Abstract
Background and Objectives
Despite notable advances in genetic understanding of stroke recovery, most studies focus only on candidate genes. To date, only 2 genome-wide association studies (GWAS) have focused on stroke outcomes, but they were limited to the modified Rankin Scale (mRS). The mRS maps poorly to biological processes. Therefore, we performed a GWAS to discover single nucleotide polymorphisms (SNPs) associated with motor recovery poststroke.
Methods
We used the Vitamin Intervention for Stroke Prevention (VISP) data set of 2,100 genotyped participants with nondisabling stroke. We included only participants who had motor impairment at randomization. Participants with a recurrent stroke during the trial were excluded. Genotyped data underwent strict quality control and imputation. The GWAS used logistic regression models with generalized estimating equations to leverage the repeated NIH Stroke Scale motor score measurements spanning 6 time points over 24 months. The primary outcome was a decrease in the motor drift score of ≥1 vs <1 at each time point. Our model estimated the odds ratio (OR) of motor improvement for each SNP after adjusting for age, sex, race, days from stroke to visit, initial motor score, VISP treatment arm, and principal components.
Results
A total of 488 (64%) participants with a mean (SD) age of 66 ± 11 years were included in the GWAS. Although no associations reached genome-wide significance (p < 5 × 10−8), our analysis detected 115 suggestive associations (p < 5 × 10−6). Notably, we found multiple SNP clusters near genes with plausible neuronal repair biology mechanisms. The CLDN23 gene had the most convincing association with rs1268196-T as its most significant SNP (OR 0.32; 95% CI 0.21–0.48; p value 6.19 × 10−7). CLDN23 affects blood-brain barrier integrity, neurodevelopment, and immune cell transmigration.
Discussion
We identified novel suggestive genetic associations with the first-ever motor-specific poststroke recovery GWAS. The results seem to describe a distinct stroke recovery phenotype compared with prior genetic stroke outcome studies that use outcome measures, such as the mRS. Replication and further mechanistic investigation are warranted. In addition, this study demonstrated a proof-of-principle approach to optimize statistical efficiency with longitudinal data sets for genetic discovery.
Introduction
A reckoning is coming to the field of stroke recovery and genomics. The research, now merging at the intersection of these fields, faces 3 major challenges. First, most studies on stroke-related genes use a candidate gene approach,1 whereas there are only 2 genome-wide association studies published to date.2,3 Therefore, understanding stroke recovery genetics is limited to an extremely small portion of the genome, encompassing only 11 associated genes.1 However, the complex and time-varying biology of stroke recovery will likely involve a much greater proportion of the genome. This suggests that study designs using genome-wide4 and epigenome-wide5 associations are well suited to discover novel recovery-associated genes and their variations. The second issue is that acute stroke treatment trials often collect blood samples useful for subsequent genetic studies. However, they tend to lack detailed measures of stroke recovery. Conversely, stroke recovery trials frequently collect these detailed and domain-specific outcome measures, but lack biospecimens for subsequent genetic analyses. The third challenge entails the issue that most studies on stroke recovery-related genes have defined their recovery phenotypes using global outcome measures that combine multiple domains of impairment (e.g. the modified Rankin Scale or total NIH Stroke Scale [NIHSS] score) rather than using domain-specific measures (e.g. the Upper Extremity Fugl-Meyer for the motor domain).6
It remains unclear whether the phenotype-genotype associations observed using multidomain measures differ from those observed using domain-specific measures of stroke recovery. For example, variants of the brain-derived neurotrophic factor (BDNF) gene have been shown to predict poor stroke outcomes defined as the 90-day modified Rankin Scale (mRS) score ≤1 for ischemic stroke or Glasgow Outcome Scale score ≤3 for hemorrhagic stroke.7 However, Cramer et al.8 recently showed that BDNF variants were not associated with a domain-specific measure of arm motor function. This suggests that change in a multidomain outcome measure may represent a different phenotype-genotype relationship than a change in a domain-specific measure. The distinction is not trivial. As noted in the Stroke Recovery and Rehabilitation Roundtable9 guidelines, “brain repair maps best onto fine-grained movement quality measures that are sensitive and specific.” In other words, using domain-specific measures of stroke recovery is better suited for studies that aim to discover genetic mechanisms of brain plasticity. Thus, genetic studies of stroke recovery using domain-specific measures are urgently needed.
In an effort to address this need, Braun et al.10 argued that changes in NIHSS subscores, which measure impairment in distinct neurologic domains, can be considered as an efficient and clinically feasible means to obtain domain-specific measures of stroke recovery. They noted that the NIHSS motor impairment subscores are comparable with the Fugl-Meyer regarding arm and leg motor function. They also have good inter-rater reliability (kappa 0.77–0.78). This study is the first effort to define a phenotype-genotype association specific to poststroke motor recovery using the change in NIHSS subscores.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
This retrospective analysis uses the clinical and genetic data obtained from the Vitamin Intervention for Stroke Prevention (VISP) clinical trial (NCT00004734 at ClinicalTrials.gov).11 The VISP trial received approval from the Institutional Review Boards of Wake Forest University School of Medicine and the University of North Carolina at Chapel Hill School of Medicine. Furthermore, all participants provided informed consent, and all enrollment sites were in accordance with the Declaration of Helsinki. This retrospective analysis falls within the informed consent of the participants in the VISP trial.
Discovery Cohort
The VISP trial investigated the effect of vitamin supplementation dosage on the risk of recurrent stroke with a randomized double-blinded design. The study enrolled participants with a nondisabling ischemic stroke (mRS ≤3) greater than or equal to 72 hours before enrollment. Participants were randomized to a high-dose or low-dose vitamin supplementation arm if they were at least 75% compliant with taking a low-dose supplementation packet for 1 month prior. All participants were reassessed every 3 months until a recurrent stroke event, but not longer than 2 years.11 The trial successfully enrolled a total of 3,680 randomized participants. However, 10 sites were not approved for genetic studies, resulting in a subset of 2,100 genotyped participants.
