LUNG CANCER IS THE LEADING CAUSE OF CANCER DEATHS IN THE UNITED States1 and worldwide. The two major forms of lung cancer are non–small-cell lung cancer (about 85% of all lung cancers) and small-cell lung cancer (about 15%). Despite advances in early detection and standard treatment, non–small-cell lung cancer is often diagnosed at an advanced stage and has a poor prognosis. The treatment and prevention of lung cancer are major unmet needs that can probably be improved by a better understanding of the molecular origins and evolution of the disease.
Non–small-cell lung cancer can be divided into three major histologic subtypes: squamous-cell carcinoma, adenocarcinoma, and large-cell lung cancer. Smoking causes all types of lung cancer but is most strongly linked with small-cell lung cancer and squamous-cell carcinoma; adenocarcinoma is the most common type in patients who have never smoked (Fig. 12-8). This review will focus on major recent advances in the molecular study of the origins and biology of squamous-cell carcinoma and adenocarcinoma, since they constitute the vast majority of diagnosed lung cancers (Table 15,8-14). These advances have been facilitated by the development of molecular techniques and biomarkers for defining cancer risk, prognosis, and optimal therapy aimed at personalized prevention and treatment of lung cancer.
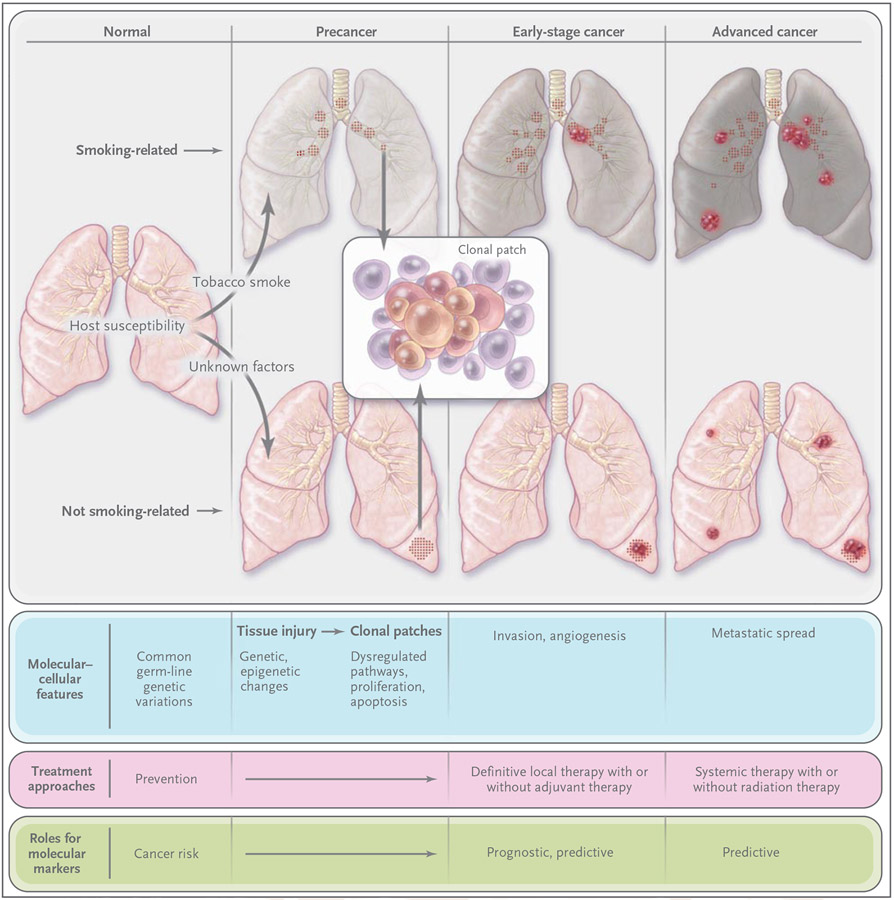
Figure 1. Molecular Evolution of Lung Cancer.
Environmental factors, such as tobacco smoke, and genetic susceptibility interact to influence carcinogenesis. Factors that are unrelated to smoking — including genetic, hormonal, and viral (e.g., human papillomavirus) factors — have been suggested.2 Tissue injury (e.g., from tobacco smoke, reflected in the discolored smoking-related lungs) initially occurs in the form of genetic and epigenetic changes (e.g., mutations, loss of heterozygosity, and promoter methylation) and global transcriptome changes (e.g., inflammation and apoptosis pathways). These changes can persist long term3,4 and eventually lead to aberrant pathway activation and cellular function (e.g., dysregulated proliferation and apoptosis) to produce premalignant changes, including dysplasia and clonal patches. Additional changes can result in angiogenesis, invasion and early-stage cancer, and advanced cancer and metastasis.5 Many molecular changes in earliest-stage cancer also occur in advanced disease.6,7 Premalignant patches contain clones and subclones (inset), which can involve loss of heterozygosity, microsatellite instability, and mutations (e.g., in p53 and epidermal growth factor receptor [EGFR]). Lung cancers unrelated and related to smoking have strikingly different molecular profiles, including those of mutations in p53, KRAS, EGFR, and HER2. Smoking-related patches and primary cancers (usually squamous-cell carcinoma and small-cell lung cancer) most often develop in the central airway.4,8 Most tumors that are not related to smoking are adenocarcinomas and develop in the peripheral airways. Molecular markers can signify risk (in people without cancer), prognosis (outcome independent of treatment), and sensitivity to treatment through predictive markers. Such stage-specific markers can span the course of disease from its early stages through its late stages. They also can help define mechanisms of resistance to therapy.
Table 1.
Genetic Abnormalities Specific in the Lung to Non–Small-Cell Lung Cancer and Small-Cell Lung Cancer.*
| Abnormality | Non–Small-Cell Lung Cancer | Small-Cell Lung Cancer | |
|---|---|---|---|
| Squamous-Cell Carcinoma |
Adenocarcinoma | ||
| Precursor | |||
| Lesion | Known (dysplasia) | Probable (atypical adenomatous hyperplasia) | Possible (neuroendocrine field)† |
| Genetic change | p53 mutation | KRAS mutation (atypical adenomatous hyperplasia in smokers), EGFR kinase domain mutation (in nonsmokers) | Overexpression of c-MET |
| Cancer | |||
| KRAS mutation | Very rare | 10 to 30%‡ | Very rare |
| BRAF mutation | 3% | 2% | Very rare |
| EGFR | |||
| Kinase domain mutation | Very rare | 10 to 40%‡ | Very rare |
| Amplification§ | 30% | 15% | Very rare |
| Variant III mutation | 5%¶ | Very rare | Very rare |
| HER2 | |||
| Kinase domain mutation | Very rare | 4% | Very rare |
| Amplification | 2% | 6% | Not known |
| ALK fusion∥ | Very rare | 7% | Not known |
| MET | |||
| Mutation | 12% | 14% | 13% |
| Amplification | 21% | 20% | Not known |
| TITF-1 amplification | 15% | 15% | Very rare |
| p53 mutation | 60 to 70% | 50 to 70%‡ | 75% |
| LKB1 mutation | 19% | 34% | Very rare |
| PIK3CA | |||
| Mutation | 2% | 2% | Very rare |
| Amplification | 33% | 6% | 4% |
Non–small-cell lung cancer includes squamous-cell carcinoma and adenocarcinoma.
Neuroendocrine fields have been detected only in tissue surrounding tumors and have been characterized by extremely high rates of allelic loss and by c-MET overexpression (Salgia R: personal communication).
Variations are based in part on smoking profiles.
