Abstract
Arabidopsis (Arabidopsis thaliana) root development is regulated by multiple dynamic growth cues that require central metabolism pathways such as β-oxidation and auxin. Loss of the pectin biosynthesizing enzyme GALACTURONOSYLTRANSFERASE 10 (GAUT10) leads to a short-root phenotype under sucrose-limited conditions. The present study focused on determining the specific contributions of GAUT10 to pectin composition in primary roots and the underlying defects associated with gaut10 roots. Using live-cell microscopy, we determined reduced root growth in gaut10 is due to a reduction in both root apical meristem size and epidermal cell elongation. In addition, GAUT10 was required for normal pectin and hemicellulose composition in primary Arabidopsis roots. Specifically, loss of GAUT10 led to a reduction in galacturonic acid and xylose in root cell walls and altered the presence of rhamnogalacturonan-I (RG-I) and homogalacturonan (HG) polymers in the root. Transcriptomic analysis of gaut10 roots compared to wild type uncovered hundreds of genes differentially expressed in the mutant, including genes related to auxin metabolism and peroxisome function. Consistent with these results, both auxin signaling and metabolism were modified in gaut10 roots. The sucrose-dependent short-root phenotype in gaut10 was linked to β-oxidation based on hypersensitivity to indole-3-butyric acid (IBA) and an epistatic interaction with TRANSPORTER OF IBA1 (TOB1). Altogether, these data support a growing body of evidence suggesting that pectin composition may influence auxin pathways and peroxisome activity.
During root development, changes in cell wall composition facilitate cell growth properties and auxin metabolism.
Introduction
Roots are a key organ in plants for water and nutrient acquisition. In angiosperms, the primary root is established during embryogenesis and further elaborated via the coordination of complex cellular processes, including cell wall remodeling and hormone pathways. The plant cell wall is a dynamic structure during root development and can exhibit distinct properties based on cellular function (Somssich et al. 2016). In addition, the plant cell wall provides physical strength to the plant cells and can protect them from internal factors like turgor pressure and external factors like pathogens (Ridley et al. 2001). Because plant cells are held in place through cell walls, the root apical meristem (RAM) must monitor the state of the cell wall to properly coordinate cell divisions (Serra and Robinson 2020; Gu and Rasmussen 2022). While the transcription factor networks regulating the root stem cell populations have been established over time with extensive genetic studies (Sozzani and Iyer-Pascuzzi 2014; Fisher and Sozzani 2016), it is essential to understand the mechanisms beyond transcriptional regulation. In the root, the transition of daughter cells through differentiation leading to their maturation involves regulated cell elongation. Currently, limited information is available regarding cell wall properties within the Arabidopsis (Arabidopsis thaliana) RAM (Somssich et al. 2016).
In most angiosperms, including eudicots and nongraminaceous monocots, ∼35% of the primary cell wall is comprised of pectin (Mohnen 2008). Pectins are a family of polysaccharides, of which ∼70% are covalently linked units of galacturonic acid (GalA) residues (Keegstra et al. 1973). Pectins are synthesized in the Golgi apparatus and subsequently transported to the cell wall (Mohnen 2008), where they regulate cell wall properties such as extensibility and thickness (Majda and Robert 2018). The 3 major classes of pectins that are extensively studied are homogalacturonan (HG), rhamnogalacturonan-I (RG-I), and substituted galacturonans that are further subclassified as rhamnogalacturonan-II (RG-II), xylogalacturonan, and apiogalacturonan (Scheller et al. 2007; Mohnen 2008; Lampugnani et al. 2018).
Currently, limited information is available on the cell wall properties of root meristem cells compared to other tissues (Jobert et al. 2023). For example, the differences in the cell wall composition of mitotically dormant QC cells, surrounding stem initials, and the differentiating cell layers are not well understood (Somssich et al. 2016). In addition, the timing and regulation of polysaccharide deposition in differentiating root cells are poorly understood, but a few emergent properties have been described to date (Sinclair et al. 2022). Differentiating plant cells have a thin primary cell wall that facilitates frequent cell division (Baluska et al. 1996). In such dividing cells, callose predominantly constitutes the early stages of the cell plate assembly (Miart et al. 2014; Drakakaki 2015). In addition, pectins and hemicellulose contribute to the cell wall structure by forming a middle lamella that supports intercellular junctions (Drakakaki 2015). Cells at the transition zone within the primary root have a unique cell wall pectin composition, with an abundance of (1→4)-β-d-galactan that demarks the transition from mitotic activity to cell elongation (McCartney et al. 2003).
Galacturonosyltransferases (GAUTs) are a conserved family of enzymes involved in pectin biosynthesis and belong to the glycosyl transferase 8 family (GT8) in the Carbohydrate-Active Enzymes (CAZy) database (Cantarel et al. 2009). Genetic and biochemical characterization of gaut mutants revealed their unique role in modulating pectin and xylan polysaccharide composition in shoot tissues and contributing to shoot phenotypes (Caffall et al. 2009; Wang et al. 2013; Lund et al. 2020; Guo et al. 2021; Engle et al. 2022). Fifteen GAUT genes annotated in Arabidopsis are phylogenetically classified into 7 clades and 10 GAUT-like (GATL) genes (Caffall and Mohnen 2009). Two GAUT members from clade B-2 have been linked to root growth and development, GAUT10 and GAUT15. GAUT15 is transcriptionally regulated by auxin and is required for root gravitropism (Lewis et al. 2013). GAUT10 is regulated by auxin posttranscriptionally and is required for primary root growth and lateral root formation (Pu et al. 2019). Loss of GAUT10 leads to short roots with reduced RAM size in the absence of exogenous sucrose (Pu et al. 2019). In addition, GAUT10 has been shown to be localized to the Golgi and is involved in pectin biosynthesis in Arabidopsis inflorescence and stem tissues (Caffall et al. 2009; Voiniciuc et al. 2018; Guo et al. 2021). GAUT10 is also important for stomata formation in combination with GAUT11, an orthologous gene (Guo et al. 2021). Altogether, these studies suggest that GAUT10 may play numerous roles in plant development.
In order to understand better the biological processes that underpin the short-root phenotype of gaut10 seedlings, we performed molecular and genetic analyses. This work focused on the gaut10-3 allele as it has been previously characterized by several research groups as a null allele of GAUT10 (Caffall et al. 2009; Voiniciuc et al. 2018; Pu et al. 2019; Guo et al. 2021). To facilitate this study, we established a permissive low sucrose growth medium that enabled sufficient growth of gaut10-3 roots for molecular analyses. Under 0.5% (15 mm) sucrose, the short-root phenotype of gaut10-3 is due to both a reduction in RAM cell number and impaired epidermal cell elongation. Furthermore, the absence of GAUT10 impacts auxin-dependent gene expression and metabolism, suggesting that the cell wall composition may influence auxin pathways indirectly. In addition, we have characterized how GAUT10 specifically contributes to root pectin and hemicellulose composition. Genetic analysis with GAUT10 and TRANSPORTER OF IBA1 (TOB1) and indole-3-butyric acid (IBA) response assays indicated that the sucrose-dependent phenotype of gaut10-3 may be due to altered peroxisome biology. This study provides a working model for GAUT10 in root development and expands our understanding of how cell wall properties can influence root growth.
Results
GAUT10 is required for root cell division and elongation
Through quantitative proteomics coupled with a reverse genetic screen, we previously identified GAUT10 as an auxin downregulated protein required for root development (Clark et al. 2019; Pu et al. 2019). Loss of function alleles of gaut10 exhibit a short RAM phenotype compared to wild-type Col-0 when grown in 0.5× MS medium lacking sucrose (Pu et al. 2019), and the gaut10-3 null allele has been well characterized (Caffall et al. 2009; Pu et al. 2019; Guo et al. 2021). Supplementing the 0.5× MS medium with 1% sucrose rescues the gaut10-3 RAM to wild-type size (Fig. 1). To determine permissive root growth conditions for analyzing gaut10-3 roots, we grew Col-0 and gaut10-3 seedlings on 0.5× MS medium supplemented with 0, 0.5%, and 1% sucrose (Fig. 1). Based on this assay, we determined that 5-d-old gaut10-3 roots are statistically shorter than Col-0 when grown on 0.5× MS supplemented with 0.5% (15 mm) sucrose but still long enough to facilitate molecular and biochemical analyses.
Figure 1.
Primary root length is shorter in gaut10-3 compared to Col-0 grown in 0 and 15 mm sucrose; supplementing the growth medium with 30 mm sucrose rescues the root length phenotype. A) Five-day-old gaut10-3 and Col-0 whole seedlings grown on 0.5× MS supplemented with 3 different sucrose concentrations: 0, 15, and 30 mm. Images were digitally extracted for comparison with all scale bars = 0.5 cm. B) Violin plots of quantified root phenotypes in Col-0 and gaut10-3. Boxplots within the violin shapes represent the 5 number summaries, where the center line is the median, box limits are the upper and lower quartiles, whiskers are 1.5× interquartile range, and points are outliers. Statistical analysis was performed using a 1-way ANOVA followed by Tukey's post hoc analysis with a P < 0.05 to assign letters (A, B, and C) to each genotype/treatment that indicates statistical significance. “n” represents the number of biological replicates quantified. Sucrose concentrations are displayed as both a percentage (weight/volume) and molarity (mm).
