Abstract
The impact of staphylococci on food poisoning and infections could be higher than previously reported. In this study, we characterised the occurrence and coexistence of antimicrobial resistance and virulence genes of staphylococci isolates in foods. Staphylococci were isolated from 236 samples of selected street-vended foods and identified. The pattern of antimicrobial resistance and virulence genes in the staphylococci were assessed using disc diffusion, PCR and analysis of next-generation sequencing data. The food samples (70.76 %) showed a high prevalence of staphylococci and differed among the food categories. Forty-five Staphylococcus species were identified and comprised coagulase-negative and positive species. Staphylococcus sciuri (now Mammaliicoccus sciuri), S. aureus, S. kloosii, S. xylosus, S. saprophyticus, S. haemolyticus and S. succinus were the most abundant species. The staphylococcal isolates exhibited resistance to tetracycline, levofloxacin, ciprofloxacin, norfloxacin, gentamicin and amikacin and susceptibility to nitrofurantoin. Antimicrobial susceptibilities were also reported for cefoperazone, ceftriaxone, cefotaxime, nalidixic acid and piperacillin-tazobactam. The antimicrobial resistance and virulence genes commonly detected consisted of tet, arl, macB, van, gyr, nor, optrA, bcrA, blaZ, taeA and S. aureus lmrS. The isolates frequently exhibited multiple resistance (30.42 %) of up to eight antimicrobial drug classes. The isolates predominantly harboured genes that express efflux pump proteins (50.53 %) for antibiotic resistance compared with inactivation (10.05 %), target alteration (26.72 %), protection (7.67 %) and replacement (3.17 %). The virulence determinants comprised genes of pyrogenic toxin superantigens (eta, etb, tst), adhesions (clf, fnbA, fnbB, cna, map, ebp, spA, vWbp, coa) and genes that express exoproteins (nuclease, metalloprotease, γ-hemolysin, hyaluronate lyase). There was a statistically significant difference in the prevalence of staphylococci isolates and their antimicrobial resistance and virulence profile as revealed by the phenotypic, PCR and next-generation sequencing techniques. The findings suggest a higher health risk for consumers. We recommend a critical need for awareness and antimicrobial susceptibility and anti-virulence strategies to ensure food safety and counteract the spread of this clinically relevant genus.
Keywords: Polymerase chain reaction, Whole-genome shotgun sequencing, Antimicrobial susceptibility, Virulence, Multiple drug resistance, Staphylococcus species
1. Introduction
Street vended foods (SVFs) are crucial in the daily activities of many individuals and urban dwellers in developed and developing countries [1]. Though these foods are easily accessible, foodborne diseases and infections of microbial origins are commonly associated with them. SVFs are mainly produced by locals with less or inappropriate conditions for personal hygiene and temperature-holding capacities. Some vendors exhibit bad handling practices and limitations in their ability to protect foods from flies and dust from roadsides or bus terminals [1,2]. Reports on the prevalence of some foodborne pathogens such as Salmonella species, Escherichia coli and Staphylococcus species have been implicated in ready-to-eat foods [3,4].
Staphylococcus species have long occurred as natural residents on the skin and mucosae of animals and humans in a once harmless relationship with beneficial effects, including the expansion of the memory of T-cells, but now they are mostly implicated in life-threatening infections [[5], [6], [7]]. The pathogen has developed a complex regulatory network to control the evasion of antibiotics and ensure survival and adaptation to different environments [8]. The plasmids and transposons including the staphylococcal cassette chromosome mec (SCCmec) of the Staphylococcus genome encode antibiotic resistance genes while its phage-related and pathogenicity islands encode toxins and other virulence determinants [9,10]. Many of the antibiotic resistance determinants in Staphylococcus species occur by horizontal transmission between species from different environments and hosts [11,12].
