Abstract
Phage Q38, a representative member of the c2 species, was purified by CsCl gradient and used to immunize BALB/c mice. Monoclonal antibodies (MAbs) were raised and then characterized by enzyme-linked immunosorbent assay. Two MAbs of isotype immunoglobulin G2a, designated 2A5 and 6G7, reacted only with phages belonging to the c2 species and not with phages of the 936 and P335 species, with a Lactococcus lactis cell extract, or with phage DNA. Immunoelectron microscopy showed that both MAbs recognized only phage head proteins. They did not react with any denatured phage proteins in Western blot assays. However, when the nitrocellulose membranes were treated with a Triton-based buffer to assist in protein renaturation, MAbs 2A5 and 6G7 recognized the two major capsid proteins with molecular masses of 80 and 170 kDa. Competitive inhibition tests showed that the two MAbs bind to overlapping epitopes. These MAbs may be a useful tool for monitoring c2 bacteriophages during dairy fermentation and in genetic studies.
Lactococcus lactis is used for the production of fermented dairy products, including cheeses, buttermilk, and sour cream. Occasionally, spontaneous fermentation failures occur during the manufacture of these products, and the main causative agents are bacteriophages (13, 22, 37). Lactococcus phages are currently classified into 12 species (6, 13, 33). However, only three species have been found to hinder industrial mesophilic milk fermentations: the prolate-headed c2 species and the isometric-headed 936 and P335 species (22). A previous study showed that the c2 species was the most common lactococcal phage species found in Canadian cheddar cheese plants (24). The c2 species is also the only lactic acid bacteriophage ranked at the genus level by the International Committee on Taxonomy of Viruses (31). In fact, c2 is one of only six genera in the family Siphoviridae. This taxonomic status emphasizes the importance of these unique and distinct phages not only to the dairy sector but also to the field of bacterial viruses.
Lactic phages are naturally found in raw milk, resist pasteurization, and have a short latent period and a large burst size. Therefore, they can spread rapidly within a cheese plant, and constant monitoring is critical (19). Two types of methods for detecting phages are already available: direct and indirect. The direct methods, which detect lytic phages or phage components, include plaque assays (10), DNA probes (25), and enzyme-linked immunosorbent assays (ELISA) (15, 21). Indirect methods detect inhibitory substances by starter activity and indicator tests, ATP measurement, or electrical impedance (9, 27, 35). Most phage-testing laboratories routinely perform starter activity tests and plaque assays because they give information on the phage sensitivity of Lactococcus strains used in the dairy plant. Both methods are reliable but time-consuming; i.e., it takes a day to obtain the results. Obviously, on-line phage detection in cheese plants is virtually impossible with such methods. Furthermore, they do not provide information on the nature of the phage species.
Biotechnology has recently provided tools for engineering phage-resistant starter cultures. These improved strains rely on the addition to phage-sensitive strains of natural plasmids encoding various phage resistance mechanisms. For industrial applications, the selection of an antiphage system to introduce into a phage-sensitive strain depends on the sensitivity of that strain to a particular phage species (22). As the next generation of phage-resistant strains and rotation strategies are being introduced into the system, a rapid method of detecting and identifying lactococcal phages would be an asset.
Several attempts have been made to develop such a detection system. The lack of, or weak, homology among the genomes of different lactococcal phage species was exploited to construct DNA probes specific to the c2 and 936 species (25). Although many samples could be processed simultaneously, the time required for performing the detection method was too long. A PCR assay was recently developed to detect phages of another lactic acid bacterium, Streptococcus thermophilus (4). In general, DNA techniques require equipment that is not readily available in dairy plants. On the other hand, the use of antibodies might be more appropriate since antibodies are easier to handle. Two ELISA systems used to detect L. lactis phages have already been described (15, 21). The first system employs polyclonal antibodies raised against native phages of the c2 and 936 species (15), whereas the second system uses monoclonal antibodies (MAbs) specific to the denatured major capsid protein (MCP) of P335 phages (21). The MCP is the most abundant protein in the phage structure and is a suitable target for detection purposes, provided it is conserved in all members of the species. Generally, members of the same phage species have similar structural protein profiles (3, 24, 29, 30).
