Abstract
Non-small cell lung cancer (NSCLC), as the main type of lung cancer, has a long history of high incidence and mortality. Despite the continuous updates to the American Joint Committee on Cancer (AJCC) staging system, which adapt to evolving treatment modalities and diagnostic advancements, it is evident that patients at the same stage exhibit varying prognoses. The heterogeneity of tumors underscores the need for molecular diagnostics to assume a pivotal role in tumor staging and patient stratification. In our investigation, we meticulously analyzed the data of the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database, incorporating clinical patients and scrutinizing pathological specimens. Through this comprehensive approach, we established a correlation between the expression of the Thymosin beta 4 X-linked (TMSB4X) gene and poorer disease-free survival (DFS) and overall survival (OS) post-surgery. Compared to the TMSB4X positive expression group, patients in the negative expression group had a better prognosis, with longer DFS (median disease-free survival (median DFS): 16.2 months vs. 11.3 months, P = 0.032) and OS (median overall survival (mOS): 29.8 months vs. 18.5 months, P = 0.033). Furthermore, our findings suggest that TMSB4X may facilitate immune evasion in non-small cell lung cancer cells by influencing the activation of infiltrating dendritic cells (DCs) in tumor infiltrating immune cells (TIICs) (R = 0.27, P = 4.8E+08). In summary, TMSB4X emerges as an unfavorable prognostic factor for NSCLC, potentially modulating the tumor immune microenvironment through its regulatory impact on dendritic cell function, thus facilitating tumor immune escape.
Keywords: TMSB4X, NSCLC, Immune microenvironment, Dendritic cell, Survival prognosis
1. Introduction
The mortality rate of lung cancer (22.5 %) ranks first among malignant tumors [1]. NSCLC is the main pathological type of lung cancer [2]. The clinical treatment of NSCLC has entered the era of precision therapy, but for patients with advanced stage III and above, the 5-year OS prognosis is still less than 20 %. For each lung cancer patient, the prognosis is different, which is closely related to the degree of malignancy of tumor and the microenvironment in which the tumor occurs and develops. The growth personality of the tumor and the tumor microenvironment interact and influence each other, which together contribute to the differences tumor ecosystem, ultimately determines the difference prognosis of every cancer patient [3].
To date, an increasing number of oncogenes and tumor suppressor genes have been unveiled, heralding a new era in cancer research [4,5]. The discovery of many genes has led to the development of drugs that can be used in the patients with NSCLC. Notably, targeted therapies tailored to specific epidermal growth factor receptor (EGFR) mutations have emerged as highly effective interventions, extending their benefits to anaplastic lymphoma kinase (ALK) mutations and beyond [6,7]. Even so, the number of oncogenes and tumor suppressor genes is still far greater than the number of targeted drugs. We need to notice that the effect of gene mutation of tumor cells on tumor microenvironment [8,9].
The immune microenvironment constitutes a crucial component within the intricate landscape of lung cancer. Notably, the immune microenvironment exhibits distinct variations across different subtypes of lung cancer [10]. The influence of individual genes on the tumor immune microenvironment has garnered escalating interest [11,12]. Molecular attributes of tumors exert a profound influence on the effectiveness of therapeutic interventions, including immunotherapy [13]. Delving into the repercussions of aberrantly expressed genes in lung cancer on the tumor immune microenvironment and prognosis shall facilitate the formulation of clinical treatment strategies and enable tailored approaches for population stratification.
TMSB4X, a versatile actin binding protein, participates in a myriad of biological phenomena, including cell migration and adhesion, wound healing, intracellular signaling and stem cell formation [14]. Its expression is notably elevated in various human malignant tumor tissues, correlating with unfavorable prognoses [15]. Most previous studies have focused on its impact on tumor cell invasion, migration, and proliferation [16,17]. However, with the advent of single-cell sequencing technology, an increasing number of TMSB4X's functions have come to light. Notably, TMSB4X acts as an actin-binding protein with increasing evidence that it may play an important role in the tumor immune microenvironment (TIME). When tumor cells die, the exposed F-actin serves as a pivotal site for DCs bind and recognize antigens [[17], [18], [19]]. In this process, F-actin plays a crucial role in enabling DCs to identify tumor antigens, thereby eliciting anti-tumor immune responses. As a key regulator of both G-actin and F-actin, TMSB4X's overexpression has been shown to induce an increase in G-actin levels and a decrease in F-actin levels, potentially facilitating immune evasion by tumors [[20], [21], [22]].
Targeted therapy and immunotherapy stand as pivotal pillars in the realm of contemporary anti-tumor therapy. Therefore, a comprehensive understanding of a single gene shall aid us in the creation of novel targeted medications, while considering the intricate interplay between tumor therapy and the microenvironment of the body, thereby optimizing the advantages bestowed upon tumor patients. Regrettably, there exists a dearth of research pertaining to TMSB4X in the realm of tumor immunity. In this study, we recruited 32 patients diagnosed with lung cancer, and their pathological sections were analyzed by immunohistochemistry to establish the correlation between TMSB4X expression and clinical prognosis of lung cancer. We observed that TMSB4X was associated with poor prognosis in patients with non-small cell lung cancer in clinical samples. We tried to explain this phenomenon from the impact of a single gene on the tumor immune microenvironment, which is necessary for drug development in this target.
