Abstract
Background and Objectives
Chronic kidney disease (CKD) increases the risk of stroke, but the extent through which this association is mediated by hypertension is unknown. We leveraged large-scale genetic data to explore causal relationships between CKD, hypertension, and cerebrovascular disease phenotypes.
Methods
We used data from genome-wide association studies of European ancestry to identify genetic proxies for kidney function (CKD diagnosis, estimated glomerular filtration rate [eGFR], and urinary albumin-to-creatinine ratio [UACR]), systolic blood pressure (SBP), and cerebrovascular disease (ischemic stroke and its subtypes and intracerebral hemorrhage). We then conducted univariable, multivariable, and mediation Mendelian randomization (MR) analyses to investigate the effect of kidney function on stroke risk and the proportion of this effect mediated through hypertension.
Results
Univariable MR revealed associations between genetically determined lower eGFR and risk of all stroke (odds ratio [OR] per 1-log decrement in eGFR, 1.77; 95% CI 1.31–2.40; p < 0.001), ischemic stroke (OR 1.81; 95% CI 1.31–2.51; p < 0.001), and most strongly with large artery stroke (LAS) (OR 3.00; 95% CI 1.33–6.75; p = 0.008). These associations remained significant in the multivariable MR analysis, controlling for SBP (OR 1.98; 95% CI 1.39–2.82; p < 0.001 for all stroke; OR 2.16; 95% CI 1.48–3.17; p < 0.001 for ischemic stroke; OR 4.35; 95% CI 1.84–10.27; p = 0.001 for LAS), with only a small proportion of the total effects mediated by SBP (6.5% [0.7%–16.8%], 6.6% [0.8%–18.3%], and 7.2% [0.5%–24.8%], respectively). Total, direct and indirect effect estimates were similar across a number of sensitivity analyses (weighted median, MR-Egger regression).
Discussion
Our results demonstrate an independent causal effect of impaired kidney function, as assessed by decreased eGFR, on stroke risk, particularly LAS, even when controlled for SBP. Targeted prevention of kidney disease could lower atherosclerotic stroke risk independent of hypertension.
Introduction
Chronic kidney disease (CKD) carries a strong epidemiologic association with stroke risk.1,2 In a large transancestry meta-analysis of 83 studies, the risk of stroke increased by 7% for every 10 mL/min/1.73 m2 decrease in estimated glomerular filtration rate (eGFR) and by 10% for every 25 mg/mmol increase in urine albumin-to-creatinine ratio (UACR).3 However, the central mechanisms underpinning the relationship between CKD and stroke risk remain unclear.4 Traditional risk factors such as hypertension, diabetes, and atrial fibrillation are all highly prevalent in patients with CKD, but nontraditional CKD-related risk factors such as chronic inflammation, uremic toxins, and oxidative stress are also purported to contribute to risk by triggering vascular injury and endothelial dysfunction.5
Because hypertension is comorbid in up to 90% of patients with CKD,6 it is not clear whether the relationship between CKD and stroke is truly independent of blood pressure (BP). We previously performed 2 systematic reviews and meta-analysis of low eGFR, proteinuria, and stroke risk, analyzing studies according to the way in which they adjusted for hypertension.7,8 For pooled studies of low eGFR and stroke risk, there was near-complete attenuation of associations after adjustment for longitudinal BP control.7 By contrast, in the meta-analysis of proteinuria, the pooled risk association did not substantially attenuate even with the extensive adjustment for BP.8
Mendelian randomization (MR) is a method that uses genetic variants associated with modifiable exposures as instrumental variables to test the potentially causal relationship between a risk factor and disease outcome.9 A previous univariable MR study found associations between genetically determined lower eGFR and higher UACR with increased risk of large artery stroke (LAS) and between genetically determined higher UACR with increased risk of intracerebral hemorrhage (ICH).10 Building on this work, multivariable MR (MVMR) offers the opportunity to further explore the role of hypertension as a potential mediator in the relationship between CKD and stroke. MVMR uses genetic variants associated with multiple, potentially related exposures to estimate the effect of each on a single outcome.11 It also allows for estimation of mediation effects.
Therefore, using MVMR, we leveraged large-scale genetic data to explore causal relationships between genetic predispositions to CKD, hypertension, and cerebrovascular disease phenotypes, testing the hypothesis that the effect of CKD on stroke risk is mediated through increased BP.
Methods
Study Design
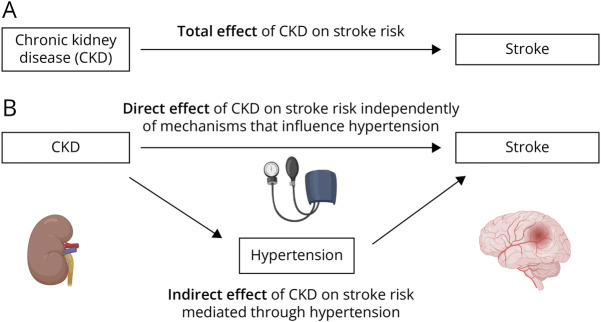
We conducted summary-level univariable, multivariable, and mediation MR analyses to investigate the effect of kidney function on stroke risk and the proportion of this effect mediated through hypertension. In MR analysis, a genetic variant is considered to be a valid instrument if certain assumptions hold, including that the genetic variant is associated with the exposure of interest, that it is not associated with the outcome through a confounding pathway and that the variant does not affect the outcome directly, only possibly indirectly through their effect on the exposure of interest.9 Figure 1 conceptually depicts our approach, and Table 1 summarizes the data sources used in each analysis.