In the present study, we report the production and characterization of the first two MAbs specific to the native MCP of lactococcal phages of the c2 species.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
The strains and phages used in the present study are listed in Table 1. L. lactis strains were grown at 30°C in M17 broth containing 0.5% glucose for strain LM0230 or 0.5% lactose for industrial strains (36). Phages were amplified, purified, and concentrated on a discontinuous CsCl gradient (11, 12, 26), followed by a one-step CsCl gradient. Ultracentrifugations were performed with a Beckman NVT65 rotor at 60,000 rpm for 18 h.
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Strain or phage | Relevant characteristics | Reference or source |
|---|---|---|
| L. lactis | ||
| LM0230 | Plasmid free, host for phages c2, c21, ml3, eb1, and p2 | 20 |
| SMQ-86 | UL8 (pSA3), Emr, host for phage ul36 | 7 |
| SMQ-189 | Industrial strain, host for phage Q7 | 22 |
| SMQ-195 | Industrial strain, host for phage Q11 | 22 |
| SMQ-196 | Industrial strain, host for phages Q38 and Q44 | This study |
| SMQ-203 | Industrial strain, host for phage Q42 | 22 |
| Bacteriophages | ||
| c2 | Prolate headed, c2 species | K. M. Polzina |
| c21 | Prolate headed, c2 species | This study |
| eb1 | Prolate headed, c2 species | L. L. McKayb |
| ml3 | Prolate headed, c2 species | W. E. Sandinec |
| Q38 | Prolate headed, c2 species | 22 |
| Q44 | Prolate headed, c2 species | 22 |
| p2 | Small isometric headed, 936 species | L. L. McKay |
| Q7 | Small isometric headed, 936 species | 22 |
| Q11 | Small isometric headed, 936 species | 22 |
| Q42 | Small isometric headed, 936 species | 22 |
| ul36 | Small isometric headed, P335 species | 24 |
New Zealand Dairy Research Institute.
University of Minnesota, Minneapolis.
Oregon State University, Corvallis.
Phage DNA analysis.
Purified phages (200 μl) were treated with proteinase K at a final concentration of 1 mg/ml for 2 h at 37°C. Then, 135 μl of 100 mM ammonium acetate (pH 7.5) was added, followed by an equal volume (335 μl) of phenol-chloroform. After centrifugation at 16,000 × g for 10 min, supernatants were taken and extraction was performed and repeated twice. DNA was precipitated with 0.7 volume of isopropanol. After centrifugation, pellets were washed with ethanol (70%) and resuspended in 50 μl of 10 mM Tris (pH 8.0). Purified DNA was treated with RNase H and digested with restriction enzymes as recommended by the manufacturer (Boehringer Mannheim, Laval, Québec, Canada). Restriction fragments were electrophoresed on a 0.8% agarose gel in TAE buffer (40 mM Tris acetate, 1 mM EDTA), stained with ethidium bromide, and visualized by UV. Agarose gels were transferred to nylon membranes (Hybond-N+; Amersham, Arlington Heights, Ill.) as described previously (26). Phage Q38 DNA digested with EcoRV was randomly labeled with a digoxigenin DNA labeling kit (Boehringer Mannheim) and used as a probe (25). Immunological detection of DNA was performed according to the manufacturer’s instructions.
Protein profiles.
Phage structural proteins were fractionated according to the method of Braun et al. (3). Protein concentrations (15 μg) were estimated by the method of Lowry et al. (16), and proteins were separated through a sodium dodecyl sulfate (SDS)–15% polyacrylamide minigel at 100 V with the Mini-Protean II apparatus (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Broad-range protein markers (New England Biolabs, Mississauga, Ontario, Canada) were used as molecular mass controls.
Production and purification of MAbs.
Female BALB/c mice were immunized four times intraperitoneally at 3-week intervals with 100 μg of phage Q38 emulsified in an equal volume of complete Freund’s adjuvant for the first injection and in incomplete Freund’s adjuvant for the subsequent injections. Three final booster injections at 1-day intervals with 100 μg of phage Q38 were administered intravenously 3 days prior to fusion. Spleen cells were fused with SP2/0 myeloma cells in the presence of polyethylene glycol 4000 and grown as described previously (8). Supernatants were screened by direct-binding ELISA according to the method of Moineau et al. (21). Immulon II microplates (Dynatech Laboratories, McLean, Va.) were coated with 100 ng of phage Q38. Hybridoma cells that produced antibodies against phage Q38 at an optical density above 0.7 were kept and retested by direct-binding ELISA against 100 ng of phages eb1, c2, ml3, Q38, and Q44 (c2 species); phages Q7, Q11, and Q42 (936 species); phage ul36 (P335 species); phage Q38 DNA; and L. lactis cell extracts. Hybrid cells that secreted antibodies reacting only with c2 species were subcloned by the limiting-dilution technique. Two stable clones, 2A5 and 6G7, were obtained. MAbs were purified through a protein G-Sepharose column, eluted with 100 mM glycine (pH 2.5), and neutralized with 1 M K2HPO4. MAbs were dialyzed against phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM K2HPO4 [pH 7.4]) and concentrated with Centricon-100 (Amicon, Oakville, Ontario, Canada).