The percentages include increased gene copy numbers from amplification or polysomy and represent percentages from resected cancers. The percentages are higher in primary tumors from patients with metastatic disease. Increased copy numbers have been reported in squamous dysplastic lesions but not in adenocarcinoma precursors.
Genomic EGFR variant III mutations have been detected only in lung squamous-cell carcinoma, and these tumors are sensitive preclinically to irreversible EGFR tyrosine kinase inhibitors. The incidence of 5% is substantially lower than that of 30 to 40% for the detection in squamous-cell carcinoma or adenocarcinoma by immunohistochemical analysis or other techniques.
The anaplastic lymphoma kinase (ALK) fusion gene (involving chromosome 2p), consisting of parts of EML4 and ALK, is transforming in fibroblasts and occurs in adenocarcinoma but not in other types of non–small-cell lung cancer or other nonlung cancers.
MOLECULAR ORIGINS
HOST SUSCEPTIBILITY
Epidemiologic studies showing an association between family history and an increased risk of lung cancer provided the first evidence of host susceptibility (Fig. 1). Lung-cancer susceptibility and risk also are increased in inherited cancer syndromes caused by rare germ-line mutations in p53,15 retinoblastoma,16 and other genes,17 as well as a germ-line mutation in the epidermal growth factor receptor (EGFR) gene.18 More recently, three large genomewide association studies identified an association between single-nucleotide polymorphism (SNP) variation at 15q24–15q25.1 and susceptibility to lung cancer. The region of the SNP variation was recently linked to lung carcinogenesis and includes two genes encoding subunits of the nicotinic acetylcholine receptor alpha, which is regulated by nicotine exposure.19-22
Lung-cancer susceptibility and risk also increase with reduced DNA repair capacity (particularly when accompanied by exposure to tobacco smoke)23 that results, for example, from germ-line alterations in nucleotide excision repair genes such as ERCC1.24 Increased expression of DNA synthesis and repair genes, including RRM1 (the regulatory subunit of ribonucleotide reductase) and ERCC1, in non–small-cell lung cancer correlates with a better prognosis overall but no benefit from platinum-based chemotherapy.25,26 Table 1 presents gene abnormalities involved in the development of different histologic types of lung cancer.
CLONAL EVOLUTION
Changes in certain genes (e.g., proinflammatory interleukin-8 [IL8] and some DNA-repair genes) occur in nonmalignant lung tissue of smokers and patients with lung cancer, a finding consistent with diffuse tissue injury.3,27-29 These changes probably precede epithelial clonal evolution, an important element of the molecular origins of lung and other cancers (Fig. 1). Patches of clonally related cells, or clonal patches containing 40,000 to 360,000 cells, have been mapped in the lung.30 The size and number of subclones in a clonal patch may contribute to the cancer risk.31 Early events in the development of non–small-cell lung cancer include loss of heterozygosity at chromosomal region 3p21.3 (site of RASSF1A, a member of the Ras association domain family, and FUS1), 3p14.2 (FHIT, a fragile histidine triad gene), 9p21 (p16), and 17p13 (p53).32 All these genes are tumor-suppressor genes. Loss-of-heterozygosity patterns in squamous-cell carcinoma and adenocarcinoma differ (e.g., chromosome 3p deletions are much more extensive in squamous-cell carcinoma). Mutations in the EGFR kinase domain occur early in the development of adenocarcinoma that is generally unrelated to smoking, and KRAS mutations occur early in the development of smoking-related adenocarcinoma.33,34 Clonal patches with methylation of promoter regions of genes (epigenetic changes), p53 mutation, EGFR mutation, c-Myc amplification, loss of heterozygosity, and microsatellite instability can occur in normal tissue surrounding non–small-cell lung tumors5,30,32,34,35 and may be associated with a greater risk of recurrence and second primary tumors. These findings suggest that in the future molecular analyses of surgical margins may help identify patients most likely to benefit from adjuvant therapy.
Methylated genes in premalignant squamous-cell lung lesions (e.g., metaplasia and dysplasia) are p16 and FHIT (frequently and very early) and O-6-methylguanine-DNA methyltransferase (MGMT), death-associated protein kinase (DAPK), and RASSF1A (less frequently or rarely and in advanced precancers).35-41 The early methylation of p16 in the development of squamous-cell lung cancer (e.g., in normal lung tissue in approximately 50% of smokers)36 exemplifies differences with that in the development of adenocarcinoma, in which p16 methylation occurs very rarely and only late in precursors (e.g., high-grade atypical adenomatous hyperplasia).37 Methylation markers in sputum are associated with the risk of lung cancer (e.g., methylated p16)38 and the recurrence of lung cancer (methylated ASC-TMS1,39 also called PYCARD). Recent data show that promoter methylation of various genes, including p16 in stage I non–small-cell lung cancer, is associated with recurrence after resection.40,41 Agents that reverse epigenetic changes have shown promise in a mouse model of lung carcinogenesis and are being tested in humans with lung cancer.42
Bronchoalveolar stem cells, which may be precursors of lung adenocarcinoma, were identified recently in studies in mice.43 The KRAS, Pten, phosphoinositide 3-kinase (PI3K), and cyclin-dependent kinase pathways have been implicated in the proliferation of these stem cells.44,45 The potential role of bronchoalveolar stem cells and other tumorigenic stem-cell populations in the development and prognosis of human lung cancer and its resistance to drugs is an important area of future investigation.
MOLECULAR EVOLUTION
EGFR FAMILY
EGFR regulates important tumorigenic processes, including proliferation, apoptosis, angiogenesis, and invasion (Fig. 2), and, along with its ligands, is frequently overexpressed in the development and progression of non–small-cell lung cancer.2,5,34,46 Clinical trials of the EGFR tyrosine kinase inhibitor erlotinib for second-line or third-line treatment of such tumors and of the monoclonal antibody against EGFR, cetuximab (combined with chemotherapy), for treatment of previously untreated, advanced disease47,48 validated EGFR as a molecular target for therapy. EGFR mutations that were discovered during clinical trials led to extensive studies of the roles of these mutations and EGFR amplification in the pathogenesis of the disease and its prognosis and sensitivity to treatment.
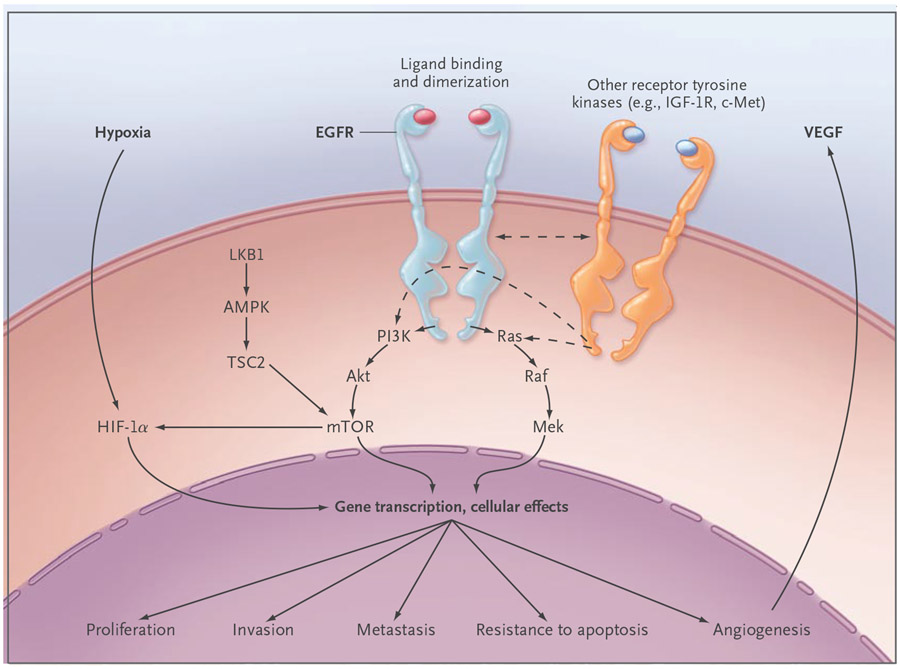
Figure 2. Epidermal Growth Factor Receptor (EGFR) Cell-Signaling Pathways.