To determine if the short-root phenotype of gaut10-3 roots was due to a reduction in cell numbers or cell elongation, we measured these properties in 5-d-old seedlings grown under 0.5% sucrose (Supplemental Table S1). The epidermal cells of gaut10-3 roots were shorter than Col-0 (Fig. 2, A, B, and I), suggesting that a defect in cell elongation may contribute to the observed short-root phenotype. In addition, we examined the RAM properties of gaut10-3 grown under 0.5% sucrose in more detail than what had been previously characterized (Pu et al. 2019). The Arabidopsis RAM has been described as consisting of “apical” and “basal” zones (Ishikawa and Evans 1993; Beemster et al. 2003; Verbelen et al. 2006; Zhang et al. 2010; Hacham et al. 2011). The apical region consists of dividing cells of uniform size, while the basal meristem, also called the “transition zone,” is comprised of slow-dividing cells that increase in size. To quantify the smaller RAM properties in gaut10-3 roots, the number of cells in the apical and basal meristem was measured using a previously described method (Hacham et al. 2011). Compared to Col-0, the number of both apical and root basal meristem (RBM) cells was reduced in gaut10-3 (Fig. 2, C to H). Altogether these data suggest that gaut10-3 roots are shorter due to both a reduction in cell number and cell elongation (Fig. 2J).
Figure 2.
The short phenotype of gaut10-3 roots is due to a smaller RAM and reduced cell elongation. Images were digitally extracted for comparison. The confocal images (20× magnification) in A, B) and C, D) show the root DZ and the root MZ of 5-d-old gaut10-3 and Col-0 seedlings, respectively, with all scale bars showing 20 μm length; the dotted double head arrows in A, B) represent the difference in the length of earliest trichoblast cells at the root DZ, and the dotted lines in C, D) demark the length of the root apical and basal meristems in gaut10-3 and Col-0 representative figures. Images are digitally extracted for comparison with all scale bars = 20 μm. Phenotypic differences observed in gaut10-3 as compared to Col-0 are quantified as box and whisker plots overlayed with scatter plots showing differences in the lengths G, H) and the number of cortical cells E, F) spanning the apical and the RBM. The meristem lengths are measured on a micrometer (μm) scale. For all phenotypic quantifications pertaining to root meristem E to I), 6 individual seedlings (biological replicates) from each genotype (Col-0 and gaut10-3) were used. In I), the average cell lengths of 6 to 10 early differentiating trichoblast cells (technical replicates) from 6 independent roots were quantified. Boxplots E to I) represent the 5 number summaries, where the center line is the median, box limits are the upper and lower quartiles, whiskers are 1.5× interquartile range, and points are outliers. The calculated P-values E to I) are from a nonparametric Wilcoxon rank-sum test used to test the statistical significance of phenotypic differences observed between the 2 genotypes. J) A cartoon schematic summarizing the short-root phenotype in gaut10 roots compared to Col-0, which is due to a smaller RAM (double-ended arrow) and shorter epidermal cells (arrows in the epidermal cells). RAM, root apical meristem; RBM, root basal meristem; MZ, meristematic zone; DZ, differentiation zone.
Epidermal and lateral root cap marker gene expression is diminished in gaut10-3
GAUT10 mRNA levels have been previously shown to be enriched in several root stem cell types, including the quiescent center, columella, and epidermis/lateral root cap (Epi/LRC) initials (Clark et al. 2019; Supplemental Fig. S1). To determine if the loss of GAUT10 impacts tissue-specific marker line expression in the RAM, we examined the expression patterns of several well-characterized GFP reporter lines for these cell types. For this experiment, wild-type and gaut10-3 5-d-old roots harboring root cell marker lines were grown in the absence (-suc) or presence of 0.5% sucrose (+suc) and imaged via confocal microscopy. In gaut10-3 roots, the SCR:GFP marker, which marks cortex/endodermal cell identity, shows diminished expression in a sucrose-dependent manner (Fig. 3A). In contrast, the QC marker WOX5:GFP is expressed normally in gaut10-3 (Fig. 3B), indicating that the stem cell organizing center is normal. In addition, marker lines for the columella (PET111:GFP) and lateral root cap (702LRC:GFP) are diminished in gaut10-3 compared to wild-type independent of sucrose (Fig. 3, C and D). An epidermal marker, WER:GFP, showed reduced expression in gaut10-3 in a sucrose-dependent manner compared to wild type (Fig. 3E). Altogether, these data suggest that GAUT10 may influence RAM marker gene expression in a sucrose-dependent manner.
Figure 3.
Root cell marker lines are altered in gaut10-3. Confocal images of Col-0 and gaut10-3 5-d-old primary roots grown in the presence of 15 mm sucrose (+suc) and absence of sucrose (−suc). A)SCR:GFP, which marks the endodermis. B)WOX5:GFP, which marks the quiescent center. C)PET111:GFP, which marks the columella. D)702LRC:GFP, which marks the lateral root cap. E)WER:GFP, which marks the lateral root cap. Scale bars = 20 µm. Images were digitally extracted for comparison.
Contributions of GAUT10 to cell wall composition
GAUT10 has been previously shown to contribute to the pectin biosynthesis of Arabidopsis shoot tissues (Caffall et al. 2009; Guo et al. 2021). To determine how the loss of GAUT10 may impact pectin composition in the root, we performed both monosaccharide composition analysis and glycome profiling (Fig. 4). The monosaccharide analysis of gaut10-3 roots compared to wild type grown under 0.5% sucrose indicated that GalA levels are reduced in the pectin-enriched fraction of gaut10-3 roots (Fig. 4A; Supplemental Table S2), which is consistent with a previous report on reduced GalA levels of gaut10 siliques (Caffall et al. 2009). The total cellulose content in gaut10-3 roots was the same as Col-0 (Supplemental Table S2). In addition, the xylose levels are significantly reduced in the hemicellulose-enriched fraction of gaut10-3 roots compared to wild type (Fig. 4B; Supplemental Table S2).
Figure 4.
Cell wall polysaccharide composition is altered in gaut10-3 roots. A, B) Concentrations of root cell wall monosaccharides are shown as mole fraction × 100 = mole percentage (Mol%) in the pectin- A) and the hemicellulose- B) enriched fractions from 3 biological replicates per genotype. The error bars show the Se of the mean (SEM). Statistical analysis was performed using a 2-sample nonparametric Wilcoxon rank-sum test; an asterisk indicates P ≤ 0.1. C) A 4D dot plot that shows differential binding of cell wall polysaccharide-specific antibodies in root cell wall extracts collected across 3 biological replicates from gaut10-3 and Col-0 measured by ELISA. The shape annotations show clades of the cell wall–binding antibodies (as defined in Pattathil et al. 2010) whereas the x axis indicates the corresponding cell wall fraction origin (i.e. pectin or hemicellulose). The size of the dots represents the log2 fold change (gaut10-3/Col-0) value calculated using the ELISA absorbance values. Significantly enriched/reduced polysaccharides (23 in total) with their binding epitopes are indicated according to Pattathil et al. (2010) annotations. Statistical significance is determined by a 2-sample nonparametric Wilcoxon rank-sum test with P ≤ 0.1. Antibody names and their binding cell wall epitope structures are annotated for each row. HG, homogalacturonan; RG-I, rhamnogalacturonan-I.
Glycome profiling was performed using a published ELISA-based method on the pectin and the hemicellulose-enriched fractions extracted from 5-d-old Col-0 and gaut10-3 roots grown under 0.5% sucrose. For this assay, a collection of 66 cell wall–specific antibodies was utilized (Pattathil et al. 2010, 2012). From these analyses, we identified 15 differentially enriched polysaccharide epitopes in the pectin-enriched fraction of gaut10-3 roots compared to the wild type and 12 in the hemicellulose-enriched fraction (Fig. 4C; Supplemental Table S2).
The epitopes present in the HG backbone (methyl esterified and demethyl esterified), RG-Is (RG-I backbone, RG-Ia, and RG-I/galactans), xylan backbone, and GlcA–xylan were enriched in the CDTA/ammonium oxalate–soluble pectin abundant fraction from gaut10-3 root cell walls in comparison with the same fraction prepared from Col-0 cell walls (Fig. 4C; Supplemental Table S2). In contrast, RG-Ic and xyloglucan (XXLG/XLLG) are reduced in the pectin-enriched fraction of gaut10-3 roots compared to Col-0 (Fig. 4C; Supplemental Table S2). In the hemicellulose-enriched fraction, 11 out of 12 significant epitopes were found to be reduced in gaut10-3 compared to Col-0, including RG-Is (RG-I backbone, RG-I/galactans, and linseed mucilage RG-I), xylans (xylan backbone, GlcA-xylan, 3Ara-xylan), and a xyloglucan (XLXG). Only one significant RG-I epitope, namely the β3-galactan backbone, was found to be enriched in the hemicellulose-enriched cell wall fraction of gaut10-3 roots in comparison to Col-0 (Fig. 4C; Supplemental Table S2). The observed reduction of xylose in the hemicellulose-enriched fraction (Fig. 4B) corresponds with the reduction of several xylan-related epitopes in glycome profiling of the same fraction (Fig. 4C). Altogether, these data demonstrate that GAUT10 influences RG and XG composition in primary roots.
