Abstract
The ability of Enterococcus faecalis to metabolically adapt to an oligotrophic environment has been analyzed. E. faecalis is able to survive for prolonged periods under conditions of complete starvation established by incubation in tap water. During incubation in this microcosm, cells developed a rippled cell surface with irregular shapes. Exponentially growing cells survived to the same extent as cells starved for glucose prior to exposure to the multiple nutrient deficient stress. Chloramphenicol treatment during incubation in tap water led to a rapid decline in plate counts for exponentially growing cells but showed progressively reduced influence on stationary-phase cells harvested after different times of glucose starvation. During incubation in the oligotrophic environment, cells from the exponential-growth phase and early-stationary phase became progressively more resistant to other environmental stresses (heat [62°C], acid [pH 3.3], UV254 nm light [180 J/m2], and sodium hypochlorite [0.05%]) until they reached a maximum of survival characteristic for each treatment. In contrast, cells starved of glucose for 24 h did not become more resistant to the different treatments during incubation in tap water. Our combined data suggest that energy starvation induces a response similar to that triggered by oligotrophy. Analysis of protein synthesis by two-dimensional gel electrophoresis revealed the enhanced synthesis of 51 proteins which were induced in the oligotrophic environment. A comparison of these oligotrophy-inducible proteins with the 42 glucose starvation-induced polypeptides (J. C. Giard, A. Hartke, S. Flahaut, P. Boutibonnes, and Y. Auffray, Res. Microbiol. 148:27–35, 1997) showed that 16 are common between the two different starvation conditions. These proteins and the corresponding genes seem to play a key role in the observed phenomena of long-term survival and development of general stress resistance of starved cultures of E. faecalis.
The natural habitat of enteric bacteria is the intestine of humans and animals. When discharged into a natural aquatic system, these allochthonous microorganisms need to adapt for survival in this hostile environment. Water systems are characterized by their oligotrophic nature (18, 19), and nutrient starvation seems to be one of the abiotic factors that negatively affects survival (2).
Recent advances in studying the physiological response of nondifferentiating bacteria following starvation of different individual nutrients has led to an understanding that such bacteria undergo a concerted rapid change in the pattern of gene expression (13). The metabolic reprogramming leads to a cellular state of enhanced resistance, compared to that in exponentially growing cells, to a great number of various stress conditions (see references 9 and 10 and references therein).
Fecal streptococci are considered to be good indicators of fecal contamination since they are present in the feces of humans and warm-blooded animals. Despite this fact, fecal streptococci have received, relative to fecal coliforms and in particular Escherichia coli, only minor attention from researchers studying the destiny of allochthonous bacteria after their release into aquatic systems. Compared to other streptococci, Enterococcus faecalis is considered to survive longer in the aquatic environment (2) and hence should be the most suitable indicator of fecal contamination in water. Furthermore, this gram-positive, nonsporulant bacterium is known as an opportunistic pathogen that causes urinary tract infection and is responsible for the majority of cases of subacute bacterial endocarditis. Because of the increasing importance of the health risks associated with exposure to contaminated water and of hospital-acquired infections, which are due to an alarming increase in resistance to various antibiotics, we have initiated studies to increase our fundamental understanding and knowledge of E. faecalis survival strategies after its release into hostile environments (3, 6–10, 15, 16, 23).
We have recently determined the starvation stress response to glucose of E. faecalis. During the first 24 h of the energy starvation period at least 42 proteins with time-dependent sequential synthesis were induced (10). The carbohydrate-starved cells showed enhanced resistance to lethal heat, oxidative, acid, ethanol (9), and NaOCl (16) stresses, compared to the growing cells, indicating that energy starvation in E. faecalis triggers development of a generally resistant phenotype. The time necessary to reach maximal resistance in the stationary phase was shown to be characteristic for each stress condition.
As stated above, starvation of only one nutrient is rarely encountered by fecal bacteria after release by the host. The objective of this study was to determine whether E. faecalis develops general stress resistance in an aquatic environment characterized by its oligotrophic nature. Furthermore, we were interested in whether the response triggered by oligotrophy overlaps with that induced in rich medium after the exhaustion of the energy source glucose. Therefore, exponentially growing and preadapted glucose-starved cells of E. faecalis were introduced into tap water, and survival rates as well as the development of resistance to different treatments were determined. Furthermore, to gain insight into the adaptation process of this fecal bacterium to an aquatic environment at the molecular level, we have analyzed changes in protein metabolism by two-dimensional gel electrophoresis.
MATERIALS AND METHODS
Bacterial strain, culture conditions, and introduction into the oligotrophic microcosm.