EGFR activates several major downstream signaling pathways, including Ras–Raf–Mek and the pathway consisting of phosphoinositide 3-kinase (PI3K), Akt, and mammalian target of rapamycin (mTOR), which in turn may have an effect on proliferation, survival, invasiveness, metastatic spread, and tumor angiogenesis through pathways that are either dependent on or independent of the hypoxia inducible factor (HIF). These pathways also may be modulated by other receptor tyrosine kinases, such as insulin-like growth factor 1 receptor (IGF-1R) and cMET, and by the LKB1–amp-activated protein kinase (AMPK) pathway, which is involved in energy sensing and cellular stress. Most of these functions depend on signaling through the kinase domain. However, kinase-independent functions, such as maintaining glucose transport, have been reported.46 TSC2 denotes tuberous sclerosis complex 2, and VEGF vascular endothelial growth factor.
Several groups of investigators independently identified somatic mutations in the kinase domain of EGFR in lung adenocarcinoma in approximately 10% of specimens from patients in the United States and in 30 to 50% of specimens from patients in Asia49 (Fig. 3). The mutations occur with increased frequency in women and nonsmokers and are tightly associated with sensitivity to the EGFR tyrosine kinase inhibitors gefitinib and erlotinib and so appear to explain most of the dramatic responses to these agents.50-52 More than 80% of these mutations in lung cancer involve in-frame deletions within exon 19 or the L858R mutant within exon 21. EGFR mutations are associated with an improved prognosis in non–small-cell lung cancer, even when treated with cytotoxic chemotherapy.53,54 EGFR amplification is detected in dysplasia (especially of a high grade), which is associated with lung-cancer risk when detected in the sputum of smokers, and is associated with a poor prognosis but also with sensitivity to EGFR inhibitors.55,56 The epidemiologic links highlight three factors — whether the patient is a nonsmoker, Asian, and female — that are associated independently and collectively with an improved response to EGFR tyrosine kinase inhibitors. However, erlotinib appears to prolong survival in virtually all subgroups of patients with non–small-cell lung cancer.48 There are major differences in clinical, pathological, sex-related, and molecular factors between smokers and lifelong nonsmokers in whom lung cancer develops (Fig. 1).
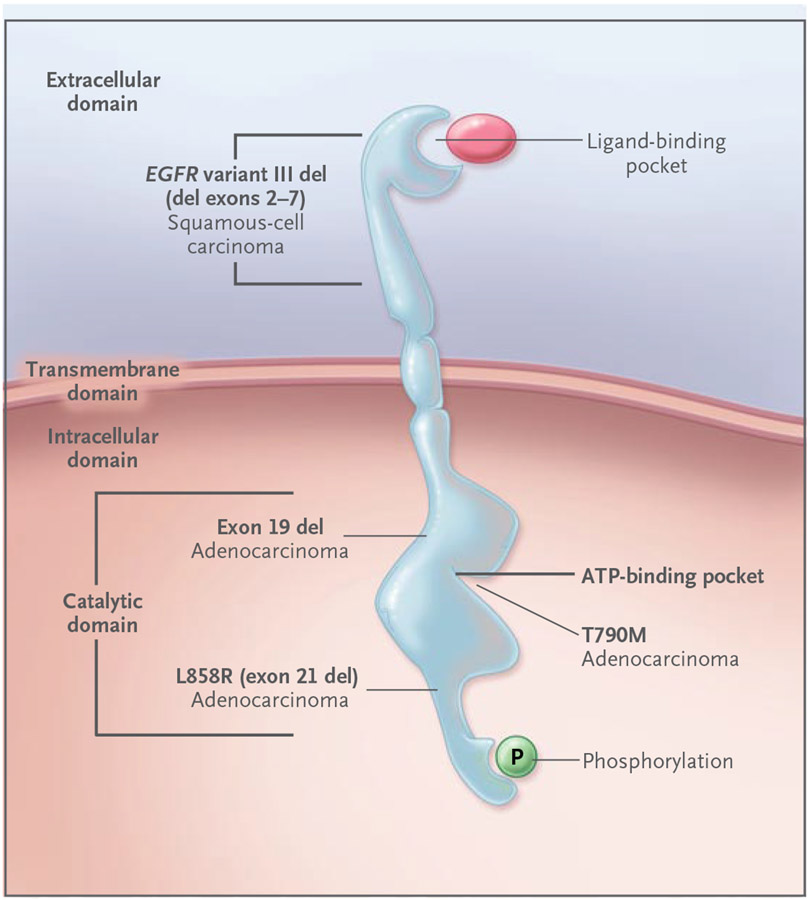
Figure 3. Effect of Deletions and Mutations in the Epidermal Growth Factor Receptor Gene (EGFR) on Disease Development and Drug Targeting.
Ligand binding to the EGFR extracellular domain results in receptor homodimerization and tyrosine phosphorylation, with the phosphate derived from ATP bound within the kinase catalytic domain. EGFR mutations have transforming potential in preclinical lung models and can occur early in human lung carcinogenesis. EGFR point mutations (e.g., L858R) and exon 19 deletions, which occur predominantly in adenocarcinoma of the lung, are located within the catalytic domain and result in constitutive EGFR activation. These mutations are associated with increased sensitivity to EGFR tyrosine kinase inhibitors, such as erlotinib and gefitinib. In contrast, mutations in T790M (an amino acid located within the ATP binding site of the EGFR kinase domain) are associated with acquired resistance to these drugs. EGFR variant III mutant deletions occur in the extracellular domain and are associated with squamous-cell cancer.
The vast majority of patients who have an initial response to erlotinib and gefitinib eventually have a relapse.49,57,58 Recent studies have identified EGFR T790M mutations (in exon 20) in tumors before drug treatment59 and in tumors of patients who had a relapse after therapy with standard reversible EGFR tyrosine kinase inhibitors.57 The binding kinetics of the mutant EGFR appear to be altered by the T790M mutation (Fig. 3). Irreversible EGFR inhibitors suppress T790M-mutant tumor cells in vitro and are promising treatments for T790M-mutant tumors.60 Amplification of the met proto-oncogene (MET), another major mechanism of acquired resistance to EGFR tyrosine kinase inhibitors, marks a poor prognosis.61-63 Other proposed resistance mechanisms include activation of other receptor tyrosine kinases, such as insulin-like growth factor 1 receptor (which can bypass EGFR to activate critical downstream signaling pathways [Fig. 2]), KRAS mutations, and the epithelial-to-mesenchymal transition.58,64,65 The epithelial-to-mesenchymal transition is a program of cell development involving loss of cell adhesion, repressed E-cadherin expression, and increased cell mobility.