Loss of GAUT10 impacts auxin and cell wall pathway gene expression
During root development, there is coordination between cell wall dynamics and gene expression (Taylor-Teeples et al. 2015). To determine how gene expression may be impacted in gaut10-3 roots, we performed transcriptomics on 5-d-old gaut10-3 and Col-0 roots grown on 0.5× MS +0.5% sucrose. Differentially expressed genes (DEGs) were identified using the DESeq2 package implemented in R (Love et al. 2014). With an adjusted P-value of <0.05, 109 upregulated and 176 downregulated genes were identified in gaut10-3 roots relative to Col-0 (Fig. 5A; Supplemental Fig. S2 and Table S3). Among these DEGs, we identified several key marker genes that are associated with root development and are consistent with the observed defects in gaut10-3 roots. For example, EXTENSIN18 (EXT18), which is required for root growth via cell elongation (Choudhary et al. 2015), is downregulated in gaut10-3 (Fig. 5B). In addition, several known auxin pathway genes are altered in gaut10-3 roots including YADOKARI 1 (YDK1), NITRILASE 1 (NIT1), MYB DOMAIN PROTEIN 34 (MYB34), CA2+-DEPENDENT MODULATOR OF ICR1 (CMI1), and INDOLE-3-ACETIC ACID INDUCIBLE 17/AUXIN RESISTANT 3 (IAA17/AXR3) (Fig. 5B). Collectively, YDK1, NIT1, and MYB34 are important regulators of auxin metabolism (Bartling et al. 1992, 1994; Bartel and Fink 1994; Takase et al. 2004; Celenza et al. 2005; González-Lamothe et al. 2012; Lehmann et al. 2017). CMI1 is an auxin-regulated gene that modulates auxin responses in the root meristem via a Ca2+-dependent pathway (Hazak et al. 2019). Loss of function of cmi1 results in a shorter primary root with reduced root meristem size (Hazak et al. 2019), which is consistent with the gaut10-3 root phenotype. IAA17/AXR3 encodes for an Aux/IAA transcription factor that represses auxin-inducible gene expression (Nagpal et al. 2000; Nakamura et al. 2006; Muto et al. 2007). Altogether, the transcriptomic analysis suggests that the altered cell wall composition in gaut10-3 may influence auxin pathway gene expression.
Figure 5.
Transcriptomic analysis of 5-d-old gaut10-3 and Col-0 roots across 3 biological replicates. A) Volcano plot showing DEGs with an adjusted Padj. value (false discovery rate) threshold of 0.05, with log2 fold change on the x axis and −log10(adjusted Padj. value) on the y axis. The upregulated genes (109) and downregulated genes (176) are above the dashed line; transcripts that are not significantly changed are below the dashed line. B) Expression values for notable key marker genes are shown in the bar plot with log2 fold change on the y axis and gene abbreviations on the x axis: YDK1, EXT18, CMI1, AUXIN-INDUCED IN ROOT CULTURES 1 (AIR1), NIT1, AXR3, and MYB34. C) The top GO terms for biological processes enriched by the DE transcripts in gaut10-3 compared to Col-0 are shown in a 4D graph, where the size and color of the puncta show the number of DEGs/proteins enriched within each GO term and their statistical significance, respectively.
In order to identify enriched biological processes among DEGs in gaut10-3 compared to the wild type, we performed Gene Ontology (GO) enrichment analysis. Overall, numerous GO terms that are typically associated with plant stress and defense are significantly enriched among DEGs in gaut10-3. Among upregulated genes, there is an enrichment for “response to hydrogen peroxide” and numerous biotic and abiotic stress terms (Fig. 5C). In addition, 2 classical defense and stress hormone GO terms are enriched (“response to salicylic acid” and “response to abscisic acid”) among the downregulated genes in gaut10 (Fig. 5C). In contrast, among the upregulated genes in gaut10-3 are GO biological process (GOBP) terms associated with “plant epidermis development,” “response to jasmonic acid,” and “flavonoid biosynthetic development” (Fig. 5C). This GO analysis suggests that loss of GAUT10 may impact numerous plant growth and stress responses.
Local auxin signaling is inhibited in the gaut10-3 root meristem
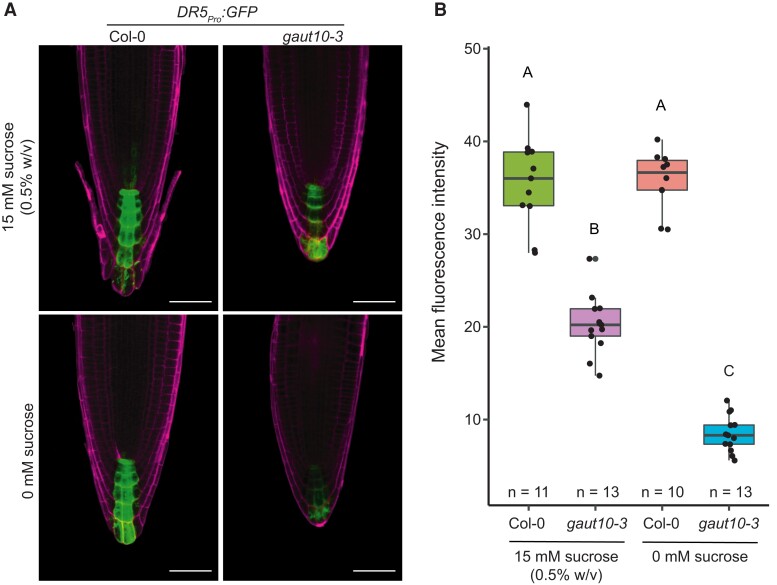
The phytohormone auxin regulates much critical growth and developmental cues in plants. Because GAUT10 abundance is regulated by auxin (Pu et al. 2019) and several auxin pathway genes are DE in gaut10-3 roots (Fig. 5), we wanted to examine auxin signaling using the DR5:GFP reporter, which is a synthetic promoter consisting of 7 tandem repeats of an auxin-responsive TCTCTC element and a minimal 35S CaMV promoter driving expression GFP (Friml et al. 2003; Heisler et al. 2005; Hayashi et al. 2014). We observed a significant reduction of DR5:GFP expression in gaut10-3 roots compared to Col-0 (Fig. 6, A and B), which was exacerbated under sucrose-deficit growth conditions (Fig. 6, A and B; Supplemental Table S4). Altogether, these results suggested a defect in local auxin signaling in the gaut10-3 root meristem.
Figure 6.
Auxin signaling defects in gaut10-3 roots compared to wild type. A) Representative 20× confocal images of DR5:GFP expression in 5-d-old Col-0 and gaut10-3 roots grown on 0 and 15 mm sucrose. Scale bars = 20 µm. Images were digitally extracted for comparison. B) Mean fluorescence intensity (MFI) quantified for each genotype/treatment in y axis. Boxplots represent the 5 number summaries, where the center line is the median, box limits are upper and lower quartiles, whiskers are 1.5× interquartile range, and points are outliers. Statistical analysis was a 1-way ANOVA followed by Tukey's post hoc analysis with a P < 0.05 to assign letters (A, B, and C) to each genotype/treatment to indicate statistical significance. “n” represents the number of biological replicates used for averaging.
Auxin metabolism is altered in gaut10-3 roots
Auxin homeostasis plays a major role in establishing postembryonic root architecture (Jones and Ljung 2012). In concert with polar auxin transport, local auxin biosynthesis forms the spatiotemporal auxin gradient within the root apex that plays an essential role in root development (Ljung et al. 2005; Petersson et al. 2009; Casanova-Sáez et al. 2021). Plants maintain auxin homeostasis through de novo auxin biosynthesis or by controlling the levels of active auxin through conjugation (primarily amino acids and simple sugars) or by degradation (Normanly 2010; Ruiz Rosquete et al. 2012; Casanova-Sáez et al. 2021). We identified several known auxin metabolite genes as DE in gaut10-3 roots (Fig. 5B), suggesting that auxin metabolism or signaling may be altered in this mutant. Specifically, YDK1 conjugates aspartate and other amino acids to IAA, consequently lowering active auxin levels in vivo in a tissue-specific manner (Takase et al. 2004; González-Lamothe et al. 2012). NIT1 encodes an enzyme that catalyzes the conversion of indole-3-acetonitrile (IAN) to indole-3-acetamide (IAM) as it modulates IAA biosynthesis (Bartling et al. 1992, 1994; Bartel and Fink 1994; Lehmann et al. 2017). NIT1 overexpression results in shorter primary roots in seedlings, explaining its role in regulating root development via auxin homeostasis (Lehmann et al. 2017). MYB34 plays a key role in the transcriptional regulation of enzymes that maintain the metabolic homeostasis of tryptophan (TRP) and indole glucosinolates in Arabidopsis (Celenza et al. 2005). To determine how the observed transcriptional changes to auxin metabolism genes in gaut10-3 roots impact auxin metabolism, we measured auxin metabolites in Col-0 and gaut10-3 roots grown in the presence of 0.5% sucrose.
