Abstract
Common purslane (Portulaca oleracea) integrates both C4 and crassulacean acid metabolism (CAM) photosynthesis pathways and is a promising model plant to explore C4-CAM plasticity. Here, we report a high-quality chromosome-level genome of nicotinamide adenine dinucleotide (NAD)-malic enzyme (ME) subtype common purslane that provides evidence for 2 rounds of whole-genome duplication (WGD) with an ancient WGD (P-β) in the common ancestor to Portulacaceae and Cactaceae around 66.30 million years ago (Mya) and another (Po-α) specific to common purslane lineage around 7.74 Mya. A larger number of gene copies encoding key enzymes/transporters involved in C4 and CAM pathways were detected in common purslane than in related species. Phylogeny, conserved functional site, and collinearity analyses revealed that the Po-α WGD produced the phosphoenolpyruvate carboxylase-encoded gene copies used for photosynthesis in common purslane, while the P-β WGD event produced 2 ancestral genes of functionally differentiated (C4- and CAM-specific) beta carbonic anhydrases involved in the C4 + CAM pathways. Additionally, cis-element enrichment analysis in the promoters showed that CAM-specific genes have recruited both evening and midnight circadian elements as well as the Abscisic acid (ABA)-independent regulatory module mediated by ethylene-response factor cis-elements. Overall, this study provides insights into the origin and evolutionary process of C4 and CAM pathways in common purslane, as well as potential targets for engineering crops by integrating C4 or CAM metabolism.
Common purslane has undergone 2 rounds of whole-genome duplication (WGD), and the ancient P-β WGD produced the ancestral genes of functional C4- and crassulacean acid metabolism (CAM)-specific gene copies involved in C4 + CAM pathways.
Introduction
Common purslane (Portulaca oleracea), a member of the Portulacaceae family in the Caryophyllales, is an annual herb that is widely dispersed over the world. Based on numerous morphological and chromosome number variations, it has been described as either a polymorphic species or a complex of subspecies (Ocampo and Columbus 2012; Rice et al. 2015; Walter et al. 2015; Ferrari et al. 2020b). Common purslane is a medicinal and edible plant, is rich in nutrients, and accumulates carotene, vitamin C, vitamin E, and ω-3 unsaturated fatty acids (Omara-Alwala et al. 1991; Simopoulos et al. 1992; Uddin et al. 2014). Notably, common purslane can tolerate extremely high temperatures combined with drought, high humidity, high salt, and low nutrient levels (Zimmerman 1976; Yang et al. 2012; D'Andrea et al. 2014). Together, these characteristics make P. oleracea an attractive model for both basic and applied research.
Common purslane has developed multiple resistance strategies to abiotic stresses (Lara et al. 2004; Jin et al. 2016; Habibi 2020). One such example is its photosynthetic pathway that is responsive to drought—although it is a canonical C4 plant, common purslane performs facultative crassulacean acid metabolism (CAM) photosynthesis under drought conditions (Koch and Kennedy 1980; Lara et al. 2004; D'Andrea et al. 2014). Such an integrated C4-CAM system has been described in another purslane, Paraguayan purslane (P. amilis) (Gilman et al. 2022), but to our knowledge, this integration is rare in land plants. C4 and CAM both facilitate high-efficiency photosynthesis by concentrating CO2 near Rubisco (Hatch 1987; Edwards and Ogburn 2012) to generate so-called CO2-concentrating mechanisms (CCMs). These 2 metabolisms share a series of biochemical reactions and enzymes (Ferrari and Freschi 2019). CO2 is incorporated into oxaloacetate by the concerted action of beta carbonic anhydrase (β-CA) and then phosphoenolpyruvate carboxylase (PEPC) during initial carboxylation, followed by its release from malate or aspartate via nicotinamide adenine dinucleotide phosphate (NAD(P))-malic enzyme (ME)–mediated decarboxylation before entering the Calvin cycle. The major difference between the C4 and CAM pathway is the spatial separation of carboxylation and decarboxylation associated with the C4 pathway—carboxylation takes place in mesophyll cells (MCs) and decarboxylation in bundle sheath cells—while in CAM, these 2 processes are separated temporally in MCs.
Recently, Moreno-Villena et al. demonstrated that CAM and C4 carbon fixation occur in the same cells and that CAM-generated metabolites are likely directly incorporated into the C4 cycle in common purslane (Hibberd 2022; Moreno-Villena et al. 2022). Based on phylogenetic analysis of PEPC, a key enzyme for C4 and CAM metabolism in the genus Portulaca, PEPC1E1 (ppc-1E1) genes were proposed to be recruited into both C4 and CAM photosynthesis in the Caryophyllales (Christin et al. 2014; Goolsby et al. 2018; Moore et al. 2018). Subsequently, several studies found that PEPC is encoded by many gene copies in Portulaca, and although it has not been possible to test the importance of all PEPC gene copies for photosynthesis, it was proposed that one copy (PPC-1E1a′) is specific for C4 and another (PPC-1E1c) for CAM in both common purslane and Paraguayan purslane (Ferrari et al. 2019; Gilman et al. 2022; Moreno-Villena et al. 2022). The PPC-1E1a′ copy is highly expressed during the daytime in well-watered plants, while the PPC-1E1c copy displays the highest expression during the day-night transition in plants under drought stress (Christin et al. 2014; Ferrari et al. 2019; Gilman et al. 2022; Moreno-Villena et al. 2022). However, it is still unknown whether and how duplication events are associated with these C4- and CAM-specific PEPC gene copies as well as others encoding essential enzymes or transporters used in C4 and CAM.
The origin of C4 and CAM in Portulaca has not been comprehensively studied. Facultative CAM is assumed to be ancestral in Portulaca because it has been observed in every major subclade as well as Portulaca's closest relatives (Anacampserotaceae and Cactaceae) (Sage 2002; Christin et al. 2014; Gilman et al. 2022). This hypothesis was supported from an analysis of the evolutionary history of PEPC genes, with CAM-specific copies of Portulaca being similar to CAM forms of other species in Cactaceae (Christin et al. 2014). However, this approach has not been possible to take for other genes important for C4 and CAM such as β-CA. A high-quality genome assembly of Portulaca species along with published genomes of Portulaca's closest relatives (e.g. dragon fruit [Hylocereus undatus] in Cactaceae) would allow us to comprehensively explore mechanisms, allowing integration of the C4 and CAM pathways. A de novo genome assembly for Paraguayan purslane from Illumina short reads was reported recently. It indeed provided a scaffold level genome and revealed evidence of coexpression networks supporting C4-CAM compatibility (Gilman et al. 2022). However, the origin process of C4 and CAM and the importance of whole-genome duplication (WGD) events in the evolutionary history of the Portulacaceae remain to be explored.
Here, we report a high-quality chromosome-level genome assembly for common purslane, which revealed 2 rounds of WGD: the ancient WGD (hereafter abbreviated as the P-β) in common purslane is shared with dragon fruit, while an independent WGD (hereafter abbreviated as the Po-α) occurred in common purslane. These WGD events and tandem duplication (TD) events produced multiple copies of genes encoding key enzymes/transporters of C4 photosynthesis and CAM. We identified 4 C4-specific PEPC genes from Po-α WGD/TD and 2 CAM-specific PEPC genes from Po-α WGD based on conserved functional sites and diurnal expression patterns. Importantly, we found that the P-β WGD event produced 2 ancestral genes of functionally differentiated (C4- and CAM-specific) β-CA genes involved in C4 + CAM pathways by phylogenetic trees and evolutionary history. Motif enrichment analysis in promoters of CAM-specific genes showed enriched motif clusters but little overlap with night or drought-specific promoters, suggesting facultative CAM results from the recruitment of complex and independent regulatory networks. Thus, this study not only provides further evidence of C4-CAM compatibility in one leaf but also sheds light on the origin and evolution of C4 and CAM in Portulaca and the integration of CAM and/or C4 metabolism, which is important for biotechnological applications of these pathways.
Results
Genome assembly and annotation of common purslane
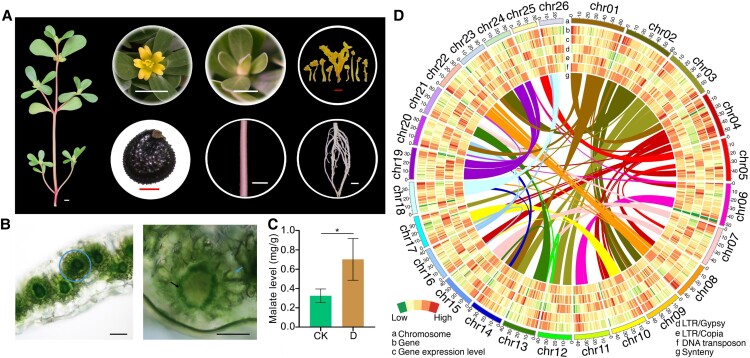
We selected a wild purslane plant growing in the field (Beijing, China) for genome sequencing and assembly. Using plant taxonomy indicators, we confirmed that this wild purslane was the common purslane (Fig. 1A). It had typical Kranz anatomy (Fig. 1B), corresponding to C4 photosynthesis. Increased level of malate accumulation was also observed at night under drought conditions (Fig. 1C), which indicates nocturnal CO2 fixation occur. Overall, these characters demonstrated that common purslane we used also performs both C4 and CAM photosynthesis pathways as other Portulaca species (Voznesenskaya et al. 2010, 2017; Gilman et al. 2022).
Figure 1.
Overview of the common purslane genome assembly and features. A) Morphology of the common purslane seedling, flowers, separated pistil and stamen, mature seeds, stem, and root. Images were digitally extracted for comparison. White scale bars in flowers, leaf, stem, and root: 0.5 cm; red scale bars in separated pistil and stamen, and mature seeds: 0.2 mm. B) Handmade leaf slice showing the C4 Kranz anatomy of common purslane leaves. Blue circle shows typical Kranz leaf anatomy. Black and blue arrows show bundle sheath and MCs, respectively. Scale bars, 0.1 mm. C) Malate accumulation in common purslane leaves under normal or drought conditions at night. Data are means ± Se of 3 biological replicates. *P < 0.05 by t-test. D) Overview of the common purslane genome assembly. Track a, 26 assembled chromosomes; Tracks b to f represent the other genomic features as indicated at the lower left of the circle plot. The colors indicate the density of genomic features in 1-Mb sliding window along chromosomes. Track g shows syntenic blocks. Band width is proportional to the size of the syntenic block. CK, control; D, drought.
To examine the karyotype of this common purslane, we utilized 2-color fluorescence in situ hybridization (FISH) with 5S rDNA and 45S rDNA as probes and detected 52 chromosomes (Supplemental Fig. S1). This result was consistent with the analysis of wild common purslane widely grown in China but different from the common purslane found in other countries used in previous studies (Ocampo and Columbus 2012; Walter et al. 2015). We estimated genome size to 1,122 Mb based on k-mer analysis of Illumina short reads (Supplemental Fig. S2), which agreed with the estimation of 1,137 Mb obtained by flow cytometry (Supplemental Fig. S2). The sequenced common purslane individual exhibited low heterozygosity, with overall genome heterozygosity of 0.059% (Supplemental Fig. S2).