Figure 1. Direct Acyclic Graph to Illustrate Total, Direct, and Indirect Effects of CKD on Stroke Risk.
Directed acyclic graphs demonstrating the hypothesized direction for the total effect of CKD on increased odds of stroke (A) and the hypothesized direction for the effect of CKD on increased hypertension (B), which may partially mediate the effect of CKD on stroke risk. CKD = chronic kidney disease.
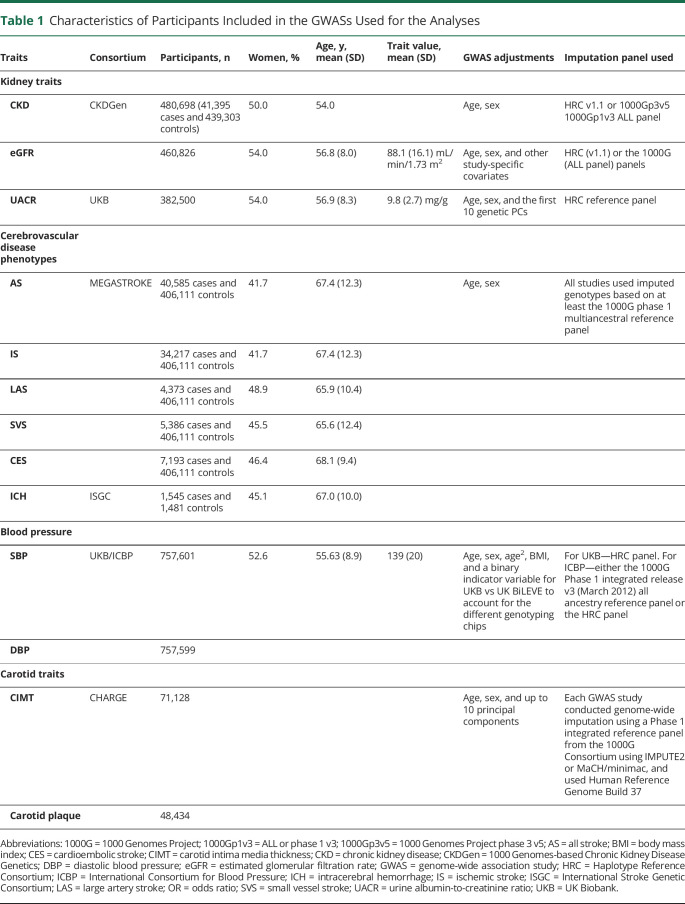
Table 1.
Characteristics of Participants Included in the GWASs Used for the Analyses
| Traits | Consortium | Participants, n | Women, % | Age, y, mean (SD) | Trait value, mean (SD) | GWAS adjustments | Imputation panel used |
| Kidney traits | |||||||
| CKD | CKDGen | 480,698 (41,395 cases and 439,303 controls) | 50.0 | 54.0 | Age, sex | HRC v1.1 or 1000Gp3v5 1000Gp1v3 ALL panel | |
| eGFR | 460,826 | 54.0 | 56.8 (8.0) | 88.1 (16.1) mL/min/1.73 m2 | Age, sex, and other study-specific covariates | HRC (v1.1) or the 1000G (ALL panel) panels | |
| UACR | UKB | 382,500 | 54.0 | 56.9 (8.3) | 9.8 (2.7) mg/g | Age, sex, and the first 10 genetic PCs | HRC reference panel |
| Cerebrovascular disease phenotypes | |||||||
| AS | MEGASTROKE | 40,585 cases and 406,111 controls | 41.7 | 67.4 (12.3) | Age, sex | All studies used imputed genotypes based on at least the 1000G phase 1 multiancestral reference panel | |
| IS | 34,217 cases and 406,111 controls | 41.7 | 67.4 (12.3) | ||||
| LAS | 4,373 cases and 406,111 controls | 48.9 | 65.9 (10.4) | ||||
| SVS | 5,386 cases and 406,111 controls | 45.5 | 65.6 (12.4) | ||||
| CES | 7,193 cases and 406,111 controls | 46.4 | 68.1 (9.4) | ||||
| ICH | ISGC | 1,545 cases and 1,481 controls | 45.1 | 67.0 (10.0) | |||
| Blood pressure | |||||||
| SBP | UKB/ICBP | 757,601 | 52.6 | 55.63 (8.9) | 139 (20) | Age, sex, age2, BMI, and a binary indicator variable for UKB vs UK BiLEVE to account for the different genotyping chips | For UKB—HRC panel. For ICBP—either the 1000G Phase 1 integrated release v3 (March 2012) all ancestry reference panel or the HRC panel |
| DBP | 757,599 | ||||||
| Carotid traits | |||||||
| CIMT | CHARGE | 71,128 | Age, sex, and up to 10 principal components | Each GWAS study conducted genome-wide imputation using a Phase 1 integrated reference panel from the 1000G Consortium using IMPUTE2 or MaCH/minimac, and used Human Reference Genome Build 37 | |||
| Carotid plaque | 48,434 |
Abbreviations: 1000G = 1000 Genomes Project; 1000Gp1v3 = ALL or phase 1 v3; 1000Gp3v5 = 1000 Genomes Project phase 3 v5; AS = all stroke; BMI = body mass index; CES = cardioembolic stroke; CIMT = carotid intima media thickness; CKD = chronic kidney disease; CKDGen = 1000 Genomes-based Chronic Kidney Disease Genetics; DBP = diastolic blood pressure; eGFR = estimated glomerular filtration rate; GWAS = genome-wide association study; HRC = Haplotype Reference Consortium; ICBP = International Consortium for Blood Pressure; ICH = intracerebral hemorrhage; IS = ischemic stroke; ISGC = International Stroke Genetic Consortium; LAS = large artery stroke; OR = odds ratio; SVS = small vessel stroke; UACR = urine albumin-to-creatinine ratio; UKB = UK Biobank.