This study was performed with E. faecalis JH2-2 obtained from the parental strain JH2 (12). Cultures were grown without shaking at 37°C in 20-ml glass tubes containing 10 ml of semisynthetic medium (Bacto-Folic AOAC Medium; Difco, Detroit, Mich.) supplemented with 0.15% glucose. Under these conditions cultures entered stationary phase at an optical density of 600 nm (OD600) of 1.1, a value which corresponds to approximately 3 × 108 CFU/ml. Cells from the exponential-growth phase were harvested at an OD600 of 0.5. Prior to introduction into tap water, the cultures were harvested by centrifugation and washed twice in an equal volume of sterile 0.9% NaCl at room temperature. Finally, the pellets were suspended in tap water (obtained from the distribution system of the city of Caen, France) to a final concentration of approximately 108 CFU/ml and incubated at 16°C without shaking in the presence or absence of chloramphenicol (100 μg/ml). Prior to its use, tap water was filtered through a 0.2-μm (pore size) cellulose filter (Millipore Corp., Bedford, Mass.) and autoclaved for 15 min at 121°C.
Challenge conditions and cell count.
For heat, acid, and NaOCl challenges the cells were harvested by centrifugation and resuspended in semisynthetic medium either prewarmed to 62°C or adjusted to pH 3.3 by HCl or containing 0.05% (vol/vol) NaOCl (Sigma Chemical Co., St. Louis, Mo.). Free and total chlorine were determined colorimetrically by the dialkyl-p-phenylenediamine method (Merck, Darmstadt, Germany). Cultures incubated in the microcosms were irradiated with UV254 nm light in a standard petri dish without any pretreatment. Control cultures were harvested prior to irradiation by centrifugation and resuspended in 10 ml of 0.9% NaCl before transfer to a petri dish. In all cases, UV irradiation was performed at a dose rate of 150 J/m2.
To determine culturable bacterial counts, serial dilutions were made in 0.9% NaCl, and the diluted cells were poured into M17 (26) agar (1.5% [wt/vol]; Difco) supplemented with 0.5% glucose. Plates were incubated at 37°C for 48 h. Survival at any time point corresponded to the ratio of CFU after incubation in tap water or after a given challenge to the number of CFU of untreated control cultures. In all experiments each point is the average of three platings. Each experiment was repeated a minimum of three times, and the data reported are replicate means.
Electron microscopy.
E. faecalis cells were fixed by the addition of glutaraldehyde to a final concentration of 2% (wt/vol) in 0.1 M sodium cacodylate buffer (pH 6.8) (SCB). After this first fixation the cells were rinsed with 0.1 M SCB and fixed for 1 h in 1% (wt/vol) osmium tetroxide in 0.1 M SCB. The samples were then washed twice with 0.1 M SCB, dehydrated with acetone, critical-point dried by the CO2 method of Anderson (1), and coated with gold. Cells were examined and photographed with a JEOL-JSM 6400F field emission scanning electron microscope operating at 5 kV.
Labeling of proteins and two-dimensional gel electrophoresis.
Culture conditions were as described above. Culture aliquots of 1 ml were pulse-labeled with 250 μCi of [35S]methionine-[35S]cysteine protein labeling mix (1,000 Ci/mmol; New England Nuclear). Exponential-growth-phase cells were labeled for 40 min at 37°C (between OD600 0.25 and 0.5) in semisynthetic medium. Labeling of cultures in tap water was done by the addition of radiolabel at the onset of oligotrophy, and the cell suspensions were incubated for different times at 16°C. Cells were harvested by centrifugation and resuspended in 500 μl of lysozyme buffer (25 mM Tris-base [pH 7.0; Millipore] containing 10 μg of lysozyme [Sigma] per ml, 1 mM phenylmethylsulfonyl fluoride [Sigma], 100 μg of chloramphenicol per ml, and 0.5 M sucrose). Control cultures from the exponential-growth phase were treated in the same manner except that the cells were washed twice in cold 0.9% NaCl before suspension in the lysozyme buffer. After 5 min at 37°C, cells were harvested by centrifugation, and lysis was performed by the addition of 200 μl of buffer I (0.3% sodium dodecyl sulfate, 200 mM dithiothreitol [Merck], 28 mM Tris-HCl [Millipore], and 22 mM Tris-base). After 5 min at 100°C, samples were chilled on ice, and 24 μl of buffer II (24 mM Tris-base, 476 mM Tris-HCl, 50 mM MgCl2, 1 mg of DNase I [Sigma] per ml, and 0.25 mg of RNase A [Sigma] per ml) was added. The reaction was stopped after 15 min at 4°C by the addition of 4 volumes of ice-cold acetone, and precipitation of proteins was allowed to continue for 20 min on ice. Proteins were collected by centrifugation at 15,000 rpm for 15 min and were suspended in 40 μl of buffer III (540 mg of urea per ml, 10 mg of dithiothreitol per ml, 2% [vol/vol] Ampholytes [pH 4 to 8; Millipore], and 0.52% Triton-X 100). High-resolution two-dimensional electrophoresis was performed according to the method of O’Farrell (21) with modifications as described previously (17) and using the Multiphor II system (Pharmacia Biotech, Uppsala, Sweden) for the first dimension and the Investigator-2D electrophoresis system (Millipore) for the second dimension. Labeled proteins were visualized by autoradiography after 4 to 12 weeks at −80°C by exposure on Hyperfilm-MP (Amersham International). Analysis of the autoradiograms was performed by visual inspection.
RESULTS
Survival of E. faecalis in an oligotrophic microcosm.