We built a de novo assembly of the common purslane genome using 4 different sequencing technologies (Supplemental Fig. S3). The first draft assembly consisted of 201 contigs, spanning 1,119 Mb, derived from 80× coverage with Nanopore long reads (read N50 = 29 kb; contig N50 = 18.23 Mb; Table 1). We then integrated optical mapping data, yielding an assembly of 1,134 Mb with 101 scaffolds (N50 of 35.10 Mb), covering 99.74% of the estimated genome size based on flow cytometry and k-mer analyses (Supplemental Table S1). Finally, we ordered and oriented the scaffolds into 26 pseudochromosomes using High-resolution chromosome conformation capture (Hi-C) data (Fig. 1D and Supplemental Fig. S4). Notably, the longest contig spanned the entire length of chromosome 16. We assessed the completeness and quality of the genome assembly with several strategies. First, we generated transcriptome deep-sequencing (RNA-seq) data from 7 different tissues and resequenced the common purslane genome via Illumina short reads. We then mapped the RNA-seq, DNA-seq data, and Nanopore long reads onto the final assembly with mapping rates of 96.47%, 99.39%, and 99.44%, respectively (Supplemental Table S2). Second, the complete benchmarking universal single copy orthologs (BUSCOs) and consensus quality value (QV) of genome assembly were 96.8% (Supplemental Table S2) and 32.4, respectively. Third, we estimated the long terminal repeat (LTR) assembly index (LAI) of the common purslane genome to be 17.96, indicating that it shows the qualities of a “reference” genome (10 < LAI < 20) (Supplemental Fig. S5). Finally, we calculated the genome coverage using Nanopore long reads and Hi-C joins, yielding results of 98.1% and 99.7%, respectively. Overall, we obtained a high-quality genome in terms of genome completeness, continuity, and accuracy.
Table 1.
Summary statistics of the common purslane genome assembly and annotation
| Genomic features | Values |
|---|---|
| Estimated genome size | 1,137 Mb |
| Assembled genome size | 1,134 Mb |
| No. of contigs | 201 |
| N50 length of contigs | 18.23 Mb |
| No. of scaffolds | 101 |
| N50 length of scaffolds | 35.10 Mb |
| No. of pseudochromosomes | 26 |
| Length of sequence assigned to chromosomes | 1,131 Mb |
| Percentage of sequence assigned to chromosomes | 99.70% |
| Percentage of repeat sequences | 67.64% |
| No. of annotated high-confidence genes | 45,250 |
| Complete BUSCOs (embryophyta_odb10) | 98.00% |
| Percentage of gene with functional annotation | 91.90% |
Using a combination of plant homology searches, transcriptome-based predictions, and ab initio gene predictions, we identified 45,250 high-confidence, protein-coding genes, corresponding to a BUSCO score of 98.00% for complete genes (single-copy and duplicated; Supplemental Table S3). We obtained potential functional annotation information for 41,585 (91.90%) genes using the EuKaryotic Orthologous Groups, Kyoto Encyclopedia of Genes and Genomes, Nonredundant Protein Sequence Database, SwissProt, and Gene Ontology (GO) databases (Supplemental Table S4). Transposons made up 63.70% of the common purslane genome sequence, with LTR retrotransposons being the largest family, accounting for about 76.61% of all transposable elements (TEs) and representing 48.80% of the genome assembly (Supplemental Table S5). Among LTR retrotransposons, Ty3/Gypsy elements represented 23.71% of the genome and were much more numerous than Ty1/Copia elements, which covered only 3.94% of the genome (Supplemental Table S5). We observed a distinct unimodal distribution for the insertion times of intact LTR/Gypsy in the common purslane genome, with a peak of amplification around 0.5 million years ago (Mya), while the LTR/Copia showed a burst about 0.8 Mya (Supplemental Fig. S6). Like other plant genomes, the common purslane genome also exhibited high Ty3/Gypsy density in regions with low protein-coding gene density (Fig. 1D).
Whole-genome duplications
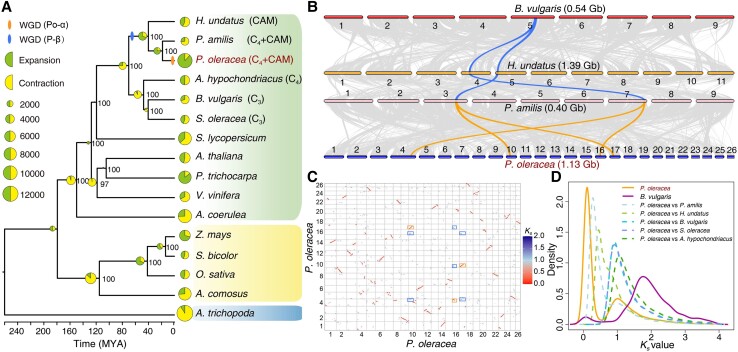
To investigate the evolution of the common purslane genome, we included another 15 genomes from major angiosperm clades, comprising 10 eudicots, 4 monocots, and Amborella as an outgroup, for comparative genomic analyses. We utilized a set of 47 single-copy gene families from these 16 species to construct a phylogenetic tree (Fig. 2A). The resulting topology was consistent with that of the Angiosperm Phylogeny Group IV (Byng et al. 2016). We observed that the Portulacaceae (P. oleracea and P. amilis) cluster with Cactaceae (dragon fruit [H. undatus]) among the Caryophyllales. Further phylogenetic dating analysis indicated that P. oleracea (Portulacaceae) diverged from H. undatus (Cactaceae) around 40 Mya, while the clade containing P. oleracea and H. undatus diverged from the Amaranthaceae (spinach [Spinacia oleracea], sugar beet [Beta vulgaris], and amaranth [Amaranthus hypochondriacus]) around 70 Mya (Fig. 2A).
Figure 2.
Evolutionary analysis of the common purslane genome. A) Phylogenetic species tree constructed based on single-copy putative orthologs. The lineage divergence times (Mya) and gene family expansion and contraction are shown. The divergence times were estimated by r8s (v1.81). The pie charts at each branch of the tree represent the proportion of gene families undergoing gain or loss events. The size of the pie charts is proportional to the number of gene families expanded (green) or contracted (yellow). The numbers at each branch represent the percentage support. B) Macrosynteny comparisons among B. vulgaris (0.54 Gb), H. undatus (1.39 Gb), P. amilis (0.40 Gb), and P. oleracea (1.13 Gb) revealing a 1:2:2:4 ratio, the region highlighted in orange provides 1 example. C) Syntenic dot plot of P. oleracea against itself. Syntenic gene pairs were colored as a function of the synonymous substitution rate (Ks) values (Ks > 1, blue; Ks < 1, red). The recent and relatively ancient WGDs are highlighted by the orange and blue boxes, respectively. D)Ks distribution in the identified syntenic putatively paralogous blocks from P. oleracea and B. vulgaris (solid lines), and P. oleracea putative orthologs with 5 other species (dashed lines).
The high percentage of complete duplicated genes in the P. oleracea genome, as indicated by BUSCO analysis, suggested widespread duplication events. We therefore explored the evolutionary history of WGD events in the P. oleracea lineage by combining evidence from synonymous substitution rate (Ks) and synteny analyses (Fig. 2, B to D). The most recent WGD event in B. vulgaris is the ancestral gamma hexaploidization (γ-WGD) shared by core eudicots (Dohm et al. 2013; Xu et al. 2017); we thus used the B. vulgaris genome as a reference to identify duplication events in the Caryophyllales. Syntenic analyses detected a clear 1:3 colinearity relationship when we compared the P. oleracea genome with itself (Fig. 2C), suggesting that 2 WGDs occurred in the evolutionary history of P. oleracea. Dotplots of syntenic gene pairs identified a 4:1 syntenic ratio between the P. oleracea and B. vulgaris genomes, a 4:2 syntenic ratio between P. oleracea and H. undatus, and a 4:2 syntenic ratio between P. oleracea and P. amilis (Supplemental Fig. S7). Macrosynteny patterns between genomic regions from B. vulgaris, H. undatus, P. amilis, and P. oleracea clearly showed a 1:2:2:4 ratio, supporting 2 rounds of WGD in P. oleracea after its divergence from Amaranthaceae and 1 WGD in the H. undatus lineage (Fig. 2B). To validate this WGD history, we plotted the Ks distribution of anchor gene pairs from intra- and intergenomic syntenic blocks. We observed 2 distinct Ks peaks in the P. oleracea genome (Fig. 2D), consistent with a genome history with 2 WGD events. Given the Ks peak of putatively orthologous gene pairs between P. oleracea and P. amilis (Ks = 0.28) and H. undatus (Ks = 0.45), we hypothesized that the P-β WGD in P. oleracea was shared with H. undatus, while the Po-α WGD occurred in P. oleracea. Together with evidence from synteny and Ks analyses, our results strongly suggested the P-β WGD be in the common ancestor to Portulacaceae and Cactaceae, which would be consistent with previous study (Yang et al. 2018; Wang et al. 2019), and the Po-α WGD in the P. oleracea lineage occurred after its divergence from P. amilis. Finally, using a synonymous substitution rate per site per year of 7.54 × 10−9 for Caryophyllales, we estimated that the 2 rounds of WGD occurred around 66.30 and 7.47 Mya, respectively (Fig. 2A).
More gene copies encoding key enzymes/transporters for C4 and CAM probably laid the foundation for the coexistence of 2 CCMs in one leaf in P. oleracea
Recently, the expression patterns of P. oleracea genes encoding some key enzymes involved in C4 or CAM metabolism were explored via RNA-seq (Christin et al. 2014; Ferrari et al. 2019; Gilman et al. 2022; Moreno-Villena et al. 2022). Yet, such analysis was still not complete due to the lack of the genome of P. oleracea, and thus, the investigation to the mechanisms of the integrated C4 and CAM photosynthesis pathways was highly limited. Here, we systematically identified the P. oleracea homologs of genes encoding key enzymes/transporters of C4 and CAM based on their functional annotation. Notably, most of these genes had a much higher gene copy number in P. oleracea than that in other species (Supplemental Table S6). Clustering and phylogenetic analyses showed that WGDs contributed much to the increased gene copy numbers, such as the genes encoding aspartate transaminase (AspAT) (all the 10 genes were from WGD), phosphate dikinase (PPDK) (all the 8 genes were from WGD), and β-carbonic anhydrase (β-CA) (9 of the 11 genes were from WGD) (Supplemental Fig. S8). We then separated these genes into day- or night-phased genes based on their expression pattern in normal and stress conditions, as well as their preferential expression in leaves resulting in a list of 29 C4-related and 9 CAM-related genes (Supplemental Figs. S9 and S10). Among the 29 C4-related genes, we noticed genes encoding 12 key enzymes or transporters, including β-CA, NAD(P)-ME, PEPC, and AspAT. These gene copies were highly expressed during the day under normal conditions but were downregulated under stress (Supplemental Fig. S9). Similarly, the list of CAM-related copies exhibits higher expression at night under stress. The 9 CAM-related copies encoded 2 β-CAs, 1 PEPC-K, 2 CAM-PEPCs, 2 aluminum-activated malate transporters, 1 PPDK, and 1 PPDK-RE. In contrast to most of the single-copy genes previously identified based on transcriptome data, those newly identified photosynthesis-related gene copies might shed comprehensive light onto future bioengineering that achieves CAM-to-C4 progression.
