Abstract
Differentiation of viable cells from nonviable cells is of considerable importance in the development of methods to detect foodborne pathogens. To study the suitability of 16S rRNA as an indicator of cell viability in nucleic acid-based detection assays, we examined rRNA stability in two representative foodborne pathogens, Escherichia coli O157:H7 and enterotoxigenic Staphylococcus aureus, which were inactivated by extreme heat, moderate heat, and UV irradiation. Cell death under all conditions was confirmed by a failure to grow in brain heart infusion broth after incubation for 48 h at 37°C. rRNA stability was monitored by a Northern blot analysis, and detection was evaluated by using reverse transcription (RT)-PCR performed with two primer sets (which produced 325- and 1,400-bp amplicons). rRNA of neither pathogen was detected by Northern blot analysis and RT-PCR after cells were killed by autoclaving at 121°C for 15 min. In contrast, intact rRNA of both pathogens were detected by Northern blotting and could be amplified by RT-PCR up to 48 h after cells were killed by heat treatment at 80°C and UV irradiation at 254 nm. rRNA was a suitable target molecule for monitoring bacterial viability under extreme heat conditions, but the presence of rRNA was not correlated with viability following moderate heat inactivation or UV irradiation of cells.
When used to detect pathogenic microorganisms in food, the PCR is a rapid, inexpensive, sensitive, and specific alternative to standard cultural procedures (4). Typically, DNA is the target molecule used for PCR-based amplification followed by DNA hybridization with internal oligonucleotide probes, which is used to confirm the identities of PCR amplicons. Recent studies, however, have brought into question the association between DNA and cellular viability (3, 9). This is particularly important in foods and the environment, where nonviable, inactivated pathogens may be present after treatments such as heating, sanitation, and irradiation. In general, investigators have found that DNA is stable enough to be amplified by PCR many days after cell viability has been lost, as determined by culturing (2, 9).
While DNA is the target nucleic acid most commonly used for amplification reactions, research has indicated that certain RNA types can be present at levels of thousands of copies per cell (21). Such high copy numbers can significantly improve detection limits, and for this reason rRNA has been a popular target molecule for commercial probe assays designed to detect foodborne pathogens (11). Although Taq polymerase cannot use RNA as a target for amplification, conversion of RNA to cDNA prior to the PCR makes the assay readily adaptable to this template.
rRNA is an appealing target for nucleic acid amplification-based detection. Since rRNA is a universal constituent of bacterial ribosomes and is present in high copy numbers (103 to 104 molecules per actively growing cell) (14), targeting this molecule increases the potential sensitivity of detection assays. Extensive bacterial rRNA sequence information is currently available (8). Finally, some studies have shown that the status of ribosomes is a sensitive biological indicator of the physiological state of cells and that rRNA may be more closely associated with cellular viability than DNA is (10, 13, 19).
To study the suitability of 16S rRNA as an indicator of cell viability in nucleic acid-based detection assays, we examined the stability of this molecule in two representative foodborne pathogens, Escherichia coli O157:H7 and enterotoxigenic Staphylococcus aureus. Stability was assessed by Northern hybridization and on the basis of the ability to support RT-PCR amplification after cells were killed by moderate heat, extreme heat, or UV irradiation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli O157:H7 strain ATCC 43895 and S. aureus ATCC 13565 were obtained from Douglas Marshall, Department of Food Science and Technology, Mississippi State University. Cultures were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) at 37°C for 16 h prior to all experiments.
Inactivation treatments.