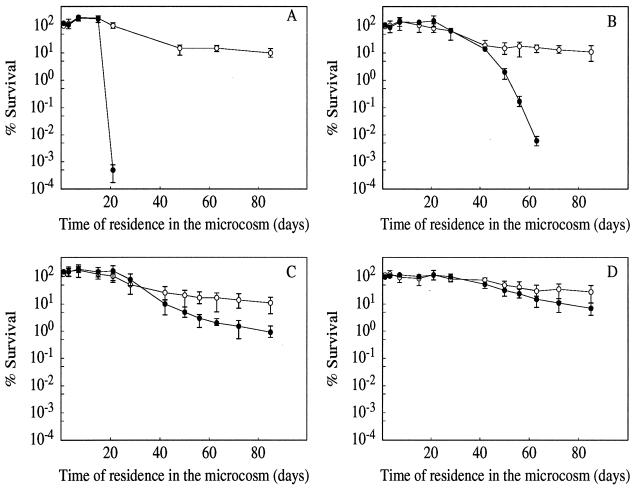
The survival of E. faecalis in tap water at 16°C is shown in Fig. 1. During the 85 days of incubation no significant differences were seen between growing cells and those harvested at the onset of glucose starvation or after 3 and 24 h in the stationary phase. In all cases 10 to 30% of cells were culturable.
FIG. 1.
Survival of E. faecalis in membrane-filtered, autoclaved tap water obtained from the distribution system of the city of Caen, France. Cells were harvested from the exponential-growth phase (A) and during a time course of the stationary phase—at onset (B), 3 h (C), and 24 h (D)—established by exhaustion of the energy source glucose. Each experiment was conducted in triplicate in the absence (○) or presence (•) of chloramphenicol (100 μg/ml). The data are means and standard deviations (bars) for two independent experiments.
To determine whether de novo protein synthesis was necessary for adapting to the oligotrophic environment, we treated the cultures during incubation in tap water with chloramphenicol. Under these conditions the culturability of the cells from the exponential-growth phase decreased rapidly after 20 days of incubation. On the other hand, cells harvested from the stationary phase showed progressively reduced sensitivity to the blockage of protein synthesis. The survival of cultures from the onset of starvation started to decline after 40 days; those harvested after 3 and 24 h were practically insensitive to the chloramphenicol treatment.
Morphological changes of E. faecalis upon residence in the nutrient-poor microcosm.
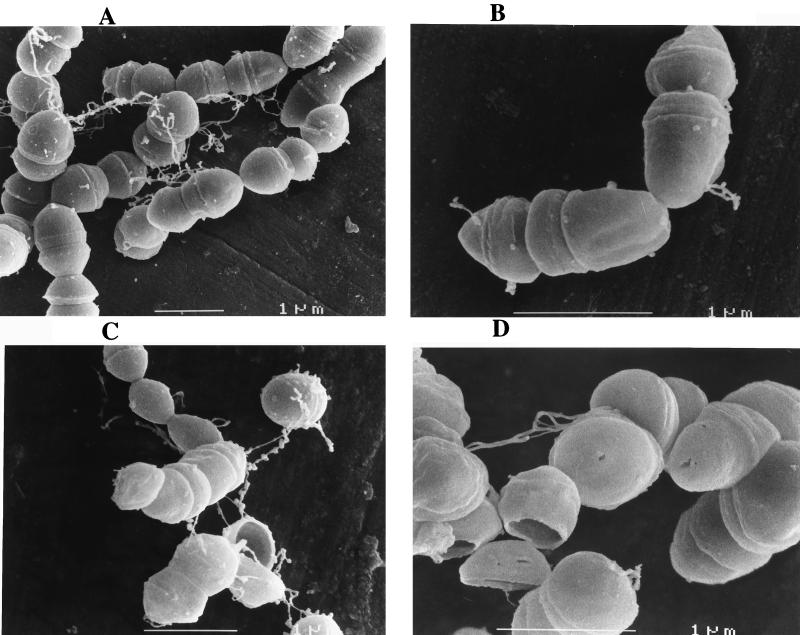
Scanning electron micrographs revealed that cells incubated for 24 h under oligotrophic conditions showed no obvious difference from growing cultures (Fig. 2). The cells had a smooth and spherical appearance and were organized mainly in chains. However, the cells developed a rippled cell surface with irregular shapes and significant alterations within 3 to 7 weeks of starvation. At these longer incubation times, cells were mainly organized as pairs; only rarely were longer chains detected. Some cells had collapsed envelopes, and other cells showed breaks in the septal regions and had surface tears (Fig. 2).
FIG. 2.
Analysis of morphological changes of E. faecalis harvested from the exponential-growth phase during incubation in tap water. Over the entire experiment some representative photographs obtained after 24 h (A), 3 weeks (B), 4 weeks (C), and 7 weeks (D) are shown.
Development of multiresistance of E. faecalis during incubation in tap water.
