Summary
Rhodopsin-1 (Rh1), the main photosensitive protein of Drosophila, is a seven-transmembrane domain protein, which is inserted co-translationally in the endoplasmic reticulum (ER) membrane. Biogenesis of Rh1 occurs in the ER, where various chaperones interact with Rh1 to aid in its folding and subsequent transport from the ER to the rhabdomere, the light-sensing organelle of the photoreceptors. Xport-A has been proposed as a chaperone/transport factor for Rh1, but the exact molecular mechanism for Xport-A activity upon Rh1 is unknown. Here, we propose a model where Xport-A functions as a chaperone during the biogenesis of Rh1 in the ER by stabilizing the first five transmembrane domains (TMDs) of Rh1.
Subject areas: Biochemistry, Biological sciences, Molecular biology, Molecular interaction
Graphical abstract
Highlights
-
•
AlphaFold2 predicts that Xport-A interacts with the first five TMDs of rhodopsin-1
-
•
Molecular dynamics simulations corroborate AlphaFold2 predictions
-
•
Xport-A may function as a transient placeholder for TMDs 6 and 7 of rhodopsin-1
-
•
The C-terminal domain of Xport-A stabilizes the N-terminal domain of rhodopsin-1
Biochemistry; Biological sciences; Molecular biology; Molecular interaction
Introduction
Rh1 functional protein comprises the apoprotein opsin and the covalently bound chromophore 11-cis-3-hydroxy-retinal.1 The opsin corresponds to the protein moiety of Rh1 and is encoded by the ninaE (neither inactivation nor afterpotential E) gene.2,3 This gene encodes an integral membrane protein composed of seven transmembrane domains (TMDs),2 which is inserted co-translationally into the endoplasmic reticulum (ER) membrane. Rh1 maturation in the ER involves post-translational modifications such as transient glycosylation, followed by de-glycosylation4,5,6 and chromophore binding. The chromophore of Drosophila Rh1 is made from beta carotene that is uptaken in the diet1 and then processed to vitamin A, which is subsequently converted into 11-cis-3-hydroxy-retinal.7,8 The lack of chromophore incorporation in Drosophila Rh1, by carotenoid dietary restriction, leads to extremely reduced protein levels of functional Rh1, resulting in glycosylated Rh1 that appears to be retained in the ER, presumably due to folding defects.1,9 Misfolded Rh1 can be degraded either by ER-associated degradation or by the lysosome.10,11 Upon successful folding and maturation, Rh1 is transported through the Golgi compartment to the rhabdomere, where phototransduction occurs.12,13
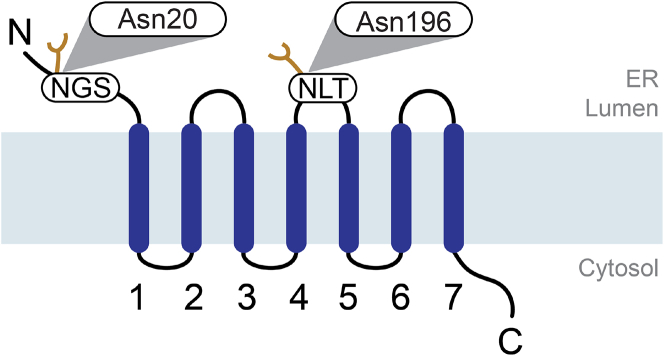
Rh1 contains two possible glycosylation sites (Figure 1): N20, within the N-terminal region and N196, in the second extracellular loop, between TMD4 and TMD5.4,5,6 Although in vitro experiments with mammalian microsomes have shown that Rh1 can be glycosylated at both sites,5 only glycosylation at N20 has been shown to occur in “wild-type” (WT) and ninaA mutant flies.4,5,6 Nonetheless, mutation of the asparagine residue at either glycosylation sites (N20 or N196) to isoleucine (N20I and N196I) interferes with the biogenesis of mutant and WT Rh1, resulting in the ER retention of Rh1.6 Consequently, Drosophila eyes expressing Rh1 N20I or Rh1 N196I present large accumulations of Rh1 in the ER and Golgi membranes.6 Furthermore, as less Rh1 reaches the rhabdomeres, these mutant (Rh1 N20I or Rh1 N196I) eyes also present late onset age-related retinal degeneration.6,14
Figure 1.
Schematic model of Drosophila rhodopsin-1 showing the two N-glycosylation sites
Rhodopsin contains two predicted glycosylation sites, NGS (asparagine-glycine-serine) in the N-terminal region, and NLT (asparagine-leucine-threonine) in the second extracellular loop region, between TMD4 and TMD5. The predicted first glycosylation site is mapped to asparagine at position 20 (N20) and the second glycosylation site is mapped to asparagine at position 196 (N196). Sugar residues are represented in yellow.
While glycosylation of Rh1 in N20 is necessary for maturation and/or transport in the secretory pathway, mature Drosophila Rh1 is completely deglycosylated.6 During maturation, Rh1 fully glycosylated 40 kDa form is first trimmed in the ER to a 39 kDa partly deglycosylated form, which is subsequently deglycosylated in the Golgi to a completely or almost completely deglycosylated protein.15,16
Rh1 requires several chaperones for proper folding and transport out of the ER, including ninaA (neither inactivation nor afterpotential A),17,18,19,20 calnexin,21,22 and exit protein of rhodopsin and TRP – Xport.23,24 The Xport locus is bicistronic; it is transcribed as a single mRNA that encodes two different proteins: Xport-A and Xport-B.24 Xport-A is a tail-anchored (TA) protein and Xport-B is predicted to be a type III membrane protein. Both proteins have homologs in insects and the bicistronic nature of the locus is also conserved.24 Xport-A was the first to be described,23 in a screening of the Zuker collection of EMS-mutagenized flies. Xport-A mutant (Xport-A1) homozygous flies presented electroretinogram (ERG) profiles similar to mutants of the TRP (transient receptor potential) channel and displayed extremely reduced levels of both Rh1 and TRP proteins. The mutation in Xport-A1 is recessive, as the heterozygotes presented normal protein levels of TRP and Rh1. When reared in the dark, Xport-A1 flies exhibited ER and Golgi expansions, but the rhabdomere morphology was preserved. Upon exposure to light, the defect worsened and the mutants presented retinal degeneration. Interestingly, overexpression of Xport-A in the Xport-B mutant, or overexpression of Xport-B in the Xport-A mutant failed to rescue the ERG defects observed in these mutants, indicating that the roles of Xport-A and Xport-B are not redundant.24
Recently, we have shown that the ER membrane protein complex (EMC) is required for the biogenesis and membrane insertion of Xport-A,25 not being directly required for the biogenesis of Rh1 and TRP, as proposed previously.26 In that manuscript, we expressed 3 truncations of Rh1 containing the first (TMD1), the first 3 (TMD1-3) or the first 5 (TMD1-5) transmembrane domains of Rh127 in the background of flies heterozygous or homozygous for Xport-A1 mutation. Surprisingly, only Rh1 TMD1-5 biogenesis was impaired in the Xport-A1 homozygous flies, with Rh1 TMD1-5 being double glycosylated in N20 and N196, in contrast to single N20 glycosylation of Rh1 TMD1-5 in the Xport-A1 heterozygous background.25 Rh1 TMD1 and TMD1-3 presented normal biogenesis in Xport-A1 mutant homozygous flies, with both constructs being glycosylated in N20, demonstrating normal insertion of the first TMD of Rh1 in the ER membrane of Xport-A mutant flies.25 Here, mostly based on AlphaFold2 (AF2) and molecular dynamics simulations, we propose a model where Xport-A functions as a chaperone by stabilizing the first 5 TMDs of Rh1, during its biogenesis in the ER.