Analysis of the origin and evolution of PEPC gene copies specific for CAM and C4 pathways at the genome-wide scale
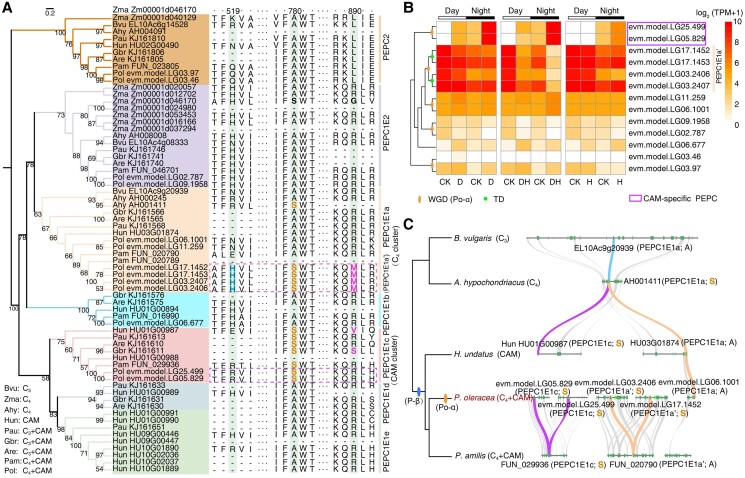
PEPC, as an essential enzyme, can catalyze the fixation of HCO3− to the receptor phosphoenolpyruvate resulting in the formation of oxaloacetate during the process of CO2 fixation. Prior studies revealed one key CAM-specific PEPC isoform, which was upregulated under drought at night, and one C4-specific PEPC, which exhibited high expression level during daytime under well-watered condition (Christin et al. 2014; Ferrari et al. 2020b). However, a general atlas of all PEPC gene copies in P. oleracea has not been explored. Taking advantage of the whole-genome gene information, we first performed a phylogenetic analysis on PEPC proteins encoded by the P. oleracea (C4 + CAM), P. amilis (C4 + CAM) (Gilman et al. 2022), H. undatus (CAM) (Chen et al. 2021; Zheng et al. 2021), B. vulgaris (C3) (Dohm et al. 2013), and A. hypochondriacus (C4) (Lightfoot et al. 2017) genomes as well as several others (C3 + CAM) that have been reported in the Caryophyllales (Christin et al. 2014) (Fig. 3A and Supplemental Table S7). Of the 13 PEPC proteins in P. oleracea, 2 were grouped into the PEPC2 cluster, another 2 into PEPC1E2, and 9 into PEPC1E1; this latter cluster consists of 6 PEPC1E1a (putative C4-specific subcluster), 1 PEPC1E1b, and 2 PEPC1E1c (putative CAM-specific subcluster) proteins. After analyzing the expression level of each PEPC gene, we obtained 4 C4-specific PoPEPC1E1a′ genes with high expression level during the day regardless of growth conditions and 2 CAM-specific PoPEPC1E1c genes being highly expressed under stress conditions at night (Fig. 3B). Strikingly, the expression levels of the 2 CAM-specific genes were hundreds to thousands of times higher under stress at night than in the control samples or stress samples during the day (Supplemental Fig. S11A) and was demonstrated by reverse transcription quantitative PCR (RT-qPCR) (Supplemental Figs. S11B and S12).
Figure 3.
Analysis of C4- and CAM-specific PEPC genes in the common purslane genome. A) Phylogenetic tree of PEPC proteins from P. oleracea (Pol, C4 + CAM), P. amilis (Pam, C4 + CAM), H. undatus (Hun, CAM), A. hypochondriacus (Ahy, C4), B. vulgaris (Bvu, C3), Z. mays (Zma, C4), and several identified PEPC proteins from Pereskia aureiflora (Pau, C3 + CAM), Anacampseros retusa (Are, C3 + CAM), and Grahamia bracteata (Gbr, C3 + CAM) in Caryophyllales. Only bootstrap values greater than 50% were shown. Branches were colored according to the PEPC subclass. A multiple sequence alignment of partially conserved amino acids was shown to the right. Sites at positions of 519 (known as position of D509 in the Kalanchoë genome), 780, and 890 (according to the numbering in Zm00001d046170) were highlighted in light-green background. Potentially functional C4- and CAM-specific PEPC proteins were highlighted in magenta and purple boxes, respectively. B) Gene expression of PEPC genes identified in P. oleracea under heat and drought treatments during the day and night. Orange ovals in the gene trees to the left represent WGD Po-α events; green dots represent TD events. CAM-specific PEPC genes were highlighted in magenta boxes. C) Synteny analysis of C4- and CAM-specific PEPC genes in the P. oleracea, P. amilis, H. undatus, A. hypochondriacus, and B. vulgaris genomes in Caryophyllales. CK, control group; D, drought group; H, heat group; DH, drought combined heat group.
According to previous studies, C4/CAM-specific PEPC proteins that are functional or exhibit high efficiency during photosynthesis generally harbor a serine (S) at residue 780 (using the amino acid numbering of the protein encoded by Zm00001d046170 in maize [Zea mays]), while other PEPC proteins carry a conserved alanine (A) at the equivalent position (Svensson et al. 2003; Christin et al. 2014). Although the above observation was not absolute (Rao et al. 2008; Christin et al. 2014), we determined that it likely holds true in P. oleracea (Fig. 3A). Indeed, protein sequence alignments with other PEPCs from classical C4 and CAM species highlighted 4 PEPC1E1a proteins in P. oleracea (PoPEPC1E1a) with an S residue at position 780 (classified as PoPEPC1E1a′; the other 2 had an A at this position), as well as 2 PoPEPC1E1c enzymes with S at position 780 (Fig. 3A). Notably, the other reported C4 + CAM plant P. amilis had only one copy each for the PEPC1E1a and PEPC1E1c subclusters, of which only PaPEPC1E1c carried the S residue. We also examined residue 890 (according to the numbering in Zm00001d046170), where an arginine (R) in PEPC was shown to support tight inhibitor binding in C3 plants, while C4-related PEPCs have a glycine (G) or other residues with lower inhibitor affinity (Paulus et al. 2013). All 4 candidate C4-specific PoPEPC1E1a′ proteins possessed a methionine (M) instead of an R at this position, which may be responsible for the high efficiency of photosynthetic carbon fixation in P. oleracea. By contrast, position 890 in the only PEPC1E1a of P. amilis harbored an R, like most C3 plants. We also detected residue 519 (according to the numbering in Zm00001d046170) known as position of D509; an aspartic acid (D) residue was demonstrated to reduce malate inhibition in the Kalanchoë genome (Yang et al. 2017). Histidine (H) and glutamic acid (E) were present at this position in PEPC1E1a′ in the P. oleracea and P. amilis genomes respectively, which was consistent with the result in the paper of the P. amilis genome (Gilman et al. 2022). Overall, these results provided further evidence at the molecular level that P. oleracea is a C4 and CAM plant and that the 4 C4-specific PoPEPC1E1a′ and 2 PoPEPC1E1c genes we identified here may be keys to its C4 and CAM photosynthesis.
In addition, to explore the origin and evolution of PEPC genes in P. oleracea, we compared the genomes among P. oleracea (C4 + CAM), P. amilis (C4 + CAM), H. undatus (CAM), A. hypochondriacus (C4), and B. vulgaris (C3) in the Caryophyllales. Synteny analyses of PEPC1E1a and PEPC1E1c genes in these species suggested that the P-β WGD event may produce the ancestral copies of C4 and CAM-specific PEPC genes (Fig. 3C). Combing with the information of phylogenetic trees and collinearity evidence (Fig. 3, A and C), we speculated that the ancestral PEPC1E1a’ copies in P. oleracea genome might be duplicated from the PEPC1E1a copy, and then 4 copies were generated by the recent Po-α WGD and TD events (Fig. 3, A and B). By contrast, the ancestral copy of 2 PEPC1E1c copies probably came from the P-β WGD event, followed by duplication after the Po-α WGD (Fig. 3). Overall, P-β WGD, Po-α WGD, and TD events contributed to the occurrence of multicopies of C4- and CAM-specific PEPC genes in P. oleracea.
Gene duplications produced functionally differentiated β-CA genes involved in C4 + CAM pathways
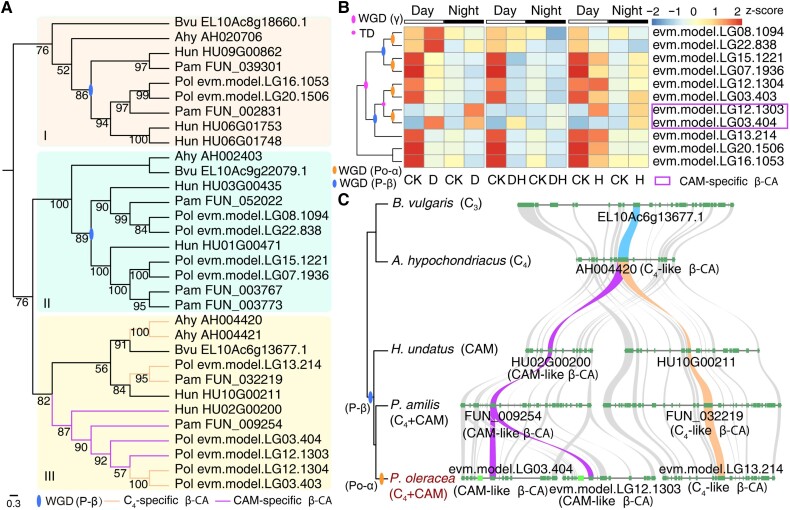
In both CCMs pathways, CO2 is converted to HCO3− by a key enzyme β-CA, which has been identified and classified into C4- and CAM-specific β-CA based on transcriptome data in P. oleracea (Ferrari et al. 2019). To explore the origin and evolutionary process of C4 and CAM pathway in Portulaca more comprehensively and accurately, we analyzed evolution of β-CA genes coding first-step key enzymes in C4 and CAM pathways in the genomes of P. oleracea (C4 + CAM), P. amilis (C4 + CAM), H. undatus (CAM), A. hypochondriacus (C4), and B. vulgaris (C3) in the Caryophyllales. Firstly, we identified 32 β-CA genes in the above 5 species including 11 in P. oleracea, 7 in P. amilis, 7 in H. undatus, 4 in A. hypochondriacus, and 3 in B. vulgaris (Fig. 4A). By constructing phylogenetic trees, we have divided these genes into 3 categories, of which clade III contained C4- (FUN_032219) and CAM-specific (FUN_009254) genes in P. amilis (Gilman et al. 2022) and the corresponding 3 C4-specific and 2 CAM-specific β-CA gene copies in P. oleracea (Fig. 4B). Synteny analyses of these genes indicated that P-β and Po-α WGD events contribute to the occurrence of multicopies of β-CA genes in P. oleracea (Fig. 4, B and C). Furthermore, collinearity analysis of the chromosomal segment where C4- and CAM-specific genes located showed that P-β WGD event produces C4-like cluster (HU10G00211 in H. undatus, FUN_032219 in P. amilis and evm.model.LG13.214 in P. oleracea) and CAM-like cluster (HU02G00200 in H. undatus, FUN_009254 in P. amilis and evm.model.LG03.404 and evm.model.LG12.1303 in P. oleracea), which indicated that P-β WGD event resulted in the origin of 2 types of β-CA genes (Fig. 4C). Interestingly, TD events prior to Po-α WGD event in P. oleracea genome led to repeated evolution of C4-specific β-CA genes, while Po-α WGD event occurring independently in P. oleracea may promote the photosynthetic efficiency by increasing the gene expression dosage (Fig. 4, B and C). Similar to the evolutionary pattern of PEPC gene, but more clearly, duplication preceded functional evolution: P-β WGD event produced 2 ancestral genes of functionally differentiated (C4- and CAM-specific) β-CA genes involved in C4 + CAM pathways.