2. Materials and methods
2.1. Source of pathological sections and immunohistochemical experiments
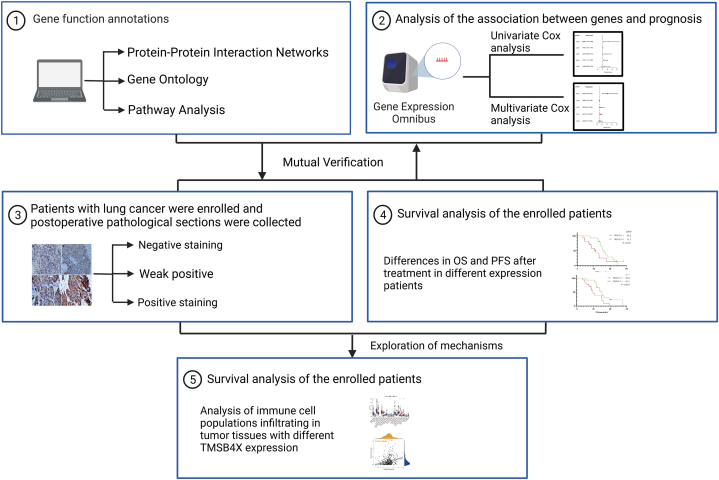
Overall workflow diagram of this study is summarized in Fig. 1 workflow diagram of this study. The study included patients with pathologically confirmed non-small cell lung cancer at the Affiliated Hospital of Zunyi Medical University from January 1, 2013 to January 31, 2018, aged >18 years and <75 years, who had undergone at least surgical treatment, and all patients were treated according to the guidelines [[23], [24], [25]]. Prior to the surgery, all patients provided their informed consent and signed consent forms for the utilization of surgically excised tumor tissues for scientific research purposes. Paraffin sections of these patients were collected. Immunohistochemical staining experiment was performed by dewaxing, antigen repair, serum blocking, addition of TMSB4X antibody (19850-1-AP, Proteintech), addition of secondary antibody (SE134, Solarbio), addition of streptavidin (SE068, Solarbio), Diaminobenzidine (DAB, DA1010, Solarbio) color development, repeat staining, dehydration, and blocking, and images were acquired by light microscopy.
Fig. 1.
Workflow diagram of this study.
Use the IHC Profiler plugin in the ImageJ software for automated quantitative analysis and scoring of IHC electron images. The software will automatically assign scores based on a formula (see Ref. [26]), with four levels: 3, 2, 1, and 0. High Positive is assigned a score of 3, Positive is assigned a score of 2, Low Positive is assigned a score of 1, and Negative is assigned a score of 0. In this study, Positive is defined as scores of 3 and 2, Weak Positive as a score of 1, and Negative as a score of 0.
2.2. Bioinformatics analysis of GEO database
The NSCLC dataset GSE42127 (case = 176) containing prognostic information was downloaded via the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The chip data file (GSE42127) was preprocessed using Perl software (version 5.32.1.1) through the annotation file of the GPL570 platform. Perl software was used to annotate the microarray data and convert gene probe IDs to gene names. Differentially expressed genes were obtained by screening the expression of all genes using P < 0.05 and |log2FC| ≥ 1 as the defined value. The clinical and prognostic data of each patient in the dataset was paired with TMSB4X by Perl software. Univariate Cox and multivariate Cox regression analyses were performed on TMSB4X in the GSE42127 dataset using R language software (version 3.6.3) and package ("Survival" package (version 3.1–11) https://cran.r-project.org/web/packages/survival/index.html) to evaluate the impact of TMSB4X and other clinical factors on NSCLC prognosis.
2.3. Analysis of immune cell infiltration in tumor tissue
We retrieve transcriptomic data for lung adenocarcinoma from the TCGA database (https://portal.gdc.cancer.gov/). Subsequently, we compute the median expression value of TMSB4X in tumor tissue across all cases. This median value in all cases was served as the designated cut-off point. Patients are then categorized into two groups: TMSB4X high expression and low expression, based on this cut-off value. To estimate the extent of immune cell infiltration in the tumor tissue, we employ the Cibersort algorithm, utilizing the "CIBERSORT package" within the R language software [[27], [28], [29]].
2.4. Survival prognostic analysis
Follow-up was conducted mainly through inpatient and outpatient data or telephone, and the last follow-up was on December 31, 2020. The primary endpoint was OS, defined as the duration from surgery to death resulting from any cause among patients diagnosed with NSCLC. The second endpoint was DFS, which was delineated as the duration from the commencement of surgical intervention to the manifestation of disease recurrence or mortality resulting from any cause, whichever comes first. Tumor progression was assessed based on Resist 1.1 criteria. The results of imaging examinations are determined by a diagnostic imaging physician and a clinician with medical licenses.
2.5. Statistical analysis
Data were organized using Microsoft Excel software (version 2016), and analyzed by IBM SPSS 22.0 software for Windows.
Given that survival time is often influenced by multiple factors, we have incorporated univariate and multivariate Cox regression analysis to ascertain whether the expression of TMSB4X and common clinical prognostic factors exert an impact on survival. In univariate and multivariate Cox regression analysis, the coefficients of the variables included in the analysis were assumed to be the exponent of e, and the resulting values are the Hazard Ratio (HR), where HR > 1 (coefficient >0) indicates a risk factor, and conversely, a protective factor. The p-value is used to indicate the significance level, with p < 0.05 considered as a significant (risk/protective) factor [30].
The patients' clinical characteristics fall under the realm of unordered classification data. To investigate whether there are differences in the expression of TMSB4X among different clinical characteristics, a chi-square test was employed. A P < 0.05 was considered statistically significant, indicating that there is a significant difference in the expression of TMSB4X among patients with different classifications of this clinical characteristic. Conversely, if the P-value is greater than 0.05, it suggests that there is no significant difference in the expression of TMSB4X.
The Kaplan-Meier method was used to describe the median survival time, median DFS time, survival rate and survival curve. The log-rank test was used to compare the differences in survival rates between different groups, where a P-value <0.05 accepted the null hypothesis, indicating a significant difference in survival rates between the two groups, and vice versa indicating no significant difference. In addition, GraphPad Prism 8 software was used to plot survival curves.