Quality Control
The Center for Inherited Disease Research at Johns Hopkins University performed genotyping on the Illumina HumanOmni1-Quad-v1 array (Illumina, Inc.). The genotyped data underwent strict quality control measures that filtered out SNPs as follows: (1) missing call rate >2%, (2) Mendelian errors in control trios, (3) deviation from Hardy-Weinburg equilibrium in controls, (4) discordant calls in duplicate samples, (5) sex differences in allele frequency or heterozygosity, and (6) minor allele frequency <0.05 in line with previously published recommendations.12 We increased the number of SNP with genetic imputation using the TOPMed Imputation server,13,14 which implements the Minimac Imputation procedure.15 The TOPMed study13 has a large cohort of 97,256 individuals with diverse backgrounds, which was preferred because of the sizeable proportion of non-European ancestry participants in the VISP genotyped cohort. After filtering out imputed SNPs with poor imputation quality (r2 < 0.80) and MAFs < 0.05, the final count of SNPs came to 6,588,085.
Phenotyping
As suggested,10 we used the motor drift subscores of the NIHSS as a measurement of motor weakness. The NIHSS subscores 5A/5B and 6A/6B defined the degree of limb weakness for the upper and lower extremities, also known as drift. The subscores rate limb weakness or drift on an ordinal scale from 0 to 5: 0 shows no drift, 1 drift is present, 2 observed some effort against gravity, 3 shows no effort against gravity, 4 there is no movement, and 5 the limb is amputated. We assumed that the enrolling stroke caused all motor drift scores between 1 and 4 of the contralateral limb to the stroke lesion. The VISP data set does not have imaging data for retrospective validation. A limb was not phenotyped if it had a motor drift score of 5 (amputation) at study randomization. Motor improvement is defined as the decrease in the initial motor drift subscore of the weakest limb from randomization to each follow-up period. We chose the upper limb if participants had equally affected upper and lower limbs. To maximize statistical power and model stability, we dichotomized motor improvement as a decrease in initial motor drift by ≥1 vs <1 for each follow-up period.
Data Analysis Plan
We implemented a logistic regression model with generalized estimating equations (GEE) with the “gee” R package.16 The GEE model allows the incorporation of repeated measurements of the motor drift subscore over 2 years,17 which provides notable statistical power gains compared with the traditional case/control GWAS study design. A priori we planned to adjust for age, sex, initial motor drift score, treatment arm, and population stratification through principle components. We calculated the top 10 principal components using KING software18 to account for population stratification in our cohort with genotyped SNPs after pruning. To determine which principle components to include in the GWAS model, we used a backward selection procedure optimizing the AIC with the “stepAIC” function from the MASS R package.19
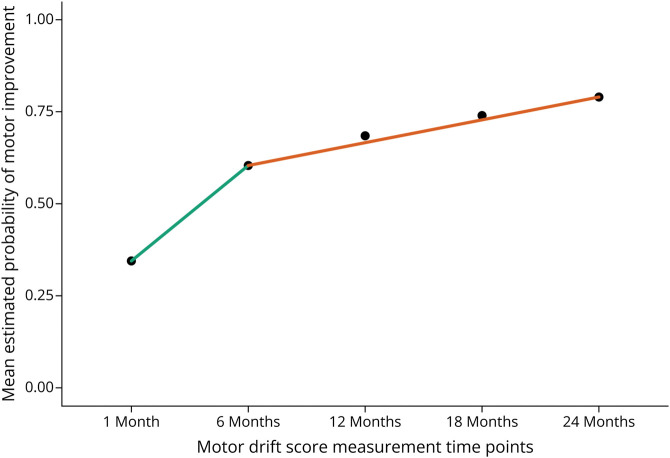
In addition to the a priori covariates, time since stroke onset is an important covariate when modeling stroke recovery because of changing rates of recovery based on well-defined time epochs (i.e. acute, early and late subacute, and chronic).20 These epochs are tied to biological processes of inflammation and scarring early on into recovery with a transition to mainly endogenous plasticity in later stages. To account for this effect in the model, we added the covariate of time from stroke onset to time of motor drift measurement in days for each follow-up period. Furthermore, we investigated the nonlinear relationship of the probability of poststroke motor improvement and time since stroke onset. Figure 1 shows the mean estimated probability of motor drift improvement over time since randomization. We used a spline of time from onset to measurement with 1 knot at 250 days to better model the nonlinear relationship and maintain the clinical. Finally, we considered possible loss to follow-up effects. We investigated which baseline characteristics predict missing motor drift scores. Any associated baseline characteristics would be added to the final GEE model with an exchangeable correlation structure as covariates.
Figure 1. Nonlinear Probability of Motor Improvement Over Time.
The nonlinear relationship of the mean probability of motor improvement for each follow-up time point since study randomization. The green and orange line segments highlight the notable change in slope from 1- to 6-month visits to 6- to 24-month visits after randomization. The nonlinear relationship of motor improvement time is well known from chronic stroke rehab trials of the upper extremity.
Sensitivity Analysis
We performed 2 sensitivity analyses. First, we evaluated the interaction of time of stroke onset to follow-up period spline with each SNP that reached a p value threshold of p < 5 × 10−6. We suspected that the effect of the SNP may change depending on the stroke recovery phase. Second, we observed a wide spread of time from stroke onset to VISP randomization (median 72 days; interquartile range [IQR] 45.75–102 days). We generated an early vs late poststroke randomization variable defined as < the median (72 days) being early and ≥ the median as late randomization and then estimated its interaction with each.
Look-Up Analysis
We investigated if the reported SNPs from the GISCOME GWAS study2 on stroke functional recovery replicate with our poststroke motor recovery associated SNPs. The GISCOME study is the largest poststroke recovery GWAS by combined sample size (n = 6,021) from 12 studies. Söderholm et al. defined good recovery as a mRS of ≤2 and a mRS of ≥3 signified poor recovery. We planned to compare our GWAS results with all SNPs with a p value <5 × 10−6 from the GISCOME study. The p values of the look-up analysis will receive a multiple comparison adjustment at an FDR of 10%.
Data Availability
The clinical VISP data are available at the NIHND Archived Clinical Research Datasets website (ninds.nih.gov/current-research/research-funded-ninds/clinical-research/archived-clinical-research-datasets). Raw genotype data for VISP are available in dbGaP (Accession phs000343.v4.p1). The genetic association results generated by this study can be found in the Cerebrovascular Disease Knowledge Portal.