The former experiments showed that E. faecalis was able to survive for long periods under oligotrophic conditions and that survival was independent of the physiological state of the cells prior to the transfer to the microcosms (Fig. 1). One phenomenon induced by starvation is that microorganisms develop general stress resistance (13). Our previous results showed that glucose starvation triggers resistance to a number of environmental stresses in E. faecalis, e.g., heat (62°C), lactic acid (pH 3.2), H2O2 (20 mM), ethanol (17% [vol/vol]) (9), and NaOCl (10−2% [vol/vol]) (16). We were therefore interested in determining whether E. faecalis could evolve into a more resistant state under conditions that were closer to situations encountered by allochthonous bacteria in the environment.
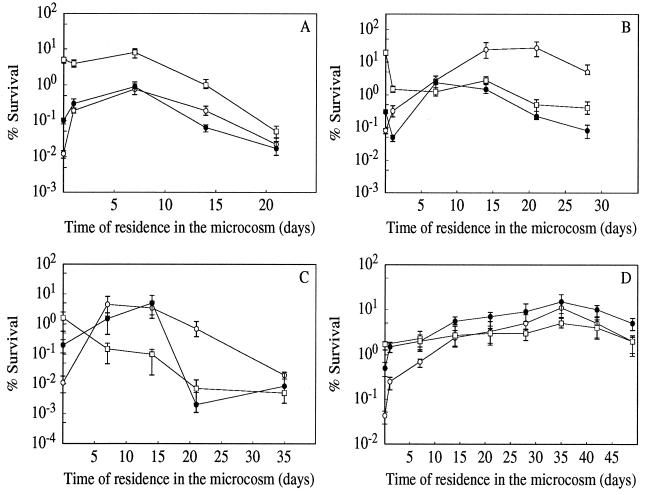
The changes in sensitivity of growing and starved cells of E. faecalis during incubation in tap water at 16°C to heat shock (62°C), acid stress (pH 3.3), UV256 nm irradiation (180 J/m2), and sodium hypochlorite treatment (0.05% [vol/vol]) are shown in Fig. 3. Cultures harvested from the stationary phase (due to glucose starvation) were, prior to the transfer into the microcosm, more resistant to all the treatments than were exponentially growing cultures. These results are in agreement with our previous findings (9, 16). Furthermore, in all cases cultures starved for 24 h were more resistant than those harvested at the onset of starvation. However, the resistance of former cultures either stagnated (in the case of heat shock and UV irradiation) or declined (in the case of acid challenge and NaOCl stress) in the first days of incubation in the oligotrophic environment. In contrast, cells harvested from the exponential-growth phase and those from the onset of starvation became progressively more resistant to these treatments during incubation in the microcosm. Whereas the kinetics of the increase of resistance against the heat challenge, NaOCl stress, and UV irradiation were comparable, cells from the exponential-growth phase developed a significantly greater resistance against the acid challenge. For each stress, resistance reached a maximum level between days 10 and 15 of incubation in the microcosm for the heat, acid, and NaOCl stresses. However, for UV irradiation the cells mounted resistance progressively for up to 35 days. The maximum tolerance factors for cultures harvested from the exponential-growth phase were 64, 345, 420, and 332 against the heat, acid, NaOCl, and UV irradiation stresses, respectively. The tolerance factor is defined as the ratio of survival after a given challenge of oligotrophy-stressed cells versus the survival of untreated control cells. After this maximum of resistance, the cells again became more sensitive in all cases. At the end of the experiments the resistances to the different treatments of exponential- or stationary-phase cells were comparable.
FIG. 3.
Development of stress resistance to heat (A; 62°C for 30 min), acid (B; pH 3.3 for 30 min), NaOCl (C; 10−2% [vol/vol] for 8 min), and UV254 nm irradiation (D; 180 J/m2) in E. faecalis during incubation in tap water. Before transfer into the aquatic system, the cultures were harvested from the exponential-growth phase (○) or at the onset (•) or after 24 h (□) of glucose starvation. Values shown on the ordinate represent the sensitivities of the control cultures prior to transfer into the microcosm. For all stresses, the resistance of preadapted glucose-starved control cultures was initially significantly higher than that of exponentially growing cells. The data are means and standard deviations (bars) for at least three independent experiments.
Analysis of modifications in protein pattern during incubation in tap water by two-dimensional gel electrophoresis.
The most spectacular development into a multiresistant cellular state during incubation in tap water was evident in cells harvested from the exponential-growth phase. In contrast, the resistance of cultures which experienced glucose starvation for 24 h prior to incubation in the oligotrophic environment was not further enhanced. We therefore began to analyze the changes in protein synthesis in naive cells (i.e., those harvested from the exponential-growth phase) during the incubation in the oligotrophic environment.