To further explore associations between genetically predicted kidney disease traits and LAS, we additionally performed MR analyses to investigate the relationship between lower eGFR, carotid intima media thickness (CIMT), and the presence of carotid plaque.
Data Sources
We used data from genome-wide association study (GWAS) of European ancestry to identify genetic proxies for kidney function (CKD diagnosis, eGFR, and UACR), blood pressure (systolic blood pressure [SBP], diastolic blood pressure [DBP]), and cerebrovascular disease (ischemic stroke and its subtypes and ICH). Similar to previous work,10 we used CKD diagnosis as well as both eGFR and UACR to explore impaired kidney function because these exposures may capture different aspects of kidney pathophysiology, and their combined assessment of function is increasingly recommended in both clinical practice and research.12,13 The cystatin-C–based formula for estimating eGFR was chosen instead of a creatinine-based one because of its superior accuracy and better cardiovascular predictive ability.14,15
CKD diagnosis and eGFR GWAS summary meta-analysis statistics were derived from the 1000 Genomes-based Chronic Kidney Disease Genetics (CKDGen) consortium effort.16 The genotypic and phenotypic procedures of the CKDGen consortium GWAS are outlined elsewhere.17,18 In brief, genetic instrumental variables for eGFR were identified from a GWAS meta-analysis from the CKDGen Consortium and UK Biobank (n = 460,826 for cystatin-based eGFR).18 For CKD diagnosis, we obtained genetic association estimates from an earlier CKDGen GWAS meta-analysis (n = 480,698 [41,395 cases and 439,303 controls]).17 Summary statistics for the UACR trait were obtained from a UK Biobank-based GWAS (n = 382,500).19
The MEGASTROKE consortium20 and the International Stroke Genetic Consortium (ISGC) GWAS for ICH were used to derive genetic association estimates for cerebrovascular phenotypes.21 The details of the study populations and stroke subtyping ascertainment are available online at the Cerebrovascular Disease Knowledge Portal.22 In brief, our analysis was based on the European ancestry GWAS summary statistics of the study (40,585 cases; 406,111 controls). The phenotypes used included (1) all stroke (AS), inclusive of all subtypes; (2) ischemic stroke (IS) regardless of subtype; the 3 available etiologic ischemic subtypes (3) LAS, (4) small vessel stroke (SVS), and (5) cardioembolic stroke (CES); and (6) ICH. IS and ICH were defined based on clinical and imaging criteria, and IS subtypes were classified based on the Trial of Org 10172 in Acute Stroke Treatment classification system.23
SBP and DBP summary meta-analysis statistics were obtained from the largest GWAS of blood pressure traits to date in which data were combined from UK Biobank and the International Consortium of Blood Pressure.24,25 The study included 757,601 individuals of European ancestry. SBP values were based on the average of 2 values and adjusted for antihypertensive therapy.
CIMT and carotid plaque summary statistics were available from a meta-analysis of GWAS data in individuals of European ancestry for CIMT (up to 71,128 participants from 31 studies) and carotid plaque (up to 48,434 participants from 17 studies; 21,540 with defined carotid plaque).26
The methodologies for genotyping and bioinformatic genetic analysis of each of the GWAS cited were consistent or comparable across the studies, and further details are available in the citations.17-21,24,26 All the summary statistics were derived from inverse-variance weighted meta-analysis restricted to participants of European ancestry after adjusting for age, sex, and principal components reflecting ancestry. eGFR and UACR were log-transformed. The meta-analysis in the BP GWAS additionally adjusted for body mass index and genotyping platform.
Statistical Analysis
We performed summary-level univariable MR analyses to investigate the total, indirect, and direct effects of CKD (clinical diagnosis, eGFR, and UACR) on each of the stroke phenotypes (Figure 1). Power calculations for these analyses are available in eTable 1 (links.lww.com/WNL/D128). The total effect is defined as the net effect of genetically predicted CKD on stroke risk irrespective of mechanism. The indirect effect is defined as the effect of genetically predicted CKD on stroke risk that is mediated through hypertension. The indirect effect was calculated using the product of coefficients method,27,28 in which we multiplied the univariable MR estimate for the effect of CKD on SBP and DBP and the multivariable MR estimate for the effect of SBP and DBP on stroke risk controlling for CKD. To test the null hypothesis of no mediation through hypertension, we calculated CIs for the indirect effect using the previously described Monte Carlo method.29 A p value for the indirect effect was derived using the propagation of error method.30 We calculated the proportion of the mediated effect by dividing the indirect effect by the total effect. The direct effect is defined as the association of genetically determined CKD on stroke risk through mechanisms independent of mediation. We used multivariable MR to estimate the direct effect.31
For MR analyses, we used as instruments genetic variants that were associated with CKD diagnosis, eGFR, UACR, SBP, and DBP at genome-wide significance (p < 5 × 10−8) after pruning based on linkage disequilibrium between variants (distance window of 10,000 kB and r2 < 0.001). The instruments for kidney traits are presented in eTable 2 (links.lww.com/WNL/D129) and those used as proxies for BP traits have been previously published.32 The genetic association estimates for the described outcomes were obtained from the MEGASTROKE and ISGC GWAS summary statistics. After extraction of the association estimates, we next performed harmonization of the direction of effects between exposure and outcome associations based on the effect allele, where palindromic single-nucleotide polymorphisms (SNPs) were aligned when minor allele frequencies were less than 0.3 or were otherwise excluded. We determined individual MR estimates for each instrument using the Wald estimator and calculated standard errors using the delta method.33 Individual MR estimates were pooled using fixed-effects inverse-variance weighted (IVW) analyses. The overall effect sizes on stroke risk were reported as odds ratios (ORs) and 95% CIs of OR. Heterogeneity across estimates was examined with the I2 and the Cochran Q test (I2 > 50% and p < 0.05 were considered statistically significant).34 We performed sensitivity analyses that are more robust than the IVW method to certain forms of pleiotropy, including the weighted median35 and MR-Egger.36 The estimated value of the intercept in MR-Egger regression can be considered to be an estimate of the average pleiotropic effect across the genetic variants. An intercept term that differs from zero is indicative of overall directional pleiotropy.36 To investigate the influence of outlying or pleiotropic genetic variants, we performed a leave-one-out analysis in which we omitted 1 SNP in turn.37 All MR analyses were performed in R (version 4.1.0; the R Foundation for Statistical Computing, Vienna, Austria) using the MendelianRandomization and TwoSampleMR packages. Finally, we also tested for the inverse association using kidney traits as outcomes and cerebrovascular disease phenotypes as exposures (bidirectional MR).29 Given the 6 cerebrovascular disease phenotypes studied for each of the 3 kidney traits, for the univariable MR analysis, we set statistical significance at an false discovery rate (FDR)–adjusted p value <0.05. We did not apply FDR adjustment for the p values for the multivariable MR analyses because we considered these to be exploratory in nature.