Results and discussion
Xport-A61-116 accommodates in Rh1 TMD1-5 but not in full-length Rh1
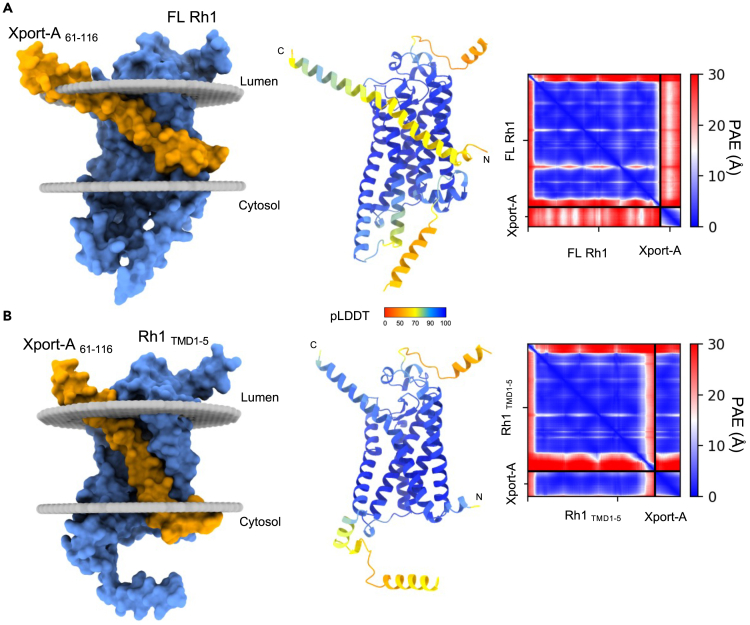
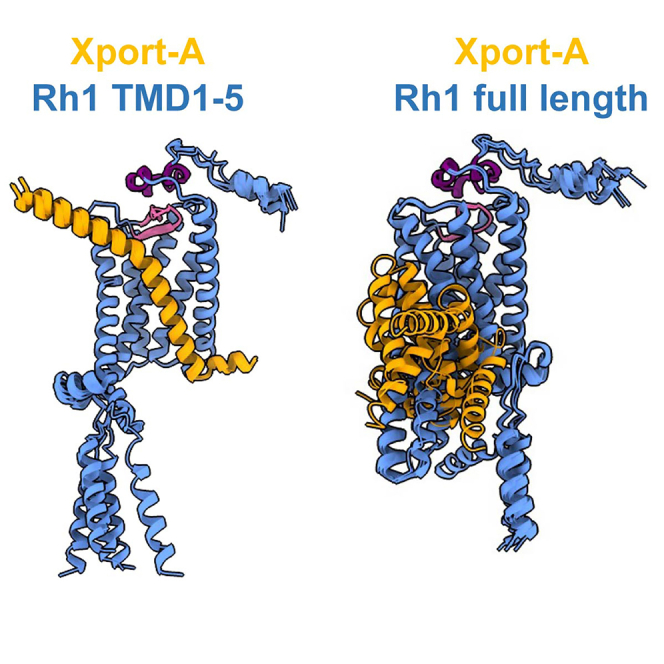
Based on our previous findings,25 we hypothesized whether Xport-A could be required for Rh1 biogenesis at the stage when the first 5 TMDs of Rh1 are inserted into ER membrane, rather than later, when all TMDs of the full-length (FL) Rh1 are inserted into the membrane. To provide insights into this issue, we resorted to AF227,28 structural predictions of Xport-A61-116 (amino acids 61–116, including the TMD and C-terminal ER luminal domain) together with FL Rh1 (Figure 2A) or Rh1 TMD1-5 (Figure 2B). While in Figure 2, we only display the top-ranked AF2 model (rank 1) for each complex, in Figure S1 we display overlays of the 5 top-ranked AF2 models. When together with FL Rh1 (Figure 2A), the predicted structure of Xport-A61-116 has low certainty standards (predicted Local Distance Difference Test - pLDDT values per residue), but an overall high pLDDT (>85) in complex with Rh1 TMD1-5 (Figure 2B). Moreover, Xport-A/FL Rh1 structural predictions also display high inter-complex predicted alignment errors and less favorable contacts, with 3 out of the 5 poses showing Xport-A61-116 (Figure S1A) in a reverse topology (N terminal in the ER lumen and C terminal in the cytosol) from what is expected (Xport-A is a TA protein, with its N terminal in the cytosol and C terminal in the ER lumen). AF2 predictions for FL Rh1/Xport-A61-116 fail to converge to defined and plausible structures, presenting an average RMSD (root-mean-square deviation) of 6.68 ± 2.02 Å (Figure S1A). In contrast, all Rh1 TMD 1-5/Xport-A61-116 predictions are very close, with an average RMSD of 0.30 ± 0.09 Å from the top-ranked model (Figure S1B).
Figure 2.
Xport-A TMD accommodates in Rh1 TMD1-5 but not in the full-length Rh1 protein
(A and B) AlphaFold2 structural predictions of Xport-A together with (A) full-length Rh1 or (B) Rh1 TMD1-5 (M1 to V241 + H333-A373). Only the top-ranked model for each complex is displayed, of the five models computed by AlphaFold2. To the left, Xport-A (amino acids 61 to 116) is represented in orange and Rh1 FL and Rh1 TMD1-5 are in light blue. In the middle, AlphaFold2 produces an estimate of confidence for each amino acid residue (pLDDT - Local Distance Difference Test), color-coded on a scale from 0 to 100. Values of pLDDT >90 (blue) are expected to be modeled with high accuracy. To the right are represented the predicted aligned errors (PAE) on a scale from 0 to 30 Å in a blue-white-red gradient for each of the structure predictions. PAE is a metric of confidence in the relative position and orientation of the different chains of the model.