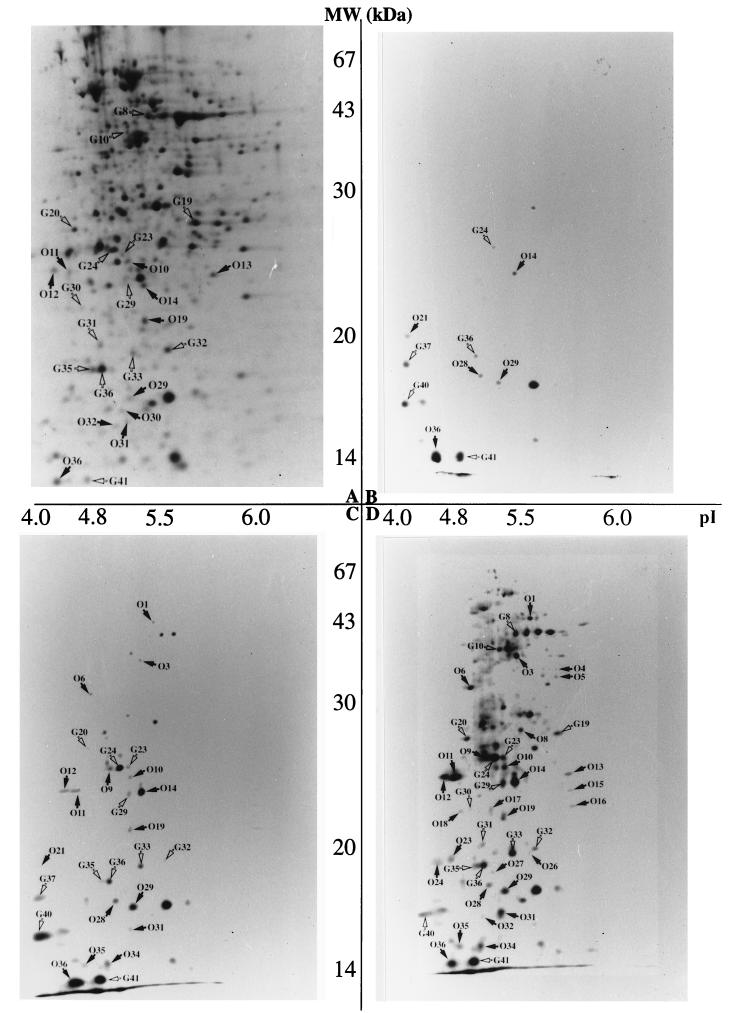
The autoradiograms obtained with cultures starved in water for different periods are shown in Fig. 4. The first 24 h of incubation in the microcosm was characterized by a shutdown of synthesis of the majority of polypeptides that are normally expressed in growing cultures. Only 40 to 50 proteins were present in the autoradiograms, with the majority at the lower limit of detection and thus not visible on the photographs. However, some protein spots, especially those in the low-molecular-weight range, were more intense and are indicated by arrows. Interestingly, a proportion of these have been identified in a previous work as glucose starvation-inducible proteins (Gls proteins) (10) and thus are indicated with a “G” in the autoradiograms. Whereas polypeptides G24 and G36 were still at the limit of detection, G41 was clearly induced over the level of synthesis in control cells, and G37 and G40 were new polypeptides not present in the growing cultures. Five other proteins not yet identified as stress proteins in E. faecalis JH2-2 (10, 16) were among the polypeptides synthesized during the first day of incubation under the oligotrophic conditions. They are indicated by the character “O.” Some of these were also present in growing cells (O14, O29, and O36), but they showed relatively enhanced synthesis in cultures incubated for 24 h in tap water. Others (O21 and O28) seem to be specifically induced under these conditions. One protein (indicated by an intense spot to the right of O29 in the autoradiogram) seems to be different from the others because its spot intensity is comparable to that in the control cells and does not change during the 4-week incubation period in the microcosm (compare Fig. 4A to D).
FIG. 4.
Autoradiograms of two-dimensional gels obtained from cultures of E. faecalis labeled with 35S during incubation in tap water. Growth, labeling, and gel running were performed in two independent experiments and led to comparable results. (A) Reference gel obtained from a culture labeled during the exponential-growth phase. (B to D) Gels from cultures incubated for 1 day, 3 days, and 2 weeks in the microcosm, respectively. Arrows indicate the proteins which are relatively induced over the level of growing cultures or specific for the starvation stress. The intense spot to the right of O29 indicates a polypeptide with a spot intensity that is comparable in growing and starved cultures and is independent of the length of incubation in the oligotrophic environment. See the text for a more detailed explanation.
After 3 days of incubation more proteins were detectable on the two-dimensional gels; of these six others were Gls proteins (G20, G23, G29, G32, G33, and G36). The intensities of most Gls and O proteins, which were low after 24 h of incubation in the microcosm, now showed significant increases in synthesis (G24, G36, G40, and G41; O14 and O29). The remaining polypeptides that became detectable at this time were those identified as induced over the level of synthesis in control cells in the oligotrophic environment at longer incubation times (O1, O3, O6, O9, O10, O11, O12, O19, O31, O34, and O35).
After 1 week in the microcosm, more proteins, including those of higher molecular weight, became detectable (data not shown). The spot intensities of most of the starvation proteins identified so far were more intense on the autoradiograms, with the most spectacular increase in the synthesis of polypeptides O1, O3, O6, and O21 and G28 and G37.