Results
Demographics
We included participants with weakness of an arm or leg defined as a greater than zero on the motor drift time from the NIH stroke scale at randomization. We excluded participants who had an incident recurrent stroke during the trial. This resulted in 488 participants in this GWAS, who provided 2,095 individual observations over the entire VISP study 2-year period from randomization to months 1, 6, 12, 18, and 24. Participants had a median (IQR) 5 (4–5) number of motor drift assessments with a minimum of 1 to a maximum of 5. Table 1 presents the demographics of this cohort. Furthermore, 170 (34.8%) left and 145 (29.7%) right upper limbs were phenotyped, while 81 (16.6%) left and 92 (18.8%) right lower limbs were phenotyped. Most of the participants had worse arm weakness (67%) than leg weakness likely because of the VISP inclusion criteria of nondisability strokes defined by a mRS ≤3. Once we phenotyped the limb of interest as motor “weakness present” (motor drift >0) vs “no weakness,” we observed a steady increase in the percentage of participants with “no weakness” over the 2 years. By 24 months after randomization, 73.89% of participants had no motor weakness (motor drift = 0) (see Table 2). It is well known that the higher mRS scores are biased toward lower extremity weakness and inability to walk compared with upper extremity weakness. Of note, the distribution of patient ancestry generally reflects the national US population. By 24 months, 26.6% of participants were lost to follow-up. We found sex and self-identified race were associated with loss to follow-up. Male participants made up 76% who were lost to follow-up vs female participants at 59% (p < 0.001). Participants who self-identified as Black were more likely to be lost to follow-up than those who self-identified as White (odds ratio [OR] 2.10; 95% CI 1.59–2.75; p < 0.001). Similarly, participants of non-European or African ancestry (collapsed into a classification “other”, due to small sample sizes) were 1.76 (OR) more likely to be lost to follow-up than White participants (95% CI 1.17–2.60; p value 0.005).
Table 1.
Vitamin Intervention for Stroke Prevention (VISP) Trial Demographics at the Time of Randomization
| VISP cohort (n = 488) | |
| Treatment Arm, n (%) | |
| High Dose | 225 (47) |
| Low Dose | 251 (53) |
| Age, y | |
| Mean (SD) | 66 (±11) |
| Sex, n (%) | |
| Male | 310 (64) |
| Weakest LIMB, n (%) | |
| Arm | 333 (67) |
| Stroke onset to randomization | |
| Days | 72.5 (±31.2) |
| Body mass index (BMI) | |
| N-Miss | 7 |
| Mean (SD) | 28.54 (6.47) |
| Hypertension, n (%) | |
| N-Miss | 1 |
| No | 114 (23) |
| Yes | 383 (77) |
| Ever smoker, n (%) | |
| N-Miss | 1 |
| No | 173 (36) |
| Yes | 314 (64) |
| Diabetes mellitus type II, n (%) | |
| No | 331 (68) |
| Yes | 157 (32) |
| Ancestry, n (%) | |
| European | 347 (73) |
| African | 92 (19) |
| Other | 37 (8) |
Information is presented as counts (%) or means (SD).
Table 2.
Number (%) of Participants With a Motor Drift Score Greater Than 0 on Their Phenotyped Limb at Each Study Time Point Since Randomization
| Motor weakness | ||
| Study visit | Present, n (%) | Absent, n (%) |
| Randomization | 488 (100) | 0 (0) |
| 1 mo | 312 (65.14) | 167 (34.86) |
| 6 mo | 194 (43.21) | 255 (56.79) |
| 12 m | 133 (31.59) | 288 (68.41) |
| 18 m | 115 (29.04) | 281 (70.96) |
| 24 mo | 94 (26.11) | 266 (73.89) |
Most study participants were in their chronic phase of poststroke recovery since they were randomized on average (SD) of 72.5 (±31.2) days from stroke onset.
GWAS
Primary Results
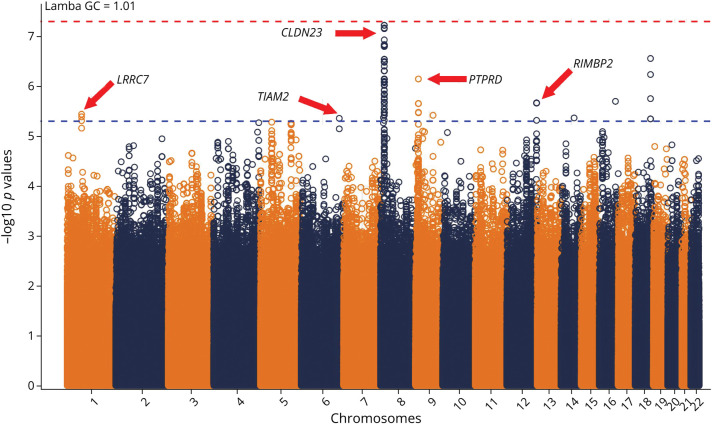
None of the SNPs reached genome-wide significance (p < 5 × 10−8). However, 115 SNPs reached suggestive associations with motor improvement (p < 5 × 10−6). Figure 2 shows the p values of the OR of motor improvement for each SNP. The GWAS's calculated genomic control factor is 1.01, which suggests no genomic inflation. The suggestive SNPs found themselves in chromosomes 1 (n = 3), 6 (n = 1), 8 (n = 92), 9 (n = 6), 12 (n = 6), 14 (n = 1), 16 (n = 1), and 18 (n = 5). The top 2 SNPs, rs12681936 and rs12680789, in chromosome 8 had the smallest p values (5.96 × 10−8), which were just shy of genome-wide significance (p < 5 × 10−8). See eTable 1 (links.lww.com/WNL/D159) for a full list of all suggestive SNP associations with annotations from Ensembl.org's variant effect predictor software.21 Figure 2 shows a strong signal on chromosome 8. This locus is better visualized by the locus zoom plot in Figure 3A. This locus is within <0.1 megabases of the CLDN23 gene.
Figure 2. Genome-wide Association Study Manhattan Plot of Poststroke Motor Recovery.
This Manhattan plot shows each SNP and its -log10 (p value) associated with poststroke motor improvement. None of the SNPs reached genome-wide significance (above the red line). However, 115 SNPs had suggestive associations (above the blue line), with 2 right under the red line. The most convincing genetic locus is the large spike in chromosome 8, near the Claudin 23 gene. This gene affects the blood-brain barrier and immune cell transmigration. Red dotted line marks the Bonferoni threshold of -log10 (5e-8). Blue dotted line marks the suggestive threshold of -log10 (5e-6). SNP = single nucleotide polymorphism.