The numbers of cells in broth cultures of E. coli and S. aureus were determined by the standard plate count method. When the cell density reached 106 CFU ml−1 (after 12 h of growth), broth-containing tubes (total volume, 3 ml) were either placed in an 80°C water bath for 20 min, autoclaved at 121°C for 15 min, or exposed to UV irradiation for 2.5 h. All treatments were performed in triplicate. For UV irradiation, 3 ml of a culture was placed in a sterile petri plate with the lid removed. The petri plate was placed in a 30.5-cm3 plastic exposure chamber, and a 254-nm light source (1,200 mW/cm2) was placed in the top of the treatment box 3 cm from the culture in the petri plate. The entire system was placed on a rotary shaker at 120 rpm, and the plate was exposed to the UV light for 2.5 h at 23°C without room lights. Immediately following the heat and UV treatments, 100-μl portions of the cell suspensions were placed into 10-ml portions of prewarmed (37°C) BHI broth (duplicate tubes) and incubated at 37°C for 48 h in order to confirm that the cells were dead by the absence of turbidity. Prior to RNA extraction at different times, this process was repeated to confirm that the cells were dead and to eliminate the possibility that injured cells had been resuscitated. A 100-μl portion of each preparation also was surface plated in duplicate onto BHI agar (Difco) and incubated at 37°C for 24 h.
RNA isolation.
Total RNA was purified from E. coli O157:H7 by using the Trizol reagent (Life Technologies, Gaithersburg, Md.) according to the manufacturer’s instructions. To isolate total RNA from S. aureus, the following method was used (a temperature of 4°C was used for all steps, unless stated otherwise). The cells in a 1-ml suspension were pelleted by centrifugation in a microcentrifuge tube and washed twice in cold sterile 0.1% (wt/vol) peptone water (Difco). After the second wash, the pellet was resuspended in 100 μl of RNase-free TBE buffer (90 mM Tris base, 90 mM boric acid, 2 mM EDTA [all obtained from Sigma Chemical Co., St. Louis, Mo.]; pH 7.8), and the tube was immersed in a boiling water bath for 1.5 min. An equal volume of phenol-chloroform (pH 4.6) (Amresco, Solon, Ohio) was immediately added, and the preparation was mixed and centrifuged at 12,000 × g for 8 min. The supernatant was removed and extracted with an equal volume of chloroform (Amresco). Following centrifugation at 10,000 × g for 10 min, the aqueous phase, which contained the total RNA, was transferred to a new tube and precipitated with 0.1 volume of RNase-free 3 M sodium acetate (Sigma) and 2.5 volumes of ice-cold 100% ethanol. The RNA was allowed to precipitate at −20°C for 4 h before it was pelleted by centrifugation at 15,000 × g for 30 min at 4°C. The pellet was dried at 37°C for 15 min and then resuspended in 10 μl of nuclease-free water (Geno Technology, St. Louis, Mo.).
To eliminate carryover DNA, DNase digestion was performed with purified RNA from both S. aureus and E. coli O157:H7. Briefly, 100 μl of a cocktail containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 5 mM MgCl2, and 1 mM CaCl2 (Calbiochem, La Jolla, Calif.) was added to the resuspended pellet along with 1 μl of RNasin, 5 μl of DNase I, and 10 μl of 0.1 M dithiothreitol (Promega, Madison, Wis.). The digest was mixed and placed in a 37°C water bath for 1 h. The acidic phenol-chloroform and chloroform extraction and precipitation steps described above were repeated, and the pelleted RNA, which was now free of DNA, was resuspended in 10 μl of nuclease-free water (Geno Technology). The amount of RNA was determined at 260 nm with a model UV-1201 spectrophotometer (Shimadzu, Inc., Kyoto, Japan).
Control RNA were purified from E. coli O157:H7 and S. aureus before heat or UV inactivation. For autoclaved cultures, RNA was extracted immediately following treatment and then 2, 6, and 24 h after treatment. For E. coli O157:H7 killed by treatment at 80°C, RNA was purified 0, 12, 24, and 48 h after treatment. RNA from E. coli O157:H7 that was killed by UV treatment was extracted 0, 24, and 48 h after treatment. For S. aureus inactivated by treatment at both UV and 80°C, RNA was purified 0, 1, 2, 4, 6, 8, 10, 24, 36, and 48 h after treatment. In all cases following inactivation treatments, the cultures were maintained at room temperature until the appropriate times for RNA isolation.