3. Results
3.1. Functional analysis of TMSB4X gene
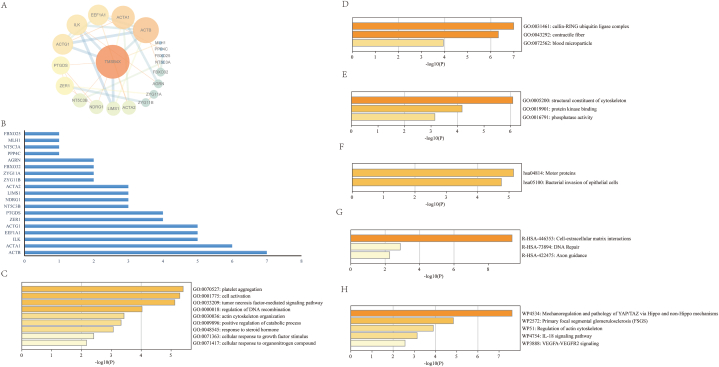
To better understand the biological function of TMSB4X, we searched for 20 target genes that interact with TMSB4X on the Genecard website (https://www.genecards.org) and created a protein-protein interaction network (PPI) network (Fig. 2 A). Based on the degree score, which reflects the strength of their interaction, we have sorted these genes (Fig. 2 B). The top five genes with the strongest association with TMSB4X were β-Actin (ACTB), α-Actin 1 (ACTA1), integrin-linked kinase (ILK), eukaryotic translation elongation factor 1 (EEF1A1), and γ-Actin 1 (ACTG1). Studying the functions of these genes can reflect the main biological functions of TMSB4X. Therefore, we conducted gene ontology (GO) and Pathway analysis. In terms of biological processes, these genes are mainly involved in platelet aggregation, cell activation, tumor necrosis factor-mediated signaling pathway, regulation of DNA recombination, and so on (Fig. 2C and Supplementary Table 1). In terms of cellular components, these genes are mainly enriched in the cullin-RING ubiquitin ligase complex, contractile fiber, blood microparticle, and other cellular components (Fig. 2 D and Supplementary Table 2). In terms of molecular function, the genes that interact with TMSB4X are mainly enriched in structural constituent of cytoskeleton, protein kinase binding, phosphatase activity, and other molecular functions (Fig. 2 E and Supplementary Table 3). Subsequently, through Pathway analysis, we found that all the related genes are enriched in Motor proteins, Bacterial invasion of epithelial cells, Cell-extracellular matrix interactions, DNA Repair, Axon guidance, Mechanoregulation and pathology of Yes-associated protein/transcriptional co-activator with PDZ-binding motif (YAP/TAZ) via Hippo and non-Hippo mechanisms, Primary focal segmental glomerulosclerosis (FSGS), Regulation of actin cytoskeleton, IL-18 signaling pathway, vascular endothelial growth factor A-vascular endothelial growth factor receptor 2 (VEGFA-VEGFR2) signaling pathway, and other pathways (Fig. 2 F, G, H and Supplementary Tables 4, 5, 6).
Fig. 2.
(A) Protein-protein interaction network of TMSB4X-interacting gene sets. (B) The 19 key genes in the PPI network. (C) Gene Ontology of Biological Processes. (D) Gene Ontology of Cellular Components. (E) Gene Ontology of Molecular Functions. (F) Enrichment Analysis of KEGG Pathway. (G) Enrichment Analysis of Reactome Gene Sets. (H) Enrichment Analysis of Wiki Pathways.
3.2. TMSB4X is associated with poorer prognosis in NSCLC by bioinformatics analysis
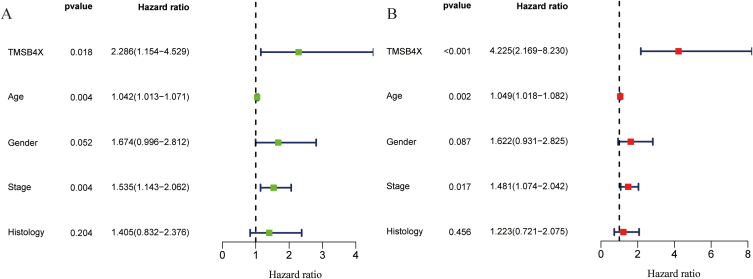
Analysis of the GSE42127 (number of cases = 176) dataset by bioinformatics methods and univariate Cox regression analysis showed that high expression of TMSB4X (HR = 2.286, 95%CI: 1.143–4.529), venerable age (HR = 1.042, 95%CI: 1.013–1.071), and later disease stage (HR = 1.535, 95%CI: 1.143–2.062) in NSCLC patients were associated with poor prognosis (p < 0.05) (Fig. 3 A). Multivariate Cox regression analysis showed that HR of TMSB4X was 4.225 (95%CI: 2.169–8.230), HR of age was 1.049 (95%CI: 1.018–1.082), and HR of stage was 1.481 (95%CI: 1.074–2.042) (Fig. 3 B).
Fig. 3.
Univariate and Multivariate Cox analysis. (A) Univariate Cox analysis (B) Multivariate Cox analysis.