IAA levels are the same in gaut10-3 compared to wild type (Supplemental Fig. S3); however, several IAA precursors and conjugates were altered in abundance (Fig. 7). Anthranilate (ANT), the primary rate-limiting precursor of TRP biosynthesis, was increased in gaut10-3 roots compared to Col-0 (Fig. 7A). In contrast, TRP levels were significantly reduced in gaut10-3 roots compared to Col-0 (Fig. 7B). IAN, which is debatably involved in indole-3-acetaldoxime (IAOx) to IAA conversion (Sugawara et al. 2009; Casanova-Sáez et al. 2021), was depleted in gaut10-3 roots (Fig. 7C). In addition, several inactive forms of IAA were significantly decreased in gaut10-3 compared to wild type, including IAA–aspartate (IAA–Asp), IAA–glutamate (IAA–Glu), IAA–glucose (IAA–Glc), and 2-oxindole-3-acetic acid–Glc (oxIAA–Glc) (Fig. 7, D to G). The observed reduction of IAN and IAA–Asp and increase in ANT are consistent with the differential expression of their respective metabolic enzymes (YDK1, NIT1, and MYB34) in gaut10-3 roots (Fig. 7H). Collectively, these data suggest that pectin composition may influence auxin metabolism within primary roots.
Figure 7.
Auxin metabolism is altered in gaut10-3 roots compared to wild type. Metabolite quantification by LC–MS/MS visualized by box and whisker plots for with the corresponding P-value (P) indicated for each metabolite across 5 biological replicates per genotype. For all metabolites quantified, a 2-sample nonparametric Wilcoxon rank-sum test followed by Benjamini–Hochberg correction for multiple testing was performed to identify significantly altered metabolites with P ≤ 0.1. A) ANT. B) TRP. C) IAN. D) IAA–Asp. E) IAA–Glu. F) IAA–Glc. G) oxIAA–Glc. Boxplots A to G) represent the 5 number summaries, where the center line is the median, box limits are the upper and lower quartiles, whiskers are 1.5× interquartile range, and the points are outliers. H) Summary model figure showing annotated auxin biosynthesis pathways. Reduced metabolites are indicated in gaut10-3 roots in red text; likewise, pathway enzymes shown in red are downregulated, whereas the genes in blue text show increased transcript levels. The solid arrows indicate pathways that have known enzymes, genes, and intermediates, while dashed arrows indicate pathways that are not well defined.
Genetic interactions between GAUT10 and TOB1
To infer possible mechanisms associated with the sucrose-dependent phenotype of gaut10-3 roots, we examined the transcriptomic data (Supplemental Table S5). Sucrose-dependent root growth in Arabidopsis has been linked to numerous pathways, including sugar signaling, sugar transport, and lipid metabolism via the peroxisome (Woodward 2005; Marzec and Kurczynska 2014; Urano et al. 2016; Wang et al. 2021). Among the DEGs in gaut10-3 roots, there were no hallmark sugar-signaling genes. However, 2 genes related to peroxisome function that were altered in gaut10-3 roots include TOB1 and PEROXIN 11C (Supplemental Table S3). Peroxisome-defective mutants exhibit hallmark sucrose-dependent short-root phenotypes (Strader et al. 2011), suggesting that such dysfunction may underpin the gaut10-3 phenotype. In gaut10-3 roots, TOB1 transcript is elevated while PEX11C expression is decreased. To examine potential genetic interactions between GAUT10 and TOB1, we generated a gaut10-3 tob1-3 double mutant. Previous work established that gaut10-3 roots respond normally to IAA (Pu et al. 2019), but the response of gaut10-3 to IBA had not been characterized in this mutant. The response to IBA and IAA was examined in gaut10-3, tob1-3, and gaut10-3 tob1-3 roots (Fig. 8). Both wild-type and gaut10-3 roots are inhibited to exogenous IAA treatment while tob1-3 roots are slightly insensitive, as previously reported (Michniewicz et al. 2019) (Fig. 8, A, H, and N). Both tob1-3 and gaut10-3 are insensitive to IBA, and the gaut10-3 tob1-3 double mutant exhibits a response like tob1-3 (Fig. 8N). In addition, the short-root phenotype of gaut10-3 is suppressed in the gaut10-3 tob1-3 background (Fig. 8M). Altogether, these data suggest an epistatic interaction between GAUT10 and TOB1.
Figure 8.
Genetic interaction between gaut10 and tob1. A, E, I) Wild-type Col-0 roots grown under 0.5% sucrose (mock), 0.5% sucrose + 10 mm IBA, or 0.5% sucrose + 10 mm IAA. B, F, J)gaut10-3 roots grown under 0.5% sucrose (mock), 0.5% sucrose + 10 mm IBA, or 0.5% sucrose + 10 mm IAA. C, G, K)tob1-3 roots grown under 0.5% sucrose (mock), 0.5% sucrose + 10 mm IBA, or 0.5% sucrose + 10 mm IAA. D, H, L)gaut10-3 tob1-3 roots grown under 0.5% sucrose (mock), 0.5% sucrose + 10 mm IBA, or 0.5% sucrose + 10 mm IAA. Images in A to L) were digitally extracted for comparison with all scale bars = 0.5 cm. M) Quantification of primary root length of Col-0, gaut10-3, tob1-3, and gaut10-3 tob1-3. Boxplots represent the 5 number summaries, where the center line is the median, box limits are the upper and lower quartiles, whiskers are 1.5× interquartile range, and points are outliers. N) IBA and IAA responses in Col-0, gaut10-3, tob1-3, and gaut10-3 tob1-3. All statistical analysis was a 1-way ANOVA followed by Tukey's post hoc analysis with a P < 0.1 to assign letters (A, B, and C) to each genotype/treatment to indicate statistical significance. “n” represents the number of biological replicates used for averaging in all cases. The error bars represent the Se.
Discussion
In the elongation zone of the primary root, cells transition from the meristematic zone (MZ) and rapidly elongate along the longitudinal axis while preventing any lateral expansion of cells that require cell wall remodeling (Ledbetter and Porter 1963; Schiefelbein and Benfey 1991; Somssich et al. 2016). Cell wall remodeling enzymes largely contribute to such unidimensional cell expansions in response to internal turgor pressure (Ledbetter and Porter 1963; Cosgrove 2005). GalA is one of the main monosaccharides constituting all pectin polysaccharides, including HG, RG-I, RG-II, and xylogalacturonan (Caffall and Mohnen 2009). Many of the characterized GAUT mutants display substantial reductions in the amount of GalA present in their cell walls prepared from various above-the-ground parts of the plants (Caffall et al. 2009; Voiniciuc et al. 2018; Guo et al. 2021); however, some mutants like gaut13 and gaut14 showed higher content of GalA in their siliques and inflorescences (Caffall et al. 2009). To our knowledge, roots have not been analyzed in any gaut mutant plants. Here, we demonstrated that gaut10-3 roots also have cell walls with reduced GalA and xylose. Our results are consistent with previous results obtained for gaut10-1 and gaut10-2 inflorescence and siliques (Caffall et al. 2009). Double gaut10 gaut11 plants also showed reduced pectin biosynthesis in stomata cells (Guo et al. 2021). Recombinant GAUT10 showed catalytic activity when given UDP–GalA as a substrate; however, the activity was very low, and it has been proposed that GAUT10 may have a unique role in synthesizing either a unique HG compared to other GAUTs (such as GAUT1) or an HG connected to RG-I (Engle et al. 2022).
Monosaccharides such as GalA and xylose are significantly reduced in the pectin and hemicellulose-enriched cell wall fractions of gaut10-3 roots, respectively, which showed a net decrease in GalA and xylose containing polysaccharides in the gaut10-3 root cell wall as compared to Col-0. The glycome analysis performed here of both cell wall fractions observed significant alterations in the levels of specific polysaccharides, which is consistent with the monosaccharide analysis. Antibodies specific to GalA containing RG-I and HG epitopes were enriched in the pectin fraction. We also observed a significant decrease in RG-I, xylan, and xyloglucan-related epitopes in the hemicellulose-enriched fraction. The increased HG and multiple RG-I pectin–related epitopes, specifically in pectin fraction, might indicate the higher extractability of polysaccharides carrying such epitopes due to their weaker binding nature in the gaut10-3 cell wall in comparison with cell walls of Col-0. For example, epitopes recognized by 4 particular cell wall antibodies (M126, M107, M72, and M150) were increased in the more soluble fraction extracted with CDTA–ammonium oxalate but consequently reduced in the following extract with a strong alkali, suggesting the increased solubility of the corresponding polysaccharide molecules within gaut10 roots. It is also possible that cell wall reinforcement in gaut10 roots is loosened, resulting in reduced cell expansion and suppressed growth that is necessary to compensate for the changes in how specific polysaccharides are integrated into cell walls. In the future, more detailed studies will be needed to clarify the impact of loss of GAUT10 on the specific polysaccharide structure/interactions and the general impact on cell wall organization in root cells.
Interestingly, gaut10 siliques and inflorescences did not show a reduction of xylose (Caffall et al. 2009); however, a reduction of both GalA and xylose was observed earlier in another gaut mutant, quasimodo 1 (qua1/gaut8) (Orfila et al. 2005). Pectic galactans have been associated with wall strengthening (McCartney et al. 2000), and the higher levels of pectin-specific epitopes in gaut10-3 mutant roots in comparison with Col-0 roots may also be a result of cell wall structural changes leading to increased cell wall rigidity in mutant root cells resulting in suppression of their growth. Altogether, we can infer that GAUT10 plays a critical role in synthesizing/maintaining pectic polysaccharides within primary root cell walls (Fig. 9).
Figure 9.