Northern blotting.
rRNA stability in viable and killed E. coli and S. aureus cultures was analyzed by using the NorthernMax system (Ambion, Austin, Tex.). The concentration of the purified RNA was determined spectrophotometrically. Purified RNA (8 μg) was hybridized to either a 323-nucleotide 16S rRNA-specific DNA probe (E. coli) or a 396-nucleotide 16S rRNA-specific DNA probe (S. aureus) (Table 1). The probes were synthesized with a MAXIscript in vitro transcription kit (Ambion) as recommended by the manufacturer. Blot detection was performed with the BrightStar BioDetect nonisotopic system (Ambion), and blots were developed with Kodak X-OMAT LS imaging film. To confirm that the method was reproducible, Northern analyses were performed in triplicate for each pathogen at different times.
TABLE 1.
Oligonucleotide primer sequences used in this studya
| Primer | Sequence (5′-3′) | Amplicon length (bp) | Use |
|---|---|---|---|
| S4F | GACAACTAGAGATAGAGCCTTCC | ||
| S4R | AGTCGAGTTGCAGACTAC | 324 (986–1310)b | RT-PCR, S. aureus |
| S6F | GGCCACACTGGAACTGAGACAC | Probe synthesis | |
| S6R | CTCTGCGCATTTCACCGCTACACATGG | 396 (291–687) | Northern blot, S. aureus |
| S8F | GCGGCGTGCCTAATACATGCAAGT | ||
| S8R | CGGTGTGTACAAGACCCGGGAA | 1,371 (14–1385) | RT-PCR, S. aureus |
| E5F | ATCCTGGCTCAGATTGAACGCT | ||
| E5R | TACAAGGCCCGGGAACGTATTCA | 1,439 (1–1439) | RT-PCR, E. coli |
| E6F | GCGGTCCGGCCGGGAACTCAAA | RT-PCR and probe | |
| E5R | TACAAGGCCCGGGAACGTATTCA | 323 (1116–1439) | Synthesis (Northern blot), E. coli |
RT-PCR.
Two microliters of RNA from each of the preparations was subjected to reverse transcription (RT)-PCR by using the primers listed in Table 1 for E. coli and S. aureus. RT was performed at 42°C for 50 min in a 25-μl (total volume) mixture containing 2 μl of purified RNA, 20 pmol of the appropriate reverse primer, 1 U of RNasin (Promega), 4 μl of 1st strand buffer (Life Technologies), 3 μl of 0.1 M dithiothreitol, 1 μl (200 U) of Superscript II reverse transcriptase (Life Technologies), 0.9 μl of a deoxynucleoside triphosphate mixture containing each deoxynucleoside triphosphate at a concentration of 800 mM, and 9 μl of nuclease-free water. For S. aureus, 2 μl of dimethyl sulfoxide was added to the reverse transcription cocktail to reduce the target secondary structure and to facilitate amplification (15, 17). Following reverse transcription, the reaction mixtures were placed in a 70°C water bath for 15 min to inactivate the reverse transcriptase; 4 μl of each reaction mixture was used in the DNA PCR along with 20 pmol of each primer and 40 μl of Supermix (Life Technologies). Dimethyl sulfoxide (2 μl) was included in the S. aureus PCR mixture.
Amplification was performed with a Power Block II cycler (Ericomp, San Diego, Calif.) for 35 cycles consisting of 94°C for 15 s, 54°C for 1 min, and 72°C for 2 min, followed by a final extension step at 72°C for 7 min. The PCR products (9 μl) were analyzed on a 1% (wt/vol) agarose gel. The controls included a no-template control, a no-RT control, and a positive DNA control for each primer set. Two sets of RT-PCR were performed for each pathogen (Table 1). One primer set amplified a small region in the 16S rRNA of S. aureus or E. coli O157:H7 (324 nucleotides for S. aureus and 323 nucleotides for E. coli). The second primer set amplified a much larger region, which spanned nearly the entire length of the 16S rRNA molecule for each microorganism (1,371 nucleotides for S. aureus and 1,439 nucleotides for E. coli). We expected that the latter primer set would better gauge the integrity of the rRNA of the two pathogens at the various times after the different treatments. RT-PCR with both primer sets for S. aureus and E. coli were performed on three different occasions to confirm the reproducibility of the method.
RESULTS
No growth was observed after 48 h of incubation at 37°C in any of the BHI broth tubes or on any of the BHI agar plates immediately following heat or UV treatment or at any time thereafter, which confirmed that the inactivation treatments were effective and that viable bacterial cells were absent.
rRNA stability as evaluated by Northern blotting.