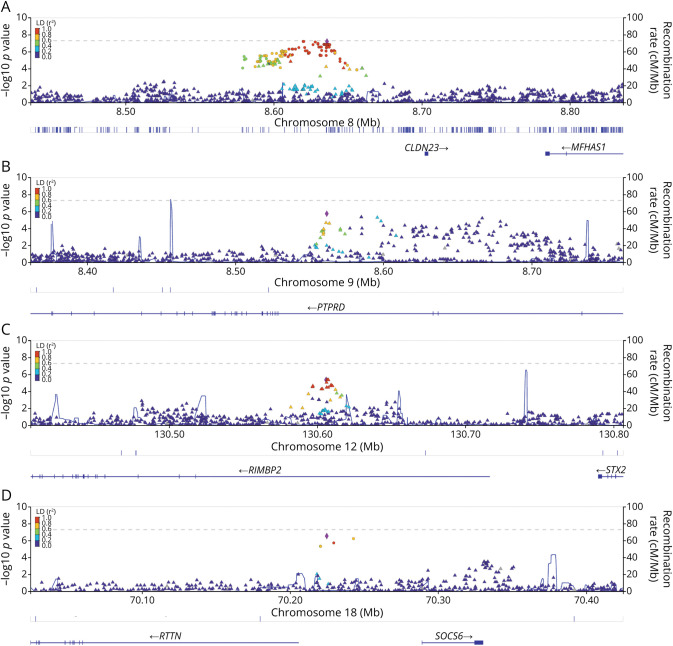
Figure 3. Suggestive Genetic Loci of Interest and Their Linkage Disequilibrium.
Panel plot of Locus Zoom figures (A–D) corresponding to genetic loci of interest. The colors refer to the correlation of each SNP to the top SNP in each panel, with red having an r2 ≥ 0.80. (A) Genetic locus near the CLDN23 gene on chromosome 8. (B) Genetic locus within the PTPRD gene on chromosome 9. (C) Genetic locus within the RIMBP2 gene on chromosome 12. (D) Genetic locus between the RTTN and SOCS6 genes on chromosome 18. SNP = single nucleotide polymorphism.
Sensitivity Analysis
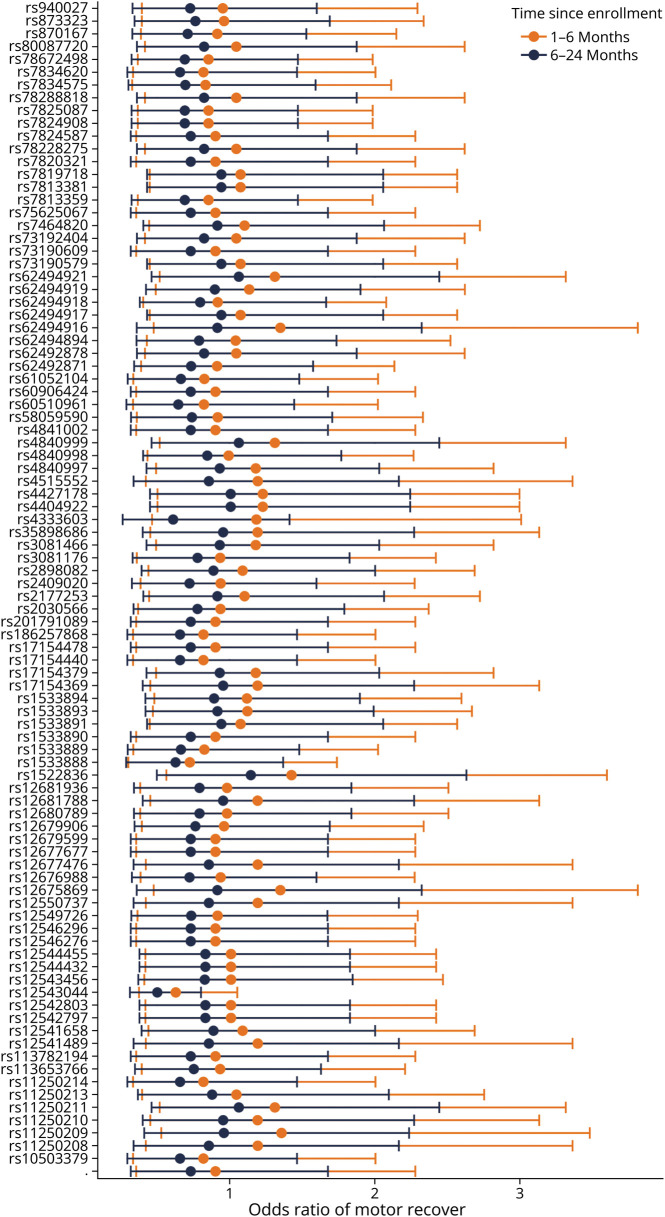
Sensitivity analysis of the interaction between the spline of stroke onset to motor drift measurement revealed that 2 SNPs had significant interactions at an FDR of 10%. They were rs113693489 in chromosome 6 and rs2967308 in chromosome 16. eTable 2 (links.lww.com/WNL/D160) contains all the interaction estimates and their q values. In general, SNP interactions with the first part of the spline (days from stroke onset to measurement <250) had a mean (±SD) OR of 0.967 (±1.33). The interactions with the second part of the spline (≥250 days) had a mean (±SD) OR of 0.806 (±1.33). Highlighting the chromosome 8 locus, Figure 4 shows the OR point estimate and their 95% confidence intervals.
Figure 4. CLDN23 Associations Have Larger Effect Sizes Within 6 Months of Enrollment vs Later.