Figure 4.
Origin and evolution of C4- and CAM-specific β-CA genes in Caryophyllales. A) Phylogenetic tree of β-CA genes from P. oleracea (Pol), P. amilis (Pam), H. undatus (Hun), A. hypochondriacus (Ahy), and B. vulgaris (Bvu) genomes in Caryophyllales. B) Gene expression of β-CA genes identified in P. oleracea under heat and drought treatments during the day and night. Magenta, blue, and orange ovals in the gene trees to the left represent γ, P-β, and Po-α WGD events, respectively; magenta dots represent TD events. CAM-specific β-CA genes were highlighted in magenta boxes. C) Synteny analysis of C4- and CAM-specific β-CA genes (clade III in Fig. 4A) in the P. oleracea, P. amilis, H. undatus, A. hypochondriacus, and B. vulgaris genomes in Caryophyllales. CK, control group; D, drought group; H, heat group; DH, drought combined heat group.
Facultative CAM likely recruits a set of cis DNA element
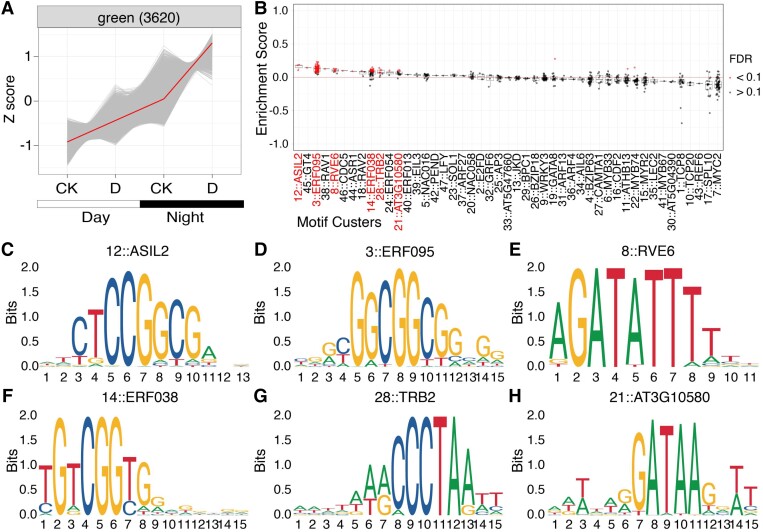
Patterns of gene expression were largely determined by interactions between cis- and trans-factors. To investigate the recruitment of cis-elements by facultative CAM associated genes, we first clustered genes using Weighted correlation network analysis (WGCNA) and identified 27 modules with distinct expression pattern (Supplemental Fig. S13). Day- or night-specific modules were defined by a lack of response to drought but preferential expression at day or night (Supplemental Figs. S14A and S14C). Two modules were defined as drought specific due to the strong induction of gene expression by drought at both day and night (Supplemental Figs. S14E and S14G). A module containing both CAM-specific PEPC1E1c′ (evm.TU.LG05.829, evm.TU.LG25.499) and β-CA (evm.TU.LG12.1303) was recognized as CAM specific and exhibited enhanced upregulation of expression at night under drought condition (Fig. 5A).
Figure 5.
CAM-specific module is enriched with motifs from clusters 12, 3, 8, 14, 28, and 21. A) Gene expression pattern of CAM-specific green module containing 3,620 genes displays more upregulated gene expression after drought treatment at nighttime; data were presented as Z-score; red line connects the mean of expression in each sample. B) Enriched TF binding sites placed in motif clusters; red dots indicate significantly enriched motifs (FDR < 0.1) compared with genomic background; motif clusters with more than 3 enriched motifs were highlighted in red text. C to H), Sequence logo of representative motifs from motif clusters 12, 3, 8, 14, 28, and 21.
Find Individual Motif Occurrences (FIMO), a software designed for scanning DNA or protein sequences with given motifs (Bailey et al. 2015), was used to scan known transcription factor (TF) binding sites from JASPAR database (656 motifs from 47 motif clusters) in upstream 2,000-bp promoter sequences, and the frequency of occurrence in each module was compared with genomic background. This identified motifs from clusters 7 (MYC2), 4 (bZIP63), 15 (MYR20), and 1 (TCP8) that were significantly enriched in genes preferentially expressed in the day (Supplemental Fig. S14B). Cluster 8 (RVE6) was enriched in genes preferentially expressed in the night, and 2 drought-specific modules were enriched with clusters 21 (AT3G10580), 7 (MYC2), and 4 (bZIP63) motifs (Supplemental Fig. S14, D, F, and H). There were few overlapping motifs between the CAM-specific module and night- or drought-specific modules except for an evening element cluster 8 (RVE6) and cluster 21 (AT3G10580); most enriched motifs belong to clusters 12 (ASIL2), 3 (ERF095), 14 (ERF038), and 28 (TRB2) (Fig. 5, B to H). Consistent with this analysis of enriched cis-elements, cognate TF genes such as REVEILLE 6 (RVE6, evm.TU.LG06.1912) and 2 TELOMERE REPEAT BINDING FACTOR 2 (TRB2) gens (evm.TU.LG13.1168 and evm.TU.LG21.800) as well as ethylene-response factor (ERF) TF genes including DREB2A/2B (evm.TU.LG19.575) and 3 cytokinin response factor genes (evm.TU.LG03.1986, evm.TU.LG04.3303, evm.TU.LG14.283) were also identified in CAM-specific module (Supplemental Tables S8 to S11). Taken together, these data suggest that facultative CAM likely results from the recruitment of a CAM-specific regulatory network using a specific set of cis-elements that are likely absent in night- or drought-specific network.
Gene duplications may contribute to common purslane stress resistance
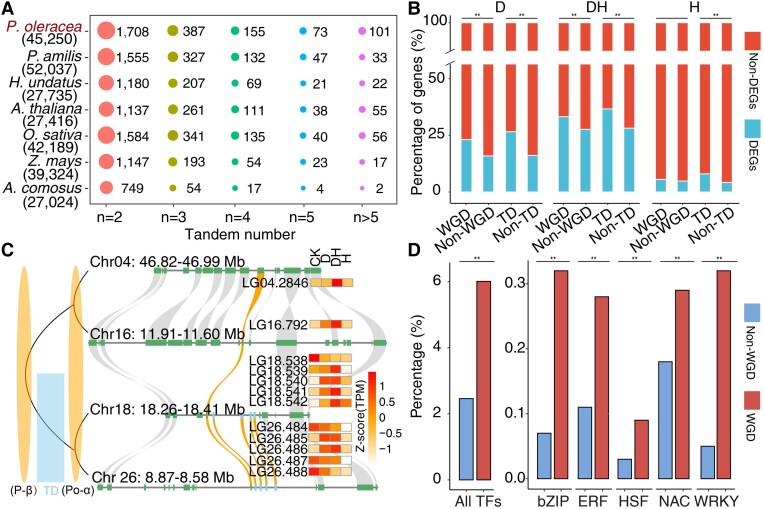
The expansion and contraction of gene families have a profound influence in shaping stress resistance and driving phenotypic diversity and adaptive evolution in flowering plants (Jiao et al. 2011; Chen et al. 2013; Wu et al. 2020; Zhang et al. 2020; Wang et al. 2022). We identified 32,605 putative orthologous gene clusters composed of 475,125 protein-coding genes from 528,054 genes across 16 plant species used in this study. Compared with the other 15 species, the P. oleracea genome possessed the largest proportion, as well as the most gene cluster expansions (9,375 gene clusters expanded) (Fig. 2A and Supplemental Table S12). The P. oleracea genome also exhibited the largest number of species-specific gene clusters (1,605) compared with its relative species (B. vulgaris, S. oleracea, and H. undatus) (Supplemental Fig. S15). Moreover, we detected 7,740 genes derived from WGD by self-synteny analysis, as they retained at least 3 copies after 2 rounds of WGD events, as well as 6,330 genes derived from TDs by Basic Local Alignment Search Tool of Protein (BLASTP) and chromosomal location analysis. We also observed that the number of tandem duplicates in P. oleracea is higher than that in other species (Fig. 6A). GO analysis revealed that WGD genes are highly enriched for the “biological regulation” and “DNA binding” categories, while TD genes tended to be enriched for the “enzyme inhibitor activity” and “response to stress” categories (Supplemental Fig. S16A). In addition, among the differentially expressed genes (DEGs) under drought and/or heat treatment, WGD and TD genes account for more than 30% (Supplemental Fig. S16B). To assess the consequence of these WGD and TD events on gene expression, we conducted an RNA-seq analysis of P. oleracea plants grown under control conditions or exposed to heat and/or drought stress. We identified more DEGs among genes having experienced WGD and TD under drought and/or heat treatment compared to non-WGD and nontandem-duplicated genes (Fig. 6B). For example, all 12 copies of a gene encoding early light-induced protein (ELIP), which underwent 2 rounds of WGD and TD events, were differentially expressed in P. oleracea seedlings exposed to heat and drought (Fig. 6C).
Figure 6.
Analyses of WGD and TD of the common purslane genome. A) Comparison of the number of tandem duplicates in different species. B) Percentage of DEGs and non-DEGs for WGD and non-WGD, TD, and non-TD genes upon heat and/or drought treatment. **Significant difference by P < 0.01 (chi-square test). C) An example of gene expansion from WGD and TD events in the P. oleracea genome. The genes encoding ELIPs, thought to act as photoprotectants, are highly upregulated upon heat and drought treatments. The highlighted orange regions are from WGD, while the light blue ones are from TD. D) TFs, especially several stress-related TF families, are significantly enriched in the WGD gene set compared with the non-WGD set. **P < 0.01 by chi-square test. CK, RNA-seq data under normal conditions; D, drought; H, heat; and DH, combined drought and heat.