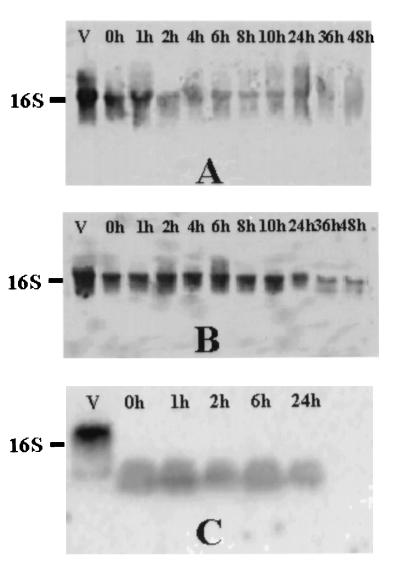
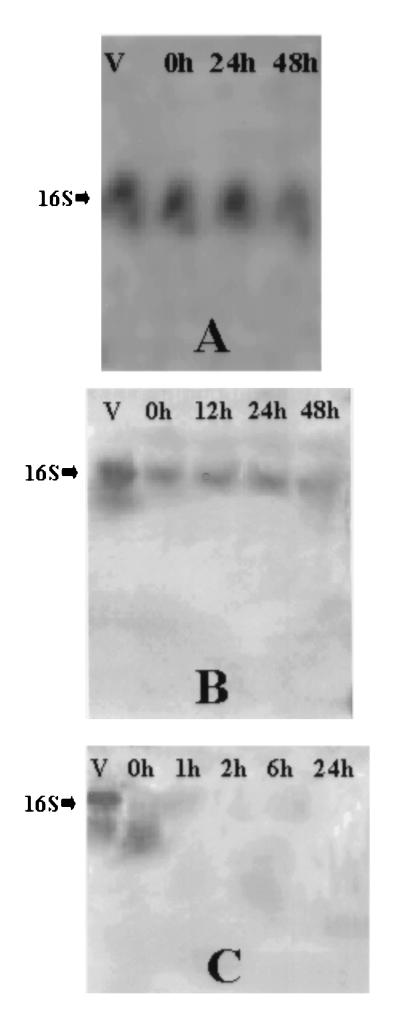
Northern blots of 16S rRNA from treated S. aureus and E. coli O157:H7 cultures are shown in Fig. 1 and 2, respectively. Autoclaved cells of both S. aureus and E. coli O157:H7 produced no signal on Northern blots, even immediately following treatment (Fig. 1C and 2C). Apparent RNA degradation products were visible throughout the 24-h holding period for autoclaved S. aureus but only immediately after autoclaving for E. coli O157:H7. In all autoclaved cells, intact rRNA was degraded. In contrast, compared to the signal intensity obtained with viable (i.e., log-phase) cells, the intensity of the 16S rRNA band decreased slightly with time following exposure to UV (Fig. 1A and 2A) or 80°C (Fig. 1B and 2B), but this band remained present through the entire 48-h holding period.
FIG. 1.
Northern blots of enterotoxigenic S. aureus cells inactivated by exposure to UV (260-nm) light for 2.5 h (A), exposure to 80°C for 20 min (B), or exposure to 121°C for 15 min (C). Total RNA (8 μg) from viable cells (lanes V) or killed cells at different times was applied to a 1% agarose–6% formaldehyde gel, transferred to a nylon membrane, and hybridized to a 323-base 16S rRNA-specific DNA probe. Blots were developed and bands were detected by using the BrightStar BioDetect nonisotopic system as described in Materials and Methods.
FIG. 2.
Northern blots of E. coli O157:H7 cells inactivated by the treatments used to kill S. aureus cells described in the legend to Fig. 1. Northern blotting and detection were performed as described in the legend to Fig. 1. Total RNA was isolated from treated E. coli at different times following exposure to UV irradiation (A), 80°C (B), or 121°C (C).
rRNA stability and detection as evaluated by RT-PCR.