Preclinical and clinical data suggest that EGFR mutations are early events in the development of non–small-cell lung cancer.66-70 EGFR mutations, including those involving exons 18, 19, and 20 and L858R, can transform fibroblasts and lung epithelial cells.67 Furthermore, in transgenic mice with lung-specific expression of exon 19 deletion or the L858R mutation, atypical adenomatous hyperplasia, which is considered to be a precursor lesion of peripheral adenocarcinoma, was followed by lesions resembling bronchioalveolar carcinoma at 5 to 6 weeks of age and invasive adenocarcinomas at 8 to 10 weeks.68 Deinduction of mutant EGFR expression led to regression of tumors, suggesting the need for persistent mutant EGFR activity for continued tumor survival. Lung tumors also developed in transgenic mice with lung-specific expression of EGFR variant III mutation (in-frame deletion of exons 2–7 from the extracellular domain) (Fig. 3).13 Mutations of the region that encodes the tyrosine kinase domain of EGFR have been detected in specimens of atypical adenomatous hyperplasia from Asian patients with no history of smoking.71 These mutations also occur in normal epithelium within and adjacent to tumors with EGFR tyrosine kinase mutations (a localized-field effect, possibly reflecting stem-cell expansion) and before EGFR amplification, a change associated with tumor progression and metastasis.72
HER2 mutations and amplification have been identified in patients with lung adenocarcinoma. The frequency of such mutations is less than 5%, and the frequency of such amplification is 5 to 10%. HER2 kinase domain mutations (in-frame insertions in exon 20) and EGFR kinase domain mutations have similar associations with female sex, nonsmoking status, and Asian background in patients with adenocarcinoma.73 HER2 amplification is associated with sensitivity to inhibitors of the EGFR tyrosine kinase74; HER2 mutations are associated with resistance to such inhibitors but also with sensitivity to HER2-targeted therapy.75 HER3 kinase domain mutations have not been detected in patients with non–small-cell lung cancer.76 Mutations in the HER4 kinase domain were found in 2 to 3% of Asian patients with this disease, with a possible association with male sex and smoking.77
Ras–Raf–Mek
The Ras–Raf–Mek pathway is involved in signaling downstream from EGFR and in other pathways leading to the growth of cancer cells and tumor progression (Fig. 2). Activating KRAS mutations are limited to non–small-cell lung cancer (predominantly adenocarcinomas), virtually mutually exclusive of mutations in the EGFR and HER2 kinase domains, and associated with resistance to EGFR inhibitors (tyrosine kinase inhibitors and cetuximab) and chemotherapy.58,78,79 Most KRAS mutations in lung adenocarcinoma are smoking-related G→T transversions (substitutions of a purine for a pyrimidine) and affect exon 12 (in 90% of patients) or exon 13.2,79 A distinct KRAS mutational profile consisting of G→A transition mutations was recently detected in patients with adenocarcinoma who had never smoked; its functional significance is unclear.79 Transversions (smokers) and transitions (non-smokers) also have been reported for p53 mutations in lung adenocarcinoma.2 KRAS mutations appear to be an early event (e.g., detectable in the preinvasive lesions of atypical adenomatous hyperplasia and bronchoalveolar carcinoma33) that precedes smoking-related lung adenocarcinoma. They generally mark a poor prognosis. Further evidence supporting this gene’s role in the pathogenesis of lung cancer comes from transgenic mice bearing a mutated KRAS and in which multifocal atypical adenomatous hyperplasia and adenocarcinoma develop.80 MET activation occurs early in KRAS-induced carcinogenesis in this model.81 BRAF mutations have also been detected in non–small-cell lung cancer9 and may be an early event in lung tumorigenesis.82
PI3K–Akt–mTOR
The pathway consisting of PI3K, Akt, and mammalian target of rapamycin (mTOR), which is downstream of EGFR, is activated early in lung carcinogenesis.83 Akt is also overexpressed in bronchial dysplasia. Inhibition of Akt can induce apoptosis of human premalignant and malignant lung cells and prevent lung carcinogenesis in an animal model. An mTOR inhibitor can block malignant progression of atypical adenomatous hyperplasia lesions in the KRas mouse model.84 Since mTOR drives tumorigenesis in part through macrophages, a prominent component of the tumor microenvironment, the antitumor effect of mTOR inhibition requires the tumor microenvironment. There is mutation or amplification of PIK3CA, which encodes the PI3K catalytic subunit, in a subgroup of non–small-cell lung tumors, especially squamous-cell carcinoma, in association with increased PI3K activity and Akt expression.85
LKB1
LKB1 (also called STK11) is frequently mutated in non–small-cell lung tumors and is thought to act as a tumor-suppressor gene through interactions with p53 and CDC42, modulating the activity of AMPK (a multifunctional protein kinase) and other possible mechanisms that are just beginning to be studied.86,87 LKB1 is thought to function in early tumorigenesis, subsequent differentiation, and the development of metastases.88 Results in transgenic KRas-mutant mice in which LKB1 was inactivated suggest that the gene plays a role in the differentiation and invasive behavior of such tumors.87 The presence of LKB1 mutations alone (i.e., without KRas mutations) was not associated with the development of lung cancer in mice. Low levels of LKB1 protein were associated with high grades of dysplasia in atypical adenomatous hyperplasia lesions, suggesting that LKB1 has an early role in the development of premalignant lesions in the lung.89 LKB1 mutations (including point mutations and deletions) were found in 34% of adenocarcinomas and 19% of squamous-cell carcinomas from 144 human specimens of non–small-cell lung cancer.87 However, much lower rates of LKB1 mutation (<5%) were found in adenocarcinomas from Asian patients.90,91 LKB1 mutations are associated with smoking and with KRAS mutations and are virtually exclusive of EGFR mutations.91
TITF1
Amplification of thyroid transcription factor 1 (TITF1, also called NKX2-1) in the 14q13.3 region was the most common focal event in a high-resolution analysis of gene copy numbers in human lung adenocarcinoma.92 This study used an array with the capacity to genotype many SNPs. As a result, the investigators also identified amplification in regions containing KRAS, Myc, vascular endothelial growth factor (VEGF), and several cell-cycle genes in the tumor specimens. TITF1 encodes a lineage-specific transcription factor that is essential for the formation of cells lining lung alveoli (type II pneumocytes). In vitro, transfection of immortalized normal human lung epithelial cells with at least two of the three genes TITF1, NKX2-8, and PAX-9 in the 14q13.3 region caused increased growth of the cells,93 suggesting that these three genes may work cooperatively in the pathogenesis of lung cancer in which there is amplification at 14q13.3 (detected by high-resolution comparative genomic hybridization array). Recent data indicate that squamous-cell carcinoma also exhibits TITF1 amplification, as detected on fluorescence in situ hybridization, but not TITF1 protein, in contrast to adenocarcinoma.94
ANGIOGENESIS
VEGF levels in bronchial epithelial cells of smokers increase in association with the progression of bronchial dysplasia from low grade to high grade.95 Bronchial hyperplasia, metaplasia, and carcinoma in situ are associated with increased microvessel density, and a distinctive pattern known as angiogenic squamous dysplasia can occur.96 Factors associated with increased tumor angiogenesis correlate with the development and prognosis of lung cancer.97-99 Circulating VEGF levels may predict the clinical benefit of VEGF inhibitors in patients with this disease. Many angiogenic factors are regulated at least in part through the hypoxia-regulated pathways, such as hypoxia-induced factor (HIF) 1α and 2α.100,101 In addition to hypoxia, VEGF and other angiogenic factors are also regulated by EGFR through HIF-dependent and independent mechanisms102 and by oncogenes such as KRAS and p53. VEGF has recently been validated as a therapeutic target on the basis of the results of a phase 3 trial, which led the Food and Drug Administration (FDA) to approve the VEGF monoclonal antibody bevacizumab in combination with standard chemotherapy for previously untreated, advanced non–small-cell lung cancer.103
Interactions between the VEGF and EGFR pathways and an association between acquired resistance to EGFR blockade and increased VEGF expression in preclinical models104 led to the hypothesis that dual blockade of VEGF and EGFR might be more effective than either approach alone. Randomized phase 2 trials of dual inhibition with bevacizumab plus erlotinib105 or the VEGF receptor–EGFR tyrosine kinase inhibitor vandetanib (combined with chemotherapy)106,107 had promising results. Phase 3 testing of both approaches in patients with platinum-resistant disease is ongoing.