Sensitivity, affinity, isotyping, and labeling of MAbs.
To determine the sensitivities of the MAbs, diluted phages were spotted onto nitrocellulose membranes (Amersham) with a Hybridot apparatus (GIBCO/BRL, Grand Island, N.Y.). Membranes were saturated with PBS–3% bovine serum albumin (BSA) for 30 min at 37°C and washed twice in TBS-Tween (25 mM Tris, 200 mM NaCl, 0.1% Tween 20 [pH 7.4]). Membranes were incubated for an hour at 37°C with the MAbs at a final concentration of 4 μg/ml diluted in PBS–0.5% BSA. After five washes with TBS-Tween, peroxidase-labeled goat anti-mouse immunoglobulin G (IgG) (diluted 1:2,000 in PBS–0.5% BSA) was added, and the membrane was incubated for 30 min at 37°C. After six washes, the colorimetric substrate (diaminobenzidine; Sigma Chemical Co., St. Louis, Mo.) was added at room temperature. The reaction was stopped with water. MAb affinity was determined by the same method, except incubation times for the MAbs were 1, 5, 10, 15, 30, and 60 min. Isotyping was performed with the Isostrip kit, and labeling was done with a peroxidase labeling kit (Boehringer Mannheim).
Competitive inhibition immunoassay.
Microplates were coated overnight at 4°C with 25, 50, 100, and 200 ng of phage Q38. After saturation with PBS–3% BSA and washing with TBS-Tween, one of the two selected MAbs was added unlabeled at various concentrations (0.1, 1, 10, and 20 μg/ml) and the mixture was incubated at 37°C for 30 min. After six washes, the remaining MAb, labeled with peroxidase, was diluted 1:2,500 in PBS–0.5% BSA and added to microplates for 30 min at 37°C. Following washing, the orthophenylenediamine colorimetric substrate was added and the optical density was measured at 450 nm. The percent inhibition was calculated as described previously (21).
Western and renaturation treatment.
After separation by SDS-polyacrylamide gel electrophoresis (PAGE), proteins were transferred with a Trans-Blot apparatus (Bio-Rad) to a nitrocellulose membrane by using Tris-glycine-methanol buffer (25 mM Tris [pH 8.3], 192 mM glycine, 20% methanol) according to the method of Sambrook et al. (32). Membranes were incubated in a renaturation buffer (0.1 M Tris-HCl [pH 7.0], 2% Triton X-100 [28]) for 30 min. After four washings (in 100 mM maleic acid, 150 mM NaCl, 0.3% Tween 20 [pH 7.5]), membranes were incubated overnight at 4°C in a 2% blocking solution (Boehringer Mannheim). Labeled MAbs diluted 1:2,500 were added and the membranes were incubated for 30 min at room temperature. Membranes were washed for 2 h (four times for 30 min), and MAb-phage protein complexes were detected by chemiluminescence with a Western blotting kit (Boehringer Mannheim).
Electron microscopy.
Specimens were observed with a Philips EM 410 electron microscope as described previously (23). Magnification was monitored with catalase crystals (18). Dimensions were measured on a photographic print at a magnification of ×135,000. Immunoelectron microscopy analysis was performed according to the method of Moineau et al. (21). MAbs were used at a final concentration of 0.1 mg/ml, and gold-labeled protein A (10 nm) was used at 3 μg/ml (Sigma).
RESULTS
Characterization of six phages from the c2 species.