Standard Protocol Approvals, Registrations, and Patient Consents
This study used publicly available, deidentified summary statistics from GWAS meta-analyses of individual studies that had already obtained ethical review board approvals and that had obtained written informed consent from all included patients or their guardians.
Data Availability
Genetic variants used are available in the eAppendix 1 (links.lww.com/WNL/D128), and the code used for all analyses is available on request.
Results
Univariable Analyses
There were 23, 195, and 38 genetic instruments for CKD diagnosis, eGFRcys, and UACR, respectively. The genetic instruments explained 0.13% of the variance in CKD diagnosis, 1.2% of the variance in eGFRcys, and 0.16% of the variance in UACR, with mean F statistics of 23, 28, and 17, respectively.
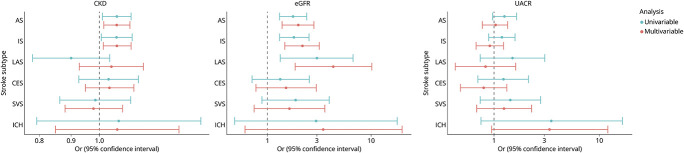
Using fixed-effects IVW MR analysis, we found genetic predisposition to CKD diagnosis to be associated with a higher risk of AS and IS subtypes (OR 1.07; 95% CI 1.01–1.12; p = 0.018 and OR 1.07; 95% CI 1.01–1.13; p = 0.031, respectively) (eFigure 1 and eTable 3, links.lww.com/WNL/D128). However, these associations did not remain significant after FDR adjustment. We found genetically determined lower eGFR to be significantly associated with AS (OR per 1-log decrement in eGFR, 1.77; 95% CI 1.31–2.40; p < 0.001), IS (OR 1.81; 95% CI 1.31–2.51; p < 0.001), and most strongly with LAS (OR 3.00; 95% CI 1.33–6.75; p = 0.008) (eFigure 1). Associations between eGFR and these stroke subtypes remained significant even after FDR adjustment of the p values (eTable 4). Genetically determined UACR was not significantly associated in univariable MR analysis with any of the cerebrovascular disease subtypes (eFigure 1). There was significant heterogeneity as defined by the Cochran Q test when exploring the associations between genetically determined eGFR, AS, and IS and between UACR and CES, but none of the intercepts from the MR-Egger regression were statistically significant, supporting a lack of significant pleiotropy in the analysis. Leave-one-out analyses did not show any indication that MR results might be driven by a single SNP (eFigure 2). Furthermore, the weighted median and the MR-Egger regression analyses provided association estimates that were directionally consistent and of similar magnitude as the ones derived from the IVW analyses, although with wider CIs, as expected given the lower statistical power of these approaches (eTables 3–5).
Multivariable Analyses
We then performed MVMR controlling for SBP and found that there was still strong evidence for an independent effect of impaired kidney function on risk of ischemic stroke (Figure 2). In particular, genetically predicted lower eGFR was still associated with risk of LAS even after controlling for SBP (OR per 1-log decrement in eGFR, 4.35; 95% CI 1.84–10.27; p = 0.001) (Figure 2 and eTable 4, links.lww.com/WNL/D128). There was also strong evidence for an independent effect of genetically predicted lower eGFR on AS (OR 1.98; 95% CI 1.39–2.82; p < 0.001) and IS (OR 2.16; 95% CI 1.48–3.17; p < 0.001) subtypes. Genetic predisposition to CKD diagnosis also remained significantly associated with AS (OR 1.07; 95% CI 1.02–1.12; p = 0.01) and IS (OR 1.07; 95% CI 1.01–1.12; p = 0.01) subtypes after controlling for SBP in the MVMR analysis (Figure 2 and eTable 3). As per the univariable analysis, the independent causal effects estimated from multivariable MR-median and MR-Egger were consistent with the IVW analysis (eTables 3–5).
Figure 2. Univariable and Multivariable Mendelian Randomization Associations Between Genetically Determined Kidney Disease Traits and Cerebrovascular Disease Phenotypes, Controlling for Systolic Blood Pressure.