Shows the estimates and 95% confidence intervals of the interaction of the study time points 1–6 months vs 6–24 months with each suggestive SNP found in chromosome 8. Interestingly, the interaction estimates of many SNPs seem to straddle one, with SNP estimates at 1–6 months having greater odds of motor recovery. In comparison, SNP estimates at 6–24 months have less odds of motor recovery. SNP = single nucleotide polymorphism
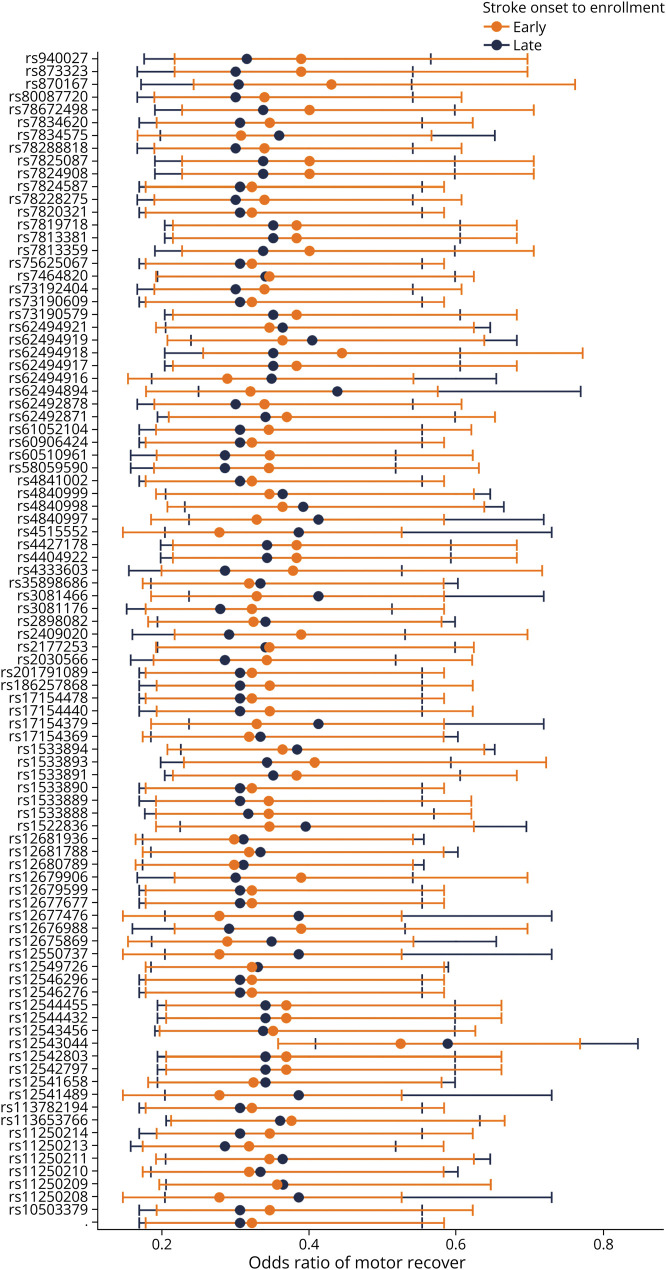
The second sensitivity analysis focused on the interaction of the suggestive SNPs based on early vs later randomization into the VISP trial from stroke onset. The SNPs' p values underwent an FDR adjustment of 10%. In contrast to the interaction analysis, all early and late randomization q values were significant; (eTable 3) (links.lww.com/WNL/D161). Early randomization interactions had a mean (±SD) OR of 0.419 (±1.68). Late randomization interaction had a similar mean (±SD) OR of 0.397 (±1.70). Figure 5 shows the estimates for each suggestive SNP interaction with early vs late randomization in the chromosome 8 locus. Although our sensitivity analysis models show that each SNP interaction is an independent predictor of motor improvement, the 95% confidence intervals have large overlaps. The overlaps suggest that ORs for each SNP interaction do not differ from early vs late randomization in the VISP trial from stroke onset.
Figure 5. CLDN23 Associations Are Robust to Time From Stroke to Study Enrollment.
Odds ratios and 95% confidence intervals of motor drift score improvement from the interaction early vs late poststroke randomization by SNP in chromosome 8. The odds ratios estimates for early vs late do not have a discernible pattern or consistency. SNP = single nucleotide polymorphism.
Look-up Analysis
Of the 500 reported SNPs (p < 5 × 10−6) from the GISCOME study,2 only 414 were present in our analysis results. After applying an FDR of 10%, none of the look-up SNPs from the GISCOME study reached significance.
Discussion
This study aimed to discover novel genetic associations of motor recovery in individuals with mildly disabling stroke, using a cohort of participants in the chronic phase of recovery. Most study participants experienced their stroke at least 30 days before study randomization. Our findings reveal that the results are more pertinent to chronic motor recovery rather than acute. Although our GWAS of poststroke motor recovery failed to show genome-wide significant associations, we found 115 novel suggestive SNPs linked to the odds of motor recovery over 2 years. These suggestive SNPs mapped to genomic loci connected with genes that are previously unknown as either candidate genes or ones from prior GWAS studies.2,3 These findings are biologically relevant and suggest that genetic factors may play a role in chronic phases of recovery for participants with mildly disabling stroke.