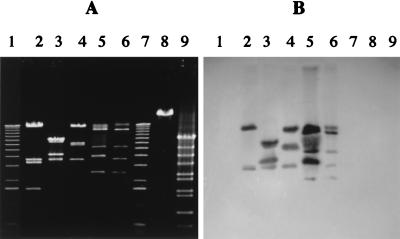
Six members of the c2 species were obtained from different international phage collections (Table 1). DNA restriction patterns showed that all six phages were different (Fig. 1A and data not shown). Southern blot assays confirmed that these phages have DNA homology (Fig. 1B). No DNA homology was found among members of the three lactococcal species (c2, 936, and P335) used in this study (Fig. 1B and data not shown). Observation under an electron microscope also revealed that the six c2 phages had the classical morphology of prolate phages (1): a 40- by 60-nm head and a 100-nm noncontractile tail (Fig. 2A).
FIG. 1.
Restriction patterns (EcoRI) of phage DNA (A) and corresponding Southern blot probed with phage Q38 DNA (B). Lanes 1 and 7, 1-kb DNA ladder (1.0, 1.6, 2.1, 3.1, 4.1, 5.1, 6.1, 7.1, 8.1, 9.1, 10.2, 11.2, and 12.2 kb) (GIBCO/BRL); lanes 2, phage eb1; lanes 3, phage c21; lanes 4, phage ml3; lanes 5, phage Q38; lanes 6, phage Q44; lanes 8, phage p2 (936 species); lanes 9, phage ul36 (P335 species).
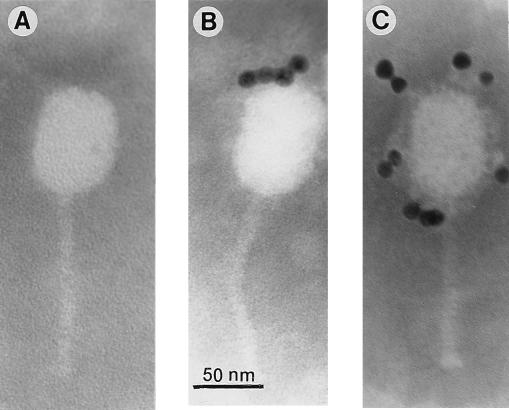
FIG. 2.
Immunoelectron microscopy with gold-labeled protein A (particle size, 10 nm). (A) Phage Q38; (B) MAb 6G7; (C) MAb 2A5.
Production and characterization of two MAbs specific to the c2 species.
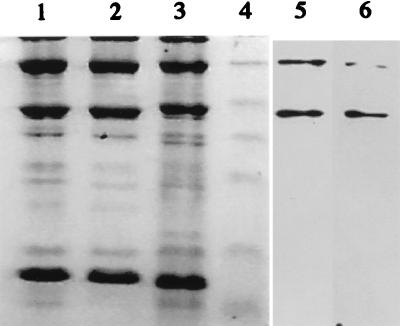
Phages of the c2 species are known to contain three major structural proteins as well as a variable number of minor proteins (3, 24, 29, 30) (Fig. 3). In this study, we estimated the sizes of the three major proteins to be 27, 80, and 170 kDa (Fig. 3). Phage Q38 was selected for mouse immunization because it could be readily amplified and purified at high concentrations. Following cell fusion, two MAbs (2A5 and 6G7) were selected for their specificity to the six native c2-like phages tested. Neither MAb reacted with phages Q7, Q11, or Q42 (936 species), ul36 (P335 species), or DNA, L. lactis cell extracts, whey, or M17 media. Both MAbs were of isotype IgG2a.
FIG. 3.
Profiles of phage structural proteins as determined by SDS–15% PAGE with the corresponding immunoblots for MAbs 2A5 and 6G7. Phage proteins were stained with Coomassie blue. Lane 1, phage Q44; lane 2, phage Q38; lane 3, phage c2; lane 4, prestained molecular mass markers (25, 32.5, 47.5, 62, 83, and 175 kDa) (New England Biolabs); lane 5, MAb 2A5 and phage c2; lane 6, MAb 6G7 and phage c2.
Binding inhibition assay.