MOLECULAR PROFILING
TECHNICAL ADVANCES
Molecular profiling, including the profiling of genes and proteins, to guide treatment may improve the clinical outcome in patients with non–small-cell lung cancer (Fig. 1). Progress in the identification of markers, mutations, and genomic signatures far outstrips the modest improvement in treatments that are based on these molecular advances. Formidable obstacles to developing effective markers include tumor heterogeneity, the highly complex interplay between the environment and host and the complexity, multiplicity, and redundancy of tumor-cell signaling networks involving genetic, epigenetic, and microenvironmental effects. Emerging high-throughput techniques for assessing genomic DNA, messenger RNA (mRNA), microRNA, methylation, and protein or phosphoprotein signaling networks should help address these obstacles (Fig. 4). The Cancer Genome Atlas is a large-scale project designed to provide a comprehensive profile of human tumors according to their gene mutations, alterations in gene copy number, and epigenetic changes. Squamous-cell carcinoma of the lung will be one of the first tumors profiled by this atlas.
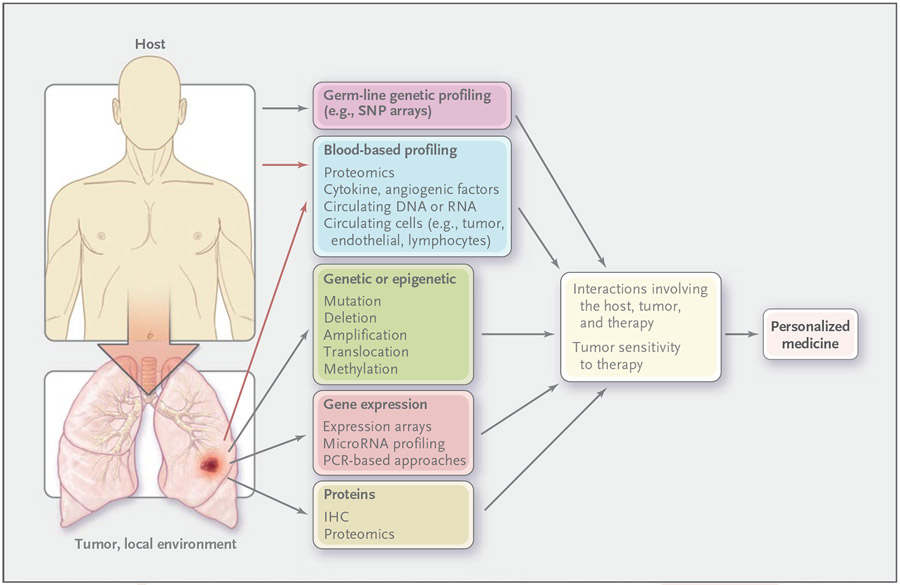
Figure 4. Molecular-Profiling Approaches to the Development of Personalized Therapy.
Host profiling involves innate characteristics of the cancer patient. All markers that are involved in profiling lung cancer can apply to the tumor or its local environment. Predictive markers identify groups of patients who are likely to have increased sensitivity or resistance to a given therapy, a critical step in personalizing treatment. It has been traditional to assess individual genetic or protein prognostic or predictive markers (e.g., HER2 for breast cancer), but emerging techniques permit global analyses of the genomic, gene-expression, epigenetic, and protein profiles of the host (innate), including markers in blood and in tumor or nonmalignant lung tissue. These methods include single-nucleotide polymorphism (SNP) arrays to assess genomic alterations, bisulfite sequencing, and methylation-specific polymerase chain reaction (PCR) to assess epigenetic changes, microarrays for assessing gene expression or microRNA levels, and proteomic methods (such as mass spectroscopy, reverse-phase protein arrays, and multiplex beads) to assess intracellular signaling in tumor tissue and cytokines and angiogenic factors in blood. Blood-based profiling includes markers derived from the host (e.g., lymphocytes) and the tumor and local environment (e.g., circulating tumor cells and tumor-derived cytokines) (red arrows). IHC denotes immunohistochemical analysis.
GENE PROFILING
Tumor molecular heterogeneity is a major reason that patients with non–small-cell lung cancer with a similar clinical stage and tumor histology can have dramatically different clinical outcomes and responses to treatment. Microarray techniques that profile the expressions of tens of thousands of genes simultaneously can measure this tumor heterogeneity at a global level. Gene-expression profiles that are associated with subtypes of non–small-cell lung cancer108,109 and with reduced recurrence-free or overall survival of patients have been identified.110-113 Combined clinical and molecular information provides better indications of cancer risk114 and prognosis.115
In a recent analysis of 672 invasion-associated genes from 125 frozen specimens of early-stage tumors,111 microarray and reverse-transcriptase–polymerase-chain-reaction (RT-PCR) analyses identified a molecular signature of five genes as an independent predictor of relapse-free and overall survival. In two validation cohorts, another recently developed gene-expression profile (metagene) predicted clinical outcome with an accuracy of 72% and 79% (greater than that for tumor stage, tumor diameter, nodal status, or other clinical measures) and predicted the outcome in patients with stage IA tumors.112 Randomized, controlled trials will need to validate these signatures and establish whether the patients with stage IA tumors who were identified as being at high risk will benefit from adjuvant therapy. One such phase 3 trial, coordinated by the Cancer and Leukemia Group B, is approved and under final review. It will evaluate a large predictive set of metagenes (or subgroups of gene-expression profiles consisting of 25 to 200 genes) in patients with stage IA tumors who are undergoing adjuvant chemotherapy.
For the majority of patients with advanced or metastatic non–small-cell lung cancer, the most important potential effect of molecular markers is likely to be in predicting the response to specific therapies with the goal of “personalizing” treatment (Fig. 4). Many exciting potential predictive markers have been developed in vitro and need validation in tumor samples and clinical trials.113 For example, gene-expression signatures have been developed for cisplatin and pemetrexed on the basis of in vitro sensitivity; the cisplatin in vitro signature predicted the likelihood of response. Recently developed in vitro profiles predicting the sensitivity of tumors to EGFR inhibitors and other therapies have yet to be assessed clinically.113
MicroRNA has recently emerged as an important regulator of gene expression. High-throughput analyses have shown that microRNA expression is commonly deregulated in lung and other cancers.116,117 Using real-time RT-PCR, investigators recently identified a five-microRNA signature that is associated with treatment outcome.116 Loss of microRNA-128b, a putative regulator of EGFR that is located on chromosome 3p, has been shown to correlate with the response to EGFR inhibition in patients with lung cancer.117
Studies suggest that information about tumor-specific genetic and epigenetic changes also may be obtained from the blood of patients with lung cancer. Circulating DNA can be detected in the plasma and serum of such patients, and levels of this DNA are associated with a poor prognosis.118,119 Tumor-specific DNA alterations (such as loss of heterozygosity), promoter methylation, and KRAS and EGFR mutations have also been detected in the blood of patients with lung cancer.120-122 New techniques for capturing circulating tumor cells allow the detection of EGFR-activating mutations and the drug-resistance allele T790M. Such techniques appear to be more sensitive than those for capturing circulating DNA. Furthermore, a decline in the number of circulating tumor cells was associated with tumor response on radiography.123 These studies suggest that blood profiling may provide useful information about genetic changes in tumors that could ultimately help detect lung cancer and guide therapy.