Shown are the results derived from inverse-variance weighted Mendelian randomization analysis. AS = all stroke; CES = cardioembolic stroke; eGFR = estimated glomerular filtration rate; ICH = intracerebral hemorrhage; IS = ischemic stroke; LAS = large artery stroke; OR = odds ratio; SVS = small vessel stroke; UACR = urine albumin-to-creatinine ratio.
In multivariable MR analysis controlling for DBP, we found consistent evidence for a causal independent effect of genetically predicted CKD on AS subtype (OR 1.06; 95% CI 1.01–1.11; p = 0.029) (eTable 6 and eFigure 3, links.lww.com/WNL/D128) and genetically predicted eGFR on AS (OR per 1-log decrement in eGFR, 1.90; 95% CI 1.33–2.72; p < 0.001), IS (OR 1.96; 95% CI 1.33–2.90; p = 0.001), and LAS (OR 2.76; 95% CI 1.16–6.58; p = 0.02) (eTable 7 and eFigure 3). Genetically determined UACR was also associated with ICH risk in the multivariable MR controlling for DBP (OR per 1 increment, 4.90; 95% CI 1.38–17.46; p = 0.014), but not SBP (eTable 8 and eFigure 3).
Mediation Analyses
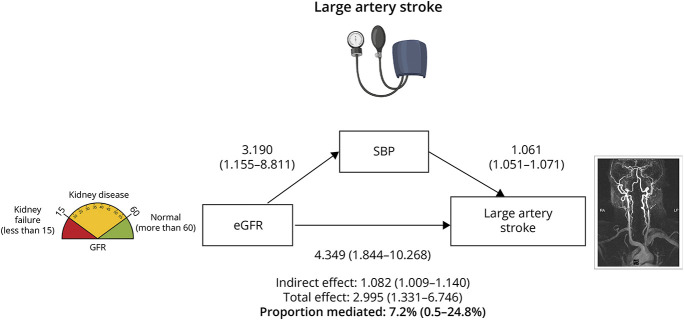
We then performed mediation MR analysis in which we calculated the proportion of the effect that was mediated by BP by dividing the indirect effect by the total effect for each of the renal traits and cerebrovascular phenotypes. The most relevant mediation analysis results are presented in Figure 3. For the effect of genetically determined lower eGFR on odds of LAS mediated through SBP, we estimated an OR of 1.08 (95% CI 1.01–1.14) and the calculated proportion mediated was 7.2% (95% CI 0.5%–24.8%) of the total effect (p = 0.03). Similarly, for the indirect effect of lower eGFR on risk of LAS mediated through DBP was 1.09 (95% CI 1.05–1.14) with DBP mediating only 7.8% (3.3%–28.6%) of the total effect (p = 0.01) (eFigure 3, links.lww.com/WNL/D128). For the genetic association between UACR and ICH risk, only 4.3% (−22.4% to 37.8%) of the proportion of the total effect was also mediated by DBP (eFigure 4).
Figure 3. Mediation Mendelian Randomization Association Between Genetically Determined Estimated Glomerular Filtration Rate and Large Artery Stroke, Controlling for Systolic Blood Pressure.
Directed acyclic graph demonstrating the total, direct, and indirect effects of estimated glomerular filtration rate on large artery stroke risk as well as the proportion of the total effect mediated by systolic blood pressure. Shown are the results derived from inverse-variance weighted Mendelian randomization analysis. eGFR = estimated glomerular filtration rate; SBP = systolic blood pressure.
To exclude reverse causation, we performed MVMR exploring the effects of BP traits on cerebrovascular phenotypes and what proportion of these effects were mediated by kidney disease traits (eFigure 5, links.lww.com/WNL/D128). There was no evidence of a bidirectional relationship between SBP and eGFR and between DBP and eGFR with little or no proportion of the total effect of BP traits on stroke subtypes mediated by lower eGFR (<1%).
To explore the association between genetically predicted lower eGFR and LAS is mediated by effects of low eGFR on atherogenesis, we examined the relationship between lower eGFR and carotid intima media thickness as well as with presence of carotid plaque (eFigure 6 and eTable 9, links.lww.com/WNL/D128). However, we found no significant associations between lower eGFR and either carotid artery trait (β for CIMT −0.004; 95% CI −0.032 to 0.025; p = 0.81 and OR per 1-log decrement in eGFR for carotid plaque 0.98; 95% CI 0.37–2.61; p = 0.60).
Discussion
Leveraging large-scale genetic data, we applied univariable and multivariable MR to determine whether impaired kidney function influences risk of stroke independently of elevated BP. Little of this effect (4%–8%) seemed to be mediated by BP. These results were largely robust to sensitivity analyses accounting for horizontal pleiotropy.
Our results build on extant efforts to leverage genetics to explore potential causal relationships between kidney disease and stroke, with the important addition of MVMR controlling for BP traits. As such, we have found strong evidence for a direct causal effect of impaired kidney function on risk of stroke, particularly between genetically determined lower eGFR and LAS. By conducting mediation analysis in the context of MR, our study adds to knowledge by formally investigating whether increased BP mediates the effect of increasing stroke risk for patients with CKD. Our study provides evidence of little indirect effect of CKD on stroke risk through BP, suggesting that other mechanisms, possibly intrinsic to kidney disease, may underlie the pathophysiologic processes leading to stroke in patients with CKD.