After 2 weeks the synthesis of higher-molecular-weight proteins increased, and among them were three additional Gls polypeptides (G8, G10, and G19). Furthermore, two low-molecular-weight Gls proteins (G30 and G31) and 13 O polypeptides (O4, O5, O8, O13, O15, O16, O17, O18, O23, O24, O26, O27, and O32) became detectable. In contrast, the intensities of the spots of some of the early starvation stress proteins, all in the low-molecular-weight range, decreased (O29, O36, and G40) or even completely disappeared (G37 and G21) in cultures incubated for 2 weeks in tap water.
Protein analysis of cultures incubated for 4 weeks in the oligotrophic environment revealed the presence of six additional O proteins (data not shown). With one exception, these polypeptides were not detectable in growing cells and thus seem to be specifically induced under these particular conditions. Furthermore, the decrease in the spot intensities observed for polypeptides O36 and G40 after 2 weeks of incubation in tap water continued.
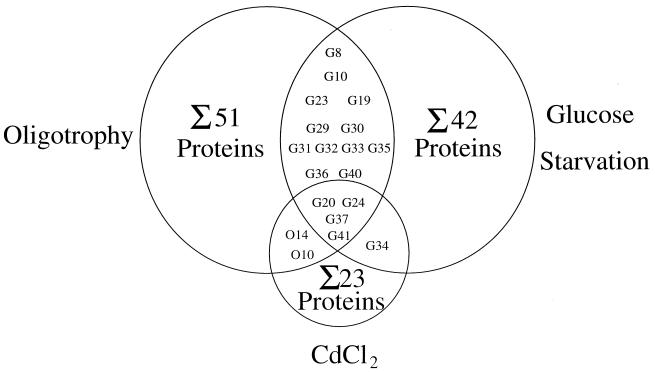
It is noteworthy that the starvation-inducible polypeptides G20, G24, G37, and G40, as well as O10 and O14, have also been found to be inducible by CdCl2 (16). A summary of the overlaps of the stress proteins induced in the oligotrophic microcosm compared to those with the glucose starvation and CdCl2 stimulons is shown in Fig. 5.
FIG. 5.
Summary of the number and overlap of the polypeptides positively regulated by oligotrophy due to incubation in tap water, starvation of energy source glucose (10), and CdCl2 stress (16). The proteins G20, G24, G34, G37, G41, O10, and O14 overlapping the CdCl2 stimulon are designated in the original study C6, C9, C17, C19, C20, C10, and C15, respectively.
DISCUSSION
In many studies the bacterial starvation stress response has been analyzed in defined in vitro systems where only one growth factor (in most this is the carbon and/or energy source) was lacking. It has been shown that these one-nutrient-starved bacteria developed a general stress resistance leading to enhanced tolerance to different, otherwise hostile environmental conditions (9, 11, 13). However, only a few investigators (20, 27) have addressed the question of whether this phenomenon also takes place with bacteria under environmental conditions where microorganisms are frequently confronted with multiple starvations. This is particularly true for enteric microorganisms [and even more for (poly)auxotrophs with complex nutritional requirements, e.g., E. faecalis], which are released directly or through wastewater into rivers and coastal areas. In the aquatic ecosystem, the survival of these organisms is affected by complex environmental stresses and killing agents that trigger cellular responses that are still poorly understood. Water systems are characterized by their oligotrophic nature, and this seems to be one of the factors that reduces the culturability of allochthonous coprotrophic bacteria (19).
We have found that E. faecalis survived well in our microcosm, as even after prolonged periods of incubation 10 to 30% could still be cultivated. Byrd et al. (4) measured the survival of several bacteria, including E. faecalis, in a drinking water system and found a rapid decrease of culturability in the first days of incubation. However, the cell density (ca. 106 CFU/ml) and incubation conditions they used were different from those used here.
Recently, it has been shown that nonculturable cells exist and exhibit various degrees of metabolic activity (24, 25). Therefore, the somewhat lower plating efficiency of cells after long-term incubation in tap water may be the result of cellular death, of a transition into a “viable but nonculturable” state, or both. However, if cellular death was mainly responsible for the decrease in CFU counts, this was not accompanied during the first weeks of incubation by significant cell lysis. This result indicates that the potential to adapt and survive in the oligotrophic environment seems not to be due to a significant cannibalism of living over dead cells but may be the result of mobilization of endogenous reserves.
The electron microscopic analysis furthermore revealed that the cells do not significantly shrink during incubation in the nutrient-poor microcosm. This finding is in contrast to the starvation response of gram-negative bacteria. During starvation E. coli become more coccoid, accompanied by a decrease in cell size (14). The shrinking process upon starvation is even more pronounced in marine Vibrio spp., where ultramicrocells as small as 0.03 μm3 can be formed (18). Furthermore, a decrease in cell length upon prolonged incubation of exponentially growing Pseudomonas fluorescens R2fRpr cells introduced into two different soils has been reported (27).