In addition, we determined the mean relative size of gene families that had been described previously by the OneKP Project (One Thousand Plant Transcriptomes Initiative 2019) in the selected genomes above. Notably, we found that most of these gene families, such as basic helix-loop-helix (bHLH), MYB, and WRKY, have more members in the P. oleracea genome than in other species (Supplemental Fig. S17). Further analysis revealed that TF genes are significantly enriched in the WGD gene set, including several reported stress-related TF gene families, such as heat shock factor (HSF), basic Leucine zipper (bZIP), NAC, WRKY, and ERF, and other important families (Fig. 6D and Supplemental Fig. S18). By contrast, we detected fewer TF genes in the TD list (Supplemental Fig. S19). For instance, a member from the HSF A (HsfA) family was present in 5 copies in the genome from TDs and WGDs; importantly, all 5 copies were highly expressed upon drought and heat stress (Supplemental Fig. S20). These results suggested that the high frequency of WGD and TD genes in the P. oleracea genome may have contributed to its adaptation to environmental stress.
Discussion
Portulaca is the only genus in the Portulacaceae family, with substantial variation in chromosome number between species (Danin et al. 1978; Ocampo and Columbus 2012; Walter et al. 2015). The base chromosome number of Portulaca was inferred to be x = 9 (Ocampo and Columbus 2012). Common purslane had been classified into several subspecies or microspecies (and as such is referred to as a P. oleracea complex) with somatic chromosome number of 2n = 18, 36, 52, and 54 in Chromosome Counts Database (Rice et al. 2015). Notably, the FISH experiment, k-mer, and genomic analysis indicated that the common purslane we used is diploid with a chromosome number of 2n = 52. There could be 2 possibilities to explain the distinct chromosome number: one is that the accession we sequenced may have been a base chromosome change prior to the Po-α WGD, and the other is that post-WGD rearrangement led to the current number (2n = 52) of chromosomes.
Plants from the Portulacaceae were reported to possess diverse photosynthesis pathways such as C4 + CAM and C3-C4 + CAM (Koch and Kennedy 1980; Voznesenskaya et al. 2010, 2017; Ferrari et al. 2020b). More than 10 common purslane genotypes were recently demonstrated to perform C4 + CAM (Ferrari et al. 2020b), making common purslane the most widely studied species for its potential as a C4/CAM model (Ferrari et al. 2020a). Our research reported a high-quality, chromosome-level genome of common purslane with 2 rounds of WGD occurring around 66.30 and 7.74 Mya, respectively. Our analysis of the common purslane genome revealed that most genes are present in multiple copies, including many that encode key enzymes or transporters involved in photosynthesis. We successfully assigned several PEPCs to C4-specific (4 enzymes) and CAM-specific (2 enzymes) groups with amino acids previously associated with these photosynthetic pathways, lending further support that common purslane is a C4 + CAM plant. Importantly, although C4- and CAM-related genes were previously reported in the related species Paraguayan purslane, such as PaPPC-1E1a′ (FUN_020790, named PaPEPC1E1a in this study) and PaPPC-1E1c (FUN_029936, named PaPEPC1E1c in this study) (Gilman et al. 2022), PaPEPC1E1a encoded an enzyme with the derived amino acids (E519 and A780) that have previously not been associated with either CAM or C4 photosynthesis. In addition, PaPEPC1E1a harbored the residue (R890) for tight inhibitor binding, much like PEPC1E1a from CAM-type dragon fruit. Further work will be required to understand the impact of these amino acids on the efficiency of PEPC activity and C4 photosynthesis in Paraguayan purslane.
Land plants have evolved C4 and/or CAM photosynthesis well over 100 times to adapt to stressful environments such as low CO2, high temperatures, and drought (Edwards 2019). Although C4 and CAM evolutionary trajectories are largely distinct, Portulaca demonstrates that they also can be compatible (Edwards 2019; Hibberd 2022; Moreno-Villena et al. 2022). A de novo genome assembly for P. amilis was reported recently. It was helpful to reveal coexpression networks that may support C4-CAM compatibility (Gilman et al. 2022). Our analysis of the common purslane genome focused more on the evolutionary contribution of gene duplication to C4-CAM compatibility. We explored this important issue through the origin and evolution of 2 key enzymes: PEPC and β-CA. Since PEPC seems to have gone through a more complex process in the evolutionary history, β-CA may be more informative than PEPC in explaining the origin and evolution of the C4 and CAM pathways. Phylogenetic trees and synteny analyses indicated that the P-β WGD produced functionally differentiated β-CA genes that become involved in the C4 + CAM pathways. However, there could be multiple possible histories of C4 related-gene evolution if the effects of convergent evolution on gene tree topology are taken into account. Interestingly, TD events prior to Po-α WGD event in common purslane genome led to repeated evolution of C4-specific β-CA genes, which provided further evidence for that C4 photosynthesis has repeatedly evolved in plants.
Upstream regulatory sequences play a critical role in determining spatial temporal gene expression. The common purslane genomic sequence provides an excellent opportunity to initiate an understanding of the cis-regulation of facultative CAM. Cis-element enrichment analysis identified G-box (7:MYC2) and TCP (1:TCP8) in day-specific genes and an evening element (8:RVE6) in night-specific genes, consistent with the involvement of these elements in diel regulation of gene expression (Wang et al. 2019). Drought-specific genes were enriched with clusters 7 (MYC2) and 4 (bZIP63), which coincide with JA or ABA associated drought response elements such as G-boxes or ABREs (Liu et al. 2018; Soma et al. 2021). It has been hypothesized that facultative CAM likely resulted from combinatorial rewiring of circadian and drought responsive networks (Gilman et al. 2022). Indeed, we discovered that the promoters of CAM-specific genes in common purslane are significantly enriched with evening-specific elements and drought-response-related cis-elements. Both the evening element RVE6 and the midnight element Telobox (TRB2, which is identical to motif cluster 28) are overrepresented in CAM-specific promoters, and this is consistent with CAM cycling genes in the C3 facultative CAM species Sedum album (Wai et al. 2019). However, S. album utilizes ABRE or ABRE-like motifs from ABA-dependent drought regulatory networks, while common purslane recruited a different set of drought-related motifs known as ERF motifs recognized by Dehydration responsive element binding protein (DREB)/ERF TFs and linked with ABA-independent drought regulatory networks. This suggests that the utilization of various drought response regulatory networks along with the conserved recruitment of evening-specific elements could be a possible mechanism for CAM-specific gene expression. Overall, these data shed light on the convergent evolution of facultative CAM and provide potential regulatory modules for incorporating C4/CAM into crops in the future.
Climate change has far-reaching and adverse effects on crop yields and human nutrition. To make matters worse, an increasing world population will require that current food production to be doubled by the year 2050. Responding to these problems will require the development of high-yield, as well as stress-tolerant crops. To achieve this, there has been an ongoing global initiative to engineer C4 or CAM pathways into C3 crops or the CAM pathway into C4 plants like maize. A key step toward engineering C4 rice was achieved through constitutive expression of maize GOLDEN2-LIKE genes, and field-grown transgenic plants resulted in a 30% to 40% increase in both vegetative biomass and grain yield (Wang et al. 2017; Li et al. 2020). Here, we accurately identified all gene copies of the key proteins involved in the C4 + CAM photosynthetic pathway. Further analysis of their diurnal expression patterns, as well as their key functional sites will allow the identification of important gene copies as the targets for engineering crops with the C4 and/or CAM photosynthetic pathway. For example, based on series analysis for PEPC gene copies, we would select evm.model.LG25.499, evm.model.LG05.829, evm.model.LG17.1452, evm.model.LG17.1453, evm.model.LG03.2406, and evm.model.LG03.2407 as the potential targets from 13 PEPC gene copies. Overall, we believe that this study will be an invaluable resource to investigate the integration of different photosynthetic pathways. Our comprehensive and in-depth analysis of a genome underpinning this complex photosynthetic pathway could provide potential targets for C4 + CAM engineering.
Materials and methods
Plant materials and DNA sequencing
In this work, we used wild common purslane (P. oleracea), which grows widely in Beijing, China. These plants were grown in an environmental chamber (10 h day/14 h night). Leaves from a 30-day-old mature plant were harvested and frozen immediately in liquid nitrogen. Genomic DNA was extracted using a QIAGEN Genomic Kit and used as material to construct sequencing libraries following the standard protocols of Oxford Nanopore Technologies. Briefly, DNA was size-selected using a BluePippin instrument (Sage Science, USA) before end-repair and adaptor ligation on the resulting blunt ends. Finally, libraries were sequenced on a PromethION platform. DNA from the same batch of samples was used to prepare Illumina libraries, following the standard manufacturer's protocol (Illumina). Libraries were sequenced on an Illumina Novaseq 6000 platform as 150-bp pair-end reads.
To assess the response of common purslane to stress, 1-mo-old plants were separated into 4 experimental groups, each being subjected to different conditions for 7 days: (i) controls in normal environment conditions (14-h-light/10-h-dark photoperiod [with lights on from 6 AM to 8 PM], with a 28 °C/22 °C day/night temperature cycle), (ii) drought stress (no water irrigation for 7 days), (iii) heat treatment (water applied regularly and temperature raised to 45 °C during the day for the last 4 days), and (iv) combined heat and drought stress (no water irrigation for 7 days and 45 °C during the day for the last 4 days). Leaves were collected after 7 days of treatment during the day (10 AM) and at night (12 AM), with 3 biological replicates.
Malate measure
After the sample was thawed and smashed, an amount of 0.05 g of the sample was mixed with 500 µL of 70% (v/v) methanol/water. The sample was vortexed for 3 min under the condition of 2,500 r/min and centrifuged at 12,000 r/min for 10 min at 4 °C. Then, 300 μL of supernatant was placed into a new centrifuge tube and into a −20 °C refrigerator for 30 min. Then, the supernatant was centrifuged again at 12,000 r/min for 10 min at 4 °C. After centrifugation, 200 μL of supernatant was transferred for further LC-MS analysis.
The sample extracts were analyzed using an LC-ESI-MS/MS system. The analytical conditions were as follows: HPLC column, ACQUITY HSS T3 (i.d. 2.1 × 100 mm, 1.8 μm); solvent system, water with 0.05% (v/v) formic acid (A), acetonitrile with 0.05% formic acid (v/v) (B); gradient started at 5% B (0 min), increased to 95% B (8 to 9.5 min), and finally ramped back to 5% B (9.6 to 12 min); flow rate, 0.35 mL/min; temperature, 40 °C; and injection volume, 2 μL.
BioNano optical maps and Hi-C sequencing
For BioNano physical mapping, DNA extracted from leaves were subject to manufacturer recommended protocols for library preparation (Bionano PrepTM Animal Tissue DNA Isolation Kit [CAT#80002]/Bionano PrepTM Plant DNA Isolation Kit [CAT#80003]) and optical scanning provided by BioNano Genomics (https://bionanogenomics.com), with the labeling enzyme Direct Label Enzyme (DLE) (Bionano PrepDLS Labeling DNA Kit, CAT#80005). Labeled DNA samples were loaded and run on the Saphyr system (BioNano Genomics) in Grandomics.