PROTEIN PROFILING
Profiling of genomic and mRNA expression provides an incomplete picture of the heterogeneity of non–small-cell lung cancer. Levels of mRNA do not always correlate with protein levels and do not provide information on protein–protein interactions or post-translational modifications such as phosphorylation that may be critical for regulating protein activity.124 Furthermore, most targeted therapeutic agents are designed to inhibit the activity of proteins such as tyrosine kinases. Therefore, protein-based profiling is likely to be essential in understanding the complexity of protein signaling networks and developing molecular signatures that predict a response to therapy.
Immunohistochemical analysis remains the most widely applied method for assessing individual proteins and may be useful for estimating prognosis and predicting the response to therapy.25,26 Emerging high-throughput proteomic techniques, such as mass spectrometry and protein microarrays, have the potential to view signal transduction networks more globally than is possible with immunohistochemical analysis. Such techniques are feasible in small amounts of tumor tissue.125 Proteomic signatures for prognosis and predicting the response to chemotherapy or EGFR inhibitors have been developed in tumors and cell lines.126,127
Proteomic profiling from blood is also under study, allowing repeated measurements during treatment without the need for tumor tissue. Serum mass spectrometry profiles can distinguish patients with non–small-cell lung cancer from normal controls124,128 and patients with better outcomes from those with worse outcomes after treatment with EGFR tyrosine kinase inhibitors.129 New techniques also permit the multiplex analysis of dozens of cytokines and angiogenic factors in small amounts of serum or plasma. This approach is being used in developing predictive markers in non–small-cell lung cancer. Although promising, blood- and tissue-based proteomic approaches remain investigational and await prospective testing and validation in large, randomized trials before they can be applied clinically.
CONCLUSIONS
The molecular origins of lung cancer lie in complex interactions between the environment and host genetic susceptibility. Lung cancer then evolves through genetic and epigenetic changes, including deregulated signaling pathways, which are potential targets for chemoprevention and therapy. Emerging techniques for genomic, gene-expression, epigenetic, and proteomic profiling92,114,125,130,131 could revolutionize clinical approaches across the spectrum of lung-cancer types and subtypes by identifying practical molecular markers of risk (in precancer), early detection and prognosis (in early-stage cancer), and treatment sensitivity (in early-stage and advanced-stage cancer). Genomewide and other molecular assessments are helping elucidate germ-line variations that may contribute to lung cancer risk,19-21 prognosis,132 and treatment sensitivity133,134 and somatic genetic alterations that occur in lung adenocarcinomas14,50-52 and in high-risk lung tissue associated with tumors or in smokers.3,28,29 Molecular targeted research has produced the recently FDA-approved EGFR and VEGF inhibitors erlotinib and bevacizumab, which have modestly improved the outcome in patients with non–small-cell lung cancer.48,103 Molecular profiling of the type described in this review has begun in clinical trials112,135-137 and promises to select patients who are most likely to benefit from therapy and to guide the development of more effective agents that will personalize standard medicine for lung cancer.138
Acknowledgments
Dr. Herbst reports receiving consulting fees and research grants from Bristol-Myers Squibb, ImClone, Genentech, Amgen, AstraZeneca, and OSI and lecture fees from Genentech; Dr. Heymach, serving on advisory boards for and receiving research grants from AstraZeneca, Pfizer, and GlaxoSmithKline and serving on an advisory board for Genentech; and Dr. Lippman, serving on advisory boards for OSI and Genentech. No other potential conflict of interest relevant to this article was reported.
We thank Lauren A. Byers, Balvindar S. Johal, and Ignacio I. Wistuba for their careful comments and other contributions regarding aspects of this work; and Bich N. Tran, Suzanne E. Davis, and Kendall Morse for their contributions to the preparation of the manuscript.
Contributor Information
Roy S. Herbst, Department of Thoracic/Head and Neck Medical Oncology, University of Texas M.D. Anderson Cancer Center, Houston.; Department of Cancer Biology, University of Texas M.D. Anderson Cancer Center, Houston.
John V. Heymach, Department of Thoracic/Head and Neck Medical Oncology, University of Texas M.D. Anderson Cancer Center, Houston.; Department of Cancer Biology, University of Texas M.D. Anderson Cancer Center, Houston.
Scott M. Lippman, Department of Thoracic/Head and Neck Medical Oncology, University of Texas M.D. Anderson Cancer Center, Houston.; Department of Clinical Cancer Prevention, University of Texas M.D. Anderson Cancer Center, Houston.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin 2008; 58:71–96. [DOI] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers — a different disease. Nat Rev Cancer 2007;7:778–90. [DOI] [PubMed] [Google Scholar]
- 3.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A 2004;101:10143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao L, Lee JS, Kurie JM, et al. Clonal genetic alterations in the lungs of current and former smokers. J Natl Cancer Inst 1997;89:857–62. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007;2:327–43. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi F, Hu J, Pelosi G, et al. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNA microarray. Clin Cancer Res 2004;10:6023–8. [DOI] [PubMed] [Google Scholar]
- 7.Zudaire I, Lozano MD, Vazquez MF, et al. Molecular characterization of small peripheral lung tumors based on the analysis of fine needle aspirates. Histol Histopathol 2008;23:33–40. [DOI] [PubMed] [Google Scholar]
- 8.Wistuba II, Berry J, Behrens C, et al. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res 2000;6:2604–10. [PMC free article] [PubMed] [Google Scholar]
- 9.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002; 62:6997–7000. [PubMed] [Google Scholar]
- 10.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283–9. [DOI] [PubMed] [Google Scholar]
- 11.Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272–81. [PubMed] [Google Scholar]
- 12.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naïve cohort. J Thorac Oncol 2008;3:331–9. [DOI] [PubMed] [Google Scholar]
- 13.Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A 2006;103:7817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. [DOI] [PubMed] [Google Scholar]
- 15.Hwang SJ, Cheng LS, Lozano G, Amos CI, Gu X, Strong LC. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum Genet 2003;113:238–43. [DOI] [PubMed] [Google Scholar]
- 16.Sanders BM, Jay M, Draper GJ, Roberts EM. Non-ocular cancer in relatives of retinoblastoma patients. Br J Cancer 1989; 60:358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet 2004;75:460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005; 37:1315–6. [DOI] [PubMed] [Google Scholar]
- 19.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor sub-unit genes on 15q25. Nature 2008;452:633–7. [DOI] [PubMed] [Google Scholar]
- 21.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam DC, Girard L, Ramirez R, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res 2007;67:4638–47. [DOI] [PubMed] [Google Scholar]
- 23.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev 2003; 12:689–98. [PubMed] [Google Scholar]
- 24.Yu D, Zhang X, Liu J, et al. Characterization of functional excision repair cross-complementation group 1 variants and their association with lung cancer risk and prognosis. Clin Cancer Res 2008;14:2878–86. [DOI] [PubMed] [Google Scholar]
- 25.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983–91. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 2007;356:800–8. [DOI] [PubMed] [Google Scholar]
- 27.Franklin WA, Gazdar AF, Haney J, et al. Widely dispersed p53 mutation in respiratory epithelium: a novel mechanism for field carcinogenesis. J Clin Invest 1997; 100:2133–7. [Erratum, J Clin Invest 1997;100:2639.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361–6. [DOI] [PubMed] [Google Scholar]