Working model for roles of GAUT10 in root morphogenesis. A) In wild-type Col-0 roots, normal cell wall composition leads to correct RAM size and DR5:GFP maxima in the QC and columella. B) In the absence of GAUT10, pectin composition is altered leading to reduced cell expansion and DR5:GFP expression. C) A molecular framework for GAUT10 function based on biochemical and genetic analyses of gaut10-3 roots. IAA represses GAUT10 protein accumulation, which is a positive regulator of pectin and hemicellulose formation in roots. GAUT10-dependent cell wall composition is required for primary root morphogenesis and for auxin metabolism.
GAUT10 was identified as an auxin downregulated protein (Clark et al. 2019; Pu et al. 2019) with roles in the regulation of RAM size by a reverse genetic study with 3 loss-of-function alleles (Pu et al. 2019). Here, we performed transcriptomics and auxin metabolomics on gaut10-3 to substantiate its role as a cell wall enzyme that interacts with auxin pathways and further delineate how GAUT10 function may be linked to auxin biology. Transcriptomic analysis of gaut10-3 roots implicated auxin metabolism and signaling associated with the short-root phenotype, which was validated by DR5:GFP expression analysis and auxin metabolite quantification. In the absence of GAUT10, both IAA precursors and inactive conjugates were altered. Within the lateral root cap, cytokinin promotes the degradation of IAA into IAA–Glu, an inactive conjugate, thereby controlling root meristem size and root growth by inducing cell differentiation (di Mambro et al. 2019). The diminished expression of 702LRC:GFP in gaut10-3 roots indicates that the lateral root cap may be altered, although the cell files appear normal, which might explain the observed decrease in IAA–Glu (along with other IAA conjugates). Previous work has shown that modulation of IAA conjugation and GH3 activity can influence root morphogenesis in Arabidopsis (Mateo-Bonmatí et al. 2021; Casanova-Sáez et al. 2022). The observed decrease of numerous IAA conjugates (IAA–Asp, IAA–Glu, IAA–Glc, and oxIAA–Glc) in gaut10-3 roots suggests that IAA homeostasis is influenced by cell wall properties. The differential expression of enzymes catalyzing the conversion of these auxin conjugates and precursors in gaut10-3 compared to wild-type roots is consistent with this working model (Fig. 7H).
The short-root phenotype in gaut10-3 manifests due to both a reduction in RAM size and reduced elongation of mature epidermal cells, suggesting that pectin composition can influence both cell proliferation and elongation during root growth. Indeed, the role of GAUT10 in influencing cell elongation properties is further supported by overexpression of genes enriched for the “plant epidermis development” GO term in the gaut10-3 roots. Root morphogenesis can be studied by visualizing the expression of cell-type–specific markers (Bargmann et al. 2013). In wild-type Col-0 roots, the expression of several cell-type–specific marker lines was not altered by the presence or absence of sucrose (Fig. 3). This result is in line with a recent report which demonstrated that sucrose can influence the relative proportion of cells within a cell type but overall does not alter cell type identities (Shulse et al. 2019). In contrast, we observed reduced expression of several root cell marker lines (SCR:GFP, WER:GFP, PET111:GFP, and 702LRC:GFP) in gaut10-3 in the absence of sucrose while expression of the QC marker WOX5:GFP was normal. However, the corresponding radial and longitudinal cell patterning in gaut10-3 roots appears normal. These results may be interpreted such that marker gene expression is diminished due to a change in pectin composition, but organ patterning remains normal in the absence of GAUT10. Given that numerous plant growth and stress genes are DE in gaut10-3 roots, these data suggest several downstream cellular processes may be impaired when pectin composition is altered. Future investigations of cell wall composition across the developing primary root may be informative for identifying which cell-type–specific contexts are associated with particular HG or RG moieties.
Sucrose positively regulates root stem cell activation and promotes root epidermal cell elongation in Arabidopsis (Wu et al. 2019; Dong et al. 2022). The sucrose-dependent root growth defects of gaut10-3 suggest a potential link between cell wall composition and sucrose-dependent growth. Notably, links between cell wall dynamics and central metabolism have been reported in studies on trehalose-6-phosphate synthase1 (tps1) (Gómez et al. 2006), glycosyl hydrolases (Lee et al. 2007), high sugar response8 (hsr8), and murus (mur) mutants (Li et al. 2007). In the absence of TPS1, which is required for the conversion of UDP-Glc to Glc-6P, pectin composition is altered, and the expression of numerous cell wall pathway genes is altered (Gómez et al. 2006). These data suggest that alteration in trehalose metabolism can impact cell wall metabolism. In addition, sugar-hypersensitive phenotypes have been reported for several enzymes that contribute to cell wall formation, namely glycosyl hydrolases and arabinose synthesis mutants, namely β-galactosidase, β-xylosidase, β-glucosidase, hsr8/mur4, mur1, and mur3 (Lee et al. 2007; Li et al. 2007). Characterization of hsr8/mur4 seedlings led to a proposed pathway from the cell wall to the nucleus that influences sugar-mediated growth (Li et al. 2007). More investigations into this observed phenomenon will be required to understand better the signal transduction pathway(s) between the cell wall and central carbon metabolism.
Several complex pathways are known to influence sugar-dependent growth in Arabidopsis including SUCROSE NON-FERMENTING 1 (SNF1)-RELATED KINASE 1 (SnRK1), TARGET OF RAPAMYCIN (TOR), HEXOKINASE 1 (HXK1), trehalose 6-phosphate (Tre6P), and β-oxidation (Rinaldi et al. 2016; Fichtner et al. 2021). It was not known which pathway(s) were impaired in gaut10-3 roots to explain the observed sugar-dependent short-root phenotype. Based on the transcriptomic analysis of gaut10-3, it appeared that alteration of β-oxidation–based processes could explain the sucrose-dependent short-root phenotype, which is a hallmark of this pathway (Rinaldi et al. 2016). Specifically, the expression of TOB1 was upregulated in gaut10-3, while PEX11C was decreased. In plant cells, the conversion of IBA to IAA requires functional peroxisomes and IBA transport via TOB1 (Damodaran and Strader 2019; Michniewicz et al. 2019). The altered response to IBA but normal response to IAA in gaut10-3 roots is consistent with altered peroxisome function. In addition, the observed epistatic interaction between TOB1 and GAUT10 supports a working model that links the cell wall composition to IAA metabolism (Fig. 9C). Thus, it appears that the short-root phenotype of gaut10-3 can be rescued by exogenous sucrose due to peroxisome dysfunction. Notably, other studies have also uncovered links between cell wall status and peroxisome function (Kanai et al. 2010; Rinaldi et al. 2016; Dai et al. 2019). Peroxisome defective mutants identified from forward genetic screens include PECTIN MEHTYLESTERASE31 (PME31) and ANTHER DEHISCENCE REPRESSOR (ADR), which both contribute to cell wall composition (Rinaldi et al. 2016; Dai et al. 2019). In addition, alteration of a peroxisomal ABC transporter PED3/PXA1/CTS can impair pectin composition in seeds (Kanai et al. 2010). Collectively, these studies suggest that cell wall composition may feedback to auxin homeostasis via several distinct pathways to influence root morphogenesis.
Materials and methods
Plant material
All seed stocks used in this study were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. Arabidopsis (A. thaliana) plants used in this study were all in the Col-0 background. SALK_092577 corresponds to the gaut10-3 null allele, which has been previously characterized (Caffall et al. 2009; Voiniciuc et al. 2018; Pu et al. 2019; Guo et al. 2021). SALK_205450 corresponds to the tob1-3 knockdown allele, previously characterized as a knockdown allele (Michniewicz et al. 2019). The WEREWOLF:GFP (epidermal cell marker), 702LRC:GFP (lateral root cap marker), SCARECROW:GFP (cortex/endodermal cell marker), PET111:GFP (columella cell marker), and DR5:GFP lines have been previously described (Friml et al. 2003; Heisler et al. 2005; Bargmann et al. 2013; Hayashi et al. 2014).
For phenotyping assays, seeds were surface sterilized using 50% bleach (v/v) and 0.01% Triton X-100 (v/v) for 10 min and then washed 5 times with sterile water. Seeds were then imbibed in sterile water for 2 d at 4 °C and then transferred to 0.5× MS plates supplemented with 15 mm sucrose and 0.8% agar (w/v). Seedlings were grown under long-day photoperiods (16 h light/8 h dark) at 23 °C in a Percival growth chamber.
Root phenotyping
Intact seedlings (5-d-old) were imaged on a flatbed scanner (Epson V600). Measurements of primary root lengths were performed using ImageJ. Roots were stained with propidium iodide (PI) and imaged under a confocal microscope for root meristem and differentiation zone (DZ) phenotyping using a 20× objective. Measurements of root meristem lengths (apical and basal) and the epidermal cells at the DZ were performed using ImageJ, and the number of cortical cells was counted across 6 biological replicates as previously described (Hacham et al. 2011). Individual cells (technical replicates) were measured from 6 independent roots per genotype (biological replicates). Two-sample nonparametric Wilcoxon rank-sum tests were performed to assess statistical significance.
Genotyping
All primers used for genotyping are provided in Supplemental Table S6 based on previously published studies (Caffall et al. 2009; Michniewicz et al. 2019; Pu et al. 2019).