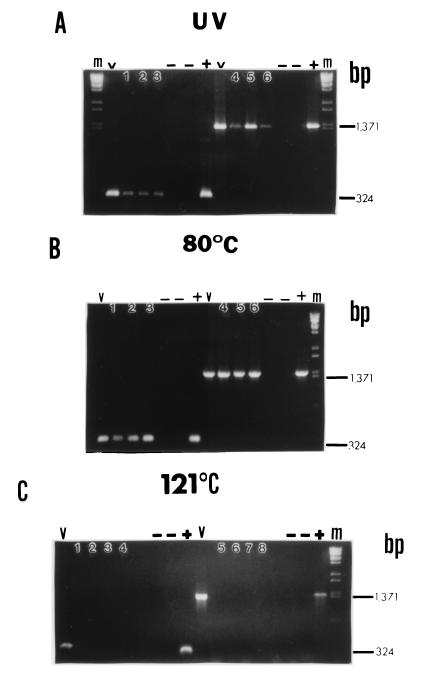
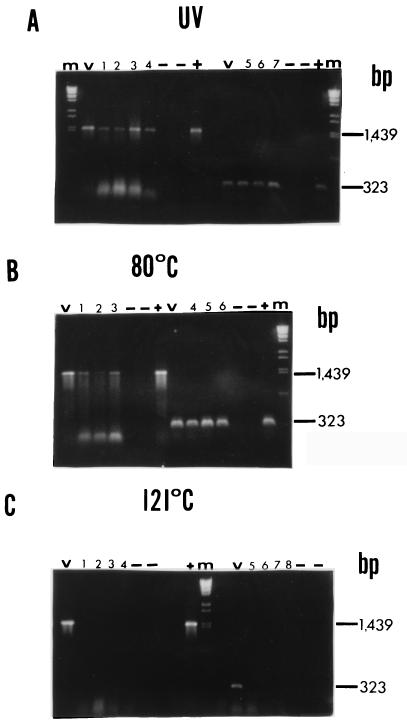
In order to corroborate the results of the Northern blotting experiments and to evaluate the ability of the rRNA in killed cells to support efficient nucleic acid amplification, RT-PCR amplification of rRNA from both S. aureus and E. coli O157:H7 was performed at various times after inactivation treatments. Two different primer sets were used (which targeted amplicons which were approximately 325 and 1,400 bp long) to aid in evaluation of rRNA integrity and the associated amplification efficiency. Fewer time points were analyzed in this experiment than in the Northern blot analyses because a spot check of selected times provided adequate information. RT-PCR products disappeared immediately after autoclaving (at zero time) for both S. aureus and E. coli O157:H7 (Fig. 3C and 4C) when preparations were analyzed with both primer sets. Although an occasional faint but spurious amplification signal was obtained at other times after autoclaving, this was not a consistent observation and is similar to the results obtained by Rosenfield (12). For UV-inactivated and 80°C-inactivated S. aureus, both primer sets amplified 16S rRNA at 0, 24, and 48 h after treatment (Fig. 3A and 3B). Similar results were obtained for UV-inactivated and 80°C-inactivated E. coli O157:H7 (Fig. 4A and 4B). There was no difference in RT-PCR amplification band intensity when the large and small amplicons were compared. However, the amplicon bands for UV-inactivated cells were almost always fainter than the amplicon bands for 80°C-inactivated cells despite the fact that the parameters for RT-PCR amplification of all samples were optimized.
FIG. 3.
RT-PCR performed with inactivated S. aureus cells. Bacteria were treated as described in the legend to Fig. 1, and purified RNA was subjected to RT-PCR with primers specific for positions 986 to 1310 of the S. aureus 16S rRNA gene (324 nucleotides, small amplicon) or primers specific for positions 14 to 1385 (1,371 nucleotides, large amplicon). Lanes m contained a λ/BstEII molecular weight standard, and lanes V contained viable cell RNA. −, no-template control or no-RT control performed with each primer set for each treatment; +, positive DNA control (1 μl of log-phase cell DNA was subjected to PCR with the appropriate primer set). (A) UV-treated cells. Lanes 1 through 3, small amplicon, 0, 24, and 48 h, respectively; lanes 4 through 6, large amplicon, 0, 24, and 48 h, respectively. (B) Cells subjected to 80°C. Lanes 1 through 3, small amplicon, 0, 24, and 48 h, respectively; lanes 4 through 6, large amplicon, 0, 24, and 48 h, respectively. (C) Cells subjected to 121°C. Lanes 1 through 4, small amplicon, 0, 2, 6, and 24 h, respectively; lanes 5 through 8, large amplicon, 0, 2, 6, and 24 h, respectively. The positions of large and small amplicon bands are indicated on the right.