Deploying more recent and larger data sets,17,18 we have also confirmed previous associations between genetically determined impaired kidney function and risk of IS demonstrated by both PRS and MR approaches.10,38 In particular, consistent with the previous univariable MR study,10 there was a strong (3-fold) association between lower eGFR and LAS. Our results extend these previous findings by additionally showing associations between lower eGFR, AS, and IS, as well as between genetic predisposition to CKD diagnosis, AS, and IS, although the latter were not robust to FDR adjustment. The consistency of these relationships across various kidney disease traits and stroke subtypes provide further support for the cerebrorenal paradigm and for the potential causal role of CKD in cerebrovascular disease. In contrast to the previous MR study,10 we did not replicate the associations between genetic predisposition to higher UACR and LAS or ICH risk. However, this discrepancy likely relates to our stricter threshold for clumping (r2 < 0.001 vs <0.1 in the earlier study) because the findings were otherwise of similar magnitude and directionally consistent.
Although several large meta-analyses that have adjusted for traditional cardiovascular risk factors support our findings of an independent effect of impaired kidney function on stroke risk,3,8,39 our results do contrast with a more recent meta-analysis in which the association was attenuated with adjustment for measures of longitudinal BP control.7 These conflicting results may relate to lack of statistical power or subtype specificity, study heterogeneity, and the inherent limitations of pooling mostly observational studies for meta-analysis. By contrast, our study benefits from the statistical power of the largest and most recent GWAS data sets available including the phenotypic characterization of stroke into its etiologic subtypes.
Several mechanisms have been suggested to explain the observed associations between CKD and stroke.4,5 One might have expected a causal link between CKD and SVS, but if hypertension mediates both of these, then it could account for the lack of causal association.40 There was instead a stronger, independent association with LAS, implicating an impact of kidney disease on the atherosclerotic disease pathways in some way. The absence of a corresponding association with CIMT suggests that CKD may not necessarily affect atherogenesis but rather that it may play a role in atheroprogression, plaque destabilization, or rupture. In observational studies, patients with CKD have been shown to have different plaque morphology with reduced total collagenous fiber content, greater levels of total calcification, and higher lesion stability and risk of rupture.41 Furthermore, serum markers of inflammation, vascular calcification, and vessel wall degradation have also been shown to be significantly higher in patients with CKD compared with those with normal kidney function including enhanced levels of fibrinogen, parathyroid hormone, fetuin-A, and matrix metalloproteinase-7, suggesting that the proinflammatory milieu of CKD may play a role.42 However, further studies are required to better understand the impact of CKD on LAS pathophysiology.
Our study had a number of strengths. Compared with conventional observational studies, MR is more robust to residual confounding and measurement error because genetic variants are randomly assigned and fixed at conception and cannot be influenced by confounding factors that act after conception.43 This study applied both univariable and multivariable MR methods, using the largest number of SNPs identified from the latest well-powered GWAS studies that could be identified in the literature. MVMR is a novel extension of MR analyses that allows exploration of causality and clarification of the direction of demonstrated associations, thus ruling out reverse causation.43 A series of pleiotropy-robust methods were rigorously applied to ensure that findings were not biased as a result of pleiotropy.
Our study also has some limitations. First, using instruments with an F-statistic >10 only reduces bias to less than a certain level, and the issues with weak instrument bias still occur.44 Second, because there was less statistical power in the analyses involving the CKD diagnosis and UACR instruments, we may not have captured all potential causal associations between impaired kidney function and the various stroke subtypes, particularly as those related to proteinuric kidney diseases. Third, there was significant heterogeneity present in some of the analyses which relate to directional pleiotropy. Confounding by cryptic pleiotropy is a known limitation of MR analyses.45 However, we addressed this possibility by quantifying pleiotropy and by using multiple MR approaches with different modeling assumptions regarding the use of pleiotropic variants in the analyses to further strengthen the validity of our MR models. Genetic variants with different functional effects on the risk factor may be an alternative explanation for some of the observed heterogeneity, whereby different biological mechanisms give rise to the association with kidney disease traits, and thus, different magnitudes of causal effect may be expected. Kidney disease itself has a very heterogenous pathobiology.46 Fourth, analyses were based on summary statistics from individuals of European ancestry, which may limit the generalizability of our findings to other ancestry groups. This is relevant to vulnerable populations such as Black and Hispanic patients where there are important phenotypic and prognostic differences in hypertension and CKD.47,48 One previous transancestry MR analysis did not demonstrate an association between eGFR and stroke risk; however, this study focused on all ischemic stroke and not individual cerebrovascular phenotypes, and they did not perform mediation analysis in the context of MR.49
In conclusion, this study used both univariable and multivariable MR analyses to demonstrate a causal effect for impaired kidney function on stroke risk that is independent of hypertension. In particular, we observed large effects for LAS risk in our multivariable MR analysis. These findings could help prioritize CKD prevention and treatment to mitigate stroke risk and motivate future studies to provide further insight into mechanisms linking kidney disease and stroke.