Confocal microscopy
Homozygous gaut10-3 plants were crossed with the following transgenic lines: WEREWOLF:GFP, 702LRC:GFP, SCARECROW:GFP, PET111:GFP, and DR5:GFP. After the initial crosses, F1 seedlings were genotyped by PCR using T-DNA and transgene-specific primers and confirmed to be heterozygous for GAUT10/gaut10-3 and carrying the correct GFP transgenes. F1 plants were selfed and then carried through to the F3 generation; genotypes were verified by PCR at each generation. F3 individuals that were homozygous for gaut10-3 and each GFP reporter were bulked for imaging. Five-day-old roots of these double transgenic and corresponding control transgenic lines were stained with PI and imaged under a 20× objective on a Zeiss LSM 700 confocal microscope. GFP was imaged with excitation at 488 nm and collection at 555 nm. PI was imaged with excitation at 555 nm and collection at 640 nm. For all experiments, the gain was set to 650, and the intensity was set to 2%. GFP fluorescence intensity was calculated in ImageJ as previously described (Lisi et al. 2012; Kim et al. 2017). In total, 6 to 10 biological replicates were imaged for each genotype under each growth condition.
Transcriptomic profiling
Transcriptomic analyses were performed as previously described (Dash et al. 2021). Col-0 and homozygous gaut10-3 seeds were surface sterilized, followed by cold stratification for 3 d in the dark at 4 °C. Next, they were plated on 0.5× MS + 15 mm sucrose plates overlaid with sterile 100-µ nylon mesh squares to facilitate tissue harvesting. Seedlings were grown under long-day photoperiods (16 h light/8 h dark) at 23 °C. Roots from 5-d-old seedlings were dissected using surgical knives and then weighed and snap frozen in liquid nitrogen; approximately 100 mg of seedling root tissue was collected per replicate/genotype. Three independent biological replicates were generated for each genotype. Snap-frozen roots were ground to a fine powder in liquid nitrogen using a mortar and pestle. Total RNA was extracted from the root samples using TRIzol, followed by column clean-up using the Quick-RNA plant kit (Zymo research). Total RNA concentration was estimated using a NanoDrop and Qubit. RNA quality was checked via Bioanalyzer at the ISU DNA Facility. QuantSeq 3′ mRNA libraries were prepared using the Lexogen 3′ mRNA-seq FWD kit and sequenced on an Illumina HiSeq 3000 as 50 bp reads at the ISU DNA Facility. QuantSeq reads were mapped to the TAIR10 genome using STAR (Dobin et al. 2013), and differential gene expression analysis was performed using DeSeq2 implemented in R (Love et al. 2014). Transcripts with an false discovery rate (FDR) cutoff of <0.05 and a log2 fold change ≥ 0.5 were defined as DE.
GO analysis
DEGs were tested for statistical overrepresentation of GOBP terms using Panther and the A. thaliana database. A standard Fisher’s exact test with FDR correction was used to calculate GO term enrichment against the total set of detected transcripts as the reference. Top GOBP terms were plotted on the y axis against their genotype/treatments on the x axis in multidimensional dot plots.
Cell wall, pectin, and hemicellulose extraction
Total cell wall material, pectin, and hemicellulose fractions from 3 biological replications of Col-0 and gaut10-3 root samples grown in 0.5× MS + 15 mm sucrose growth medium were prepared as previously described (Zabotina et al. 2012). Briefly, the root samples were ground to a fine powder in liquid nitrogen, and the cell wall was extracted by using successive organic solvents, starting with 80% (v/v) ethanol, followed by 80% (v/v) acetone, chloroform:methanol (1:1, v/v), and finally with 100% acetone. The resultant cell wall material was air dried, and the pectin and hemicellulose polysaccharides were extracted successively by using 50 mm CDTA:50 mm ammonium oxalate (1:1) buffer (v:v) and 4 m KOH, respectively. The extracts were neutralized with acetic acid, dialyzed against water, and dried by lyophilization. The dried pectin-enriched and hemicellulose-enriched extracts were stored at 4 °C for further analysis.
Glycome profiling of epitopes of pectin and hemicellulose extracts
Glycome profiling was performed as previously described (Pattathil et al. 2010, 2012). The amount of sugar in the pectin and hemicellulose extracts was determined by the phenol–sulfuric acid method, and each ELISA well was loaded with an equal amount of polysaccharide sample dissolved in water (50 µL/well from a 60 ng/µL solution). Glycome profiling was carried out using 66 different mouse primary glycome antibodies purchased from the Complex Carbohydrate Research Center (University of Georgia) as previously described (Pattathil et al. 2010, 2012; Zabotina et al. 2012). The color development was detected at 450 nm wavelength using a plate reader, and each sample’s optical density (OD) reading was statistically analyzed and compared. The ELISA assays were repeated in triplicate for both the pectin and hemicellulose extracts. Statistical analysis was performed using a nonparametric Wilcoxon rank-sum test with a P ≤ 0.1 to compare mean OD450 absorbances between Col-0 and gaut10-3.
Monosaccharide composition analysis
Monosaccharide composition was determined according to previously described methods (Brenner et al. 2012). One milligram of cell wall was hydrolyzed with 2 m trifluoroacetic acid (TFA) and analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC–PAD) (Dionex, Sunnyvale, CA) using a CarboPac PA20 column. The column was calibrated using monosaccharide standards purchased from Sigma-Aldrich, which included L254fucose (l-Fuc), l-rhamnose (l-Rha), l-arabinose (l-Ara), d-galactose (d-Gal), D255Glc (d-Glc), d-xylose (d-Xyl), d-mannose (d-Man), d-GalA, and d-glucuronic acid (D256GlcA) (Brenner et al. 2012). The content of different monosaccharides in the hydrolysates analyzed was estimated as a molecular fraction (mol%). Statistical analysis was performed using a nonparametric Wilcoxon rank-sum test with a P ≤ 0.1 to compare mean monosaccharide concentrations between Col-0 and gaut10-3.
Cellulose content measurements
The cellulose content present in the cell wall of Col-0 and gaut10-3 roots was estimated by a modified Updegraff reagent method (Updegraff 1969; Kumar and Turner 2015). From each sample, 75 to 80 mg of cell wall was placed in a preweighed screw cap glass tube, and 2 mL of Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 v/v) was added. The glass tubes were tightly closed with polytetrafluoroethylene (PTFE) seal caps. Subsequently, the tubes were incubated in a boiling water bath for 30 min, cooled to room temperature, and centrifuged at 1750 × g rpm for 10 min to pellet down the cellulose. The supernatant was removed by aspiration. The cellulose pellet was washed 5 times with water and twice with 100% acetone by repeated centrifugation and aspiration. The resultant cellulose was dried at 37° C in an oven. The dried cellulose pellet was weighed, and percent content was calculated.
Auxin metabolite profiling
Auxin metabolite profiling was performed as previously described (Novák et al. 2012). Five-day-old gaut10-3 and Col-0 roots grown on 0.5× MS + 0.5% (15 mm) sucrose were harvested in pools of 25 mg and flash frozen; 5 biological replicates were analyzed for each genotype. Extraction and LC–MS were subsequently performed as published. Statistical analysis was performed using a nonparametric Wilcoxon rank-sum test with a P ≤ 0.1 to compare mean auxin metabolite concentrations between Col-0 and gaut10-3.
IAA and IBA treatment assays
Homozygous gaut10-3 (gaut10 −/−) and tob1-3 (tob1 −/−) mutant lines were crossed to produce F1 individuals, which were verified by PCR to be heterozygous for both alleles (gaut10 +/−; tob1 +/−). F1 plants were selfed to obtain an F2 population containing double homozygous mutant seedlings of the PCR-verified genotype: gaut10 −/− and tob1 −/−. F2 individuals were selfed to obtain F3 seeds of the desired genotypes. Genotyping for the GAUT10, TOB1, gaut10-3, and tob1-3 alleles was conducted using PCR primers (Supplemental Table S6). Col-0, gaut10-3, tob1-3, and gaut10-3 tob1-3 seeds obtained from the F3 individuals were surface sterilized using 50% bleach for 10 min, followed by 5 sterile water washes. After 3 d of imbibition in sterile water and cold stratification in the dark at 4 °C, the seeds were plated on 0.5 MS + 15 mm sucrose medium. Seeds were scored for germination under a long-day photoperiod (16 h light, 8 h dark) at 23 °C. Three days postgermination, the seedlings were treated with 10 μm IAA and 10 μm IBA and both dissolved in 95% ethanol or an equivalent volume of 95% ethanol (“mock”) for 72 h by transferring the seedlings to square Petri dishes containing fresh 0.5× MS + 15 mm sucrose supplemented with IAA/IBA or mock solvent. After 72 h after transfer, the seedlings were imaged on a flatbed scanner (Epson V600). Measurements of primary root lengths were performed using ImageJ for 70 to 80 replicates per genotype/treatment. A 1-way ANOVA followed by Tukey’s post hoc analysis was performed to test the statistical significance of the root length variations observed between these 4 genotypes.
Accession numbers
Raw RNA sequence read data from this article can be found at the NCBI BioProject database under accession PRJNA694693 and ID 694693. All accession numbers for Arabidopsis genes mentioned in this study are listed in Supplemental Table S3.
Supplementary Material
Acknowledgments
We wish to thank Bastiaan Bargmann for sharing seed stocks. We also acknowledge the Swedish Metabolomics Centre for technical support.
Contributor Information
Linkan Dash, Department of Genetics, Development and Cell Biology, Iowa State University, Iowa City, IA 50011, USA.