Xport-A61-116 interacts poorly with Rh1 TMD1 and TMD1-3
We have also made AF2 predictions for Xport-A61-116 together with Rh1 TMD1 (Figures S2A–S2C) or Rh1 TMD1-3 (Figures S2D–S2F), which yielded undefined structures with low local confidence metrics. Nevertheless, for these AF2 predictions, the confidence at the binding interface between Xport-A61-116 and Rh1 TMD1 or Rh1 TMD1-3 is higher than in the AF2 predictions for Xport-A61-116 together with FL Rh1. Moreover, in the complex with TMD1-3, Xport-A61-116 shows the expected topology (N terminal in the cytosol and C terminal in the ER lumen). We observe contacts between Xport-A61-116 and the TMDs of the truncated Rh1 constructs for all predicted complexes, which become more defined when the number of Rh1 TMDs increases from TMD1 to TMD1-3 and TMD1-5. These contacts between Xport-A61-116 and the TMDs of the Rh1 are not present in the AF2 prediction of Xport-A61-116 together with the FL Rh1 protein.
AF2 predictions for FL Xport-A (Figures S3A–S3C) bear a disordered N-terminal intracellular region (amino acid residues 1–60), which is followed by the better defined short amphipathic helical segment (α1, residues 61–68), the TMD (residues 70–92), and the ER lumen-localized C-terminal domain (residues 93–116). The AF2 models for unbound Rh1 (Figures S3D–S3F) have a high-confidence pLDDT score (87.6 ± 17.3) and the only disordered regions are the intracellular loop 3 and the cytoplasmic C-terminal domain. The AF2 predictions for FL Xport-A together with Rh1 TMD1-5 (Figure S3G) and Xport-A61-116 with Rh1 TMD1-5 without the C-terminal H333-A373 (Figure S3H) gave similar results to Xport-A61-116 with Rh1 TMD1-5. Hence, we decided to omit Xport-A1-60 and use only Xport-A61-116 in our analysis, as the N-terminal disorder region of Xport-A could be a source of noise and unnecessary sampling during the prediction cycles and subsequent molecular dynamics simulations.
Xport-A may function as a transient placeholder for TMD 6 and 7 of Rh1
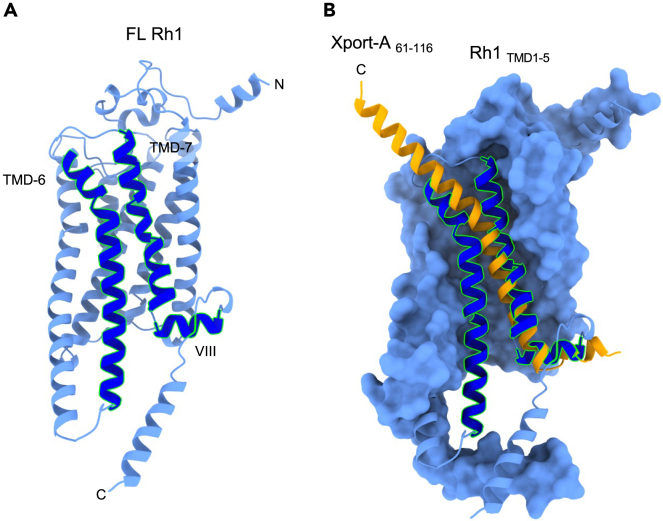
Overall, the AF2 models are therefore consistent with Xport-A binding to Rh1 TMD1-5 rather than to FL Rh1. This is supported by comparing the predicted structure of FL Rh1 by itself (Figure 3A) with the pose of Xport-A61-116 and Rh1 TMD1-5 (Figure 3B); one can visualize that Xport-A61-116 partially overlaps with the would-be locations of TMD6, TMD7, and α-helix 8 of Rh1. Hence, Xport-A could function as a chaperone by becoming a transient placeholder, by mimicking the structural features of Rh1 TMD6, TMD7, and α-helix 8, when only the first 5 TMDs of Rh1 are inserted into the membrane.
Figure 3.
Xport-A overlaps with TMD6, TMD7, and α-helix 8 of Rh1
(A) Representation of FL Rh1 with TMD6, TMD7, and α-helix 8 highlighted in darker blue with green outlines.
(B) Superimposition of Xport-A (amino acids 61 to 116 - in orange) with TMD6, TMD7, and α-helix 8 of Rh1, with Rh1 TMD1-5 surface in light blue.
Structural interactions between the TMDs of Xport-A and Rh1
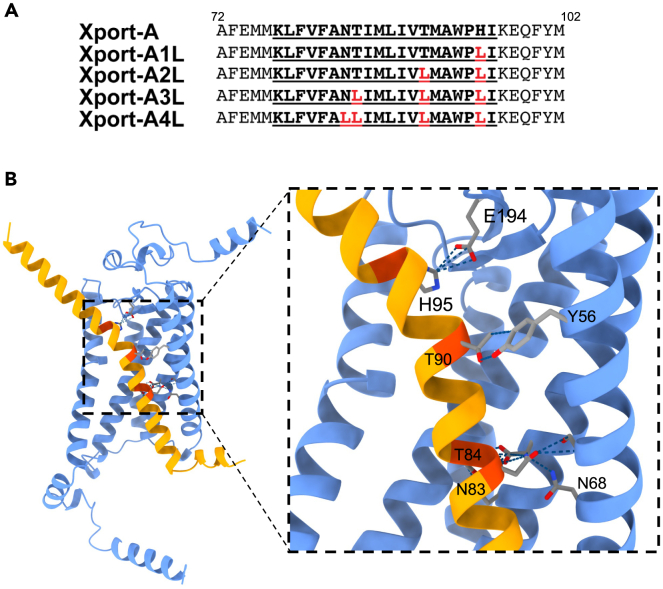
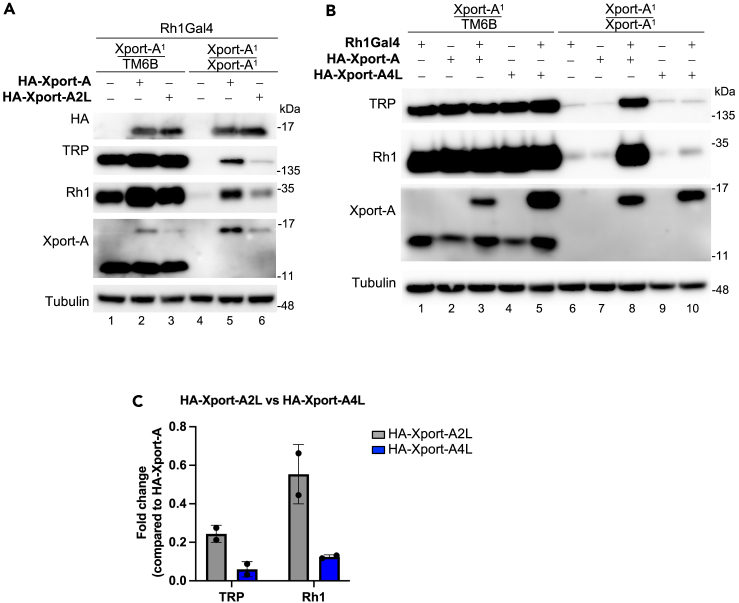
Next, we focus on some possible interactions between amino acid residues of Xport-A and Rh1, based on the most likely pose that we described in Figure 2B. Of particular interest, are the positions of the amino acid residues that we previously mutated to leucine.25 Xport-A 1L, 2L, 3L, and 4L (Figure 4A) are a series of mutants which lead to an increasingly more hydrophobic TMD of Xport-A, progressively bypassing the EMC requirement for membrane insertion of Xport-A.25 We have also shown that Xport-A 2L and 4L rescue the expression of Rh1 in EMC mutant cells,25 but in both cases these rescues were only partial, suggesting that the 2L and 4L mutants have reduced function, although they are inserted into the membrane, even in EMC mutant cells.25 In Figure 4B, we show the position of the 4 amino acid residues (N83, T84, T90, and H95) that were mutated to leucine in Xport-A 4L. All of them are oriented into the interface, in a position to interact with amino acid residues in Rh1 TMD1-5. In fact, we could observe that Xport-A 2L is worse than Xport-A (WT) but better than Xport-A 4L at rescuing the expression of Rh1 (and TRP) in fly eyes homozygous for the Xport-A1 mutation (Figure 5). This result suggests that the biological functionality of Xport-A 2L is less compromised than that of Xport-A 4L. Overall, these results support a role for Xport-A as a polarity shield to Rh1 TMD1-5, which is compromised when its own TMD’s polar residues (N83, T84, and T90) become mutated to leucine.