To anchor hybrid scaffolds onto the chromosome, genomic DNA was extracted for the Hi-C library from samples. Then, we constructed the Hi-C library and obtained sequencing data via the Illumina Novaseq/MGI-2000 platform. In brief, freshly harvested leaves were cut into 2 cm pieces and vacuum infiltrated in nuclei isolation buffer supplemented with 2% (v/v) formaldehyde. Crosslinking was stopped by adding glycine and additional vacuum infiltration. Fixed tissue was then ground to powder before resuspending in nuclei isolation buffer to obtain a suspension of nuclei. The purified nuclei were digested with 100 units of DpnII and marked by incubating with biotin-14-dATP. Biotin-14-dATP from nonligated DNA ends was removed owing to the exonuclease activity of T4 DNA polymerase. The ligated DNA was sheared into 300 to 600 bp fragments and then was blunt-end repaired and A-tailed, followed by purification through biotin-streptavidin–mediated pull down. Finally, the Hi-C libraries were quantified and sequenced using the Illumina Novaseq/MGI-2000 platform.
Transcriptome sequencing
Total RNA was isolated from higher leaves, lower leaves, higher stems, lower stems, roots, immature flowers, and mature flowers of common purslane using TRIzol reagent. mRNA-seq libraries were constructed using a TruSeq RNA Library Preparation Kit (Illumina, USA) following the manufacturer's recommendations, and 150-bp paired-end sequencing was performed on a Novaseq 6000 platform to assist gene prediction. Samples collected during the stress treatment experiments described above were also subjected to total RNA extraction using a Promega ReliaPrep RNA Tissue Miniprep System Kit. Sequencing libraries were then prepared and sequenced as above.
Preparation of chromosome spreads and FISH
Preparation of chromosome spreads and FISH were performed according to Huang et al. (2021). Briefly, 1-cm root tip segments were pretreated in 0.05% (w/v) 8-hydroxyquinoline for 2 h at 25 °C, fixed in 3:1 ethanol:acetic acid (v/v) fixative overnight, and kept at −20 °C until use. An enzymatic solution consisting of 2% (w/v) cellulase and 1% (w/v) pectolyase was used to digest the root tips at 37 °C for 3 h. Then, 10 µL of 1:3 ice-cold acetic acid:methanol mixture (v/v) was added, and the root tips were broken with tweezers, mounted onto a glass slide and allowed to air-dry. FISH was performed using a hybridization mixture (10 µL) containing 50% (w/v) formamide, 10% (w/v) dextran sulfate in 2× SSC (saline sodium citrate), and 40 ng of biotin-labeled 45S rDNA and digoxigenin-labeled 5S rDNA probes. Hybridization was carried out for 16 h at 37 °C. Digoxigenin-labeled and biotin-labeled probes were detected using rhodamine-conjugated antidigoxigenin and fluorescein-conjugated avidin, respectively. Chromosomes were counterstained with DAPI (4′,6-diamidino-2-phenylindole) in antifade solution (Vector Laboratories, USA) under a coverslip. The slides were examined with an Axio Imager Z.2 Zeiss microscope (Zeiss, Oberkochen, Germany) equipped with a Cool Cube 1 camera (Metasystems, Altlussheim, Germany) and appropriate optical filters. Final image adjustments were performed with Adobe Photoshop CC.
Genome assembly and quality assessment
The raw Nanopore data were corrected by NextDenovo software (seed_cutoff = 25k; reads_cutoff = 1k) (https://github.com/Nextomics/NextDenovo). The corrected reads were then assembled using smartdenovo software (-k 21, -j 3,000) (https://github.com/ruanjue/smartdenovo) to obtain contigs for the preliminary assembled genome. Contig sequences were polished with the Nanopore reads and Illumina reads and used as input for the Nextpolish software (default) (Hu et al. 2020). BioNano data adopt single-enzyme digestion technology, with the DLE-1 enzyme used to digest genomic DNA to obtain raw data. We constructed longer super-scaffolds by anchoring the polished contig assembly to the BioNano optical map. Then, unique Hi-C read pairs were identified through alignment to the scaffolds by bowtie2 (−very-sensitive -L 30) (Langmead and Salzberg 2012). The DpnII restriction sites were identified along the scaffolds, and the Hi-C interaction signal intensity was used to assign each read to different scaffolds. Finally, the scaffold sequences were clustered into 26 pseudo-chromosome groups by agglomerative hierarchical clustering (bottom-up hierarchical clustering) using LACHESIS software (Burton et al. 2013).
The quality and completeness of the common purslane genome assembly were assessed from 3 aspects. First, the mapping rates of the clean reads obtained from the transcriptomes and genomic DNA were mapped back to the genome assembly by Hisat2 (Kim et al. 2019) and BWA-MEM (Li 2013) with default parameters. Second, the BUSCO score was determined for all predicted genes in the final assembly against the gene list for Embryophyta_odb10 (Simão et al. 2015; Manni et al. 2021). Third, the LAI was employed to infer assembly continuity with default parameters (Ou et al. 2018). Finally, we used Merqury (Rhie et al. 2020) software to estim-ate the consensus QV of the assembly.
Gene prediction and functional annotation
Gene structure predictions adopted a combination of de novo prediction (Augustus) (Hoff and Stanke 2018), homology prediction (GeMoMa) (Keilwagen et al. 2019), and transcriptome prediction (PASA) (Xu et al. 2006). All 3 approaches were integrated by EvidenceModeler software (Haas et al. 2008). TransposonPSI (http://transposonpsi.sourceforge.net) was used to align and remove genes containing TEs to obtain the final structural genome annotation.
Repeat annotation and TE analyses
The repetitive sequences were identified using a combination of repeat homology searches and ab initio prediction. For homology searches, Repbase (2018) (Bao et al. 2015) was employed to search the genome using RepeatMasker (Tarailo-Graovac and Chen 2009) with default parameters. For ab initio predictions, a consensus sequence library was built using RepeatModeler (http://repeatmasker.org/RepeatModeler/) with the parameter “-engine ncbi”. Then, LTR_harvest (Ellinghaus et al. 2008), LTR_finder (Xu and Wang 2007), and LTR_retriever (Ou and Jiang 2018) were used to build an LTR library with default parameters. Both libraries were then used for annotating the genome using RepeatMasker, and the detected TEs were combined to obtain the final TE annotation.
Transcriptome analyses
RNA-seq raw reads were processed using Trimmomatic (Bolger et al. 2014) to remove adaptor sequences and low-quality reads. The clean reads were then mapped to the reference genome using HISAT2 (Kim et al. 2019) with default parameters. The expression abundance values were calculated using Stringtie (Pertea et al. 2016), and we averaged the abundance values from the 3 biological replicates of each sample to obtain levels of gene expression. Finally, we performed differential expression analysis between the corresponding samples by DESeq2 (Love et al. 2014).
Gene family inference and phylogenomic analysis
The nucleotide and amino acid sequences of 15 representative plant species were downloaded from various sources: Paraguayan purslane (P. amilis), amaranth (A. hypochondriacus), spinach (S. oleracea), sugar beet (B. vulgaris), tomato (Solanum lycopersicum), Arabidopsis thaliana, Populus trichocarpa, grape (Vitis vinifera), Aquilegia coerulea, pineapple (Ananas comosus), maize (Z. mays), rice (Oryza sativa), Sorghum bicolor, and Amborella trichopoda from Phytozome (https://phytozome-next.jgi.doe.gov/), and dragon fruit (H. undatus) from the Pitaya Genome Database (http://www.pitayagenomic.com/). Gene clusters of putative gene families for these species and common purslane were identified by OrthoFinder (v2.4.0) (Emms and Kelly 2019). Venn diagrams of the selected taxa were generated using Venn diagram (Chen and Boutros 2011). MAFFT (v7.471) (Katoh et al. 2002) was used to align 58 single-copy putatively orthologous gene families. Poorly aligned regions were removed using trimAL (v1.4) (Capella-Gutierrez et al. 2009) with default parameters. The concatenated amino acid alignments were used to construct a species tree by the maximum likelihood method in RAxML (v8.2.12) (Stamatakis 2014) under the “PROTGAMMAAUTO” model with 100 bootstrap replicates. Divergence time for each tree node was inferred using r8s (v1.81) (Sanderson 2003) and gene family expansion and contraction by CAFE (v4.2.1) (De Bie et al. 2006).
Genomic synteny analyses
Synteny searches were performed to identify syntenic blocks within common purslane and between common purslane and dragon fruit using MCScanX (Wang et al. 2012) by default parameter settings. Dotplots and macrosynteny patterns were drawn by JCVI (https://github.com/tanghaibao/jcvi) and R scripts, respectively.
Synonymous substitution (Ks) analysis
For each pair of homologous genes, the predicted protein sequences were used for multiple sequence alignment by MUSCLE (Edgar 2004) with default parameters, after which the nucleotide sequences were forced to fit the amino acid alignments by PAL2NAL (Suyama et al. 2006). Ks values were calculated using the Nei-Gojobori algorithm (Nei and Gojobori 1986) implemented in the codeml package of PAML (Yang 1997).
Estimate of whole genome duplication timing
To time the Portulacineae WGD, we used the methods described in Supplemental material for the opium poppy genome (Guo et al. 2018). Briefly, we estimated the average evolutionary rate for Caryophyllales using common purslane, a Portulacineae and A. hypochondriacus, an Amaranthaceae. Given the mean Ks value of common purslane-A. hypochondriacus and their divergence date T, we calculated the synonymous substitutions per site per year (r) for Caryophyllales (T = Ks/2r). The r value and Ks peak values of WGD were applied to time the common purslane WGD.
Identification of C4-/CAM-specific PEPC genes
To identify PEPC genes in common purslane, we performed a BLASTP analysis using well-annotated PEPC genes in A. thaliana and maize as queries, as well as hidden Markov model searches using the profile PF00311 from the Pfam database as seed, against the genome-wide amino acid sequences of common purslane, employing BLASTP (e-value <1e−5) and the hmmsearch in HMMER (v3.1b2) (e-value <1e−03, –domE 0.001) (Potter et al. 2018), respectively. Then, multiple sequence alignment was conducted using MAFFT (v7.471) (Katoh et al. 2002), and gene trees were constructed using the maximum likelihood method in RAxML (v8.2.12) (Stamatakis 2014) with the “PROTGAMMAAUTO” model with 100 bootstrap replicates. Based on the previous classification studies in Caryophyllales (Christin et al. 2014), PEPC genes were divided into 7 categories.