Sivakumar Swaminathan, Roy J Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Iowa City, IA 50011, USA.
Jan Šimura, Department of Forest Genetics and Plant Physiology, Umeå Plant Science Centre, Swedish University of Agricultural Sciences, Umeå 901 83, Sweden.
Caitlin Leigh P Gonzales, Department of Genetics, Development and Cell Biology, Iowa State University, Iowa City, IA 50011, USA.
Christian Montes, Department of Plant Pathology, Entomology, and Microbiology, Iowa State University, Iowa City, IA 50011, USA.
Neel Solanki, Department of Genetics, Development and Cell Biology, Iowa State University, Iowa City, IA 50011, USA.
Ludvin Mejia, Department of Genetics, Development and Cell Biology, Iowa State University, Iowa City, IA 50011, USA.
Karin Ljung, Department of Forest Genetics and Plant Physiology, Umeå Plant Science Centre, Swedish University of Agricultural Sciences, Umeå 901 83, Sweden.
Olga A Zabotina, Roy J Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Iowa City, IA 50011, USA.
Dior R Kelley, Department of Genetics, Development and Cell Biology, Iowa State University, Iowa City, IA 50011, USA.
Author contributions
L.D., S.S., J.Š., K.L., O.A.Z., and D.R.K. were involved in design of the research. L.D., C.L.P.G., S.S., J.Š., C.M., N.S., and L.M. performed the research. Data analysis, collection, and interpretation were performed by L.D., S.S., J.Š., O.A.Z., and D.R.K. The manuscript was written by L.D., S.S., O.A.Z., and D.R.K. with input from the other authors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript abundance of GAUT10 in different cell types of A. thaliana root meristem.
Supplemental Figure S2. Transcriptomic analysis of 5-d-old gaut10-3 and Col-0 roots across 3 biological replicates.
Supplemental Figure S3. Additional auxin metabolite measurements.
Supplemental Table S1. Root phenotyping data of 5-d-old gaut10-3 and Col-0 roots.
Supplemental Table S2. Glycome and monosaccharide composition data of 5-d-old gaut10-3 and Col-0 roots.
Supplemental Table S3. Transcriptomic analysis data from 5-d-old gaut10-3 and Col-0 roots with gene expressions averaged across 3 biological replicates.
Supplemental Table S4. DR5:GFP fluorescence quantification data.
Supplemental Table S5. Auxin metabolite concentrations of 5-d-old gaut10-3 and Col-0 roots.
Supplemental Table S6. Primers used in this study.
Funding
This study was supported by the National Science Foundation Award Number 2118253 to D.R.K. and O.A.Z. K.L. and J.Š. were funded by the Swedish Research Council (VR 2018-04235), the Knut and Alice Wallenberg Foundation (KAW 2016.0341 and KAW 2016.0352), and the Swedish Governmental Agency for Innovation Systems (VINNOVA 2016-00504).
Data availability
The transcriptome data that support the findings of this study are openly available at the NCBI BioProject database under accession PRJNA694693 and ID 694693. The phenotyping, glycome, and auxin metabolite data are available within the supplementary material. All seed stocks are available upon request.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Baluska F, Volkmann D, Barlow PW. Specialized zones of development in roots: view from the cellular level. Plant Physiol. 1996:112(1):3–4. 10.1104/pp.112.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BOR, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, et al. A map of cell type-specific auxin responses. Mol Syst Biol. 2013:9(1):688. 10.1038/msb.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994:91(14):6649–6653. 10.1073/pnas.91.14.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Mithofer A, Weiler EW. Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur J Biochem. 1992:205(1):417–424. 10.1111/j.1432-1033.1992.tb16795.x [DOI] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler EW. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc Natl Acad Sci USA. 1994:91(13):6021–6025. 10.1073/pnas.91.13.6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Fiorani F, Inzé D. Cell cycle: the key to plant growth control? Trends Plant Sci. 2003:8(4):154–158. 10.1016/S1360-1385(03)00046-3 [DOI] [PubMed] [Google Scholar]
- Brenner EA, Salazar AM, Zabotina OA, Lübberstedt T. Characterization of European forage maize lines for stover composition and associations with polymorphisms within O-methyltransferase genes. Plant Sci. 2012:185–186:281–287. 10.1016/j.plantsci.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009:344(14):1879–1900. 10.1016/j.carres.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D. Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant. 2009:2(5):1000–1014. 10.1093/mp/ssp062 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009:37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Sáez R, Mateo-Bonmatí E, Ljung K. Auxin metabolism in plants. Cold Spring Harb Perspect Biol. 2021:13(3):a039867. 10.1101/cshperspect.a039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005:137(1):253–262. 10.1104/pp.104.054395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary P, Saha P, Ray T, Tang Y, Yang D, Cannon MC. EXTENSIN18 is required for full male fertility as well as normal vegetative growth in arabidopsis. Front Plant Sci. 2015:6:553. 10.3389/fpls.2015.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Buckner E, Fisher AP, Nelson EC, Nguyen TT, Simmons AR, de Luis Balaguer MA, Butler-Smith T, Sheldon PJ, Bergmann DC, et al. Stem-cell-ubiquitous genes spatiotemporally coordinate division through regulation of stem-cell-specific gene networks. Nat Commun. 2019:10(1):5574. 10.1038/s41467-019-13132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005:6(11):850–861. 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Dai S-Y, Hsu W-H, Yang C-H. The gene ANTHER DEHISCENCE REPRESSOR (ADR) controls male fertility by suppressing the ROS accumulation and anther cell wall thickening in Arabidopsis. Sci Rep. 2019:9(1):5112. 10.1038/s41598-019-41382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damodaran S, Strader LC. Indole 3-butyric acid metabolism and transport in Arabidopsis thaliana. Front Plant Sci. 2019:10:851. 10.3389/fpls.2019.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash L, McEwan RE, Montes C, Mejia L, Walley JW, Dilkes BP, Kelley DR. Slim shady is a novel allele of PHYTOCHROME B present in the T-DNA line SALK_015201. Plant Direct 2021:5(6):e00326. 10.1002/pld3.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Mambro R, Svolacchia N, dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, et al. The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol. 2019:29(7):1199–1205.e4. 10.1016/j.cub.2019.02.022 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aref R, Forieri I, Schiel D, Leemhuis W, Meyer C, Hell R, Wirtz M. The plant TOR kinase tunes autophagy and meristem activity for nutrient stress-induced developmental plasticity. Plant Cell 2022:34(10):3814–3829. 10.1093/plcell/koac201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G. Polysaccharide deposition during cytokinesis: challenges and future perspectives. Plant Sci. 2015:236:177–184. 10.1016/j.plantsci.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Engle KA, Amos RA, Yang J, Glushka J, Atmodjo MA, Tan L, Huang C, Moremen KW, Mohnen D. Multiple Arabidopsis galacturonosyltransferases synthesize polymeric homogalacturonan by oligosaccharide acceptor-dependent or de novo synthesis. Plant J. 2022:109(6):1441–1456. 10.1111/tpj.15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner F, Dissanayake IM, Lacombe B, Barbier F. Sugar and nitrate sensing: a multi-billion-year story. Trends Plant Sci. 2021:26(4):352–374. 10.1016/j.tplants.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Fisher AP, Sozzani R. Uncovering the networks involved in stem cell maintenance and asymmetric cell division in the Arabidopsis root. Curr Opin Plant Biol. 2016:29:38–43. 10.1016/j.pbi.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 2003:426(6963):147–153. 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA. Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 2006:46(1):69–84. 10.1111/j.1365-313X.2006.02662.x [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, el Oirdi M, Brisson N, Bouarab K. The conjugated auxin indole-3-acetic acid–aspartic acid promotes plant disease development. Plant Cell 2012:24(2):762–777. 10.1105/tpc.111.095190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Rasmussen CG. Cell biology of primary cell wall synthesis in plants. Plant Cell 2022:34(1):103–128. 10.1093/plcell/koab249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Xiao C, Liu Q, Li R, Yan Z, Yao X, Hu H. Two galacturonosyltransferases function in plant growth, stomatal development, and dynamics. Plant Physiol. 2021:187(4):2820–2836. 10.1093/plphys/kiab432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011:138(5):839–848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Nakamura S, Fukunaga S, Nishimura T, Jenness MK, Murphy AS, Motose H, Nozaki H, Furutani M, Aoyama T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc Natl Acad Sci USA. 2014:111(31):11557–11562. 10.1073/pnas.1408960111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Mamon E, Lavy M, Sternberg H, Behera S, Schmitz-Thom I, Bloch D, Dementiev O, Gutman I, Danziger T, et al. A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Biol. 2019:17(7):e3000085. 10.1371/journal.pbio.3000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005:15(21):1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. The role of the distal elongation zone in the response of maize roots to auxin and gravity. Plant Physiol. 1993:102(4):1203–1210. 10.1104/pp.102.4.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert F, Yadav S, Robert S. Auxin as an architect of the pectin matrix. J Exp Bot. 2023. 10.1093/jxb/erad174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Ljung K. Subterranean space exploration: the development of root system architecture. Curr Opin Plant Biol. 2012:15(1):97–102. 10.1016/j.pbi.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Kanai M, Nishimura M, Hayashi M. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 2010:62(6):936–947. 10.1111/j.1365-313X.2010.04205.x [DOI] [PubMed] [Google Scholar]