To determine if the two selected MAbs bind to different epitopes, competitive binding inhibition assays were performed with native phage Q38 as an antigen. If the two MAbs recognized the same epitope, the binding of the first MAb to the phage Q38-coated well would prevent the binding of the second MAb and consequently the percent inhibition would be high. If the two MAbs recognized distinct epitopes, the percent inhibition would be low. When MAb 2A5 was used as the first and second antibody, the inhibition was 92% ± 1% (mean ± standard deviation for three experiments). Inhibition was only 25% ± 1% when MAb 6G7 was used in both incubations and at the same concentration as MAb 2A5. Increasing the concentration of MAb 6G7 in the first incubation or decreasing the antigen concentration in the well resulted in increased binding inhibition (results not shown). These results suggest that the first incubation did not saturate all 6G7 epitopes available on the native phage. It also suggests the presence of more 6G7 than 2A5 epitopes on the phage structure. Alternatively, MAb 6G7 might have less avidity or unbind more easily than MAb 2A5. When 2A5 was used as the first antibody and 6G7 as the second, the inhibition was 88% ± 3%. Thus, the prior incubation of 2A5 prevented the binding of 6G7. However, when 6G7 was incubated before 2A5, the binding inhibition was only 4% ± 5%. These results indicate that the 2A5 epitope overlaps the 6G7 epitope but that the 6G7 epitope does not span the 2A5 epitope. Based on all the above results, the two MAbs are different.
Immunoelectron microscopy and Western blotting analysis.
Since protein A has a high affinity for the heavy chain of IgG, gold particles linked to protein A were used to localize the antibody binding site on the native phage structure. Gold particles were found only along the phage head (Fig. 2), indicating that both MAbs reacted with capsid proteins. Neither MAb reacted with any of the c2 phage structural proteins in classical Western blot assays (data not shown). However, when the nitrocellulose was soaked in a protein renaturation buffer prior to the antibody reaction, both MAbs recognized the 80- and 170-kDa major structural phage proteins (Fig. 3). It appears that in both cases epitopes can be renatured after exposure to the denaturing conditions of SDS-PAGE. The above results indicated that the two different MAbs are specific to the native MCPs of the c2 phages and are most likely conformation dependent.
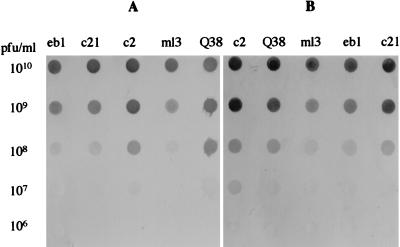
Dot immunoassay.
The sensitivity of each MAb to several phages of the c2 species was tested by dot immunoassay (Fig. 4). For both MAbs, 107 to 108 PFU of phages per ml diluted in phage buffer or in M17 media was detected within 1 min in the presence of 4 μg of peroxidase-conjugated antibody per ml (Fig. 4). These results testify to a strong affinity of both MAbs for phages of the c2 species.
FIG. 4.
Results of dot immunoassays showing sensitivities of MAbs 2A5 (A) and 6G7 (B).
DISCUSSION
Many systems using polyclonal antibodies or MAbs are available for the rapid detection and identification of microorganisms in food (5). One of our objectives is to develop a similar strategy for detection of lactococcal phages. Numerous epidemiological studies have clearly shown the diversity of the phage population in dairy plants. At least three genetically distinct phage species are routinely isolated from milk-derived samples. Due to the starter system used in Canada, the c2 species is the most predominant.
The first step in developing a more rapid method of detecting lactococcal phages involved the development of MAbs that were specific to c2-like phages. Immunoelectron microscopy and Western blot assays showed that the two MAbs 2A5 and 6G7, were raised against the native form of the MCP (80 and 170 kDa) of c2-like phages. Both MAbs recognized the six different members of the c2 species tested. They had a strong affinity for these phages, since the detection signals could be obtained within 1 min of contact between phages and MAbs. The two MAbs appeared to recognize overlapping epitopes of the MCP. Competitive inhibition assays showed that MAb 2A5 inhibited the binding of MAb 6G7, whereas MAb 6G7 did not prevent the binding of MAb 2A5. Therefore, these two MAbs could be used to develop an ELISA-based system in which MAb 6G7 would represent the capture antibody and MAb 2A5 would represent the detection antibody.
The complete nucleotide sequences of two prolate-headed phage genomes have already been determined; these phage genomes have strong homology at the nucleotide and amino acid levels (17, 34). Amino acid comparison of five deduced MCPs of prolate phages showed 95% identity, indicating that the MCP is highly conserved in this phage species (14). Furthermore, its high concentration in the phage structure makes the MCP an ideal target for immunoassays. For all the lactococcal phages of the c2 species studied so far, three major structural proteins (estimated at 27, 80, and 170 kDa) as well as numerous minor proteins have been reported (3, 24, 29, 30). N-terminal sequencing and immunoelectron microscopy analysis with polyclonal antibodies revealed that the 27-kDa protein is the major tail protein whereas the 80- and the 170-kDa proteins are the MCPs (17). The MCPs have identical N-terminal sequences and appear to be encoded by the same gene. A shift in the translational reading frame ahead of the termination codon is probably responsible for the production of the 170-kDa MCP. Lubbers et al. (17) suggested that these head proteins may also form multimers.