Figure 4.
Interactions between Xport-A TMD and Rh1 TMD1-5
(A) Amino acid sequence alignments of the TMDs (bold, underlined) of Xport-A, Xport-A1L, Xport-A2L, Xport-A3L, and Xport-A4L.
(B) Prediction of interactions between the Xport-A amino acid residues that were mutagenized to L (N83, T84, T90, and H95) and Rh1.
Figure 5.
Rescue of Rh1 and TRP in Xport-A1 homozygous mutant eyes by Xport-A, Xport-A2L, and Xport-A4L
(A) Immunoblot of fly heads expressing HA-Xport-A or HA-Xport-A2L in Xport-A heterozygous (Xport-A1/TM6B) or homozygous mutant flies (Xport-A1/Xport-A1), (B) Immunoblot of fly heads expressing HA-Xport-A or HA-Xport-A4L in Xport-A heterozygous (Xport-A1/TM6B) or homozygous mutant flies (Xport-A1/Xport-A1). In (A) and (B), the blots were probed with antibodies against HA, TRP, Rh1, Xport-A (detecting both endogenous Xport-A and overexpressed HA-Xport-A), and tubulin. HA-Xport-A, HA-Xport-A2L, and HA-Xport-A4L25 were expressed under the control of Rh1-GAL4 (BDSC 8688) and each lane was loaded with protein extracts from approximately 2.7 fly heads.
(C) Relative quantification of TRP and Rh1 levels upon the expression of HA-Xport-A2L (lane 6 in A) or HA-Xport-A4L (lane 10 in B), compared to the expression of these proteins when the HA-Xport-A construct is expressed (lane 5 in A and lane 8 in B). For quantification, two biological replicates were used (N = 2). Error bars correspond to SD.
Molecular dynamics simulations corroborate the interactions between Rh1 and Xport-A from AF2 models
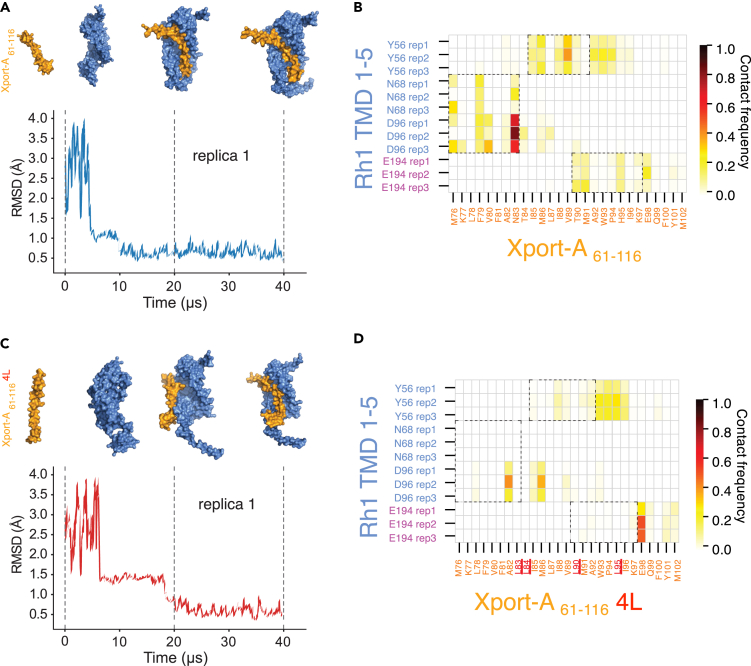
In order to provide an independent confirmation of the interactions predicted previously with AF2, we performed molecular dynamics (MD) simulations, using the Martini 3 coarse-grained model.29 Rh1 TMD1-5 and Xport-A61-116 were simulated in an ER membrane mimic, as separate proteins in the same membrane at the beginning of each simulation. As shown in Figures 6A, S4A, and S4B, all three independently performed replicates converged to a Xport-A61-116/Rh1 TMD1-5 complex analogous to the pose predicted by AF2 (Figure 2B). The molecular dynamics simulations showed Rh1 TMD1-5/Xport-A61-116 complexes with an RMSD value below 1 Å, after 10 μs (Figures 6A, S4A, and S4B), with the complexes being stable for the remaining simulation time. From the MD simulations, we could corroborate the AF2-predicted contacts between amino acid residues of Xport-A and Rh1 (Figure 4B), with the highest contact frequencies being observed between Xport-A N83 and Rh1 D96, among other pairs (Figure 6B). This result highlights that Xport-A could be important to shield Rh1 D96, a charged amino acid residue which otherwise would be unfavorably exposed to the membrane hydrophobic core.
Figure 6.