3.3. Basic information and TMSB4X expression in patients enrolled for IHC experimental verification
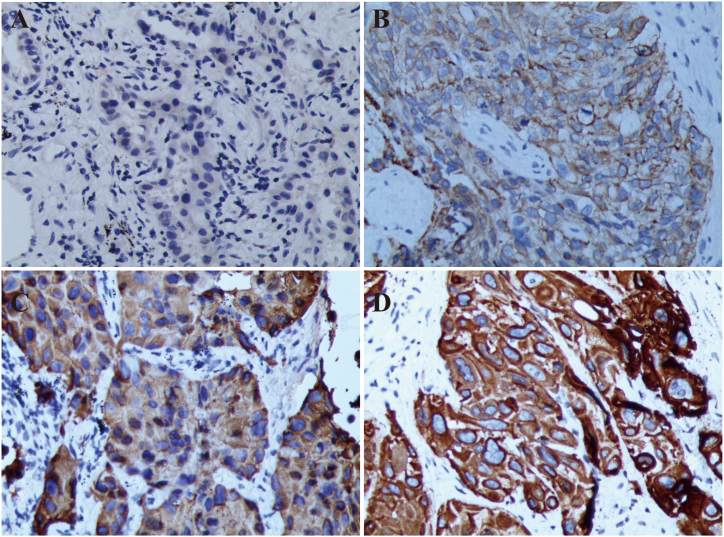
A total of 32 eligible NSCLC patients were included in this study (Supplementary Fig. 1, Supplement table 7). The maximum duration of follow-up in this study was 8 years, with a median follow-up time of 23.1 months (95 % CI: 19.5–26.7). Due to the retrospective nature of this study, data from lost-to-follow-up patients were not included in the data analysis. The patients’ clinical characteristics was shown in Table 1. Among them, 15 patients were negative expressed of TMSB4X (Fig. 4 A), and 17 patients were positive (Fig. 4 B and Fig. 4C), with a positive expression rate of 53.1 % (17/32). TMSB4X was mainly located in the cytoplasm and membrane of tumor cells (Fig. 4 D). Analysis of the relationship between TMSB4X expression and clinical characteristics of NSCLC patients showed that there was no significant differences in gender, smoking index, histological type, primary tumor site, disease stage and so on (P > 0.05) (Table 1).
Table 1.
The clinical characteristics of patients.
| Characteristic | Patients, n (%)* | TMSB4X (−) | TMSB4X (+) | P |
|---|---|---|---|---|
| gender | 0.337 | |||
| Male | 23 (72.0) | 12 | 11 | |
| Female | 9 (28.0) | 3 | 6 | |
| Age | 0.907 | |||
| >60 years | 11 (34.4) | 5 | 6 | |
| ≤60 years | 21 (65.6) | 10 | 11 | |
| smoking index | 0.755 | |||
| SI# > 400 | 18 (56.3) | 8 | 10 | |
| SI# ≤ 400 | 14 (43.7) | 7 | 7 | |
| pathological pattern | 0.288 | |||
| squamous carcinoma | 16 (50.0) | 6 | 10 | |
| Adenocarcinoma | 16 (50.0) | 9 | 7 | |
| clinical classification | 0.723 | |||
| central type | 16 (50.0) | 7 | 9 | |
| Peripheral | 16 (50.0) | 8 | 8 | |
| primary tumor site | 0.288 | |||
| left lung | 16 (50.0) | 6 | 10 | |
| right lung | 16 (50.0) | 9 | 7 | |
| T stage | 0.728 | |||
| T1 | 12 (37.5) | 7 | 5 | |
| T2 | 11 (34.4) | 5 | 6 | |
| T3 | 6 (18.7) | 2 | 4 | |
| T4 | 3 (9.4) | 1 | 2 | |
| N stage | 0.564 | |||
| N0 | 9 (28.2) | 4 | 5 | |
| N1 | 21 (65.6) | 10 | 11 | |
| N2 | 1 (3.1) | 1 | 0 | |
| N3 | 1 (3.1) | 0 | 1 | |
| AJCC stage | 0.871 | |||
| I | 6 (18.8) | 3 | 3 | |
| II | 16 (50.0) | 8 | 8 | |
| III | 10 (31.2) | 4 | 6 |
Abbreviations: SI = Smoking index; AJCC = American Joint Committee on Cancer.
#Smoking index (SI) = number of cigarettes smoked per day × number of years smoked.
*Data are presented as n (%) unless otherwise indicated.
Fig. 4.
Representative immunohistochemical image of TMSB4X expression in NSCLC tissue. (A) Negative staining (B) Weak positive (C) Positive staining (D) Positive staining; All images have been 200 times magnified.
3.4. Prognostic value of TMSB4X expression among NSCLC patients
Clinical factors commonly affecting patients' OS and the expression of TMSB4X were included in univariate and multifactorial analyses to see the effect of these factors on patients' OS. Univariate Cox analysis showed that TMSB4X is an important factor affecting patients' OS (HR = 1.762, 95%CI: 1.128–2.752; P = 0.013), but not other clinical features (Table 2). Multifactorial regression analysis likewise suggested a significant impact of TMSB4X on patient prognosis (HR = 2.222, 95 % CI: 1.160–4.258; P = 0.016), while age was also a factor influencing patient prognosis (HR = 2.856, 95%CI: 1.002–8.139; P = 0.049) (Table 3).
Table 2.
Univariate Cox analysis of clinical cases (n = 32) (endpoint: OS).
| clinical features | HR (95%CI) | P |
|---|---|---|
| Age | 2.302 (0.899–5.893) | 0.082 |
| Gender | 0.986 (0.389–2.500) | 0.976 |
| AJCC stage | 1.196 (0.677–2.111) | 0.538 |
| smoking index | 1.329 (0.576–3.063) | 0.504 |
| pathological pattern | 0.724 (0.322–1.627) | 0.434 |
| TMSB4X | 1.762 (1.128–2.752) | 0.013* |
Note: HR: risk ratio; CI: confidence interval; Smoking index (SI)= Number of cigarettes smoked per day × number of years smoked.
Table 3.
Multivariate Cox analysis of clinical cases (n = 32) (endpoint: OS).
| clinical features | HR (95%CI) | P |
|---|---|---|
| Age | 2.856 (1.002–8.139) | 0.049* |
| Gender | 0.967 (0.211–4.443) | 0.966 |
| AJCC stage | 1.853 (0.912–3.767) | 0.088 |
| smoking index | 1.192 (0.275–5.168) | 0.814 |
| pathological pattern | 1.508 (0.517–4.394) | 0.452 |
| TMSB4X | 2.222 (1.160–4.258) | 0.016* |
Note: HR: risk ratio; CI: confidence interval; Smoking index (SI)= Number of cigarettes smoked per day × number of years smoked.