As stated above, starved cells are generally considered to be in a more resistant state than their growing counterparts. However, we have not observed a significant difference between growing and preadapted glucose-starved cultures in survival oligotrophic stress. The survival of the 3- and 24-h glucose-starved cells was independent of protein synthesis during their incubation in tap water. In contrast, the persistence in the oligotrophic environment of their growing counterparts and also, but to a lesser extent, of cells from the onset of glucose starvation was dependent on an active protein metabolism. Thus, the system(s) responsible for long-term survival under oligotrophic conditions seems to be already present in cultures starved of glucose for 3 and 24 h prior to transfer into tap water. In contrast, growing cultures and cells from the onset of glucose starvation were not preconditioned to survive in the oligotrophic environment. However, they can adapt to survive this complex starvation stress by synthesizing specific proteins during incubation in the microcosm that may already be present in preadapted glucose-starved cells. These results suggests a close relationship between the stress responses triggered by glucose starvation and oligotrophy. It is noteworthy that we have obtained a very similar result in seawater (10a), showing that the observed long-term survival and dependence on protein synthesis was not specific for tap water but may reflect a more global adaptation to aquatic environments.
The suggestion of an overlap of the two different starvation responses was furthermore supported by the analysis of the evolution of resistance to heat, acid, NaOCl, and UV irradiation. Cultures which have experienced glucose starvation for 24 h prior to incubation in the oligotrophic environment do not undergo a further increase in resistance. In contrast, cells harvested from the exponential-growth phase, which were initially more sensitive to these stresses, developed resistance progressively during incubation in tap water. This shows that complete starvation does not have a synergistic effect on the multiresistance induced by energy starvation, confirming the suggestion of a molecular link between the two starvation responses in E. faecalis.
Comparable results have been obtained with P. fluorescens (27). Cells inoculated into soils mounted a general stress resistance, and preadaptation to carbon starvation of inoculant cells leads to no further enhancement of the general resistance following incubation in soil. In that study it was suggested that carbon starvation could largely induce a response similar to that triggered by the residence of inoculant cells in soil, but no molecular analysis to strengthen this suggestion was presented.
We have conducted these molecular studies by analyzing changes in protein synthesis during incubation in the oligotrophic microcosm. Incubation of E. faecalis cells in tap water leads to an enhanced synthesis of at least 51 proteins. These polypeptides showed different kinetics of induction, indicating that adaptation to oligotrophy is a highly ordered process. Interestingly, the majority of proteins induced early during incubation in tap water were of low molecular weight. Oligotrophy provoked a shutdown in synthesis of higher-molecular-weight polypeptides at the beginning of the incubation period, and synthesis restarted only after longer incubation times. It is premature to speculate whether some of the early low-molecular-weight proteins play a key role in the adaptation process which enables the cellular protein synthesis machinery to finally regain metabolism of the higher-molecular-weight proteins. However, our experimental conditions did not permit us to determine whether polypeptides which become detectable only after prolonged incubation times in tap water are subject to a delayed switching-on of expression or have lower rates of synthesis relative to those detected earlier.
At least 42 polypeptides were induced in E. faecalis after entrance into the stationary phase provoked by glucose exhaustion (10). Comparison with the proteins induced by oligotrophy revealed an overlap of 16 polypeptides. This finding shows that glucose and multiple nutrient starvation trigger similar responses. Until now only one of these polypeptides, the protein Gls24, has been analyzed at the molecular level (10). The corresponding gene showed homology to a hypothetical open reading frame of Lactococcus lactis (5), and the E. faecalis protein seems to be implicated in morphological changes in the stationary phase (8a).
The other polypeptides induced in tap water may be implicated in the starvation response to other nutrients, i.e., phosphorous, nitrogen, or amino acids. Furthermore, due to the difference in incubation temperatures used to analyze glucose and complete starvation (37 and 16°C, respectively), it is possible that some proteins belong to the cold-shock regulon. Induction of cold-shock proteins in E. faecalis JH2-2 at 8°C has recently been demonstrated (23).
Because carbon and complete starvation trigger similar physiological responses, it seems reasonable to assume that the overlapping polypeptides are responsible at the molecular level for the observed phenomena of long-term survival and the development of general resistance. Interestingly, four of these general starvation proteins have also been identified as CdCl2 inducible. Work is in progress in our laboratory to identify these proteins and their corresponding genes by reverse genetics.
ACKNOWLEDGMENTS
The expert technical assistance of Annick Blandin and Beatrice Cheval was greatly appreciated. We thank Sylviane Lemarinier for the electron microscopic studies and T. N. Ledger for the help with the English.
This work was supported with financial aid from the Agence de l’Eau Seine Normandie.