Molecular dynamics simulations of complexes between Rh1 TMD 1–5 with Xport-A61-116 or Xport-A61-116 4L
(A) RMSD (root-mean-square deviation - in Å) for the molecular dynamics simulation (replica 1) between Rh1 TMD 1–5 and Xport-A61-116. The reference frames are depicted above the respective time points at 0, 20, and 40 μs, in surface representation for coarse-grained Xport-A61-116 (orange) and Rh1 TMD 1–5 (light blue).
(B) Contact frequency between amino acid residues of Rh1 TMD 1–5 (y axis) and Xport-A61-116 (x axis), represented with a color gradient, where 1 (oxblood red) represents a persistent contact throughout the 40 μs trajectories (replicas 1, 2, and 3) and 0 (pale yellow), no contact being observed.
(C) RMSD (in Å) for the molecular dynamics simulation (replica 1) between Rh1 TMD 1–5 and Xport-A61-116 4L. The reference frames are depicted above the respective time points at 0, 20, and 40 μs, in surface representation for coarse-grained Xport-A61-116 4L (orange) and Rh1 TMD 1–5 (light blue).
(D) Contact frequency between amino acid residues of Rh1 TMD 1–5 (y axis) and Xport-A61-116 4L (x axis), represented with a color gradient, as in (B). In red are the 4 residues (82, 84, 90, and 95) mutated to L. The dashed parallelograms represent contacts observed in (B), that are absent or diminished in (D).
We have also performed the molecular dynamics simulations for Rh1 TMD1-5 together with Xport-A61-116 4L (Figures 6C, S4C, and S4D), in which replica 1 (Figure 6C) showed a different binding interface compared to the one of Rh1 TMD1-5/Xport-A61-116, taking around 20 μs to reach an RMSD value below 1 Å. Although replicas 2 and 3 (Figures S4C, and S4D) presented a similar binding interface to the one obtained for Rh1 TMD1-5/Xport-A61-116 and the RMSD eventually converged to similar values in all 3 replicas (Figure 6C), the different behaviors between replica 1 and 2, 3 suggest more unstable complexes than the ones obtained in the simulations for Rh1 TMD1-5/Xport-A61-116. We also analyzed the frequencies of contacts between the amino acid residues of Rh1 TMD1-5 and Xport-A61-116 4L, with most of the specific contacts observed for Rh1 TMD1-5/Xport-A61-116 being absent or with diminished frequencies (dashed parallelograms in Figure 6D). These results were consistent in the 3 replicas. Finally, we performed RMSD analysis between the AF2 model with the highest score (rank 1) for Rh1 TMD1-5/Xport-A61-116 and the respective complexes obtained by MD simulations. After 10 μs of simulation, the AF2 model did not deviate more than 1.5 Å from the MD simulations (Figure S4E), indicating very similar binding poses and further corroborating the consistency of the results obtained by MD simulations with the AF2 model.
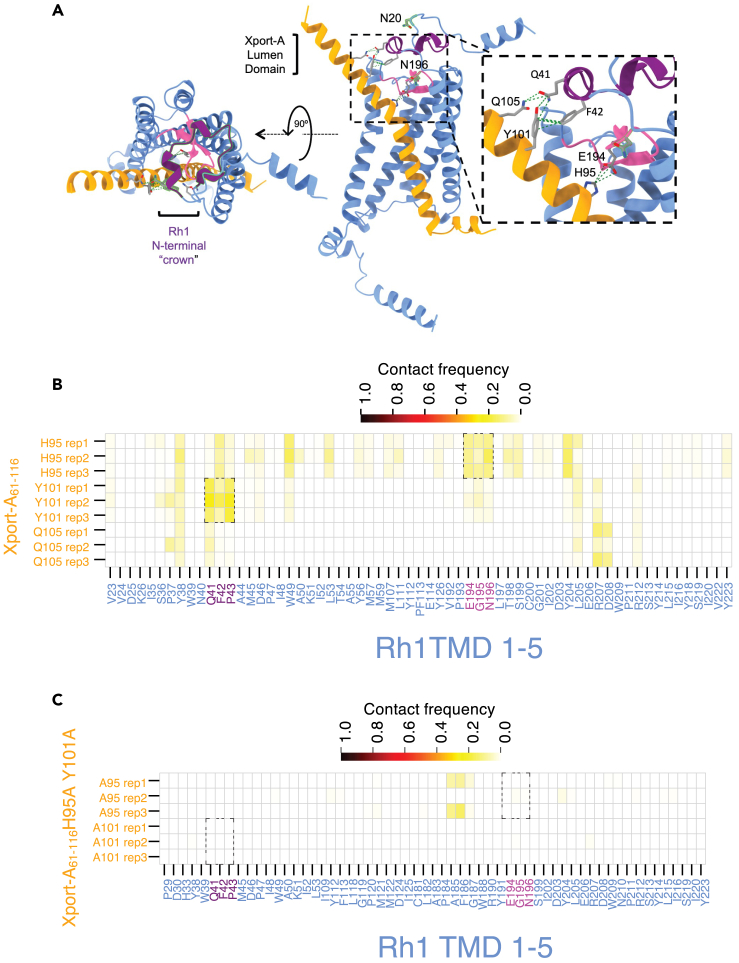
Structural interactions between the ER luminal domain of Xport-A and Rh1
We also identified possible additional interactions between amino acid residues in Rh1 and the C-terminal domain of Xport-A, which projects into the lumen of the ER (Figure 7A). For example, H95 of Xport-A interacts with E194 of Rh1 (Figures 6B and 7A, and 7B), which could be important to keep the pink beta-loop-beta (Rh1 Y191 to I202) motif deep in the plane of the membrane and protected by the purple N-terminal “crown” of 3 small α-helices (Rh1 S22 to Q41), which also interacts with amino acids residues (Y101 and Q105) in the ER luminal C-terminal domain of Xport-A (Figure 7A, inset). This N-terminal “crown” is present in the crystal structure of jumping spider (Hasarius adansoni) rhodopsin 130 but not in bovine rhodopsin.31,32,33 In fact, the AF2 model for Drosophila Rh1 is very similar to the crystal structure of jumping spider rhodopsin-1, with a backbone RMSD of 0.729 Å and 53.7% of identical amino acid residues (Figure S5). Xport-A is insect specific and is not conserved in Arachnida. In jumping spider rhodopsin 1, the N-terminal “crown” is anchored to the transmembrane bundle by contacts made by H36 and H38,30 which are not conserved in Drosophila Rh1 (Figure S5C). Interestingly, H38 is conserved in Drosophila Rh3, Rh4, and Rh5 (the rhodopsins that are expressed in the R7 and R8 photoreceptors to mediate color vision), which do not require Xport-A for their biogenesis.23 Therefore, Xport-A could substitute for the stabilization conferred by H36 and/or H38, in the case Rh1, where these residues are not conserved.