- Keegstra K, Talmadge KW, Bauer WD, Albersheim P. The structure of plant cell walls. Plant Physiol. 1973:51(1):188–197. 10.1104/pp.51.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim D-I, Kim EK, Kim C-W. CXCR4 overexpression in human adipose tissue-derived stem cells improves homing and engraftment in an animal limb ischemia model. Cell Transplant 2017:26(2):191–204. 10.3727/096368916X692708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Turner S. Protocol: a medium-throughput method for determination of cellulose content from single stem pieces of Arabidopsis thaliana. Plant Methods 2015:11(1):46. 10.1186/s13007-015-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Khan GA, Somssich M, Persson S. Building a plant cell wall at a glance. J Cell Sci. 2018:131(2):jcs207373. 10.1242/jcs.207373 [DOI] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR. A “microtubule” in plant cell fine structure. J Cell Biol. 1963:19(1):239–250. 10.1083/jcb.19.1.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Matsumura Y, Soga K, Hoson T, Koizumi N. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 2007:48(3):405–413. 10.1093/pcp/pcm009 [DOI] [PubMed] [Google Scholar]
- Lehmann T, Janowitz T, Sánchez-Parra B, Alonso M-MP, Trompetter I, Piotrowski M, Pollmann S. Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front Plant Sci. 2017:8:36. 10.3389/fpls.2017.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell 2013:25(9):3329–3346. 10.1105/tpc.11553.114868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW. Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 2007:19(8):2500–2515. 10.1105/tpc.106.049965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi A, Briganti E, Ledda M, Losi P, Grimaldi S, Marchese R, Soldani G. A combined synthetic-fibrin scaffold supports growth and cardiomyogenic commitment of human placental derived stem cells. PLoS One 2012:7(4):e34284. 10.1371/journal.pone.0034284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. Sites and regulation of auxin biosynthesis in arabidopsis roots. Plant Cell 2005:17(4):1090–1104. 10.1105/tpc.104.029272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund CH, Stenbæk A, Atmodjo MA, Rasmussen RE, Moller IE, Erstad SM, Biswal AK, Mohnen D, Mravec J, Sakuragi Y. Pectin synthesis and pollen tube growth in arabidopsis involves three GAUT1 Golgi-anchoring proteins: gAUT5, GAUT6, and GAUT7. Front Plant Sci. 2020:11:585774. 10.3389/fpls.2020.585774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majda M, Robert S. The role of auxin in cell wall expansion. Int J Mol Sci. 2018:19(4):951. 10.3390/ijms19040951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M, Kurczynska E. Importance of symplasmic communication in cell differentiation. Plant Signal Behav. 2014:9(1):e27931. 10.4161/psb.27931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo-Bonmatí E, Casanova-Sáez R, Šimura J, Ljung K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021:232(2):642–654. 10.1111/nph.17633 [DOI] [PubMed] [Google Scholar]
- McCartney L, Ormerod andrew P, Gidley MJ, Knox JP. Temporal and spatial regulation of pectic (14)-beta-D-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J. 2000:22(2):105–113. 10.1046/j.1365-313x.2000.00719.x [DOI] [PubMed] [Google Scholar]
- McCartney L, Steele-King CG, Jordan E, Knox JP (2003) Cell wall pectic (1→4)-β-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33(3): 447–454 10.1046/j.1365-313X.2003.01640.x [DOI] [PubMed] [Google Scholar]
- Miart F, Desprez T, Biot E, Morin H, Belcram K, Höfte H, Gonneau M, Vernhettes S. Spatio-temporal analysis of cellulose synthesis during cell plate formation in arabidopsis. Plant J. 2014:77(1):71–84. 10.1111/tpj.12362 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Ho C-H, Enders TA, Floro E, Damodaran S, Gunther LK, Powers SK, Frick EM, Topp CN, Frommer WB, et al. TRANSPORTER OF IBA1 links auxin and cytokinin to influence root architecture. Dev Cell. 2019:50(5):599–609.e4. 10.1016/j.devcel.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008:11(3):266–277. 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 2007:144(1):187–196. 10.1104/pp.107.096628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000:123(2):563–574. 10.1104/pp.123.2.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 2006:45(2):193–205. 10.1111/j.1365-313X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol. 2010:2(1):a001594–a001594. 10.1101/cshperspect.a001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012:72(3):523–536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong H-N, Reid JSG, Knox JP, Scheller HV. QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 2005:222(4):613–622. 10.1007/s00425-005-0008-z [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010:153(2):514–525. 10.1104/pp.109.151985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Miller JS, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: glycome profiling. Biomass conversion. Totowa (NJ): Humana Press; 2012. p. 61–72 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009:21(6):1659–1668. 10.1105/tpc.109.066480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Walley JW, Shen Z, Lang MG, Briggs SP, Estelle M, Kelley DR. Quantitative early auxin root proteomics identifies GAUT10, a galacturonosyltransferase, as a novel regulator of root meristem maintenance. Mol Cell Proteomics. 2019:18(6):1157–1170. 10.1074/mcp.RA119.001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley BL, O’Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001:57(6):929–967. 10.1016/S0031-9422(01)00113-3 [DOI] [PubMed] [Google Scholar]
- Rinaldi MA, Patel AB, Park J, Lee K, Strader LC, Bartel B. The roles of β-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics 2016:204(3):1089–1115. 10.1534/genetics.116.193169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Rosquete M, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol Plant. 2012:5(4):772–786. 10.1093/mp/ssr109 [DOI] [PubMed] [Google Scholar]
- Scheller RM, Domingo JB, Sturtevant BR, Williams JS, Rudy A, Gustafson EJ, Mladenoff DJ. Design, development, and application of LANDIS-II, a spatial landscape simulation model with flexible temporal and spatial resolution. Ecol Modell. 2007:201(3–4):409–419. 10.1016/j.ecolmodel.2006.10.009 [DOI] [Google Scholar]
- Schiefelbein JW, Benfey PN. The development of plant roots: new approaches to underground problems. Plant Cell 1991:3(11):1147–1154. 10.1105/tpc.3.11.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra L, Robinson S. Plant cell divisions: variations from the shortest symmetric path. Biochem Soc Trans. 2020:48(6):2743–2752. 10.1042/BST20200529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M, Turco GM, Zhu Y, O’Malley RC, Brady SM, et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019:27(7):2241–2247.e4. 10.1016/j.celrep.2019.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R, Hsu G, Davis D, Chang M, Rosquete M, Iwasa JH, Drakakaki G. Plant cytokinesis and the construction of new cell wall. FEBS Lett. 2022:596(17):2243–2255. 10.1002/1873-3468.14426 [DOI] [PubMed] [Google Scholar]
- Somssich M, Khan GA, Persson S. Cell wall heterogeneity in root development of Arabidopsis. Front Plant Sci. 2016:7:1242. 10.3389/fpls.2016.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Iyer-Pascuzzi A. Postembryonic control of root meristem growth and development. Curr Opin Plant Biol. 2014:17:7–12. 10.1016/j.pbi.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid–derived auxin. Plant Cell 2011:23(3):984–999. 10.1105/tpc.111.083071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA. 2009:106(13):5430–5435. 10.1073/pnas.0811226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004:37(4):471–483. 10.1046/j.1365-313X.2003.01973.x [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015:517(7536):571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. Semimicro determination of cellulose inbiological materials. Anal Biochem. 1969:32(3):420–424. 10.1016/S0003-2697(69)80009-6 [DOI] [PubMed] [Google Scholar]
- Urano D, Miura K, Wu Q, Iwasaki Y, Jackson D, Jones AM. Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol. 2016:57(3):437–445. 10.1093/pcp/pcw002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen J-P, De CT, Le J, Vissenberg K, Baluška F. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities. Plant Signal Behav. 2006:1(6):296–304. 10.4161/psb.1.6.3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Engle KA, Günl M, Dieluweit S, Schmidt MH-W, Yang J-Y, Moremen KW, Mohnen D, Usadel B. Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 2018:178(3):1045–1064. 10.1104/pp.18.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Le Gourrierec J, Jiao F, Demotes-Mainard S, Perez-Garcia M-D, Ogé L, Hamama L, Crespel L, Bertheloot J, Chen J, et al. Convergence and divergence of sugar and cytokinin signaling in plant development. Int J Mol Sci. 2021:22(3):1282. 10.3390/ijms22031282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang W, Wang Y-Q, Liu Y-Y, Wang J-X, Zhang X-Q, Ye D, Chen L-Q. Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Mol Plant. 2013:6(4):1131–1148. 10.1093/mp/sst084 [DOI] [PubMed] [Google Scholar]
- Woodward AW. Auxin: regulation, action, and interaction. Ann Bot. 2005:95(5):707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Shi L, Li L, Fu L, Liu Y, Xiong Y, Sheen J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J Exp Bot. 2019:70(8):2227–2238. 10.1093/jxb/erz028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou Y-H, Eberhard S, Danhof L, Keegstra K, Hahn MG. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 2012:159(4):1367–1384. 10.1104/pp.112.198119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jürgens G, Paul Knox J, Wang M-H. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010:64(5):764–774. 10.1111/j.1365-313X.2010.04367.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data that support the findings of this study are openly available at the NCBI BioProject database under accession PRJNA694693 and ID 694693. The phenotyping, glycome, and auxin metabolite data are available within the supplementary material. All seed stocks are available upon request.