- 29.Seike M, Yanaihara N, Bowman ED, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst 2007;99:1257–69. [DOI] [PubMed] [Google Scholar]
- 30.Park IW, Wistuba II, Maitra A, et al. Multiple clonal abnormalities in the bronchial epithelium of patients with lung cancer. J Natl Cancer Inst 1999;91:1863–8. [DOI] [PubMed] [Google Scholar]
- 31.Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 2006;38:468–73. [DOI] [PubMed] [Google Scholar]
- 32.Wistuba II, Mao L, Gazdar AF. Smoking molecular damage in bronchial epithelium. Oncogene 2002;21:7298–306. [DOI] [PubMed] [Google Scholar]
- 33.Westra WH. Early glandular neoplasia of the lung. Respir Res 2000;1:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang X, Shigematsu H, Bekele BN, et al. EGFR tyrosine kinase domain mutations are detected in histologically normal respiratory epithelium in lung cancer patients. Cancer Res 2005;65:7568–72. [DOI] [PubMed] [Google Scholar]
- 35.Guo M, House MG, Hooker C, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res 2004;10:5131–6. [DOI] [PubMed] [Google Scholar]
- 36.Bhutani M, Pathak AK, Fan Y-H, et al. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prev Res 2008;1:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licchesi JD, Westra WH, Hooker CM, Herman JG. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res 2008;14:2570–8. [DOI] [PubMed] [Google Scholar]
- 38.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 2006;66:3338–44. [DOI] [PubMed] [Google Scholar]
- 39.Machida EO, Brock MV, Hooker CM, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res 2006;66:6210–8. [Erratum, Cancer Res 2007;67:427.] [DOI] [PubMed] [Google Scholar]
- 40.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118–28. [DOI] [PubMed] [Google Scholar]
- 41.Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH. Cohypermethylation of p16 and FHIT promoters as a prognostic factor of recurrence in surgically resected stage I non-small cell lung cancer. Cancer Res 2006;66:4049–54. [DOI] [PubMed] [Google Scholar]
- 42.Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res 2003;63:7089–93. [PubMed] [Google Scholar]
- 43.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005;121:823–35. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Iwanaga K, Raso MG, et al. Phosphatidylinositol 3-kinase mediates bronchioalveolar stem cell expansion in mouse models of oncogenic K-ras-induced lung cancer. PLoS ONE 2008;3(5):e2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanagi S, Kishimoto H, Kawahara K, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest 2007;117:2929–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weihua Z, Tsan R, Huang WC, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 2008;13:385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirker R, Szczesna A, von Pawel J, et al. FLEX: a randomized, multicenter, phase III study of cetuximab in combination with cisplatin/vinorelbine (CV) versus CV alone in the first-line treatment of patients with advanced non-small cell lung cancer (NSCLC). J Clin Oncol 2008;26:Suppl:1006s. abstract. [Google Scholar]
- 48.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non–small-cell lung cancer. N Engl J Med 2005;353:123–32. [DOI] [PubMed] [Google Scholar]
- 49.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 2007;25:587–95. [DOI] [PubMed] [Google Scholar]
- 50.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 51.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 52.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160–74. [DOI] [PubMed] [Google Scholar]
- 54.Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer: a randomized phase III trial (INTEREST). Lancet 2008. (in press). [DOI] [PubMed] [Google Scholar]
- 55.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol 2008;26:3351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsao M-S, Sakurada A, Cutz J-C, et al. Erlotinib in lung cancer — molecular and clinical predictors of outcome. N Engl J Med 2005;353:133–44. [Erratum, N Engl J Med 2006;355:1746.] [DOI] [PubMed] [Google Scholar]
- 57.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2(3):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2(1):e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res 2006;66:7854–8. [DOI] [PubMed] [Google Scholar]
- 60.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A 2005;102:7665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 63.Cappuzzo F, Skokan M, Gajapathy S, et al. Effect of increased MET gene copy number on survival of surgically resected non-small cell lung cancer (NSCLC) patients. J Clin Oncol 2008;26:Suppl:589s. abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res 2007;13:2795–803. [DOI] [PubMed] [Google Scholar]
- 65.Yauch RL, Januario T, Eberhard DA, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res 2005;11:8686–98. [DOI] [PubMed] [Google Scholar]
- 66.Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res 2007;67:7319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med 2005;2(11):e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485–95. [DOI] [PubMed] [Google Scholar]
- 69.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 2006;20:1496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vikis H, Sato M, James M, et al. EGFRT-790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res 2007;67:4665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakuma Y, Matsukuma S, Yoshihara M, et al. Epidermal growth factor receptor gene mutations in atypical adenomatous hyperplasias of the lung. Mod Pathol 2007; 20:967–73. [DOI] [PubMed] [Google Scholar]
- 72.Tang X, Varella-Garcia M, Xavier AC, et al. Epidermal growth factor receptor abnormalities in the pathogenesis and progression of lung adenocarcinomas. Cancer Prev Res 2008;1:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642–6. [DOI] [PubMed] [Google Scholar]
- 74.Cappuzzo F, Varella-Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol 2005;23:5007–18. [DOI] [PubMed] [Google Scholar]
- 75.Wang SE, Narasanna A, Perez-Torres M, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 2006;10:25–38. [DOI] [PubMed] [Google Scholar]
- 76.Jeong EG, Soung YH, Lee JW, et al. ERBB3 kinase domain mutations are rare in lung, breast and colon carcinomas. Int J Cancer 2006;119:2986–7. [DOI] [PubMed] [Google Scholar]
- 77.Soung YH, Lee JW, Kim SY, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer 2006; 118:1426–9. [DOI] [PubMed] [Google Scholar]
- 78.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005;23:5900–9. [DOI] [PubMed] [Google Scholar]
- 79.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001;410:1111–6. [DOI] [PubMed] [Google Scholar]
- 81.Yang Y, Wislez M, Fujimoto N, et al. A selective small molecule inhibitor of c-Met, PHA-665752, reverses lung premalignancy induced by mutant K-ras. Mol Cancer Ther 2008;7:952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 2007; 21:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.West KA, Linnoila IR, Belinsky SA, Harris CC, Dennis PA. Tobacco carcinogen-induced cellular transformation increases activation of the phosphatidylinositol 3′-kinase/Akt pathway in vitro and in vivo. Cancer Res 2004;64:446–51. [DOI] [PubMed] [Google Scholar]
- 84.Wislez M, Spencer ML, Izzo JG, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res 2005;65:3226–35. [DOI] [PubMed] [Google Scholar]
- 85.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and gene copy number in human lung cancers. Cancer Res (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang S, Schafer-Hales K, Khuri FR, Zhou W, Vertino PM, Marcus AI. The tumor suppressor LKB1 regulates lung cancer cell polarity by mediating cdc42 recruitment and activity. Cancer Res 2008;68:740–8. [DOI] [PubMed] [Google Scholar]
- 87.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807–10. [DOI] [PubMed] [Google Scholar]
- 88.Shah U, Sharpless NE, Hayes DN. LKB1 and lung cancer: more than the usual suspects. Cancer Res 2008;68:3562–5. [DOI] [PubMed] [Google Scholar]
- 89.Ghaffar H, Sahin F, Sanchez-Cepedes M, et al. LKB1 protein expression in the evolution of glandular neoplasia of the lung. Clin Cancer Res 2003;9:2998–3003. [PubMed] [Google Scholar]