Figure 7.
Interactions between Xport-A ER lumen domain and Rh1 TMD1-5
(A) AF2 model of Xport-A (amino acids 61 to 116 - in orange) together with Rh1 TMD1-5 (light blue). The N-terminal “crown” of Rh1 (S22 to Q41) is in purple. The beta-loop-beta of Rh1 (Y191 to I202) is in pink.
(B) Contact frequency between amino acid residues of Rh1 TMD 1–5 (x axis) and Xport-A61-116 (y axis), represented with a color gradient, where 1 (oxblood red) represents a persistent contact throughout the 40 μs trajectories (replicas 1, 2, and 3) and 0 (pale yellow), no contact being observed.
(C) Contact frequency between amino acid residues of Rh1 TMD 1–5 (x axis) and Xport-A61-116 H95A, Y101A (y axis), represented with a color gradient, as in (B). The dashed parallelograms represent contacts observed in (B), that are absent or diminished in (C).
From the MD simulations, we observed that the contact frequencies between Xport-A H95 and Rh1 E194, G195, and N196 and Xport-A Y101 and Rh1 Q41, F42, and P43 (Figure 7B) are reduced or absent in simulations where H95 and Y101 are mutated to alanine (dashed parallelograms in Figure 7C). We also identified interactions between Xport-A Q105 and several amino acids in Rh1, including P37 and Q41, although with lower frequency of contacts than for H95 or Y101 (Figure 7B). Interestingly, in the replicas 1 and 3 of the simulation between Rh1 TMD1-5 and Xport-A61-116 H95A, Y101A, this Xport-A mutant localized to different regions of Rh1 from the binding pocket of “wild-type” Xport-A (transparent orange ellipsoid in Figures S6A, and S6C).
Of note, Rh1 N20 is well exposed to the glycosylating enzymes of the ER lumen, while N196 is not, since it is within the protected beta-loop-beta motif below the “crown” N-terminal domain of Rh1. So, in order for N196 to be accessible for glycosylation in Xport-A mutants, as shown,25 some dramatic misfolding of the N-terminal “crown” and the beta-loop-beta motif must occur. The chaperone role of Xport-A might therefore extend to the protection of the beta-loop-beta motif at this intermediate step of Rh1 biogenesis, when the first 5 TMDs of Rh1 are inserted into the ER membrane. Finally, we would like to highlight that the structure of the beta-loop-beta motif is important for Rh1 biogenesis, since at least 4 mutations in this motif (E194Q, E194K, G195S, and C200Y) have been reported to cause reduced biogenesis of Rh1 and retinal degeneration.34,35,36
Based on our previous results25 and the results described here, we favor a model where Xport-A acts as a chaperone during the biogenesis of Rh1, at a transient step, when the TMDs 1–5 of Rh1 are already inserted in the ER membrane, but TMDs 6–7 are not yet inserted or at least not yet present in Rh1 structure. Interactions of Xport-A with Rh1 TMD1-5 must be essential to stabilize the TMDs of Rh1 but also to stabilize the N-terminal ER luminal domain of Rh1 and the beta-loop-beta motif between TMD4 and TMD5, allowing for the correct folding and biogenesis of Rh1. Finally, our results provide an example of usefulness of AF2-based approaches to study protein complexes; in this case, the interactions between a chaperone (Xport-A) and its client (Rh1).
Limitations of the study
We were unsuccessful in producing high-confidence AF2 models for the interactions of Xport-A with the TMDs of monomeric TRP (Figure S7), Xport-A with Xport-B, or Xport-B with Xport-A/Rh1 TMD 1–5 (not shown).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Rh1 | DSHB (4C5) | RRID:AB_528451 |
| rat anti-Xport-A | kind gift of Craig Montell | Ref. Chen et al.24 |
| mouse anti-TRP | DSHB (Mab83F6) | RRID:AB_528496 |
| mouse anti-tubulin | DSHB (AA4.3) | RRID:AB_579793 |
| rat anti-HA | Chromotek (7C9) | RRID:AB_2631399 |
| Experimental models: Organisms/strains | ||
| XportA1 | BDSC | BL 60679 |
| UAS-3xHA-Xport-A | Domingos lab | Ref. Gaspar et al.25 |
| UAS-3xHA-Xport-A2L | Domingos lab | Ref. Gaspar et al.25 |
| UAS-3xHA-Xport-A4L | Domingos lab | Ref.Gaspar et al. 25 |
| Software and algorithms | ||
| GROMACS | https://www.gromacs.org/ | Ref. Abraham et al. 37 |
| ColabFold | https://github.com/sokrypton/ColabFold | Ref. Evans et al. and Mirdita et al.38,39 |
| MDTraj | https://www.mdtraj.org/1.9.8.dev0/index.html | Ref. McGibbon et al.40 |
| MD-TASK | https://github.com/RUBi-ZA/MD-TASK | Ref. Brown et al.41 |
| VMD | https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName = VMD | Ref. Humphrey et al.42 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pedro M. Domingos (domingp@itqb.unl.pt).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Drosophila stocks
Flies and crosses were raised with standard cornmeal fly food, at 25°C under 12 h light/12 h dark cycles. HA-Xport-A, HA-Xport-A2L and HA-Xport-A4L include an N terminal 3xHA tag and were described in.25 Xport-A1 mutation was described in.23
Methods details
AlphaFold modeling of Xport-A Rh1 complex
We used AlphaFold-Multimer38 to predict binding interfaces, a method that is a refined version of AlphaFold227 for complex prediction. As a first stage, we used the sequences of Rh1 and Xport-A as input to predict the 3D structure of Xport-A bound to Rh1. Next, we run independent predictions replacing the full length Rh1 with the sequences of Rh1 TMD1 (M1 to I74), Rh1 TMD1-3 (M1 to L146) and Rh1 TMD1-5 (M1 to V241), as described,43 all including the cytosolic C-terminal domain of Rh1 (H333-A373), but not the C-terminal V5 tag. We did not use template structures for the predictions iterated for up to 48 recycles, followed by energy refinement with AMBER using default settings implemented in ColabFold39 and using MMseqs2 for creating multiple sequences alignments.44 Model confidence was assessed by the predicted Local Distance Difference Test (pLDDT) and inter-complex predicted alignment error (PAE), i.e., the uncertainty about the interface. pLDDT is closely related to the pre-existing metric lDDT-Cα45 that measures the local accuracy of a prediction by determining the fraction of preserved local distances (higher is better). As a superposition-free method, lDDT is insensitive to relative domain orientation and correctly identifies segments in the full-length model deviating from the reference structure. pLDDT is given on a scale from 0 to 100. Regions with pLDDT >90 are expected to have high accuracy. PAE is not an inter-residue distance map or a contact map but the expected distance error in Å. It indicates the expected positional error at residue x if the predicted and actual structures are aligned on residue y (using the C, N and C atoms). PAEs are measured in Å and capped at 30 Å. PAE helps to assess the confidence in the relative position and orientation of the model parts (e.g., two domains or chains). For residues x and y in two chains, if the PAE values (x, y) are low, AlphaFold2 predicts the chains to have well-defined relative positions and orientations.28 Regions with pLDDT >90 are expected to have high accuracy.
