Aabo S, Hill W, Notermans S, Wernars K, Granum P E, Popovic T, Rasmussen H N, Olsvik O. Probes and polymerase chain reaction for detection of food-borne bacterial pathogens. Int J Food Microbiol. 1995;28:1–78. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 25.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petric M, Karmali M A, Richardson S, Cheung R. Purification and biological properties of Escherichia coli vero-cytotoxin. FEMS Microbiol Lett. 1987;41:63–68. [Google Scholar]
- 27.Ramotar K, Waldhart B, Church D, Szumski R, Louie T J. Direct detection of verotoxin-producing Escherichia coli in stool samples by PCR. J Clin Microbiol. 1995;33:519–524. doi: 10.1128/jcm.33.3.519-524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read S C, Clarke R C, Martin A, De Grandis S A, Hii J, McEwen S, Gyles C L. Polymerase chain reaction for detection of verotoxigenic Escherichia coli isolated from animal and food sources. Mol Cell Probes. 1992;6:153–161. doi: 10.1016/0890-8508(92)90060-b. [DOI] [PubMed] [Google Scholar]
- 29.Scheu P M, Berghof K, Stahl U. Detection of pathogenic and spoilage micro-organisms in food with the polymerase chain reaction. Food Microbiol. 1998;15:13–31. [Google Scholar]
- 30.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt C K, McKee M, O’Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scotland S M, Smith H R, Rowe B. Two distinct toxins active on Vero cells from Escherichia coli O157. Lancet. 1985;ii:885–886. doi: 10.1016/s0140-6736(85)90146-1. [DOI] [PubMed] [Google Scholar]
- 33.Steele M L, McNab W B, Poppe C, Griffiths M W, Chen S, De Grandis S A, Fruhner L C, Larkin C A, Lynch J A, Odumeru J A. A survey of Ontario bulk tank raw milk for food-borne pathogens. J Food Prot. 1997;60:1341–1346. doi: 10.4315/0362-028X-60.11.1341. [DOI] [PubMed] [Google Scholar]
- 34.Strockbine N A, Marques L R M, Newland J W, Smith W H, Holmes R K, O’Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biological activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tseng S Y, Gandhi S. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Development of a homogeneous temperature-dependent fluorescence-PCR assay for the detection of Escherichia coli O157:H7, abstr. P-72; p. 415. [Google Scholar]
- 36.Wang C N, Wu K Y, Wang H-T. Quantitative PCR using the AmpliSensor assay. In: Dieffennach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 193–202. [Google Scholar]
- 37.Witham P K, Yamashiro C, Livak K J, Batt C A. A PCR-based assay for the detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl Environ Microbiol. 1996;62:1347–1353. doi: 10.1128/aem.62.4.1347-1353.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Cooms J, Malmstrom S, Glass M. Simultaneous multianalyte nucleic acid detection for gastrointestinal bacterial pathogens using GENESTAR technology. DNA Technol Clin Lab. 1997;17:129–145. [PubMed] [Google Scholar]
- 39.Yee A J, De Grandis S A, Gyles C L. Mitomycin-induced synthesis of a Shiga-like toxin from enteropathogenic Escherichia coli H.I.8. Infect Immun. 1993;61:4510–4513. doi: 10.1128/iai.61.10.4510-4513.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yee A J, Martin A, Rozwadowski M, Read S, Todd E C D, Alves D, Johnson P, Gyles C L. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. A prevalence survey of verotoxigenic Escherichia coli in raw and ready-to-eat meat products, abstr. P-71; p. 394. [Google Scholar]