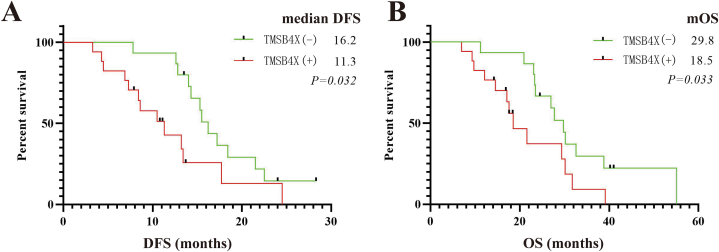
Based on the immunohistochemical results of TMSB4X, the patients were divided into negative and positive groups. Compared to the TMSB4X positive expression group, patients in the negative expression group had a better prognosis, with longer DFS (median DFS: 16.2 months vs. 11.3 months, P = 0.032) and OS (mOS: 29.8 months vs. 18.5 months, P = 0.033) (Fig. 5 A, B).
Fig. 5.
DFS and OS by tumor TMSB4X expression. (A) DFS in different expression of TMSB4X (B) OS in different expression of TMSB4X.
3.5. Shortened overall survival with high expression of TMSB4X in young adenocarcinoma patients
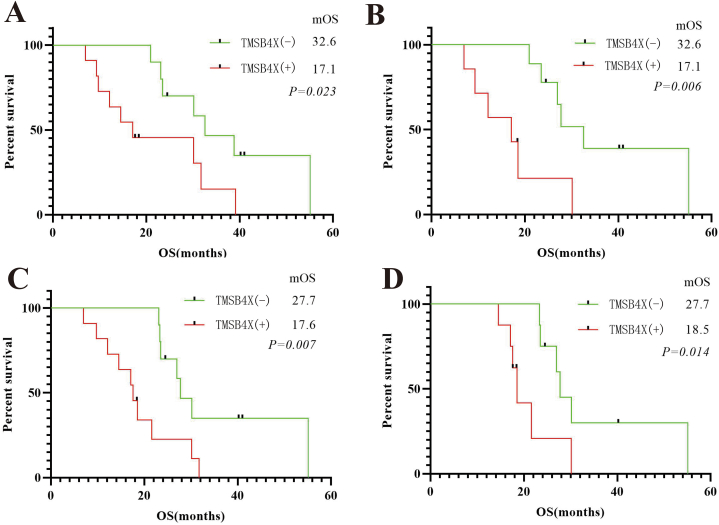
To further investigate whether the OS of each subgroup was affected by the different expression status of TMSB4X, we performed a survival analysis of each subgroup using the Kaplan-Meier method. In the age group, age ≤60 years, high expression of TMSB4X remains an important factor affecting patient OS (mOS: 32.6 months vs. 17.1 months, P = 0.023). In the lung adenocarcinoma subgroup, the high expression of TMSB4X also suggests a poor prognosis (mOS: 32.6 months vs. 17.1 months, P = 0.006). In addition, in N1 stage subgroup (mOS: 27.7 months vs. 17.6 months, P = 0.007) and stage II subgroup (mOS: 27.7 months vs. 18.5 months, P = 0.014), the expression of TMSB4X all appeared as a marker of poor prognosis (Fig. 6 A, B, C, D).
Fig. 6.
(A) aged ≤60 years subgroup, OS in different TMSB4X expressions (B) adenocarcinoma subgroup, OS in different TMSB4X expressions (C) N1 stage subgroup, OS in different TMSB4X expressions (D) stage II subgroup, OS in different TMSB4X expressions.
3.6. Effect of TMSB4X on immune cell infiltration in tumor tissues
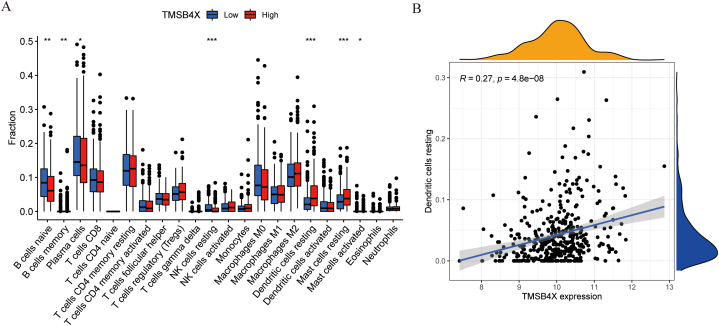
A total of 501 lung adenocarcinoma samples were downloaded from the TCGA database. Calculate the mean of TMSB4X expression in all samples, and the value was used as the cut-off value (cut-off value = 10.014). Significant differences in the expression of B cells naive, B cells memory, Plasma cells, NK cells resting, DCs resting, Mast cells resting, and Mast cells activated could be seen in the high and low expression groups based on cut-off values (Fig. 7 A). Given that our focus in tumor-infiltrating immune cells (TIICs) is mainly on DCs. We noted that the expression of TMSB4X was proportional to the number of resting DC cells in TIICs (R = 0.27, P = 4.8E+08) (Fig. 7 B).
Fig. 7.
Expression of TMSB4X and TIICs. (A) The high or low expression of TMSB4X and TIICs (B) The relevance between the expression of TMSB4X and DC cells resting.
4. Discussion
Treatment strategies for NSCLC are shifting to a precision model of care. In this model, accurate diagnosis is the first and most critical step. AJCC staging is the most commonly used staging model in tumor treatment and an important reference for implementing treatment strategies for tumor patients. However, AJCC staging is not perfect and it is often seen that patients with the same staging have different prognosis. The search for prognosis-related biomarkers will be a long-term task for achieving precise diagnosis and treatment. Immunotherapy, as an important therapeutic method in the 21st century, has been successful in the clinical application of antibody immunotherapy, which has greatly improved the clinical treatment efficacy of patients with NSCLC. However, immunotherapy is not limited to antibodies, cellular immunotherapy is also an important approach to improve the efficacy of treatment for tumor patients.