The chromosome 8 locus's apex, as seen in Figure 3A, is <0.1 megabases from the CLDN23 gene. CLDN23 (Claudin 23) is a protein-encoding gene part of the Claudin gene family, which are integral membrane proteins and components to tight junction strands.22 CLDN23 has related pathways affecting the blood-brain barrier and immune cell transmigration according to genecards.org's pathway unification database.23
In addition, CLDN23 variants are associated with blood cholesterol, triglyceride, and lipid measurements.24-26 In our study, rs62494916 located in an enhancer region near CLDN23 has an estimated OR of 0.33 (beta −1.11; standard deviation 0.23; p value 1.8 × 10−6). For each allele copy of rs62494917 (G), the odds of improving motor weakness decreased by a factor of 0.33. It is possible that certain SNPs that affect CLDN23 gene expression may also influence the dynamic stability of the blood-brain barrier resulting in a less hospitable environment for neuroplasticity and recovery to occur poststroke. Unlike the chromosome 8 locus, the chromosome 9 locus is within the PTPRD gene, part of the protein tyrosine phosphate (PTP) family. The PTPRD gene has a protein-to-protein interaction at the neuronal synapse located at the presynaptic terminal surface. It has related pathways of cell growth, differentiation, mitotic cycle, and oncogenic transformation.27,28 Interestingly, PTPRD has an association with glioblastoma.29 Figure 3C shows the chromosome 12 locus within the RIMS-binding protein (RIMBP2) gene. As the name suggests, this gene produces a binding protein. The function of this protein is predicted to involve neuromuscular synaptic transmission. It is also highly expressed in brain tissue. In 2021, Butola et al. reported that the role of RIM-BP2 is to link voltage-gated Ca2+ channels and release the sites of synaptic vesicles.30 They explain that RIMBP2 disruption leads to alterations in Cav2.1 channel topography at active zones. These active zones affect neurotransmitter release. This locus's top SNP (rs73156962) has a direct biological interpretation (p = 0.034) in the nucleus accumbens located in the basal ganglia, a highly dense interconnected neuronal tissue.31
The chromosome 18 locus sits almost equally between 2 genes, RTTN and SOCS6, each within 0.1 megabases (Figure 3D). RTTN (Rotatin) encodes a large protein without a known specific function. However, knockout mice models result in neural tube defects.32 In humans, RTTN pathologic variants lead to microcephaly and polymicrogyria with seizures.33,34 Although RTTN is linked to neurologic structure and disorder in humans, there remains a notable lack of published literature on this gene and its biological mechanisms. However, SOCS6 (suppressor of cytokine signaling 6) is part of the suppressor cytokine signaling protein family, which plays a key role in inflammation regulation and insulin signaling in human brain tissue, especially brain tissue affected by a neurodegenerative disease.35
Söderholm et al.2 performed the largest GWAS of stroke functional recovery to date. Their analysis consisted of 12 studies which totaled 6,021 participants. They defined functional stroke recovery as obtaining a mRS score ≤2 as “Good Recovery” vs ≥3 as “Poor Recovery” in their case/control approach. This study found only 1 SNP (rs1842681) significant at the genome-wide level (p < 5 × 10−8) located in the LOC105372028 gene. The LOC105372028 gene has no known biological function. They also found 33 suggestive SNPs among 12 different loci. When they used the mRS scores as an ordinal response instead of a binary one, the number of suggestive SNPs increased to 75 spread over 17 distinct loci without an increase in genome-wide significant SNPs.
When comparing the study by Söderholm et al. with ours, there are 2 notable distinctions. First, the set of associated genes of each study is unique. None of our associated genes replicated theirs. This fact is intriguing because each study's unique set of genes may be due to the functional recovery measures used. Our GWAS analysis used the motor drift scores from the NIHSS as a specific motor behavior marker of the participants in our discovery cohort who had the greatest weakness in the upper extremity instead of the lower, which suggests that motor drift score changes may not correlate with changes in the mRS. Unlike the motor drift score, the mRS encompasses multiple phenotypic domains such as cognition, motor strength, balance, and mortality. The mRS measure is likely to associate with genes that have general or systemic biological effects and include genes expressed in other tissues that interact with brain tissue such as cardiovascular and lymphatic tissues.
The second distinction is the clear difference in the number of participants in each study, ours n = 488 and Söderholm et al. n = 6,201. We capitalized on the repeated motor drift score measurements. The logistic regression model with GEE greatly enhanced the statistical efficiency in finding SNPs of interest associated with poststroke motor recovery. In fact, this study had about one-tenth of the minimum recommended sample size for GWAS studies.36,37 Thus, our analysis is a proof-of-principle that longitudinal observational studies can be a strong design for future stroke recovery genomic studies.
To note, the genetic loci of Söderholm et al. and ours did not include well-published candidate stroke recovery genes of APOE, BDNF, or COX-2.1,38-41 This has particular interest because one would imagine that at least one of these genes would present themselves in either our results or those of Söderholm et al. Even more so in line with Söderholm et al. because of the use of the mRS as a recovery measure like previous candidate gene studies. One possible explanation is that GWAS studies remain too underpowered to detect the effect size of known candidate genes. The effect sizes of the candidate genes may be smaller than anticipated. To address the issue of being underpowered, the stroke recovery community needs more genetic data linked to specific stroke recovery phenotypes of interest or at least capitalize on observational stroke outcome studies with longitudinal designs overlaid on relevant recovery milestones.
Unfortunately, our study does not have a replication cohort, despite searching internationally for other cohorts with NIHSS subscores and genetic data. For example, the National Institute of Neurological Disorders and Stroke archived that clinical research database has 22 publicly available stroke study data sets, but none of them have NIHSS subscores and genetic data. However, we performed a look-up analysis based on the findings by Söderholm et al. 2 as a reasonable surrogate replication cohort. Another limitation of this study is related to the VISP enrollment criteria. Participants enrolled in the VISP trial must have had a stroke due to atheroembolic mechanisms. Potential participants were excluded if their stroke was the result of a cardioembolic source. It is possible that our discovery cohort of participants had smaller vessel strokes compared with large artery and other stroke types. The biological mechanisms deployed and their effect on stroke recovery may differ among these subtypes, especially because large artery and cardioembolic strokes tend to have larger stroke lesion volumes than small vessel strokes.
Small vessel stroke may relate more to chronic inflammatory or hypertension exposures, which may explain CLDN23 as the most promising finding. Unfortunately, the VISP trial did not collect stroke subtype data such as the TOAST criteria.42 We are unable to investigate how the genetic associations may differ among stroke subtypes. Finally, it is important to note that most VISP cohort already experienced their acute stroke recovery which occurs within the first 30 days after stroke. The study participants included in our analysis reflect stroke survivors that have residual motor deficits and are mildly disabled. Our genetic associations should be interpreted in the context of chronic motor recovery and its relevant biological mechanisms, but not in the context of hyperacute and acute recovery phases. Although this limits the generalizability of our results to other phases of poststroke motor recovery, our findings may highlight important genetic mechanisms that influence neuroplasticity changes over the long term.
We demonstrated the first-ever use of repeated measurements and a domain-specific phenotype in a stroke recovery GWAS. This resulted in the discovery of new gene associations. As a proof-of-principle, this GWAS repurposed the NIHSS in a rich stroke clinical trial data set in line with the recommendations from Braun et al.10 and the Stroke Recovery and Rehabilitation Roundtable.20 This study's approach may greatly affect future genetic stroke study design. The longitudinal design allows one to investigate whether the SNP effect is associated with changes over time, which we believe is critical in stroke recovery genetics research.