In earlier studies, MAbs were raised against the MCPs of isometric-headed phages of the P335 species (21). In this particular case, MCPs were extracted from SDS-PAGE gels and used to immunize mice. These MAbs had a much higher affinity and avidity for the denatured form of the MCP. Consequently, for an optimal phage detection, dairy samples had to undergo a denaturation step before the assay was performed. In this study, complete phage particles were used for mouse immunizations. Lembke and Teuber also used native phages but to develop polyclonal antibodies against P008 (936 species) and P001 (c2 species) (15). Interestingly, both studies used an ELISA-based system and obtained a detection limit of only 107 phages per ml. Although it was not the objective of this study, a dot immunoassay was used to evaluate the two MAbs for detection purposes. A detection limit of 107 to 108 phages per ml was also obtained. Obviously, before a system for on-line monitoring of c2-like phages in dairy plants is developed, this sensitivity will need to be substantially improved. One way to potentially overcome this problem would be to concentrate phages. Virus precipitation by polyethylene glycol has been used to enhance the efficiency of hepatitis A virus and rotavirus detection (38). Immunomagnetic separation has also been used to increase the sensitivity of detection methods (2). These techniques could also be adapted for detection of lactococcal phages to reduce volumes and remove potential inhibitors of the antigen-antibody reaction.
Nevertheless, MAbs 2A5 and 6G7 already have useful applications in industrial and research laboratories. For example, in dairy laboratories, whey samples are tested on a day-to-day basis for the presence of phages by plaque assay or the starter activity test. If a sample tests positive, the phage concentration is likely to be high. These MAbs could then be used to rapidly identify the presence of c2-like phages in the sample. Consequently, the c2-sensitive starter would be replaced by a non-c2-related culture. These MAbs could also be used in genetic studies. The P335 MAbs have been previously used to study the mode of action of abortive infection mechanisms (23). They might also have applications in monitoring the production of MCP during the lytic cycle of c2-like phages (7).
ACKNOWLEDGMENTS
We gratefully acknowledge Eric Emond for helpful suggestions. We are also thankful to Diane Montpetit (CRDA, AAC, St.-Hyacinthe, Québec, Canada) for the electron micrographs and Immunova for assistance in MAb production.
This work was supported by a systemic grant from CORPAQ to R.E.S. and by a strategic grant from NSERC to S.M.
REFERENCES
- 1.Ackermann H W, Dubow M S. Viruses of prokaryotes. Vol. 1. Boca Raton, Fla: CRC Press; 1987. [Google Scholar]
- 2.Blake M R, Weimer B C. Immunomagnetic detection of Bacillus stearothermophilus spores in food and environmental samples. Appl Environ Microbiol. 1997;63:1643–1646. doi: 10.1128/aem.63.5.1643-1646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun V, Hertwig S, Neve H, Geis A, Teuber M. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization and protein profiles. J Gen Microbiol. 1989;135:2551–2560. [Google Scholar]
- 4.Brüssow H, Frémont M, Bruttin A, Sidoti J, Constable A, Fryder V. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl Environ Microbiol. 1994;60:4537–4543. doi: 10.1128/aem.60.12.4537-4543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candlish A A G. Immunological methods in food microbiology. Food Microbiol. 1991;8:1–14. [Google Scholar]
- 6.Casey C N, Morgan E, Daly C, Fitzgerald G F. Characterization and classification of virulent lactococcal bacteriophages isolated from a cheddar cheese plant. J Appl Bacteriol. 1993;74:268–275. [Google Scholar]
- 7.Emond E, Holler B J, Boucher I, Vandenbergh P A, Vedamuthu E R, Kondo J K, Moineau S. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl Environ Microbiol. 1997;63:1274–1283. doi: 10.1128/aem.63.4.1274-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fliss I, St. Laurent M, Emond E, Lemieux R, Simard R E, Ettriki A, Pandian S. Production and characterization of anti-DNA-RNA monoclonal antibodies and their application in Listeria detection. Appl Environ Microbiol. 1993;59:2698–2705. doi: 10.1128/aem.59.8.2698-2705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heap H A, Lawrence R C. The selection of starter strains for cheesemaking. N Z J Dairy Sci Technol. 1976;11:16–20. [Google Scholar]
- 10.Hull R R. Methods for monitoring bacteriophage in cheese factories. Aust J Dairy Technol. 1977;32:63–64. [Google Scholar]
- 11.Jarvis A W. Serological studies of a host range mutant of a lactic streptococcal bacteriophage. Appl Environ Microbiol. 1978;36:785–789. doi: 10.1128/aem.36.6.785-789.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis A W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984;47:343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I B, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 14.Labrie, S., and S. Moineau. Personal communication.