Coagulase-positive staphylococci (CoPS) such as Staphylococcus aureus strains have been reported by several researchers to often exhibit high pathogenicity, produce superantigenic enterotoxins that cause food poisoning and display multiple drug resistance [10,13,14]. Even though coagulase-negative staphylococci (CoNS) are widespread in food and have enterotoxigenic ability, their implication in the aetiology of foodborne illnesses has gained little attention [4,15]. The group has recently shown increased pathogenicity and antibiotic resistance and could be associated with a variety of infections when under-exposed primarily to infection-facilitating factors such as indwelling medical devices, skin trauma and immunocompromised hosts [[16], [17], [18]]. The growing inappropriate and indiscriminate use of antibiotics has brought about multidrug resistance (MDR), extensively drug resistance (XDR) and pan-drug resistance (PDR) in bacterial pathogens [19]. Currently, the increasing rate of spread of antimicrobial-resistant Staphylococcus species in foods threatens the ability to effectively treat staphylococcal infections and remains a global health and socio-economic concern [6,20].
The identification of staphylococcal species by phenotypic methods such as laboratory testing and the use of commercial kits, automated systems (including BD Phoenix and Vitek 2) and the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) is quite laborious, limited to only some species and infrequently reliable because of the variability in the expression of some phenotypic characteristics by the species [15,21]. In this regard, molecular approaches such as PCR primer identification and next-generation sequencing (NGS) have been successfully and reliably employed to identify staphylococcal species [21,22]. The development of bioinformatics tools has similarly ensured the efficient characterization of target genes as well as their mechanisms of action [7,23]. This study was designed to narrow the knowledge gap on the diversity of Staphylococcus species recovered from SVFs in Ghana and their corresponding determinants of virulence and resistance to antimicrobial agents of therapeutic relevance. The study further assessed the potential associations between the observed phenotypes and genotypes relative to the parameters studied.
2. Materials and methods
2.1. Sample collection and staphylococcal isolation
A total of 236 most popular and easily accessible cooked foods (n = 148), fruits (n = 49) and vegetables (n = 39) were randomly collected from street food joints, and fruits and vegetable markets in the Cape Coast metropolitan and Elmina municipality of Ghana. Each food sample was collected three times at the same vendor from January to April 2021. The initial culturing and identification of staphylococci was done based on the methods previously described [24,25]. The cultures were serially diluted in buffered peptone water up to the 103 factor, cultured in Mannitol Salt Agar (MSA) medium (Oxoid, UK) using the pour plate technique and incubated at 37 °C for 48 h. All presumptive staphylococci colonies (yellow or red coloured colonies of 2–5 mm size) were subcultured and further analysed for microscopic characteristics, gram staining reaction, catalase production and coagulase activity. A single colony of each presumptive staphylococci isolate was subcultured in a nutrient broth medium (Oxoid, UK) and preserved in 10 % glycerol at 4 °C for further use.
2.2. Staphylococci isolates genomic DNA extraction
A 24 h culture of staphylococcal isolates was prepared in a nutrient broth medium and centrifuged for genomic DNA extraction according to the procedure described by Agyirifo et al. [26]. The extracted DNA was resolved in ethidium bromide-stained 1 % agarose gel electrophoresis at 90 V for 45 min and visualised in a GelDoc Go Imaging System (BIO-RAD, USA) to assess their quality. The concentration and purity of each DNA were determined in the 7415 nano spectrophotometer (Jenway, UK). The genomic DNA was then used for PCR analyses and next-generation sequencing (NGS) at the Beijing Genomic Institute (BGI), Hong Kong. The genomic DNA from all isolates was pooled and a replicate was obtained. The pooled genomic DNA and its replicate were sequenced using the whole-genome shotgun (WGS) sequencing method to obtain the NGS data.
The two cleaned sequence reads were submitted to the MG-RAST public database. The sequences were assigned the IDs 4963321.3 and 4963322.3. These sequence reads were used for taxonomy, antibiotic resistance and virulence factors profiling.
2.3. Identification of Staphylococcus species based on PCR
The identities of the isolates as staphylococci were confirmed at the genus level and the presence of S. aureus and CoNS in each food type was identified in a monoplex PCR. The primer sequences, annealing temperatures, and reaction volume and conditions used are shown in Table S1. The reaction mix comprised 10 μl 2× Accurate Taq master mix (Accurate Biotechnology Co. Ltd., China), 2 μl genomic DNA, 2 μl primer (reverse plus forward) and 6 μl PCR-grade water in a 0.2 ml tube. The PCR was done in a thermocycler (BIORAD, Singapore) and the products were resolved in ethidium bromide-stained 2 % agarose gel at 80 V for 40 min and visualised in a GelDoc Go Imaging System (BIORAD, USA). The reference strains used in this study are shown in Table S2. PCR-grade water was used as a negative control. The amplicons were scored against a 50 bp DNA ladder (Takara Bio Inc., China).