FIG. 4.
RT-PCR performed with inactivated E. coli O157:H7 cells. Bacteria were treated as described in the legend to Fig. 1, and purified RNA was subjected to RT-PCR with primers specific for positions 1116 to 1439 of the E. coli 16S rRNA gene (323 nucleotides, short amplicon) or primers specific for positions 1 to 1439 (large amplicon). Lanes m contained a λ/BstEII molecular weight standard, and lanes V contained the products of a RT-PCR performed with viable cell RNA. −, no-template control or no-RT control performed with each primer set for each treatment; +, positive DNA control (1 μl of log-phase cell DNA was subjected to PCR with appropriate primer set). (A) UV-treated cells. Lanes 1 through 4, large amplicon, 0, 12, 24, and 48 h, respectively; lanes 5 through 7, small amplicon, 0, 24, and 48 h, respectively. (B) Cells subjected to 80°C. Lanes 1 through 3, large amplicon, 0, 24, and 48 h, respectively; lanes 4 through 6, small amplicon, 0, 24, and 48 h, respectively. (C) Cells subjected to 121°C. Lanes 1 through 4, large amplicon, 0, 2, 6, and 24 h, respectively; lanes 5 through 8, small amplicon, 0, 2, 6, and 24 h, respectively. The positions of large and small amplicon bands are indicated on the right.
DISCUSSION
PCR-based techniques have proven to be adaptable for detection of an assortment of pathogens in a variety of food samples. Because DNA from dead cells may be amplified by PCR, targeting genomic sequences may not be adequate for determining the viability of bacteria. For instance, when Masters et al. (9) evaluated the effect of several stress treatments (starvation, heating, hydrogen peroxide, acid, and desiccation) on PCR detection of Listeria monocytogenes and E. coli, they found that DNA was detectable after plate counts had declined to <3 CFU regardless of the treatment. In a similar study, Herman (3) concluded that L. monocytogenes DNA could be detected by PCR for more than 30 days after cell viability was lost due to various inactivation treatments (disinfectants, pH adjustments, ethanol, and moderate heat). In environmental systems, Dupray et al. (2) found that the DNA of dead Salmonella spp. could be detected for up to 55 days in seawater stored at 10°C and that free DNA could be detected for 2 days in seawater stored at 20°C.
A number of workers have investigated the usefulness of various mRNA transcripts for determining viability in pathogenic bacteria. However, mRNA may be an unsuitable indicator of bacterial viability due to the difficulty of finding a transcript that is constitutively expressed and species specific. Moreover, mRNA is typically present in low copy numbers and is unstable. Sheridan et al. (16) used RT-PCR to evaluate the stability of mRNA from one constitutively expressed E. coli housekeeping gene (tufA, a gene encoding an elongation factor) and two genes of a stress response regulon (rpoH and groEL) and found in each case that the mRNA was degraded more quickly in heat-killed cells (killed at 100 and 80°C) than in bacteria killed by either 50 or 67% ethanol. These authors suggested that mRNA disappears more rapidly from bacteria killed by treatments that do not inactivate the degradative RNase enzymes and that the overall persistence of mRNA in dead cells depends on the method used to kill the bacteria.