Gene coexpression and motif enrichment analysis
Transcription abundance from drought-treated and control samples were used to define condition-specific gene expression clusters. Genes with Transcripts Per Million (TPM) > 1 in at least 2 biological replicates from at least one condition were defined as expressed genes (30833 genes) and then log2-transformed TPM were used for coexpression analysis using the Weighted Gene Coexpression Network Analysis (WGCNA v1.70) (Langfelder and Horvath 2008) with signed network approach and a soft power threshold of 18. Minimal module size was set as 100 genes, initial module eigengenes with correlation coefficient >0.85 were merged, and 27 unique color-coded coexpression modules were produced. Cis-element enrichment analysis was performed as previously described (Rhie et al. 2020) with modifications. Briefly, genes with module membership >0.8 were selected, and 2000-bp upstream sequences from transcription start site were extracted. Occurrence of frequencies of the 656 known plant nonredundant motifs from JASPAR database (Castro-Mondragon et al. 2021) were determined by FIMO (v5.4.0) (Bailey et al. 2015) in each module as previously reported. The frequency of occurrence of motifs in promoters of all expressed genes was used as background. Enrichment analysis was performed using a Fisher's exact test with false discovery rate (FDR) correction (Yang et al. 2018), and motifs with FDR < 0.1 were defined as enriched individual motif. Enrichment score was calculated as log2 transformed ratio of occurrence frequency between coexpression module and genomic background. To reduce the redundancy of similar motifs, individual motifs were placed in 47 motif clusters determined by RSAT matrix-clustering (Castro-Mondragon et al. 2017), which were deposited on JASPAR website (https://jaspar.genereg.net/matrix-clusters/plants/).
Accession numbers
The data generated in this study has been uploaded to the NCBI database and can be retrieved under accession numbers PRJNA978934 and PRJNA868526. The genome assembly and annotation have also been deposited in the Genome Warehouse in National Genomics Data Center under accession number GWHCBIU00000000 that is accessible at https://ngdc.cncb.ac.cn/gwh.
Supplementary Material
Acknowledgments
We thank Prof Yalong Guo, Prof Baichen Wang, and Dr. Suhua Yang from Institute of Botany, Chinese Academy of Sciences for invaluable input. For open access, the authors have applied a Creative Commons Attribution (CC BY) license to any Author Accepted Manuscript version arising from this submission.
Contributor Information
Xiaoliang Wang, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; State Key Laboratory of Plant Diversity and Specialty Crops, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; China National Botanical Garden, Beijing 100093, China.
Xuxu Ma, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; China National Botanical Garden, Beijing 100093, China; Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Ge Yan, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; China National Botanical Garden, Beijing 100093, China; Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Lei Hua, Department of Plant Sciences, University of Cambridge, Cambridge CB2 3EA, UK.
Han Liu, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Wei Huang, National Maize Improvement Center, China Agricultural University, Beijing 100193, China.
Zhikai Liang, Department of Plant and Microbial Biology, University of Minnesota, Saint Paul, MN 55108, USA.
Qing Chao, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Julian M Hibberd, Department of Plant Sciences, University of Cambridge, Cambridge CB2 3EA, UK.
Yuannian Jiao, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; State Key Laboratory of Plant Diversity and Specialty Crops, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; China National Botanical Garden, Beijing 100093, China.
Mei Zhang, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; China National Botanical Garden, Beijing 100093, China; Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Author contributions
M.Z. and Y.J. initiated and conceived the genome project. X.M. collected materials and performed some data analyses.; X.W. performed most data analyses.; G.Y. performed most of the benchwork. W.H. performed the experiments of FISH. L.H. performed motif analyses. L.H. and J.M.H. provided valuable advice. X.W., X.M., and L.H. wrote the manuscript. M.Z., Y.J., and J.M.H. revised the manuscript. All the authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fluorescence in situ hybridization (FISH) of common purslane chromosomes with 45S rDNA and 5S rDNA.
Supplemental Figure S2. Estimation of common purslane genome size.
Supplemental Figure S3. Workflow of the de novo assembly of the common purslane genome.
Supplemental Figure S4. Hi-C contact matrix of the 26 pseudo-chromosomes for the common purslane assembly.
Supplemental Figure S5. LTR assembly index (LAI) assessment for each assembled common purslane chromosome.
Supplemental Figure S6. Estimation of the burst time of transposable elements in the common purslane genome.
Supplemental Figure S7. Syntenic blocks among the B. vulgaris, H. undatus, P. amilis, and P. oleracea genomes.
Supplemental Figure S8. Gene expression patterns of several key genes encoding enzymes or transporters identified in common purslane under drought and heat treatments during the day or night.
Supplemental Figure S9. Diagram depicting the gene copies encoding the main enzymes and transporters in the C4/CAM pathway and their expression levels under normal or stressful conditions.
Supplemental Figure S10. Expression patterns of C4-related copies and CAM-related gene copies in different tissues.
Supplemental Figure S11. Gene expression and differential expression patterns of PEPC genes identified in common purslane under heat and drought treatments during the day and night.
Supplemental Figure S12. RT-qPCR analysis of CAM-specific PEPC genes under control or stress treatments during the day (A) and at night (B).
Supplemental Figure S13. Heatmap of gene expression patterns of the 27 color-coded coexpression modules identified by WGCNA.
Supplemental Figure S14. Enrichment of cis-elements in day, night and drought-specific modules.
Supplemental Figure S15. Venn diagrams showing the numbers of shared and species-specific gene clusters in the 4 selected species.
Supplemental Figure S16. Analysis of whole-genome duplication and tandem duplication of the common purslane genome.
Supplemental Figure S17. Variation in gene copy number of several important gene families.
Supplemental Figure S18. Percentage of transcription factor genes within WGD and non-WGD genes.
Supplemental Figure S19. Percentage of transcription factor genes within TD and non-TD genes.
Supplemental Figure S20. An example of HSF genes resulting from WGD and TD in the common purslane genome that are upregulated upon stress treatment.
Supplemental Table S1. Summary statistics of contigs and scaffolds of the common purslane genome.
Supplemental Table S2. Mapping rates of RNA-seq and DNA-seq data to the assembled genome.
Supplemental Table S3. Benchmarking Universal Single Copy Orthologs (BUSCO) scores to assess the quality of the assembly and annotation.
Supplemental Table S4. Summary of the functional annotation for the annotated protein-coding genes.
Supplemental Table S5. Repeat contents in the common purslane genome.
Supplemental Table S6. Number of photosynthesis-related genes in different species.
Supplemental Table S7. Gene ID, species, and data source for all PEPC genes used in this study.
Supplemental Table S8. Summary of motif enrichment.
Supplemental Table S9. Assignment of genes into color-coded modules identified by WGCNA.
Supplemental Table S10. Genes with module membership > 0.8 in each module.
Supplemental Table S11. Significantly enriched motifs (FDR < 0.1) in each coexpression module.
Supplemental Table S12. Number of expanded and contracted gene families in the 16 species genomes analyzed in this study.
Funding
This work was funded by the National Key Research and Development Program of China (2020YFE0202300) to M.Z. and the National Natural Science Foundation of China (32221001, received by Y.J.). L.H. and J.M.H. were funded by a C4 Rice project grant from the Bill and Melinda Gates Foundation to the University of Oxford.
Competing interests: The authors declare no competing interests.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015:43(W1):W39–W49. 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015:6(1):11. 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013:31(12):1119–1125. 10.1038/nbt.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byng J, Chase M, Christenhusz M, Fay M, Judd W, Mabberley D, Sennikov A, Soltis D, Soltis P, Stevens P. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016:181(1):1–20. 10.1111/boj.12385 [DOI] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009:25(15):1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mondragon JA, Jaeger S, Thieffry D, Thomas-Chollier M, van Helden J. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res. 2017:45(13):e119. 10.1093/nar/gkx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mondragon J, Riudavets Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Pérez N, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2021:50(D1):D165–D173. 10.1093/nar/gkab1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. Venndiagram: a package for the generation of highly-customizable venn and Euler diagrams in R. BMC Bioinformatics. 2011:12(1):35. 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. New genes as drivers of phenotypic evolution. Nat Rev Genet. 2013:14(9):645–660. 10.1038/nrg3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-Y, Xie F-F, Cui Y-Z, Chen C-B, Lu W-J, Hu X-D, Hua Q-Z, Zhao J, Wu Z-J, Gao D, et al. A chromosome-scale genome sequence of pitaya (Hylocereus undatus) provides novel insights into the genome evolution and regulation of betalain biosynthesis. Hortic Res. 2021:8(1):164. 10.1038/s41438-021-00612-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Arakaki M, Osborne CP, Bräutigam A, Sage RF, Hibberd JM, Kelly S, Covshoff S, Wong GK-S, Hancock L, et al. Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. J Exp Bot. 2014:65(13):3609–3621. 10.1093/jxb/eru087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea RM, Andreo CS, Lara MV. Deciphering the mechanisms involved in Portulaca oleracea (C4) response to drought: metabolic changes including crassulacean acid-like metabolism induction and reversal upon re-watering. Physiol Plant. 2014:152(3):414–430. 10.1111/ppl.12194 [DOI] [PubMed] [Google Scholar]
- Danin A, Baker I, Baker HG. Cytogeography and taxonomy of the Portulaca oleracea L. Polyploid complex. Israel J Bot. 1978:27(3/4):177–211. [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006:22(10):1269–1271. 10.1093/bioinformatics/btl097 [DOI] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 2013:505(7484):546–549. 10.1038/nature12817 [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004:32(5):1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ. Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytol. 2019:223(4):1742–1755. 10.1111/nph.15851 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. Int J Plant Sci. 2012:173(6):724–733. 10.1086/666098 [DOI] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008:9(1):18. 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019:20(1):238. 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari RC, Bittencourt PP, Nagano PY, Oliveira WS, Freschi L. Developing Portulaca oleracea as a model system for functional genomics analysis of C4/CAM photosynthesis. Funct Plant Biol. 2020a:48(7):666–682. 10.1071/FP20202 [DOI] [PubMed] [Google Scholar]
- Ferrari RC, Bittencourt PP, Rodrigues MA, Moreno-Villena JJ, Alves FRR, Gastaldi VD, Boxall SF, Dever LV, Demarco D, Andrade SCS, et al. C4 and crassulacean acid metabolism within a single leaf: deciphering key components behind a rare photosynthetic adaptation. New Phytol. 2019:225(4):1699–1714. 10.1111/nph.16265 [DOI] [PubMed] [Google Scholar]