2.4. Antimicrobial susceptibility testing of staphylococci isolates
The susceptibility of the staphylococcal isolates to antibiotics was tested on Mueller-Hinton Agar (MHA) medium (HiMedia, India) using the Kirby-Bauer disk diffusion method and the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [27]. The bacterial cells were standardised in sterile physiological Saline at 0.5 McFarland and Axiom multidisc (Axiom Laboratories, India) consisting of 12 antibiotics used as therapy for staphylococcal infections were used for the test. The antibiotics comprised amikacin (30 μg), ciprofloxacin (5 μg), tetracycline (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), gentamicin (10 μg), levofloxacin (5 μg), norfloxacin (10 μg), nalidixic acid (30 μg), nitrofurantoin (300 μg), piperacillin-tazobactam (75/10 μg) and cefoperazone (30 μg). The MHA plate cultures containing the antibiotic discs were allowed to stand for 30 min and incubated at 37 °C for 24 h. Staphylococcus aureus ATCC 29213 strain was used for quality control. Double distilled water (ddH20) was used as a negative control. The testing was repeated three times and the zones of inhibition were measured. The isolates were categorised as susceptible or resistant to seven antibiotics according to the interpretative criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [28]. Currently, there is no susceptibility breakpoint applicable to staphylococcal species for cefoperazone, ceftriaxone, cefotaxime, nalidixic acid and piperacillin-tazobactam.
2.5. Detection of antibiotic resistance and virulence determinants based on PCR
The staphylococcal isolates were screened for antibiotic resistance and virulence genes using PCR. The genes considered for antibiotic resistance were acrA, aac(3)-I, tetA, tetB, ermA, ermB, strA, vanA, mecA and blaTEM. Even though blaTEM (extended-spectrum beta-lactamase) is normally produced by Gram-negative bacteria, recent studies have detected the enzyme in Staphylococcus species [29,30]. The virulence comprised genes for adherence factors (icaA, cna and sdrE), exfoliative toxins (eta and etb), toxic shock syndrome toxin (tst), hemolysin (hlg) and enterotoxins (sea, seb, sec, sed and see). The PCR was done in a thermocycler and the products were resolved, visualised and scored as described earlier. PCR-grade water was used as a negative control. The primer sequences, annealing temperatures, and reaction volume and conditions used are provided in Table S1.
2.6. Detection of Staphylococcus species composition and antibiotic resistance and virulence determinants
The total composition of the staphylococcal species and their abundance were determined by loading the assembled sequences into the Kraken2 program [31]. Kranken2 uses exact k-mer matches for taxonomic classification. The antibiotic resistance genes and proteins together with associated antimicrobials were determined in the Comprehensive Antibiotic Resistance Database (CARD) [32]. The detection of major virulence factors in the Staphylococcus sequences was also performed in the MG-RAST [33] and the virulence factor database (VFDB) [34].
2.7. Statistical analysis
The proportions and frequencies of all variables were calculated using descriptive statistics in Excel. Categorical variables were compared using chi-square and Fischer's exact test at a 5 % (p < 0.05) statistical significance level. The relationship between the resistance profile of all antibiotics was tested using the Spearman correlation. Only strong associations (−0.8 ≥ r ≥ 0.8) were considered [35]. The statistical analyses were done using SPSS version 26 statistical software (SPSS Inc., USA).