REFERENCES
- 1.Anderson T F. Techniques for the preservation of three-dimensional structure in preparing specimens for the electron microscope. Trans N Y Acad Sci. 1951;13:130–134. [Google Scholar]
- 2.Barcina I, Lebaron P, Vives-Rego J. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol Ecol. 1997;23:1–9. [Google Scholar]
- 3.Boutibonnes P, Giard J C, Hartke A, Thammavongs B, Auffray Y. Characterization of the heat-shock response of Enterococcus faecalis. Antonie Leeuwenhoek. 1993;64:47–55. doi: 10.1007/BF00870921. [DOI] [PubMed] [Google Scholar]
- 4.Byrd J J, Xu H S, Colwell R R. Viable but nonculturable bacteria in drinking water. Appl Environ Microbiol. 1991;57:875–878. doi: 10.1128/aem.57.3.875-878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donkersloot J A, Thompson J. Cloning, expression, sequence analysis, and site-directed mutagenesis of the Tn5306-encoded N5-(carboxyethyl)ornithine synthase from Lactococcus lactis K1. J Biol Chem. 1995;270:12226–12234. doi: 10.1074/jbc.270.20.12226. [DOI] [PubMed] [Google Scholar]
- 6.Flahaut S, Frère J, Boutibonnes P, Auffray Y. Comparison of the bile salts and sodium dodecyl sulfate stress response in Enterococcus faecalis ATCC 19433. Appl Environ Microbiol. 1996;62:2416–2420. doi: 10.1128/aem.62.7.2416-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flahaut S, Hartke A, Giard J C, Benachour A, Boutibonnes P, Auffray Y. Relationship between heat shock, bile salts and ethanol treatments in Enterococcus faecalis. FEMS Microbiol Lett. 1996;138:49–54. doi: 10.1111/j.1574-6968.1996.tb08133.x. [DOI] [PubMed] [Google Scholar]
- 8.Flahaut S, Benachour A, Giard J C, Boutibonnes P, Auffray Y. Defense against lethal treatments and de novo protein synthesis induced by NaCl in Enterococcus faecalis ATCC 19433. Arch Microbiol. 1996;165:317–324. doi: 10.1007/s002030050333. [DOI] [PubMed] [Google Scholar]
- 8a.Giard, J. C. Unpublished data.
- 9.Giard J C, Hartke A, Flahaut S, Benachour A, Boutibonnes P, Auffray Y. Starvation-induced multiresistance in Enterococcus faecalis JH2-2. Curr Microbiol. 1996;32:264–271. doi: 10.1007/s002849900048. [DOI] [PubMed] [Google Scholar]
- 10.Giard J C, Hartke A, Flahaut S, Boutibonnes P, Auffray Y. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res Microbiol. 1997;148:27–35. doi: 10.1016/S0923-2508(97)81897-9. [DOI] [PubMed] [Google Scholar]
- 10a.Hartke, A. Unpublished data.
- 11.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL 1403. Appl Environ Microbiol. 1994;60:3474–3478. doi: 10.1128/aem.60.9.3474-3478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kjelleberg S. Starvation in bacteria. New York, N.Y: Plenum Press, Inc.; 1993. [Google Scholar]
- 14.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary phase Escherichia coli cells is controlled by the novel sigma factor ςs (rpoS) J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laplace J M, Boutibonnes P, Auffray Y. Unusual resistance and acquired tolerance to cadmium chloride in Enterococcus faecalis. J Basic Microbiol. 1996;36:311–317. doi: 10.1002/jobm.3620360504. [DOI] [PubMed] [Google Scholar]
- 16.Laplace J M, Thuault M, Hartke A, Boutibonnes P, Auffray Y. Sodium hypochlorite stress in Enterococcus faecalis: influence of antecedent growth conditions and induced proteins. Curr Microbiol. 1997;34:284–289. doi: 10.1007/s002849900183. [DOI] [PubMed] [Google Scholar]
- 17.Lopez M F, Patton W F, Utterback B L, Chung-Welch N, Barry P, Skea W M, Cambria R P. Effect of various detergents on protein migration in the second dimension of two-dimensional gels. Anal Biochem. 1991;199:35–44. doi: 10.1016/0003-2697(91)90266-v. [DOI] [PubMed] [Google Scholar]
- 18.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press, Inc.; 1993. pp. 8–16. [Google Scholar]
- 19.Morita R Y. Encyclopedia of microbiology. Vol. 2. New York, N.Y: Academic Press, Inc.; 1992. Low-nutrient environments; pp. 617–624. [Google Scholar]
- 20.Nyström T, Olsson R M, Kjelleberg S. Survival, stress resistance, and alteration in protein expression in the marine Vibrio sp. strain S14 during starvation for different individual nutrients. Appl Environ Microbiol. 1992;58:55–65. doi: 10.1128/aem.58.1.55-65.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Farrell P H. High two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver J D. Formation of viable but non-culturable cells. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press, Inc.; 1993. pp. 239–272. [Google Scholar]
- 23.Panoff J M, Corroler D, Thammavongs B, Boutibonnes P. Differentiation between cold shock proteins and cold acclimation proteins in a mesophilic gram-positive bacterium, Enterococcus faecalis JH2-2. J Bacteriol. 1997;179:4451–4454. doi: 10.1128/jb.179.13.4451-4454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roszak D B, Colwell R R. Metabolic activity of bacterial cells enumerated by direct viable count. Appl Environ Microbiol. 1987;53:2889–2893. doi: 10.1128/aem.53.12.2889-2893.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terzaghi B E, Sandine W. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;2:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Overbeek L S, Eberl L, Givskov M, Molin S, Van Elsas J D. Survival of, and induced stress resistance in, carbon-starved Pseudomonas fluorescens cells residing in soil. Appl Environ Microbiol. 1995;61:4202–4208. doi: 10.1128/aem.61.12.4202-4208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]