As a next step, the CLDN23 gene represents a strong candidate for further research into its potential impact on stroke recovery. We recognize that our most convincing finding, the CLDN23 gene, was not replicated in an independent cohort; however, the biological role of this gene is very relevant to stroke outcomes through its expression in the blood-brain barrier and role in the immune cell migration pathway. This pathway is dynamic and a known important structure and interface in the context of CNS diseases.43 Although the biological relevance is high for poststroke recovery, it is important that these findings are replicated in an independent cohort focused on chronic motor recovery after stroke. Mendelian randomization approaches may be good intermediate steps to test causal relationships of blood-brain barrier dysfunction or disruption and poststroke recovery before initiating animal models.
Acknowledgment
We thank all the participants in the Vitamin Intervention for Stroke Prevention trial that made this study possible. The authors acknowledge Research Computing at The University of Virginia for providing computational resources and technical support that have contributed to the results reported within this publication. URL: rc.virginia.edu.
Glossary
- BDNF
brain-derived neurotrophic factor
- GEE
generalized estimating equations
- GWAS
genome-wide association studies
- IQR
interquartile range
- mRS
modified Rankin Scale
- NIHSS
NIH Stroke Scale
- PTP
protein tyrosine phosphate
- SNP
single nucleotide polymorphism
- VISP
Vitamin Intervention for Stroke Prevention
Appendix. Authors

| Name | Location | Contribution |
| Chad M. Aldridge, DPT, MS-CR | Department of Neurology, University of Virginia, Charlottesville, VA | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Robynne Braun, MD, PhD | Department of Neurology, University of Maryland, Baltimore, MD | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
| Keith L. Keene, PhD | Department of Biology and Center for Health Disparities, Brody School of Medicine, East Carolina University Greenville, NC; Center for Public Health Genomics, University of Virginia, Charlottesville, VA | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
| Fang-Chi Hsu, PhD | Department of Biostatistics and Data Science, Wake Forest University School of Medicine, Winston-Salem, NC | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Bradford B. Worrall, MD, MSc | Department of Neurology and Center for Public Health Genomics, University of Virginia, Charlottesville, VA | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design |
Study Funding
The GWAS component of the VISP study was supported by the US National Human Genome Research Institute (NHGRI), Grant U01 HG005160 (PI Michèle Sale & Bradford Worrall), as part of the Genomics and Randomized Trials Network (GARNET). Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to the Johns Hopkins University. Assistance with data cleaning was provided by the GARNET Coordinating Center (U01 HG005157; PI Bruce S Weir). Study recruitment and collection of data sets for the VISP clinical trial were supported by an investigator-initiated research grant (R01 NS34447; PI James Toole) from the US Public Health Service, NINDS, Bethesda, Maryland. Control data for comparison with European ancestry VISP stroke cases were obtained through the database of genotypes and phenotypes (dbGAP) High-Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation (phs000187.v1.p1; R01CA100264,3P50CA093459, 5P50CA097007, 5R01ES011740, 5R01CA133996, HHSN268200782096C; PIs Christopher Amos, Qingyi Wei, Jeffrey E. Lee). For VISP stroke cases of African ancestry, a subset of the Healthy Aging in Neighborhoods of Diversity across the Life Span study (HANDLS) were used as stroke free controls. HANDLS is funded by the National Institute of Aging (1Z01AG000513; PI Michele K. Evans).
Disclosure
B.B. Worrall serves as the Deputy Editor for the journal Neurology. All other authors report no relevant disclosures. Go to Neurology.org/N for full disclosures.
References
- 1.Lindgren A, Maguire J. Stroke recovery genetics. Stroke. 2016;47(9):2427-2434. doi: 10.1161/STROKEAHA.116.010648 [DOI] [PubMed] [Google Scholar]
- 2.Söderholm M, Pedersen A, Lorentzen E, et al. . Genome-wide association meta-analysis of functional outcome after ischemic stroke. Neurology. 2019;92(12):E1271-E1283. doi: 10.1212/WNL.0000000000007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mola-Caminal M, Carrera C, Soriano-Tárraga C, et al. . PATJ low frequency variants are associated with worse ischemic stroke functional outcome. Circ Res. 2019;124(1):114-120. doi: 10.1161/CIRCRESAHA.118.313533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20(8):467-484. doi: 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- 5.Campagna MP, Xavier A, Lechner-Scott J, et al. . Epigenome-wide association studies: current knowledge, strategies and recommendations. Clin Epigenetics. 2021;13(1):214. doi: 10.1186/s13148-021-01200-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fugl-Meyer AR, Jääskö L, Leyman I, Olsson S, Steglind S. The post-stroke hemiplegic patient. 1. A method for evaluation of physical performance. J Rehabil Med. 1975;7(1):13-31. doi: 10.2340/1650197771331 [DOI] [PubMed] [Google Scholar]
- 7.Siironen J, Juvela S, Kanarek K, Vilkki J, Hernesniemi J, Lappalainen J. The Met allele of the BDNF Val66Met polymorphism predicts poor outcome among survivors of aneurysmal subarachnoid hemorrhage. Stroke. 2007;38(10):2858-2860. doi: 10.1161/STROKEAHA.107.485441 [DOI] [PubMed] [Google Scholar]
- 8.Cramer SC, See J, Liu B, et al. . Genetic factors, brain atrophy, and response to rehabilitation therapy after stroke. Neurorehabil Neural Repair. 2022;36(2):131-139. doi: 10.1177/15459683211062899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwakkel G, Van Wegen EEH, Burridge JH, et al. . Standardized measurement of quality of upper limb movement after stroke: consensus-based core recommendations from the Second Stroke Recovery and Rehabilitation Roundtable. Int J Stroke. 2019;14(8):783-791. doi: 10.1177/1747493019873519 [DOI] [PubMed] [Google Scholar]
- 10.Braun RG, Heitsch L, Cole JW, et al. . Domain-specific outcomes for stroke clinical trials: what the modified rankin isn't ranking. Neurology. 2021;97(8):367-377. doi: 10.1212/WNL.0000000000012231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toole JF, Malinow MR, Chambless LE, et al. . Lowering homocysteine in patients with ischemic stroke to prevent recurrent stroke, myocardial infarction, and death: the Vitamin Intervention for Stroke Prevention (VISP) randomized controlled trial. JAMA. 2004;291(5):565-575. doi: 10.1001/jama.291.5.565 [DOI] [PubMed] [Google Scholar]