Most previous explorations of prognostic markers for tumor patients have been limited to oncogenes and oncogenes that are abnormally expressed on tumor cells and their effects on tumor proliferation, apoptosis, invasion, and metastasis. With the continuous progress of immunotherapy and cellular immunotherapy, it will be more beneficial to understand the factors affecting the efficacy of tumor therapy, especially immunotherapy, by revealing the effects of abnormally expressed genes on tumor cells on the tumor immune microenvironment.
In this study, we screened tumor tissues for the aberrantly expressed gene-TMSB4X from samples in publicly available databases and found that its overexpression was highly correlated with poor prognosis in tumor patients (Fig. 3 A and B). For the first time, TMSB4X was identified as an important factor contributing to poor prognosis in NSCLC. Immunohistochemistry of clinical specimens in the Chinese population also suggested that TMSB4X-positive NSCLC patients had poor DFS and OS (Fig. 5 A, B and Fig. 6 A, B, C, D). This deepens our understanding of TMSB4X as a poor prognostic factor. This suggests that in the clinical practice of lung cancer diagnosis and treatment, immunohistochemical staining of TMSB4X is necessary, which will help us stratify patients and predict the treatment effect of patients. In addition, since it is mainly expressed in the cytoplasm and cell membrane of lung cancer cells, further research on this gene will provide ideas and help for the development of antibodies and cell therapy products for NSCLC treatment. In our clinical trial, the expression status of TMSB4X in each patient was determined by immunohistochemical staining, and the positive rate was 53.1 %. The dyeing results of this study was visible TMSB4X expressed mainly in the cytoplasm and cell membrane, tumor cells increased obviously presents the nucleus and deformity, and cytoplasm reduced as a result, increasing the extruded nucleus cell abnormity, also gathered more visible malignant tumor cells, easy to identify with the surrounding normal cells, with other similar studies always similar [31,32]. This subcellular localization study facilitates the development of drugs with corresponding targets, including Chimeric Antigen Receptor T-Cell Immunotherapy (CAR-T) as well as T-cell receptor-transduced T cell (TCR-T) products.
In addition, subsequent analysis of clinical samples from the TCGA database revealed the effect of TMSB4X on DC activation from the perspective of the tumor immune microenvironment - which is critical for cellular immunity against tumors (Fig. 7 A and B). DCs loaded with tumor antigens are critical in activating effector T cells for cellular immunity [33,34]. A study suggests that tumor antigen-loaded DCs combined with tumor-infiltrating lymphocytes can achieve high objective remission rates in advanced melanoma [35]. In the field of oncology therapy, extensive researches have been conducted on DCs, such as: NCT03671720, NCT03803397, NCT02033616, NCT03410732 etc. This study suggests that with the further study of TMSB4X, DCs loaded with TMSB4X antigen can activate effector T cells and exert anti-lung cancer therapeutic effect, which may be a very good therapeutic strategy in the near future.
Although this present study did not explore the impact of TMSB4X overexpression in NSCLC cells on DC function and its role in promoting tumor growth, We are eager to publish this phenomenon due to the identification of a potential mechanism of TMSB4X in tumor progression, which differs from previous research. Previous studies have primarily focused on the effects of TMSB4X on tumor cell invasion, metastasis, and proliferation capacity [22,[36], [37], [38]]. In contrast, our study delves deeper into comprehending the impact of tumor gene expression on the immune microenvironment within tumors and the continuous adaptation of the tumors’ survival environment throughout its development. Ultimately, this leads to the establishment of an ecological environment conducive to tumor survival [3].
Certainly, this study possesses several limitations. Primarily, it should be noted that this study adopts a retrospective design. Consequently, due to the absence of follow-up data, certain patients were excluded from the cohort. This may introduce bias to the overall findings, thereby highlighting the necessity of incorporating a larger sample size in subsequent prospective investigations. Secondly, in addition to the bioinformatics analysis employed, the method employed for immunohistochemical detection of TMSB4X expression is relatively simplistic. To address this, we intend to incorporate more comprehensive techniques such as Western blotting (WB) or polymerase chain reaction (PCR) experiments in our forthcoming, more in-depth study. Moreover, TMSB4X, being a tumor-associated antigen, bioinformatics analysis suggests its potential recognition and uptake by dendritic cells (DCs), thereby promoting the transition of immature DCs (resting) to mature cells (activated). However, it is important to acknowledge that this study has not yet experimentally confirmed this phenomenon, thus representing a limitation. Consequently, in future research endeavors, we aim to further validate and explore the underlying mechanisms associated with this intriguing characteristic. Finally, being a retrospective study conducted at a single center, it is important to acknowledge that the findings of this study might be influenced by the specific attributes of the local patient population. This aspect stands as one of the inherent limitations of this investigation.
Considering the ubiquitous expression of TMSB4X in NSCLC and its profound influence on tumor cell proliferation, invasion, migration and the tumor immune microenvironment, we believe that the development of targeted therapies for TMSB4X will bring certain therapeutic benefits for NSCLC.
5. Conclusion
The results of our study showed that TMSB4X expression status was related to the prognosis of NSCLC patients through the analysis of network tumor database and the mutual verification of clinical case collection and follow-up, and the high expression of TMSB4X significantly reduced the OS and DFS of patients, and TMSB4X is expected to become an effective prognostic biomarker for NSCLC patients.
Declarations
Ethics approval and consent to participate
This study involving human participants was reviewed and approved by the Medical Ethics Committee of the Affiliated Hospital of Zunyi Medical University (approval number: KLL-2019-006).