3. Results
3.1. Prevalence of Staphylococcus species based on phenotypes and PCR
A total of 167 representing 70.76 % of the 236 food samples had staphylococci contamination (Table 1). A minimum of 3 out of the total samples collected for each food showed the presence of staphylococci. The prevalence of staphylococci was more common in vegetables (82.05 %) and cooked foods (75 %) than in fruits (48.98 %). The highest prevalence of staphylococci was found in beans and salad (100 %), carrots (100 %) and apples (72.73 %) for each food type. A high prevalence of the species (≥76.47 %) was recorded for five food samples (fish, fufu, pepper sauce, waakye and cabbage). On the contrary, tomato (61.54 %), banana (23.08 %) and banku (30 %) had the lowest percentage of staphylococci contamination among the food types. There was a statistically significant difference (p < 0.05) in the prevalence of staphylococcal isolates between food samples. Similarly, a statistically significant difference (p < 0.05) was observed in the prevalence of staphylococcal isolates between food types. All the staphylococci isolates had an amplicon for the Staphylococcus genus-specific tuf gene based on analysis with the TstaG422 and Tstag765 primers. The Staphylococcus species-specific PCR-based assay identified nine species (Table 2). Staphylococcus haemolyticus was the most prevalent (98.80 %) in the foods followed by S. saprophyticus (95.21 %) and S. xylosus (91.62 %). A high prevalence of S. aureus as well as S. epidermidis (77.25 %) was also shown in the foods. The prevalence of S. capitis was similarly high (71.26 %) whereas S. caprae (44.91 %), S. warneri (38.92 %) and S. pasteuri (14.37 %) were relatively low. Nevertheless, there was a total absence of S. pasteuri in three foods (waakye, cabbage and tomato). There was no statistically significant difference (p > 0.05) in the prevalence of the staphylococcal species between the food types as well as the food samples. However, the abundance of the staphylococci significantly differed (p < 0.05) between food samples.
Table 1.
Prevalence of staphylococci isolates from various food samples based on phenotypic analysis.
| Food type | Food Sample | Number of samples collected | Number of samples with staphylococci |
|---|---|---|---|
| Cooked | Banku | 10 | 3 (30) |
| Beans | 15 | 15 (100) | |
| Fish | 18 | 16 (88.89) | |
| Fufu | 15 | 14 (93.33) | |
| Kenkey | 16 | 6 (37.50) | |
| Pepper sauce | 17 | 13 (76.47) | |
| Salad | 14 | 14 (100) | |
| Soup | 27 | 16 (59.26) | |
| Waakye | 16 | 14 (87.50) | |
| Total | 148 | 111 (75) | |
| Vegetable | Cabbage | 12 | 10 (83.33) |
| Carrot | 14 | 14 (100) | |
| Tomato | 13 | 8 (61.54) | |
| Total | 39 | 32 (82.05) | |
| Fruit | Apple | 11 | 8 (72.73) |
| Banana | 13 | 3 (23.08) | |
| Orange | 12 | 7 (58.33) | |
| Pineapple | 13 | 6 (46.15) | |
| Total | 49 | 24 (48.98) | |
| Total | 236 | 167 (70.76) |
Cooked foods are local cuisines in Ghana. Values in brackets represent the percentage of the number of samples with staphylococci relative to the total number of each sample collected.
Table 2.
Prevalence of Staphylococcus species from various food samples based on PCR.
| Food type | Food sample | Number of Staphylococcus species |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | S. epidermidis | S. haemolyticus | S. pasteuri | S. saprophyticus | S. xylosus | S. warneri | S. caprae | S. capitis | ||
| Cooked | Banku (3) | 3 (100) | 3 (100) | 3 (100) | 1 (33.33) | 3 (100) | 3 (100) | 1 (33.33) | 3 (100) | 3 (100) |
| Beans (15) | 13 (86.67) | 11 (73.33) | 15 (100) | 2 (13.33) | 15 (100) | 13 (86.67) | 3 (20) | 3 (20) | 11 (73.33) | |