Immunoblotting of fly heads
Heads from 1-day old flies were homogenized in 2xLDS buffer + DTT (0.5 M Tris, 1 mM EDTA, 4% Lithium dodecyl sulfate, 5% glycerol, 0,0375% SERVA Blue G250, 0,0125% Phenol Red and 0.1 M DTT (Dithiothreitol)) with a pellet pestle, and then diluted with equal volume of MilliQ water. Protein denaturation was performed by incubating extracts at 65°C (15 min). Samples were run in SDS-PAGE, transferred to PVDF membranes (Amersham Hybond) and probed with the following antibodies: mouse anti-V5 (1:1,000) (R960-25, Invitrogen), mouse anti-TRP (1:300) (Mab83F6, DSHB), mouse anti-Rh1 (1:200) (4C5, DSHB), rat anti-Xport-A antibody (1:400) (kind gift of Craig Montell), and mouse anti-tubulin (1:1,000) (AA4.3, DSHB).
Molecular dynamics simulation of the interaction between Rh1 and XportA
The Martini 3 CG model was used to simulate protein interactions in a membrane setting.29 The CG topology and structure of the Rh1 TMD1-5, Xport-A and Xport-A 4L proteins was built using the martinize2 script (https://github.com/marrink-lab/vermouth-martinize), employing elastic network restraints for maintaining the Rh1 TMD1-5 structures, but only main-chain angle/torsion potentials in maintaining Xport-A’s structure. Xport-A’s secondary structure was assumed to be entirely α-helical with the exception of termini, proline, and proline-flanking residues; this yielded three helical segments: a 23-residue central segment compatible with a TM span, and N-terminal and C-terminal segments compatible with an interfacial anchoring role. Membranes were built and proteins inserted using the insane script,46 approximating the lipid composition to that of the endoplasmic reticulum membrane,47 totaling 702 lipid molecules. The number of individual lipid molecules was the following: palmitoyl-oleoyl-phosphatidylcholine: 138, palmitoyl-linoleoyl-phosphatidylcholine: 157, stearoyl-arachidonoyl-phosphatidylcholine: 237, palmitoyl-oleoyl-phosphatidylethanolamine: 42, palmitoyl-stearoyl-phosphatidylethanolamine: 68, stearoyl-arachidonoyl-phosphatidylethanolamine: 60. Lipids models were used as published with the Martini 3 release,29 or when unavailable there, adapted following the same tail-headgroup combination rules. The box size was 150 × 150 × 150 Å3. Rh1 TMD1-5 and Xport-A were initially placed with centers of mass 73 Å apart along the x axis, corresponding approximately to an equidistant separation along that box vector when considering periodicity. The xy box size was chosen so that i) proteins never simultaneously interact with two periodic images of one another, but also so that ii) at their greatest separation along either x or y axis (analogously to their starting position) no lipid simultaneously interacts with the two proteins. The box size and the starting distance allow sufficient protein separation for diffusion not to favor specific binding poses, while maintaining a tractable membrane size and computational cost.
Rh1 TMD1-5 and Xport-A were inserted separately in the membrane and three separate replicates were performed for at least 40 μs each. Molecular dynamics analysis was done using the MDTraj40 and MD-TASK41 packages. The Root-Mean-Square Deviation (RMSD) was performed by aligning the last frame of the trajectory against all other frames, for each replica. For contact frequency map calculations, a 6 Å contact threshold between residues was employed, implemented in MD-TASK. We used the GROMACS simulation package version 2020.37 Lennard-Jones interactions were cut-off at 1.1 nm; Coulombic interactions were treated, with the same cut-off, using reaction-field electrostatics with a dielectric constant of 15 and an infinite reaction-field dielectric constant. Temperature was kept at 300 K by a v-rescale thermostat with a coupling time of 4.0 ps. Pressure was coupled semi-isotropically at 1.0 bar to a Parrinello-Rahman barostat, with a relaxation time of 16.0 ps. Simulations were run at a 20 fs time step. Visualization and rendering of the simulations were performed with the molecular graphics viewer VMD version 1.9.3.47
Acknowledgments
The authors acknowledge Fundação para a Ciência e a Tecnologia, I.P. (FCT) for funding project MOSTMICRO-ITQB, with references UIDB/04612/2020 and UIDP/04612/2020. M.N.M. further acknowledges FCT for fellowship CEECIND/04124/2017 and T.N.C. is the recipient of the grant CEECIND/01443/2017. The project leading to these results was funded by the grants LCF/PR/HR17/52150018 ('la Caixa' Foundation), FCT AGA-KHAN/541141368/2019 (FCT and Aga Khan Foundation), and SR&TD project (PTDC/BIA-BFS/0391/2021).
Author contributions
C.J.G. conceived the study, did the immunoblots, and wrote the manuscript. T.G. did the MD simulation analysis. J.C.M. did the MD simulations. M.N.M. supervised the MD simulations. C.A. conceived the study. T.N.C. conceived the study and did the AF2 analysis. P.M.D. conceived the study and wrote the manuscript. All authors read and edited the manuscript.
Declaration of interests
C.J.G. is now an employee of Genentech, Inc., a member of the Roche Group.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108309.
Contributor Information
Colin Adrain, Email: c.adrain@qub.ac.uk.
Tiago N. Cordeiro, Email: tiago.cordeiro@itqb.unl.pt.
Pedro M. Domingos, Email: domingp@itqb.unl.pt.