- Ferrari RC, Cruz BC, Gastaldi VD, Storl T, Ferrari EC, Boxall SF, Hartwell J, Freschi L. Exploring C4–CAM plasticity within the Portulaca oleracea complex. Sci Rep. 2020b:10(1):14237. 10.1038/s41598-020-71012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari RC, Freschi L. C4/CAM facultative photosynthesis as a means to improve plant sustainable productivity under abiotic–stressed conditions: regulatory mechanisms and biotechnological implications. In: Plant signaling molecules. Chennai, India: Woodhead Publishing; 2019. p. 517–532 [Google Scholar]
- Gilman IS, Moreno-Villena JJ, Lewis ZR, Goolsby EW, Edwards EJ. Gene co-expression reveals the modularity and integration of C4 and CAM in Portulaca. Plant Physiol. 2022:189(2):735–753. 10.1093/plphys/kiac116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby EW, Moore AJ, Hancock LP, De Vos JM, Edwards EJ. Molecular evolution of key metabolic genes during transitions to C4 and CAM photosynthesis. Am J Bot. 2018:105(3):602–613. 10.1002/ajb2.1051 [DOI] [PubMed] [Google Scholar]
- Guo L, Winzer T, Yang X, Li Y, Ning Z, He Z, Teodor R, Lu Y, Bowser TA, Graham IA, et al. The opium poppy genome and morphinan production. Science. 2018:362(6412):343–347. 10.1126/science.aat4096 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008:9(1):R7. 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi G. Comparison of CAM expression, photochemistry and antioxidant responses in Sedum album and Portulaca oleracea under combined stress. Physiol Plant. 2020:170(4):550–568. 10.1111/ppl.13187 [DOI] [PubMed] [Google Scholar]
- Hatch MD. C4 photosynthesis: a unique elend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta (BBA)—Rev Bioenerget. 1987:895(2):81–106. 10.1016/S0304-4173(87)80009-5 [DOI] [Google Scholar]
- Hibberd JM. Photosynthesis: compatibility between incompatible pathways explained. Curr Biol. 2022:32(20):R1035–R1036. 10.1016/j.cub.2022.08.078 [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics. 2018:65(1):e57. 10.1002/cpbi.57 [DOI] [PubMed] [Google Scholar]
- Hu J, Fan J, Sun Z, Liu S. Nextpolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020:36(7):2253–2255. 10.1093/bioinformatics/btz891 [DOI] [PubMed] [Google Scholar]
- Huang Y, Ding W, Zhang M, Han J, Jing Y, Yao W, Hasterok R, Wang Z, Wang K. The formation and evolution of centromeric satellite repeats in Saccharum species. Plant J. 2021:106(3):616–629. 10.1111/tpj.15186 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011:473(7345):97–100. 10.1038/nature09916 [DOI] [PubMed] [Google Scholar]
- Jin R, Wang Y, Liu R, Gou J, Chan Z. Physiological and metabolic changes of purslane (Portulaca oleracea L.) in response to drought, heat, and combined stresses. Front Plant Sci. 2016:6:1123. 10.3389/fpls.2015.01123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002:30(14):3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Hartung F, Grau J. Gemoma: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol. 2019:1962:161–177. 10.1007/978-1-4939-9173-0_9 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K, Kennedy RA. Characteristics of crassulacean acid metabolism in the succulent C(4) Dicot, Portulaca oleracea L. Plant Physiol. 1980:65(2):193–197. 10.1104/pp.65.2.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008:9(1):559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MV, Drincovich MF, Andreo CS. Induction of a crassulacean acid-like metabolism in the C(4) succulent plant, Portulaca oleracea L: study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell Physiol. 2004:45(5):618–626. 10.1093/pcp/pch073 [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics2013. 10.48550/arXiv.1303.3997, preprint: not peer reviewed. [DOI]
- Li X, Wang P, Li J, Wei S, Yan Y, Yang J, Zhao M, Langdale JA, Zhou W. Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun Biol. 2020:3(1):151. 10.1038/s42003-020-0887-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DJ, Jarvis DE, Ramaraj T, Lee R, Jellen EN, Maughan PJ. Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 2017:15(1):74. 10.1186/s12915-017-0412-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lv Z, Liu Y, Li L, Zhang L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet Mol Biol. 2018:41(3):624–637. 10.1590/1678-4685-gmb-2017-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021:38(10):4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AJ, Vos JM, Hancock LP, Goolsby E, Edwards EJ. Targeted enrichment of large gene families for phylogenetic inference: phylogeny and molecular evolution of photosynthesis genes in the portullugo clade (Caryophyllales). Syst Biol. 2018:67(3):367–383. 10.1093/sysbio/syx078 [DOI] [PubMed] [Google Scholar]
- Moreno-Villena JJ, Zhou H, Gilman IS, Tausta SL, Cheung CYM, Edwards EJ. Spatial resolution of an integrated C4 + CAM photosynthetic metabolism. Sci Adv. 2022:8(31):eabn2349. 10.1126/sciadv.abn2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986:3(5):418–426. 10.1093/oxfordjournals.molbev.a040410 [DOI] [PubMed] [Google Scholar]
- Ocampo G, Columbus JT. Molecular phylogenetics, historical biogeography, and chromosome number evolution of Portulaca (Portulacaceae). Mol Phylogenet Evol. 2012:63(1):97–112. 10.1016/j.ympev.2011.12.017 [DOI] [PubMed] [Google Scholar]
- Omara-Alwala TR, Mebrahtu T, Prior DE, Ezekwe MO. Omega-three fatty acids in purslane (Portulaca oleracea) tissues. J Am Oil Chem Soc. 1991:68(3):198–199. 10.1007/BF02657769 [DOI] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019:574(7780):679–685. 10.1038/s41586-019-1693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Chen J, Jiang N. Assessing genome assembly quality using the LTR assembly index (LAI). Nucleic Acids Res. 2018:46(21):e126. 10.1093/nar/gky730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N. LTR_Retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018:176(2):1410–1422. 10.1104/pp.17.01310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus JK, Schlieper D, Groth G. Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat Commun. 2013:4(1):1518. 10.1038/ncomms2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-Seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016:11(9):1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018:46(W1):W200–W204. 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SK, Reiskind JB, Bowes G. Kinetic analyses of recombinant isoforms of phosphoenolpyruvate carboxylase from Hydrilla verticillata leaves and the impact of substituting a C4-signature serine. Plant Sci. 2008:174(4):475–483. 10.1016/j.plantsci.2008.01.010 [DOI] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020:21(1):245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I. The chromosome counts database (CCDB)—a community resource of plant chromosome numbers. New Phytol. 2015:206(1):19–26. 10.1111/nph.13191 [DOI] [PubMed] [Google Scholar]
- Sage RF. Are crassulacean acid metabolism and C4 photosynthesis incompatible? Funct Plant Biol. 2002:29(6):775–785. 10.1071/PP01217 [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003:19(2):301–302. 10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015:31(19):3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Simopoulos AP, Norman HA, Gillaspy JE, Duke JA. Common purslane: a source of omega-3 fatty acids and antioxidants. J Am Coll Nutr. 1992:11(4):374–382. 10.1080/07315724.1992.10718240 [DOI] [PubMed] [Google Scholar]
- Soma F, Takahashi F, Yamaguchi-Shinozaki K, Shinozaki K. Cellular phosphorylation signaling and gene expression in drought stress responses: ABA-dependent and ABA-independent regulatory systems. Plants. 2021:10(4):756. 10.3390/plants10040756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014:30(9):1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006:34(Web Server):W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. Evolution of C4 phosphoenolpyruvate carboxylase. Arch Biochem Biophys. 2003:414(2):180–188. 10.1016/S0003-9861(03)00165-6 [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009:4:4.10.1–4.10.14. 10.1002/0471250953.bi0410s25 [DOI] [PubMed] [Google Scholar]
- Uddin MK, Juraimi AS, Hossain MS, Nahar MA, Ali ME, Rahman MM. Purslane weed (Portulaca oleracea): a prospective plant source of nutrition, omega-3 fatty acid, and antioxidant attributes. ScientificWorldJournal. 2014:2014:951019. 10.1155/2014/951019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Edward GE, Ocampo G. Unique photosynthetic phenotypes in Portulaca (Portulacaceae): C3–C4 intermediates and NAD-ME C4 species with pilosoid-type Kranz anatomy. J Exp Bot. 2017:68(2):225–239. 10.1093/jxb/erw393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G. Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3–C4 intermediate in genus Portulaca L. (Portulacaceae). J Exp Bot. 2010:61(13):3647–3662. 10.1093/jxb/erq178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai CM, Weise SE, Ozersky P, Mockler TC, Michael TP, VanBuren R. Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album. PLoS Genet. 2019:15(6):e1008209. 10.1371/journal.pgen.1008209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Vekslyarska T, Dobeš C. Flow cytometric, chromosomal and morphometric analyses challenge current taxonomic concepts in the Portulaca oleracea complex (Portulacaeae, Caryophyllales). Bot J Linn Soc. 2015:179(1):144–156. 10.1111/boj.12309 [DOI] [Google Scholar]
- Wang P, Khoshravesh R, Karki S, Tapia R, Balahadia CP, Bandyopadhyay A, Quick WP, Furbank R, Sage TL, Langdale JA. Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr Biol. 2017:27(21):3278–3287. 10.1016/j.cub.2017.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012:40(7):e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yan X, Hu Y, Qin L, Wang D, Jia J, Jiao Y. A recent burst of gene duplications in triticeae. Plant Commun. 2022:3(2):100268. 10.1016/j.xplc.2021.100268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J, et al. Evolution of portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Mol Biol Evol. 2019:36(1):112–126. 10.1093/molbev/msy200 [DOI] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant. 2020:13(1):59–71. 10.1016/j.molp.2019.10.012 [DOI] [PubMed] [Google Scholar]
- Xu C, Jiao C, Sun H, Cai X, Wang X, Ge C, Zheng Y, Liu W, Sun X, Xu Y, et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun. 2017:8(1):15275. 10.1038/ncomms15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007:35(Web Server):W265–W268. 10.1093/nar/gkm286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang X, Yang J, Vaynberg J, Qin J. PASA—a program for automated protein NMR backbone signal assignment by pattern-filtering approach. J Biomol NMR. 2006:34(1):41–56. 10.1007/s10858-005-5358-0 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997:13(5):555–556. 10.1093/bioinformatics/13.5.555 [DOI] [PubMed] [Google Scholar]
- Yang Y, Chen J, Liu Q, Ben C, Todd CD, Shi J, Yang Y, Hu X. Comparative proteomic analysis of the thermotolerant plant Portulaca oleracea acclimation to combined high temperature and humidity stress. J Proteome Res. 2012:11(7):3605–3623. 10.1021/pr300027a [DOI] [PubMed] [Google Scholar]
- Yang X, Hu R, Yin H, Jenkins J, Shu S, Tang H, Liu D, Weighill DA, Yim WC, Ha J, et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat Commun. 2017:8(1):1899. 10.1038/s41467-017-01491-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Moore MJ, Brockington SF, Mikenas J, Olivieri J, Walker JF, Smith SA. Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytol. 2018:217(2):855–870. 10.1111/nph.14812 [DOI] [PubMed] [Google Scholar]
- Zhang X, Li X, Zhao R. Evolutionary strategies drive a balance of the interacting gene products for the CBL and CIPK gene families. New Phytol. 2020:226(5):1506–1516. 10.1111/nph.16445 [DOI] [PubMed] [Google Scholar]
- Zheng J, Meinhardt LW, Goenaga R, Zhang D, Yin Y. The chromosome-level genome of dragon fruit reveals whole-genome duplication and chromosomal co-localization of betacyanin biosynthetic genes. Hortic Res. 2021:8(1):63. 10.1038/s41438-021-00501-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman CA. Growth characteristics of weediness in Portulaca Oleracea L. Ecology. 1976:57(5):964–974. 10.2307/1941061 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.