Previous studies have suggested that rRNA may be a better indicator of viability when bacteria are subjected to certain stresses. For instance, it has been reported that carbon or phosphate starvation is correlated with degradation of 16S rRNA (1, 5, 18). Other studies in which the effects of heat on cells were examined revealed that thermal injury resulted in degradation of the 30S subunit (10, 13, 17). The shortwave UV irradiation which we used to kill S. aureus and E. coli was a less traumatic means of cell inactivation than either of the heat treatments that we employed. Neither exposure to UV irradiation at room temperature nor heat treatment at 80°C would be expected to significantly impede the activity of endogenous RNases following cell death. However, we noticed no marked differences in the overall integrity of rRNA in cells exposed to UV irradiation and cells killed by exposure to 80°C, perhaps because the ribosomes retained their secondary structure and associated proteins, thereby avoiding RNase degradation. After both treatments, the rRNA was structurally sound in the Northern blots through 48 h after treatment, and rRNA could serve as a template for RT-PCR even when a primer set spanning nearly the entire length of the 16S rRNA molecule was employed. We expected that the latter study would yield more valuable information regarding ribosomal integrity after bacterial death as a function of time, as any damage to rRNA at the primary sequence level could potentially prevent full-length amplicon production during RT-PCR. These observations were consistent for both of the pathogens which we studied.
Destruction of bacterial rRNA during moderate heating has been studied in several bacterial species. Uyttendaele et al. (20) targeted the 16S rRNA of Campylobacter jejuni by using the nucleic acid sequence-based amplification procedure and found that rRNA was stable for 5 h following treatment at 100°C. Sheridan et al. (16) monitored E. coli 16S rRNA by RT-PCR following thermal inactivation at three different temperatures (100, 80, and 60°C). A positive signal was obtained through 16 h after treatment, which is consistent with our findings. Khalil and Villota (6) studied the effect of sublethal heating on the rRNA of S. aureus and found that rRNA damage occurred following exposure to 50°C for 30 min, as determined by sucrose density gradient centrifugation and corresponding measurements of absorbance at 260 nm. However, these authors were for the most part investigating the rate of ribosomal recovery from damage (i.e., denaturation) by heating, while other studies, including our study, have focused more on the presence or absence of an intact rRNA template after a lethal heat or UV treatment and on how such treatments dictate the extent to which the rRNA may be used as a determinant of bacterial viability.
In this study, a combination of high temperature and pressure (autoclaving at 121°C for 15 min) degraded S. aureus and E. coli O157:H7 rRNA enough so that signals were lost during Northern hybridization, and rRNA could not be used as a template in subsequent RT-PCR. Rosenfield (12) studied the effects of autoclaving Salmonella enteriditis on the subsequent ability to amplify 16S rRNA by RT-PCR. rRNA could be detected by RT-PCR only sporadically and at very low levels after autoclaving. Rosenfield concluded that the presence of rRNA can be correlated with cellular viability following inactivation of the bacteria with extreme temperatures (i.e., autoclaving). Our data are consistent with this conclusion.
We hypothesized that in the case of extreme heat (i.e., autoclaving) changes in the secondary structure of rRNA and ribosomal proteins affected the overall integrity of the ribosomes, exposing sequences not normally exposed to the intracellular RNases. The loss of ribosomal integrity observed on Northern blots supports this theory. Alternatively, or in combination, heat may disrupt the bacterial cell wall, releasing the cellular contents and exposing the rRNA to RNases. We hypothesized that under less extreme conditions (80°C or UV irradiation) the ribosomal unit remained intact and the combination of nucleic acid secondary structure and associated ribosomal proteins protected the rRNA from degradation by endogenous or exogenous nucleases. Perhaps a more likely explanation is that the shear heat and pressure of autoclaving simply degraded the ribosomes, preventing them from serving as either target molecules in Northern blots or as templates for RT-PCR. In other words, the extreme physical conditions of autoclaving, not the presence of RNases, destroyed the ribosomes.
rRNA is a good choice for a target amplification molecule because it is present in high copy numbers, is constitutively expressed, and is less stable than DNA. Our study indicated that rRNA is a good indicator of viability under extreme heat conditions. However, many food-processing heat treatments involve lower temperatures and longer times, and our results indicate that detection of rRNA may not be associated with viability under these conditions. When a food is subjected to heat, however, exposed bacterial rRNA may be degraded by food-associated RNases. Therefore, it would be beneficial to evaluate the relationship among cell viability, rRNA integrity, and RT-PCR detection in model food systems. Such studies are currently underway in our laboratory.