Supplemental information
References
- 1.Ozaki K., Nagatani H., Ozaki M., Tokunaga F. Maturation of major drosophila rhodopsin, ninaE, requires chromophore 3-hydroxyretinal. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-Z. [DOI] [PubMed] [Google Scholar]
- 2.O’Tousa J.E., Baehr W., Martin R.L., Hirsh J., Pak W.L., Applebury M.L. The Drosophila nina E gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 3.Zuker C.S., Cowman A.F., Rubin G.M. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 4.O’Tousa J.E. Requirement of N-linked glycosylation site in Drosophila rhodopsin. Vis. Neurosci. 1992;8:385–390. doi: 10.1017/s0952523800004910. [DOI] [PubMed] [Google Scholar]
- 5.Katanosaka K., Tokunaga F., Kawamura S., Ozaki K. N-Linked glycosylation of Drosophila rhodopsin occurs exclusively in the amino-terminal domain and functions in rhodopsin maturation. FEBS Lett. 1998;424:149–154. doi: 10.1016/S0014-5793(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 6.Webel R., Menon I., O’Tousa J.E., Colley N.J. Role of asparagine-linked oligosaccharides in rhodopsin maturation and association with its molecular chaperone. J. Biol. Chem. 2000;275:24752–24759. doi: 10.1074/jbc.M002668200. [DOI] [PubMed] [Google Scholar]
- 7.Wang T., Jiao Y., Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J. Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T., Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 9.Huber A., Wolfrum U., Paulsen R. Opsin maturation and targeting to rhabdomeral photoreceptor membranes requires the retinal chromophore. Eur. J. Cell Biol. 1994;63:219–229. [PubMed] [Google Scholar]
- 10.Kang M.-J., Ryoo H.D. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl. Acad. Sci. USA. 2009;106:17043–17048. doi: 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H.-W., Brown B., Chung J., Domingos P.M., Ryoo H.D. highroad Is a Carboxypetidase Induced by Retinoids to Clear Mutant Rhodopsin-1 in Drosophila Retinitis Pigmentosa Models. Cell Rep. 2018;22:1384–1391. doi: 10.1016/j.celrep.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colley N.J., Baker E.K., Stamnes M.A., Zuker C.S. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-Z. [DOI] [PubMed] [Google Scholar]
- 13.Wolff T., Ready D.F. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Dev. Camb. Engl. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- 14.Kumar J.P., Ready D.F. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Dev. Camb. Engl. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- 15.Satoh A., Tokunaga F., Kawamura S., Ozaki K. In situ inhibition of vesicle transport and protein processing in the dominant negative Rab1 mutant of Drosophila. J. Cell Sci. 1997;110:2943–2953. doi: 10.1242/jcs.110.23.2943. [DOI] [PubMed] [Google Scholar]
- 16.Rosenbaum E.E., Vasiljevic E., Brehm K.S., Colley N.J. Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in Drosophila melanogaster. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pak W.L., Grossfield J., Arnold K.S. Mutants of the Visual Pathway of Drosophila melanogaster. Nat. Publ. Group. 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- 18.Schneuwly S., Shortridge R.D., Larrivee D.C., Ono T., Ozaki M., Pak W.L. Drosophila ninaA gene encodes an eye-specific cyclophilin (cyclosporine A binding protein) Proc. Natl. Acad. Sci. USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker E.K., Colley N.J., Zuker C.S. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong B., Bellen H.J. Rhodopsin homeostasis and retinal degeneration: lessons from the fly. Trends Neurosci. 2013;36:652–660. doi: 10.1016/j.tins.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenbaum E.E., Hardie R.C., Colley N.J. Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron. 2006;49:229–241. doi: 10.1016/j.neuron.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearse B.R., Hebert D.N. Lectin chaperones help direct the maturation of glycoproteins in the endoplasmic reticulum. Biochim. Biophys. Acta. 2010;1803:684–693. doi: 10.1016/j.bbamcr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum E.E., Brehm K.S., Vasiljevic E., Liu C.H., Hardie R.C., Colley N.J. XPORT-Dependent Transport of TRP and Rhodopsin. Neuron. 2011;72:602–615. doi: 10.1016/j.neuron.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z., Chen H.C., Montell C. TRP and Rhodopsin Transport Depends on Dual XPORT ER Chaperones Encoded by an Operon. Cell Rep. 2015;13:573–584. doi: 10.1016/j.celrep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaspar C.J., Vieira L.C., Santos C.C., Christianson J.C., Jakubec D., Strisovsky K., Adrain C., Domingos P.M. EMC is required for biogenesis of Xport-A, an essential chaperone of Rhodopsin-1 and the TRP channel. EMBO Rep. 2022;23 doi: 10.15252/embr.202153210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh T., Ohba A., Liu Z., Inagaki T., Satoh A.K. dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. Elife. 2015;4 doi: 10.7554/eLife.06306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souza P.C.T., Alessandri R., Barnoud J., Thallmair S., Faustino I., Grünewald F., Patmanidis I., Abdizadeh H., Bruininks B.M.H., Wassenaar T.A., et al. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods. 2021;18:382–388. doi: 10.1038/s41592-021-01098-3. [DOI] [PubMed] [Google Scholar]
- 30.Varma N., Mutt E., Mühle J., Panneels V., Terakita A., Deupi X., Nogly P., Schertler G.F.X., Lesca E. Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR. Proc. Natl. Acad. Sci. USA. 2019;116:14547–14556. doi: 10.1073/pnas.1902192116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palczewski K. G Protein–Coupled Receptor Rhodopsin. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Edwards P.C., Burghammer M., Villa C., Schertler G.F.X. Structure of Bovine Rhodopsin in a Trigonal Crystal Form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 33.Okada T., Sugihara M., Bondar A.-N., Elstner M., Entel P., Buss V. The Retinal Conformation and its Environment in Rhodopsin in Light of a New 2.2Å Crystal Structure††This paper is dedicated to Dr Yoshimasa Kyogoku. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 34.Colley N.J., Cassill J.A., Baker E.K., Zuker C.S. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc. Natl. Acad. Sci. USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurada P., O’Tousa J.E. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 36.Zheng L., Farrell D.M., Fulton R.M., Bagg E.E., Salcedo E., Manino M., Britt S.G. Analysis of Conserved Glutamate and Aspartate Residues in Drosophila Rhodopsin 1 and Their Influence on Spectral Tuning. J. Biol. Chem. 2015;290:21951–21961. doi: 10.1074/jbc.M115.677765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 38.Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J., et al. Protein Complex Prediction with AlphaFold-Multimer. biorxiv. 2022;1 doi: 10.1101/2021.10.04.463034. Preprint at. [DOI] [Google Scholar]
- 39.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernández C.X., Schwantes C.R., Wang L.-P., Lane T.J., Pande V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown D.K., Penkler D.L., Sheik Amamuddy O., Ross C., Atilgan A.R., Atilgan C., Tastan Bishop Ö. MD-TASK: a software suite for analyzing molecular dynamics trajectories. Bioinformatics. 2017;33:2768–2771. doi: 10.1093/bioinformatics/btx349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 43.Hiramatsu N., Tago T., Satoh T., Satoh A.K. ER membrane protein complex is required for the insertions of late-synthesized transmembrane helices of Rh1 in Drosophila photoreceptors. Mol. Biol. Cell. 2019;30:2890–2900. doi: 10.1091/mbc.E19-08-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 45.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassenaar T.A., Ingólfsson H.I., Böckmann R.A., Tieleman D.P., Marrink S.J. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theor. Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 47.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.