- 90.Matsumoto S, Iwakawa R, Takahashi K, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene 2007;26:5911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer 2008;99:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A 2007;104:16663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang X, Sun M, Behrens C, et al. TITF-1 gene amplification and protein expression pattern identify adenocarcinoma of lung with worse prognosis. Presented at the American Association for Cancer Research Annual Meeting, San Diego, CA, April 12–16, 2008. abstract. [Google Scholar]
- 95.Merrick DT, Haney J, Petrunich S, et al. Overexpression of vascular endothelial growth factor and its receptors in bronchial dypslasia demonstrated by quantitative RT-PCR analysis. Lung Cancer 2005; 48:31–45. [DOI] [PubMed] [Google Scholar]
- 96.Keith RL, Miller YE, Gemmill RM, et al. Angiogenic squamous dysplasia in bronchi of individuals at high risk for lung cancer. Clin Cancer Res 2000;6:1616–25. [PubMed] [Google Scholar]
- 97.Stricter RM. Out of the shadows: CXC chemokines in promoting aberrant lung angiogenesis. Cancer Prev Pres 2008. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dowlati A, Gray R, Sandler AB, Schiller JH, Johnson DH. Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non-small cell lung cancer treated with chemotherapy with or without bevacizumab — an Eastern Cooperative Oncology Group study. Clin Cancer Res 2008;14:1407–12. [DOI] [PubMed] [Google Scholar]
- 99.Heymach JV, Hanrahan EO, Mann H, et al. Baseline VEGF as a potential predictive biomarker of vandetanib clinical benefit in patients with advanced NSCLC. J Clin Oncol 2008;26:Suppl:426s. abstract. [Google Scholar]
- 100.Giatromanolaki A, Koukourakis MI, Sivridis E, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer 2001;85:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoogsteen IJ, Marres HA, van der Kogel AJ, Kaanders JH. The hypoxic tumour microenvironment, patient selection and hypoxia-modifying treatments. Clin Oncol (R Coll Radiol) 2007;19:385–96. [DOI] [PubMed] [Google Scholar]
- 102.Pore N, Jiang Z, Gupta A, Cerniglia G, Kao GD, Maity A. EGFR tyrosine kinase inhibitors decrease VEGF expression by both hypoxia-inducible factor (HIF)-1-independent and HIF-1-dependent mechanisms. Cancer Res 2006;66:3197–204. [DOI] [PubMed] [Google Scholar]
- 103.Sandler A, Gray R, Perry MC, et al. Paclitaxel–carboplatin alone or with bevacizumab for non–small-cell lung cancer. N Engl J Med 2006;355:2542–50. [Erratum, N Engl J Med 2007;356:318.] [DOI] [PubMed] [Google Scholar]
- 104.Viloria-Petit A, Crombet T, Jothy S, et al. Acquired resistance to the antitumor effect of epidermal growth factor receptor-blocking antibodies in vivo: a role for altered tumor angiogenesis. Cancer Res 2001;61:5090–101. [PubMed] [Google Scholar]
- 105.Herbst RS, O’Neill VJ, Fehrenbacher L, et al. Phase II study of efficacy and safety of bevacizumab in combination with chemotherapy or erlotinib compared with chemotherapy alone for treatment of recurrent or refractory non small-cell lung cancer. J Clin Oncol 2007;25:4743–50. [DOI] [PubMed] [Google Scholar]
- 106.Heymach JV, Johnson BE, Prager D, et al. Randomized, placebo-controlled phase II study of vandetanib plus docetaxel in previously treated non small-cell lung cancer. J Clin Oncol 2007;25:4270–7. [Erratum, J Clin Oncol 2008;26:165-6.] [DOI] [PubMed] [Google Scholar]
- 107.Heymach J, Paz-Ares L, De Braud F, et al. Vandetanib alone or with paclitaxel and carboplatin as first-line treatment for advanced NSCLC: a randomized phase II study. J Clin Oncol (in press). [DOI] [PubMed] [Google Scholar]
- 108.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meyerson M, Carbone D. Genomic and proteomic profiling of lung cancers: lung cancer classification in the age of targeted therapy. J Clin Oncol 2005;23:3219–26. [DOI] [PubMed] [Google Scholar]
- 110.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816–24. [DOI] [PubMed] [Google Scholar]
- 111.Chen H-Y, Yu S-L, Chen C-H, et al. A five-gene signature and clinical outcome in non–small-cell lung cancer. N Engl J Med 2007;356:11–20. [DOI] [PubMed] [Google Scholar]
- 112.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non–small-cell lung cancer. N Engl J Med 2006;355:570–80. [Erratum, N Engl J Med 2007;356:201-2.] [DOI] [PubMed] [Google Scholar]
- 113.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med 2006;12:1294–300. [Erratum, Nat Med 2007;13:1388.] [DOI] [PubMed] [Google Scholar]
- 114.Beane J, Sebastiani P, Whitfield TH, et al. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res 2008;1:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multisite, blinded validation study. Nat Med 2008;14:822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008; 13:48–57. [DOI] [PubMed] [Google Scholar]
- 117.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol 2008;19:1053–9. [DOI] [PubMed] [Google Scholar]
- 118.Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004;22:4157–64. [DOI] [PubMed] [Google Scholar]
- 119.Ramirez JL, Rosell R, Taron M, et al. 14-3-3sigma Methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival. J Clin Oncol 2005;23:9105–12. [DOI] [PubMed] [Google Scholar]
- 120.Chen XQ, Stroun M, Magnenat JL, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med 1996;2:1033–5. [DOI] [PubMed] [Google Scholar]
- 121.Belinsky SA, Grimes MJ, Casas E, et al. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer 2007; 96:1278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ludovini V, Pistola L, Gregorc V, et al. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the Perugia multidisciplinary team for thoracic oncology. J Thorac Oncol 2008;3:365–73. [DOI] [PubMed] [Google Scholar]
- 123.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating tumor lung-cancer cells. N Engl J Med 2008;359:366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yildiz PB, Shyr Y, Rahman JS, et al. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol 2007;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo A, Villén J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A 2008;105:692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Byers L, Nanjundan M, Girard L, et al. Reverse-phase protein array (RPPA) profiling of response to taxanes and epidermal growth factor receptor (EGFR) inhibitors identifies an inverse correlation between markers of sensitivity to docetaxel and erlotinib in non-small cell lung cancer lines. Presented at the American Association for Cancer Research Annual Meeting, San Diego, CA, April 12–16, 2008. abstract. [Google Scholar]
- 127.Yanagisawa K, Tomida S, Shimada Y, Yatabe Y, Mitsudomi T, Takahashi T. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst 2007;99:858–67. [DOI] [PubMed] [Google Scholar]
- 128.Patz EF Jr, Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE II. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol 2007; 25:5578–83. [DOI] [PubMed] [Google Scholar]
- 129.Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst 2007;99:838–46. [DOI] [PubMed] [Google Scholar]
- 130.Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148–59. [DOI] [PubMed] [Google Scholar]
- 131.Tsou JA, Galler JS, Siegmund KD, et al. Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer 2007;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heist RS, Zhai R, Liu G, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol 2008;26:856–62. [DOI] [PubMed] [Google Scholar]
- 133.Gregorc V, Hidalgo M, Spreafico A, et al. Germline polymorphisms in EGFR and survival in patients with lung cancer receiving gefitinib. Clin Pharmacol Ther 2008;83:477–84. [DOI] [PubMed] [Google Scholar]
- 134.Liu G, Gurubhagavatula S, Zhou W, et al. Epidermal growth factor receptor polymorphisms and clinical outcomes in non-small-cell lung cancer patients treated with gefitinib. Pharmacogenomics J 2008; 8:129–38. [DOI] [PubMed] [Google Scholar]
- 135.Simon G, Sharma A, Li X, et al. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J Clin Oncol 2007;25:2741–6. [DOI] [PubMed] [Google Scholar]
- 136.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol 2007;25:2747–54. [DOI] [PubMed] [Google Scholar]
- 137.Zhou X, Liu S, Kim ES, Herbst RS, Lee JJ. Bayesian adaptive design for targeted therapy development in lung cancer — a step toward personalized medicine. Clin Trials 2008;5:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Herbst RS, Lippman SM. Molecular signatures of lung cancer — toward personalized therapy. N Engl J Med 2007;356:76–8. [DOI] [PubMed] [Google Scholar]