Glossary
- AS
all stroke
- BP
blood pressure
- CES
cardioembolic stroke
- CIMT
carotid intima media thickness
- CKD
chronic kidney disease
- CKDGen
1000 Genomes-based Chronic Kidney Disease Genetics
- DBP
diastolic blood pressure
- eGFR
estimated glomerular filtration rate
- FDR
false discovery rate
- GWAS
genome-wide association study
- ICH
intracerebral hemorrhage
- IS
ischemic stroke
- ISGC
International Stroke Genetic Consortium
- IVW
inverse-variance weighted
- LAS
large artery stroke
- MVMR
multivariable MR
- MR
Mendelian randomization
- OR
odds ratio
- SBP
systolic blood pressure
- SNP
single-nucleotide polymorphism
- SVS
small vessel stroke
- UACR
urine albumin-to-creatinine ratio
Appendix. Authors

| Name | Location | Contribution |
| Dearbhla M. Kelly, MB BCh, BAO, MSc, DPhil, MRCPI | J. Philip Kistler Stroke Research Center, Department of Neurology, Massachusetts General Hospital, Harvard Medical School; Program in Medical and Population Genetics, Broad Institute of Harvard and the Massachusetts Institute of Technology, Boston | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design; analysis or interpretation of data |
| Marios K. Georgakis, MD, PhD | Program in Medical and Population Genetics, Broad Institute of Harvard and the Massachusetts Institute of Technology, Boston; Institute for Stroke and Dementia Research, University Hospital of LMU Munich, Germany; McCance Center for Brain Health, Massachusetts General Hospital, Boston | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design; analysis or interpretation of data |
| Nora Franceschini, MD, MPH | Department of Epidemiology, University of North Carolina, Chapel Hill | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; analysis or interpretation of data |
| Deborah Blacker, MD, ScD | Department of Psychiatry, Massachusetts General Hospital, Harvard Medical School; Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA | Drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data |
| Anand Viswanathan, MD, PhD | J. Philip Kistler Stroke Research Center, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston | Drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data |
| Christopher D. Anderson, MD, MMSc | Program in Medical and Population Genetics, Broad Institute of Harvard and the Massachusetts Institute of Technology; McCance Center for Brain Health, Massachusetts General Hospital; Department of Neurology, Brigham and Women's Hospital, Boston, MA | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design; analysis or interpretation of data |
Study Funding
No targeted funding reported.
Disclosure
D.M. Kelly is an Atlantic Fellow for Equity in Brain Health at the Global Brain Health Institute (GBHI) and is supported with funding from GBHI, Alzheimer's Association, and Alzheimer's Society (GBHI ALZ UK-22-868940) and is the recipient of an NIH StrokeNet Fellowship. M.K. Georgakis is supported by a Walter-Benjamin fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft [DFG], GZ: GE 3461/1-1) and the FöFoLe program of Ludwig-Maximilians-University Munich (FöFoLe-Forschungsprojekt Reg.-Nr. 1120). This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy: ID 390857198 to MKG). N. Franceschini is supported by NIH R01 HL163972. A. Viswanathan is supported by NIH P50 AG005134 NIH AG047975 R01 NS104130. C.D. Anderson is supported by NIH R01NS103924, U01NS069673, AHA 18SFRN34250007, and AHA-Bugher 21SFRN812095 for this work and receives sponsored research support from Bayer AG and has consulted for ApoPharma, Inc. Go to Neurology.org/N for full disclosures.
References
- 1.Abramson JL, Jurkovitz CT, Vaccarino V, Weintraub WS, McClellan W. Chronic kidney disease, anemia, and incident stroke in a middle-aged, community-based population: the ARIC study. Kidney Int. 2003;64(2):610-615. doi: 10.1046/j.1523-1755.2003.00109.x [DOI] [PubMed] [Google Scholar]
- 2.Murray AM, Seliger S, Lakshminarayan K, Herzog CA, Solid CA. Incidence of stroke before and after dialysis initiation in older patients. J Am Soc Nephrol. 2013;24(7):1166-1173. doi: 10.1681/asn.2012080841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masson P, Webster AC, Hong M, Turner R, Lindley RI, Craig JC. Chronic kidney disease and the risk of stroke: a systematic review and meta-analysis. Nephrol Dial Transplant. 2015;30(7):1162-1169. doi: 10.1093/ndt/gfv009 [DOI] [PubMed] [Google Scholar]
- 4.Kelly D, Rothwell PM. Disentangling the multiple links between renal dysfunction and cerebrovascular disease. J Neurol Neurosurg Psychiatry. 2020;91(1):88-97. doi: 10.1136/jnnp-2019-320526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyoda K, Ninomiya T. Stroke and cerebrovascular diseases in patients with chronic kidney disease. Lancet Neurol. 2014;13(8):823-833. doi: 10.1016/s1474-4422(14)70026-2 [DOI] [PubMed] [Google Scholar]
- 6.Muntner P, Anderson A, Charleston J, et al. Hypertension awareness, treatment, and control in adults with CKD: results from the Chronic Renal Insufficiency Cohort (CRIC) study. Am J Kidney Dis. 2010;55(3):441-451. doi: 10.1053/j.ajkd.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly DM, Rothwell PM. Does chronic kidney disease predict stroke risk independent of blood pressure? A systematic review and meta-regression. Stroke. 2019;50(11):3085-3092. doi: 10.1161/strokeaha.119.025442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly DM, Rothwell PM. Proteinuria as an independent predictor of stroke: systematic review and meta-analysis. Int J Stroke. 2020;15(1):29-38. doi: 10.1177/1747493019895206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133-1163. doi: 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 10.Marini S, Georgakis MK, Chung J, et al. Genetic overlap and causal inferences between kidney function and cerebrovascular disease. Neurology. 2020;94(24):e2581-e2591. doi: 10.1212/wnl.0000000000009642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coresh J, Grams ME, Chen TK. Using GFR, albuminuria, and their changes in clinical trials and clinical care. Am J Kidney Dis. 2021;78(3):333-334. doi: 10.1053/j.ajkd.2021.04.003 [DOI] [PubMed] [Google Scholar]
- 13.National Institute for Health and Care Excellence. Chronic kidney disease in adults: assessment and management. Clinical guideline [online]. 2021. [PubMed] [Google Scholar]
- 14.Stevens LA, Coresh J, Schmid CH, et al. Estimating GFR using serum cystatin C alone and in combination with serum creatinine: a pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis. 2008;51(3):395-406. doi: 10.1053/j.ajkd.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lees JS, Welsh CE, Celis-Morales CA, et al. Glomerular filtration rate by differing measures, albuminuria and prediction of cardiovascular disease, mortality and end-stage kidney disease. Nat Med. 2019;25(11):1753-1760. doi: 10.1038/s41591-019-0627-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ckdgen.imbi.uni-freiburg.de/.