No animal ethics were involved in this research project.
Consent for publication
Not applicable.
Availability of data and material
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
Ze Yang, Jihang Luo, Mengmei Zhang, Meixiao Zhan, Yuju Bai, Yi Yang, Wei Wang and Ligong Lu declare that they have no conflict of interest.
Funding
This study was supported by Joint Zunyi City Science and Technology Plan Project (Grant No. Zunyi City Kehe HZ Zi (2020) No. 65); Zunyi Medical University 2019 Special Project for Innovation of Academic Seedling Medium (Grant No.: Qianke Platform Talent [2019−004]); Master Start-up Fund of Zunyi Medical University (Grant No. F-925) and Lianyungang Huilan Charity Foundation (Grant No. HX-2021-02).
Data availability statement
All relevant data are included in article and its supplementary material.
CRediT authorship contribution statement
Ze Yang: Data curation, Funding acquisition, Writing – original draft. Jihang Luo: Methodology. Mengmei Zhang: Data curation, Investigation. Meixiao Zhan: Data curation. Yuju Bai: Data curation, Investigation. Yi Yang: Data curation. Wei Wang: Data curation, Software. Ligong Lu: Project administration, Supervision, Validation, Writing – review & editing.
Declaration of competing interest
All authors declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21505.
Contributor Information
Wei Wang, Email: intensivecare@foxmail.com.
Ligong Lu, Email: lu_ligong@163.com.
Abbreviations
- TMSB4X
Thymosin beta 4 X-linked
- NSCLC
Non-small cell lung cancer
- AJCC
American Joint Committee on Cancer
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- DC
Dendritic Cell
- OS
Overall Survival
- DFS
Disease-free survival
- TIICs
tumor infiltrating immune cells
- TIME
tumor immune microenvironment
- HR
Hazard ratio
- CI
Confidence interval
- DAB
3,3-N-Diaminobenzidine Tertrahydrochloride
- GO
Gene Ontology
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Qu Y., Emoto K., Eguchi T., Aly R.G., Zheng H., Chaft J.E., et al. Pathologic assessment after neoadjuvant chemotherapy for NSCLC: importance and implications of distinguishing adenocarcinoma from squamous cell carcinoma. J. Thorac. Oncol. 2019;14:482–493. doi: 10.1016/j.jtho.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Song E. The theory of tumor ecosystem. Cancer Commun. 2022;42:587–608. doi: 10.1002/cac2.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Li X., Zhang L., Wong S.H., Wang M.H.T., Tse G., et al. Oncogenes expand during evolution to withstand somatic amplification. Ann. Oncol. 2018;29:2254–2260. doi: 10.1093/annonc/mdy397. [DOI] [PubMed] [Google Scholar]
- 5.Kussainova A., Bulgakova O., Aripova A., Khalid Z., Bersimbaev R., Izzotti A. The role of mitochondrial miRNAs in the development of radon-induced lung cancer. Biomedicines. 2022;10 doi: 10.3390/biomedicines10020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu S., Dong X., Jian H., Chen J., Chen G., Sun Y., et al. AENEAS: a randomized phase III trial of aumolertinib versus gefitinib as first-line therapy for locally advanced or MetastaticNon-small-cell lung cancer with EGFR exon 19 deletion or L858R mutations. J. Clin. Oncol. 2022;40:3162–3171. doi: 10.1200/jco.21.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson S., Gupta A., Scheuer N., Mackay E., Arora P., Thorlund K., et al. Assessment of alectinib vs ceritinib in ALK-positive non-small cell lung cancer in phase 2 trials and in real-world data. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.26306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim A., Lim S.M., Kim J.H., Seo J.S. Integrative genomic and transcriptomic analyses of tumor suppressor genes and their role on tumor microenvironment and immunity in lung squamous cell carcinoma. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.598671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen J.B.E., Medema J.P., Gootjes E.C., Tauriello D.V.F., Verheul H.M.W. Mutant RAS and the tumor microenvironment as dual therapeutic targets for advanced colorectal cancer. Cancer Treat Rev. 2022;109 doi: 10.1016/j.ctrv.2022.102433. [DOI] [PubMed] [Google Scholar]
- 10.Busch S.E., Hanke M.L., Kargl J., Metz H.E., MacPherson D., Houghton A.M. Lung cancer subtypes generate unique immune responses. J. Immunol. 2016;197:4493–4503. doi: 10.4049/jimmunol.1600576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang H., Sheng S., Chen B., Wang J., Mao D., Han Y., et al. A pan-cancer analysis of the oncogenic role of cell division cycle-associated protein 4 (CDCA4) in human tumors. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.826337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Launonen I.M., Lyytikäinen N., Casado J., Anttila E.A., Szabó A., Haltia U.M., et al. Single-cell tumor-immune microenvironment of BRCA1/2 mutated high-grade serous ovarian cancer. Nat. Commun. 2022;13:835. doi: 10.1038/s41467-022-28389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogino S., Galon J., Fuchs C.S., Dranoff G. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat. Rev. Clin. Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shomali N., Baradaran B., Deljavanghodrati M., Akbari M., Hemmatzadeh M., Mohammadi H., et al. A new insight into thymosin β4, a promising therapeutic approach for neurodegenerative disorders. J. Cell. Physiol. 2020;235:3270–3279. doi: 10.1002/jcp.29293. [DOI] [PubMed] [Google Scholar]