- 15.Lembke J, Teuber M. Detection of bacteriophages in whey by an enzyme-linked immunosorbent assay. Milchwissenschaft. 1979;34:457–458. [Google Scholar]
- 16.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 17.Lubbers M W, Waterfield N R, Beresford T P J, Le Page R W F, Jarvis A W. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl Environ Microbiol. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luftig R. An accurate measurement of the catalase crystal period and its use as an internal marker for electron microscopy. J Ultrastruct Res. 1967;20:91–102. doi: 10.1016/s0022-5320(67)80038-8. [DOI] [PubMed] [Google Scholar]
- 19.McIntyre K, Heap H A, Davey G P, Limsowtin G K Y. The distribution of lactococcal bacteriophages in the environment of a cheese manufacturing plant. Int Dairy J. 1991;1:183–197. [Google Scholar]
- 20.McKay L L, Baldwin K A, Zottola E A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972;23:1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moineau S, Bernier D, Jobin M, Hébert J, Klaenhammer T R, Pandian S. Production of monoclonal antibodies against the major capsid protein of the Lactococcus bacteriophage ul36 and development of an enzyme-linked immunosorbent assay for direct phage detection in whey and milk. Appl Environ Microbiol. 1993;59:2034–2040. doi: 10.1128/aem.59.7.2034-2040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moineau S, Borkaev M, Holler B J, Walker S A, Kondo J K, Vedamuthu E R, Vandenbergh P A. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J Dairy Sci. 1996;79:2104–2111. [Google Scholar]
- 23.Moineau S, Durmaz E, Pandian S, Klaenhammer T R. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl Environ Microbiol. 1993;59:208–212. doi: 10.1128/aem.59.1.208-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moineau S, Fortier J, Ackermann H W, Pandian S. Characterization of lactococcal phages from Québec cheese plants. Can J Microbiol. 1992;38:875–882. [Google Scholar]
- 25.Moineau S, Fortier J, Pandian S. Direct detection of lactococcal bacteriophages in cheese whey using DNA probes. FEMS Microbiol Lett. 1992;92:169–174. [Google Scholar]
- 26.Moineau S, Pandian S, Klaenhammer T R. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol. 1994;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce L E. Starters and phage activity tests for cheese starter cultures. N Z J Dairy Sci Technol. 1969;4:246–247. [Google Scholar]
- 28.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214:241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 29.Powell I B, Arnold P M, Hillier A J, Davidson B E. Molecular comparison of prolate and isometric headed bacteriophages of lactococci. Can J Microbiol. 1989;35:860–866. [Google Scholar]
- 30.Prevots F, Mata M, Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990;56:2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pringle C R. Virus taxonomy 1996—a bulletin from the Xth International Congress of Virology in Jerusalem. Arch Virol. 1996;141:2251–2256. doi: 10.1007/BF01718231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schouler C. Genomic organization of lactic acid bacteria phages. Lait. 1996;76:81–89. [Google Scholar]
- 34.Schouler C, Ehrlich S D, Chopin M C. Sequence and organization of lactococcal prolate headed bIL67 phage genome. Microbiology (United Kingdom) 1994;140:3061–3069. doi: 10.1099/13500872-140-11-3061. [DOI] [PubMed] [Google Scholar]
- 35.Svensson U K, Christiansson A. Methods for phage monitoring. Bull Int Dairy Fed. 1991;263:29–39. [Google Scholar]
- 36.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehead H R, Cox G A. The occurrence of bacteriophages in starter cultures of lactic streptococci. N Z J Sci Technol. 1935;16:319–320. [Google Scholar]
- 38.Zhou Y-J, Estes M K, Jiang X, Metcalf T G. Concentration and detection of hepatitis A virus and rotavirus from shellfish by hybridization tests. Appl Environ Microbiol. 1991;57:2963–2968. doi: 10.1128/aem.57.10.2963-2968.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]