- 12.International HapMap 3 Consortium, Altshuler DM, Gibbs RA, et al. . Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52-58. doi: 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taliun D, Harris DN, Kessler MD, et al. . Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290-299. doi: 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Forer L, Schönherr S, et al. . Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31(5):782-784. doi: 10.1093/bioinformatics/btu704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey V, Lumley T, Ripley B. gee: Generalized Estimation Equation Solver. R package version 4.13-19. R Foundation for Statistical Computing; 2015. [Google Scholar]
- 17.Zeger SL, Liang K-Y. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121-130. doi: 10.2307/2531248 [DOI] [PubMed] [Google Scholar]
- 18.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867-2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venables WN, Ripley BD. Modern Applied Statistics with S [online]. Springer; 2002. Accessed January 20, 2022. stats.ox.ac.uk/pub/MASS4/ [Google Scholar]
- 20.Bernhardt J, Hayward KS, Kwakkel G, et al. . Agreed definitions and a shared vision for new standards in stroke recovery research: the Stroke Recovery and Rehabilitation Roundtable Taskforce. Neurorehabil Neural Repair. 2017;31(9):793-799. doi: 10.1177/1545968317732668 [DOI] [PubMed] [Google Scholar]
- 21.McLaren W, Gil L, Hunt SE, et al. . The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh M, Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med. 2003;11(6):683-689. doi: 10.3892/ijmm.11.6.683 [DOI] [PubMed] [Google Scholar]
- 23.PathCards. Accessed November 15, 2022. pathcards.genecards.org/
- 24.Sinnott-Armstrong N, Tanigawa Y, Amar D, et al. . Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53(2):185-194. doi: 10.1038/s41588-020-00757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson TG, Leyden GM, Wang Q, et al. . Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022;20(2):e3001547. doi: 10.1371/journal.pbio.3001547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adewuyi EO, Mehta D, International Endogene Consortium IEC, 23andMe Research Team, Nyholt DR. Genetic overlap analysis of endometriosis and asthma identifies shared loci implicating sex hormones and thyroid signalling pathways. Hum Reprod. 2022;37(2):366-383. doi: 10.1093/humrep/deab254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pulido R, Krueger NX, Serra-Pagès C, Saito H, Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem. 1995;270(12):6722-6728. doi: 10.1074/jbc.270.12.6722 [DOI] [PubMed] [Google Scholar]
- 28.Mizuno K, Hasegawa K, Katagiri T, Ogimoto M, Ichikawa T, Yakura H. MPTP delta, a putative murine homolog of HPTP delta, is expressed in specialized regions of the brain and in the B-cell lineage. Mol Cell Biol. 1993;13(9):5513-5523. doi: 10.1128/mcb.13.9.5513-5523.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Diaz AK, Paugh BS, et al. . The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444-450. doi: 10.1038/ng.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butola T, Alvanos T, Hintze A, et al. . RIM-binding protein 2 Organizes Ca2+ channel topography and Regulates release probability and vesicle Replenishment at a Fast central synapse. J Neurosci. 2021;41(37):7742-7767. doi: 10.1523/JNEUROSCI.0586-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. doi: 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kheradmand Kia S, Verbeek E, Engelen E, et al. . RTTN mutations link primary cilia function to organization of the human cerebral cortex. Am J Hum Genet. 2012;91(3):533-540. doi: 10.1016/j.ajhg.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavallin M, Bery A, Maillard C, et al. . Recurrent RTTN mutation leading to severe microcephaly, polymicrogyria and growth restriction. Eur J Med Genet. 2018;61(12):755-758. doi: 10.1016/j.ejmg.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Stouffs K, Moortgat S, Vanderhasselt T, et al. . Biallelic mutations in RTTN are associated with microcephaly, short stature and a wide range of brain malformations. Eur J Med Genet. 2018;61(12):733-737. doi: 10.1016/j.ejmg.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 35.Walker DG, Whetzel AM, Lue LF. Expression of suppressor of cytokine signaling genes in human elderly and Alzheimer's disease brains and human microglia. Neuroscience. 2015;302:121-137. doi: 10.1016/j.neuroscience.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziyatdinov A, Kim J, Prokopenko D, et al. . Estimating the effective sample size in association studies of quantitative traits. G3 (Bethesda). 2021;11(6):jkab057. doi: 10.1093/g3journal/jkab057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore CM, Jacobson SA, Fingerlin TE. Power and Sample Size Calculations for Genetic Association Studies in the Presence of Genetic Model Misspecification; 2020. Accessed April 11, 2022. karger.com/hhe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DY, Quinlan EB, Gramer R, Cramer SC. BDNF Val 66 Met Polymorphism Is Related to Motor System Function After Stroke [online]; 2016. Accessed November 18, 2021. academic.oup.com/ptj/article/96/4/533/2686517 [DOI] [PMC free article] [PubMed]
- 39.Kim JM, Stewart R, Park MS, et al. . Associations of BDNF genotype and promoter methylation with acute and long-term stroke outcomes in an East Asian cohort. PLoS ONE 2012;7(12):e51280. doi: 10.1371/journal.pone.0051280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan A, Yan J, Csurhes P, Greer J, McCombe P. Circulating brain derived neurotrophic factor (BDNF) and frequency of BDNF positive T cells in peripheral blood in human ischemic stroke: effect on outcome. J Neuroimmunol. 2015;286:42-47. doi: 10.1016/j.jneuroim.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 41.Gómez-Pinilla F, Ying Z, Roy RR, Molteni R, Edgerton VR. Voluntary exercise induces a BDNF-mediated mechanism that promotes neuroplasticity. J Neurophysiol. 2002;88(5):2187-2195. doi: 10.1152/jn.00152.2002 [DOI] [PubMed] [Google Scholar]
- 42.Adams HP, Bendixen BH, Kappelle LJ, et al. . Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35-41. doi: 10.1161/01.str.24.1.35 [DOI] [PubMed] [Google Scholar]
- 43.Erickson MA, Dohi K, Banks WA. Neuroinflammation: a common pathway in CNS diseases as mediated at the blood-brain barrier. Neuroimmunomodulation. 2012;19(2):121-130. doi: 10.1159/000330247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The clinical VISP data are available at the NIHND Archived Clinical Research Datasets website (ninds.nih.gov/current-research/research-funded-ninds/clinical-research/archived-clinical-research-datasets). Raw genotype data for VISP are available in dbGaP (Accession phs000343.v4.p1). The genetic association results generated by this study can be found in the Cerebrovascular Disease Knowledge Portal.