- 15.Chi L.H., Chang W.M., Chang Y.C., Chan Y.C., Tai C.C., Leung K.W., et al. Global proteomics-based identification and validation of thymosin beta-4 X-linked as a prognostic marker for head and neck squamous cell carcinoma. Sci. Rep. 2017;7:9031. doi: 10.1038/s41598-017-09539-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makowiecka A., Malek N., Mazurkiewicz E., Mrówczyńska E., Nowak D., Mazur A.J. Thymosin β4 regulates focal adhesion formation in human melanoma cells and affects their migration and invasion. Front. Cell Dev. Biol. 2019;7:304. doi: 10.3389/fcell.2019.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita T., Hayashi K. Tumor progression is mediated by thymosin-β4 through a TGFβ/MRTF signaling Axis. Mol. Cancer Res. 2018;16:880–893. doi: 10.1158/1541-7786.Mcr-17-0715. [DOI] [PubMed] [Google Scholar]
- 18.Hanč P., Fujii T., Iborra S., Yamada Y., Huotari J., Schulz O., et al. Structure of the complex of F-actin and DNGR-1, a C-type lectin receptor involved in dendritic cell cross-presentation of dead cell-associated antigens. Immunity. 2015;42:839–849. doi: 10.1016/j.immuni.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanč P., Schulz O., Fischbach H., Martin S.R., Kjær S., Reis e Sousa C. A pH- and ionic strength-dependent conformational change in the neck region regulates DNGR-1 function in dendritic cells. EMBO J. 2016;35:2484–2497. doi: 10.15252/embj.201694695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheller I., Beck S., Göb V., Gross C., Neagoe R.A.I., Aurbach K., et al. Thymosin β4 is essential for thrombus formation by controlling the G-actin/F-actin equilibrium in platelets. Haematologica. 2021 doi: 10.3324/haematol.2021.278537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padmanabhan K., Grobe H., Cohen J., Soffer A., Mahly A., Adir O., et al. Development; 2020. Thymosin β4 Is Essential for Adherens Junction Stability and Epidermal Planar Cell Polarity; p. 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia C., He Z., Cai Y. Quantitative proteomics analysis of differentially expressed proteins induced by astragaloside IV in cervical cancer cell invasion. Cell. Mol. Biol. Lett. 2020;25:25. doi: 10.1186/s11658-020-00218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Standards for the diagnosis and treatment of primary lung cancer (2018 Version) electronic journal of comprehensive tumor therapy. 2019;5:100–120. doi: 10.12151/JMCM.2019.03-16. [DOI] [Google Scholar]
- 24.Xiuyi Z., Yuankai S., Jinming Y. Chinese guidelines on the diagnosis and treatment of primary lung cancer (2015 version) Chin. J. Oncol. 2015;37:67–78. doi: 10.3760/cma.j.issn.0253-3766.2015.01.014. [DOI] [Google Scholar]
- 25.Xiuyi Z., Yilong W., Shenglin M., Tianyou W., Changli W., Yuankai S., et al. Chinese guidelines on the diagnosis and treatment of primary lung cancer (2011 version) Chin. J. Lung Cancer. 2012;15:677–688. doi: 10.3779/j.issn.1009-3419.2012.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varghese F., Bukhari A.B., Malhotra R., De A. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emura T., Matsui S., Chen H.Y. compound.Cox: univariate feature selection and compound covariate for predicting survival. Comput. Methods Progr. Biomed. 2019;168:21–37. doi: 10.1016/j.cmpb.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Grote H.J., Feng Z., Schlichting M., Helwig C., Ruisi M., Jin H., et al. Programmed death-ligand 1 immunohistochemistry assay comparison studies in NSCLC: characterization of the 73-10 assay. J. Thorac. Oncol. 2020;15:1306–1316. doi: 10.1016/j.jtho.2020.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Wu C.E., Chang C.F., Kou-Sheng L., Chiang J., Lee S.W., Chiu Y.C. PD-L1 immunohistochemistry comparability and their correlation with clinical characteristics in NSCLC. Anal. Cell Pathol. 2020;2020 doi: 10.1155/2020/3286139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau S.P., van Montfoort N., Kinderman P., Lukkes M., Klaase L., van Nimwegen M., et al. Dendritic cell vaccination and CD40-agonist combination therapy licenses T cell-dependent antitumor immunity in a pancreatic carcinoma murine model. J Immunother Cancer. 2020:8. doi: 10.1136/jitc-2020-000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reap E.A., Suryadevara C.M., Batich K.A., Sanchez-Perez L., Archer G.E., Schmittling R.J., et al. Dendritic cells enhance polyfunctionality of adoptively transferred T cells that target cytomegalovirus in glioblastoma. Cancer Res. 2018;78:256–264. doi: 10.1158/0008-5472.Can-17-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saberian C., Amaria R.N., Najjar A.M., Radvanyi L.G., Haymaker C.L., Forget M.A., et al. Randomized phase II trial of lymphodepletion plus adoptive cell transfer of tumor-infiltrating lymphocytes, with or without dendritic cell vaccination, in patients with metastatic melanoma. J Immunother Cancer. 2021:9. doi: 10.1136/jitc-2021-002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An H.W., Kim S.Y., Kwon J.W., Seok S.H., Woo S.H., Kim D.Y., et al. In vivo CRISPR-Cas9 knockout screening using quantitative PCR identifies thymosin beta-4 X-linked that promotes diffuse-type gastric cancer metastasis. Mol. Carcinog. 2021;60:597–606. doi: 10.1002/mc.23326. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y., Clark K.C., Nguyen E.V., Niranjan B., Horvath L.G., Taylor R.A., et al. Cell Oncol (Dordr); 2022. Proteomic Characterisation of Prostate Cancer Intercellular Communication Reveals Cell Type-Selective Signalling and TMSB4X-dependent Fibroblast Reprogramming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D., Wang S., Wang A., Chen X., Zhang H. Thymosin beta 4 silencing suppresses proliferation and invasion of non-small cell lung cancer cells by repressing Notch 1 activation. Acta Biochim. Biophys. Sin. 2016;48:788–794. doi: 10.1093/abbs/gmw070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
All relevant data are included in article and its supplementary material.