- 17.Wuttke M, Li Y, Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957-972. doi: 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanzick KJ, Li Y, Schlosser P, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun. 2021;12(1):4350. doi: 10.1038/s41467-021-24491-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas ME, Aragam KG, Emdin CA, et al. Genetic association of albuminuria with cardiometabolic disease and blood pressure. Am J Hum Genet. 2018;103(4):461-473. doi: 10.1016/j.ajhg.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. doi: 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo D, Falcone GJ, Devan WJ, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94(4):511-521. doi: 10.1016/j.ajhg.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CKDGen Meta-Analysis Data. Accessed October 1, 2021. cd.hugeamp.org/.
- 23.Adams HP Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35-41. doi: 10.1161/01.str.24.1.35 [DOI] [PubMed] [Google Scholar]
- 24.Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412-1425. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Accessed October 1, 2021. grasp.nhlbi.nih.gov/FullResults.aspx.
- 26.Franceschini N, Giambartolomei C, de Vries PS, et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun. 2018;9(1):5141. doi: 10.1038/s41467-018-07340-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Daniel RM, Butterworth AS, Thompson SG. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 2015;44(2):484-495. doi: 10.1093/ije/dyu176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter AR, Sanderson E, Hammerton G, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021;36(5):465-478. doi: 10.1007/s10654-021-00757-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Thompson DJ, Rees JMB, Day FR, Perry JR, Ong KK. Dissecting causal pathways using Mendelian randomization with summarized genetic data: application to age at menarche and risk of breast cancer. Genetics. 2017;207(2):481-487. doi: 10.1534/genetics.117.300191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter AR, Gill D, Davies NM, et al. Understanding the consequences of education inequality on cardiovascular disease: Mendelian randomisation study. BMJ. 2019;365:l1855. doi: 10.1136/bmj.l1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanderson E. Multivariable Mendelian randomization and mediation. Cold Spring Harb Perspect Med. 2021;11(2):a038984. doi: 10.1101/cshperspect.a038984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgakis MK, Gill D, Webb AJS, et al. Genetically determined blood pressure, antihypertensive drug classes, and risk of stroke subtypes. Neurology. 2020;95(4):e353-e361. doi: 10.1212/wnl.0000000000009814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teumer A. Common methods for performing Mendelian randomization. Front Cardiovasc Med. 2018;5:51. doi: 10.3389/fcvm.2018.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J, Hemani G, Davey Smith G. Invited commentary: detecting individual and global horizontal pleiotropy in Mendelian randomization: a job for the humble heterogeneity statistic? Am J Epidemiol. 2018;187(12):2681-2685. doi: 10.1093/aje/kwy185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389. doi: 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holliday EG, Traylor M, Malik R, et al. Polygenic overlap between kidney function and large artery atherosclerotic stroke. Stroke. 2014;45(12):3508-3513. doi: 10.1161/strokeaha.114.006609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M, Saver JL, Chang KH, Liao HW, Chang SC, Ovbiagele B. Low glomerular filtration rate and risk of stroke: meta-analysis. BMJ. 2010;341(1):c4249. doi: 10.1136/bmj.c4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S, Nagasawa T, Abe M, Mori T. Strain vessel hypothesis: a viewpoint for linkage of albuminuria and cerebro-cardiovascular risk. Hypertens Res. 2009;32(2):115-121. doi: 10.1038/hr.2008.27 [DOI] [PubMed] [Google Scholar]
- 41.Pelisek J, Assadian A, Sarkar O, Eckstein HH, Frank H. Carotid plaque composition in chronic kidney disease: a retrospective analysis of patients undergoing carotid endarterectomy. Eur J Vasc Endovasc Surg. 2010;39(1):11-16. doi: 10.1016/j.ejvs.2009.09.024 [DOI] [PubMed] [Google Scholar]
- 42.Pelisek J, Hahntow IN, Eckstein HH, et al. Impact of chronic kidney disease on carotid plaque vulnerability. J Vasc Surg. 2011;54(6):1643-1649. doi: 10.1016/j.jvs.2011.05.049 [DOI] [PubMed] [Google Scholar]
- 43.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764. doi: 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 45.Bennett DA, Holmes MV. Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart. 2017;103(18):1400-1407. doi: 10.1136/heartjnl-2016-310605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess SBJ, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30-42. doi: 10.1097/ede.0000000000000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duru OK, Li S, Jurkovitz C, et al. Race and sex differences in hypertension control in CKD: results from the Kidney Early Evaluation Program (KEEP). Am J Kidney Dis. 2008;51(2):192-198. doi: 10.1053/j.ajkd.2007.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hounkpatin HO, Fraser SDS, Honney R, Dreyer G, Brettle A, Roderick PJ. Ethnic minority disparities in progression and mortality of pre-dialysis chronic kidney disease: a systematic scoping review. BMC Nephrol. 2020;21(1):217. doi: 10.1186/s12882-020-01852-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris AP, Le TH, Wu H, et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun. 2019;10(1):29. doi: 10.1038/s41467-018-07867-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genetic variants used are available in the eAppendix 1 (links.lww.com/WNL/D128), and the code used for all analyses is available on request.