ACKNOWLEDGMENTS
This work was funded by Dairy Management, Inc., and the Mississippi Agricultural and Forestry Experiment Station.
Footnotes
This is Mississippi Agricultural and Forestry Experiment Station project number J9387.
REFERENCES
- 1.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupray E, Caprais M P, Derrien A, Fach P. Salmonella DNA persistence in natural seawaters using PCR analysis. J Appl Microbiol. 1997;82:507–510. doi: 10.1046/j.1365-2672.1997.00143.x. [DOI] [PubMed] [Google Scholar]
- 3.Herman L. Detection of viable and dead Listeria monocytogenes by PCR. Food Microbiol. 1997;14:103–110. [Google Scholar]
- 4.Hill W E. The polymerase chain reaction: applications for the detection of food-borne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan R, Apirion D. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J Biol Chem. 1975;250:1854–1863. [PubMed] [Google Scholar]
- 6.Khalil H, Villota R. The effect of microwave sublethal heating on the ribonucleic acids of Staphylococcus aureus. J Food Prot. 1989;52:544–548. doi: 10.4315/0362-028X-52.8.544. [DOI] [PubMed] [Google Scholar]
- 7.King T C, Schlessinger D. Processing of RNA transcripts. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 703–718. [Google Scholar]
- 8.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S rRNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masters C I, Shallcross J A, Mackey B M. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J Appl Bacteriol. 1994;77:73–79. doi: 10.1111/j.1365-2672.1994.tb03047.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller L L, Ordal Z J. Thermal injury and recovery of Bacillus subtilis. Appl Microbiol. 1972;24:878–884. doi: 10.1128/am.24.6.878-884.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons G. Development of DNA probe-based commercial assays. J Clin Immunoassay. 1988;11:152–160. [Google Scholar]
- 12.Rosenfield S. Development of reverse transcription polymerase chain reaction (RT-PCR) approaches to the detection of bacterial and viral foodborne disease agents. M.S. thesis. Raleigh: North Carolina State University; 1998. [Google Scholar]
- 13.Rosenthal L J, Iandolo J J. Thermally induced intracellular alteration of ribosomal ribonucleic acid. J Bacteriol. 1970;103:833–835. doi: 10.1128/jb.103.3.833-835.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheu P M, Berghof K, Stahl U. Detection of pathogenic and spoilage microorganisms in food with the polymerase chain reaction. Food Microbiol. 1998;15:13–31. [Google Scholar]
- 15.Shen W H, Hohn B. DMSO improves PCR amplification of DNA with complex secondary structure. Trends Genet. 1992;8:227. [Google Scholar]
- 16.Sheridan G E C, Masters C I, Shallcross J A, Mackey B M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidhu M K, Liao M-J, Rashidbaigi A. Dimethyl sulfoxide improves RNA amplification. BioTechniques. 1996;21:44–47. doi: 10.2144/96211bm08. [DOI] [PubMed] [Google Scholar]
- 18.Tolker-Nielsen T, Larsen M H, Kyed H, Molin S. Effects of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int J Food Microbiol. 1997;35:251–258. doi: 10.1016/s0168-1605(97)01242-7. [DOI] [PubMed] [Google Scholar]
- 19.Tomlins R I, Ordal Z J. Precursor ribosomal ribonucleic acid and ribosome accumulation in vivo during the recovery of Salmonella typhimurium from thermal injury. J Bacteriol. 1971;107:134–142. doi: 10.1128/jb.107.1.134-142.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uyttendaele M, Bastiaansen A, Debevere J. Evaluation of the NASBA® nucleic acid amplification system for assessment of the viability of Campylobacter jejuni. Int J Food Microbiol. 1997;37:13–20. doi: 10.1016/s0168-1605(97)00039-1. [DOI] [PubMed] [Google Scholar]
- 21.Waters A P, McCatchan T F. Ribosomal RNA: nature’s own polymerase-amplified target for diagnosis. Parasitol Today. 1990;6:56–59. doi: 10.1016/0169-4758(90)90071-b. [DOI] [PubMed] [Google Scholar]