| Fish (16) | 11 (68.75) | 15 (93.75) | 15 (93.75) | 3 (18.75) | 15 (93.75) | 15 (93.75) | 7 (43.75) | 9 (56.25) | 14 (87.50) | |
| Fufu (14) | 13 (92.86) | 12 (85.71) | 14 (100) | 2 (14.29) | 14 (100) | 14 (100) | 6 (42.86) | 7 (50) | 11 (78.57) | |
| Kenkey (6) | 5 (83.33) | 5 (83.33) | 6 (100) | 1 (16.67) | 6 (100) | 6 (100) | 3 (50) | 2 (33.33) | 5 (83.33) | |
| Pepper sauce (13) | 12 (92.31) | 12 (69.23) | 13 (100) | 3 (23.08) | 13 (100) | 13 (100) | 6 (46.15) | 7 (53.85) | 8 (61.54) | |
| Salad (14) | 11 (78.57) | 10 (71.43) | 14 (100) | 2 (14.29) | 13 (92.86) | 12 (85.71) | 6 (42.86) | 5 (35.71) | 11 (78.57) | |
| Soup (16) | 10 (62.50) | 11 (68.75) | 15 (93.75) | 3 (18.75) | 14 (87.50) | 12 (75) | 3 (18.75) | 5 (31.25) | 11 (75) | |
| Waakye (14) | 11 (78.57) | 11 (78.57) | 14 (100) | 0 (0) | 13 (92.86) | 13 (92.86) | 8 (57.14) | 6 (42.86) | 9 (64.29) | |
| Total (111) | 89 (80.18) | 87 (78.38) | 109 (98.20) | 17 (15.32) | 106 (95.50) | 101 (90.99) | 43 (38.74) | 47 (42.34) | 84 (75.68) | |
| Vegetable | Cabbage (10) | 6 (60) | 7 (70) | 10 (100) | 0 (0) | 9 (90) | 9 (90) | 4 (40) | 4 (40) | 5 (50) |
| Carrot (14) | 6 (42.86) | 7 (71.43) | 14 (100) | 2 (14.29) | 14 (100) | 13 (92.86) | 5 (35.71) | 6 (42.86) | 7 (50) | |
| Tomato (8) | 8 (100) | 7 (87.50) | 8 (100) | 0 (0) | 8 (100) | 8 (100) | 4 (50) | 5 (62.50) | 6 (75) | |
| Total (32) | 20 (62.50) | 24 (75) | 32 (100) | 2 (6.25) | 31 (96.88) | 30 (93.75) | 13 (40.63) | 15 (46.88) | 18 (56.25) | |
| Fruit | Apple (8) | 6 (75) | 6 (75) | 8 (100) | 2 (25) | 8 (100) | 7 (87.50) | 3 (37.50) | 4 (50) | 5 (62.50) |
| Banana (3) | 2 (66.67) | 2 (66.67) | 3 (100) | 1 (33.33) | 2 (66.67) | 2 (66.67) | 1 (33.33) | 1 (33.33) | 2 (66.67) | |
| Orange (7) | 6 (85.71) | 5 (71.43) | 7 (100) | 1 (14.29) | 6 (85.71) | 7 (100) | 3 (42.86) | 5 (71.43) | 5 (71.43) | |
| Pineapple (6) | 6 (100) | 5 (83.33) | 6 (100) | 1 (16.67) | 6 (100) | 6 (100) | 2 (33.33) | 3 (50) | 5 (83.33) | |
| Total (24) | 20 (83.33) | 18 (75) | 24 (100) | 5 (20.83) | 22 (91.67) | 22 (91.67) | 9 (37.50) | 13 (54.17) | 17 (70.83) | |
| Total (167) | 129 (77.25) | 129 (77.25) | 165 (98.80) | 24 (14.37) | 159 (95.21) | 153 (91.62) | 65 (38.92) | 75 (44.91) | 119 (71.26) | |
Cooked foods are local cuisines in Ghana, Food sample: Value in bracket represent the food samples with staphylococci isolates, Number of Staphylococcus species: Value in bracket represent the percentage of the Staphylococcus species present in each food sample.
3.2. Patterns of antimicrobial resistance of staphylococci based on phenotypes and PCR
The staphylococci from all the foods showed phenotypic resistance to tetracycline, gentamicin, levofloxacin, ciprofloxacin, amikacin and norfloxacin, and susceptibility to nitrofurantoin (Table S3). A strong association was found between nitrofurantoin and levofloxacin (r = 0.863, p = 0.000) and nitrofurantoin and amikacin resistance (r = 0.827, p = 0.001). There was a statistically significant difference (p < 0.05) in the antibiotic resistance pattern between the food types. For the five antibiotics without breakpoints, staphylococci from vegetables and cooked foods showed the highest inhibition zones for cefoperazone (20.75 mm), cefotaxime (15 mm), and piperacillin-tazobactam (14 mm), ceftriaxone (15.75 mm) and nalidixic acid (13 mm), respectively, compared with inhibition zones showed by staphylococci from fruits.
The PCR-based analysis indicated several antimicrobial resistance encoded genes in the staphylococci isolates, confirming the phenotypic assay (Table 3). Genes encoding for resistance to a broad spectrum of antimicrobials such as beta-lactams and fluoroquinolones (acrA), tetracycline (tetA) and streptomycin (strA) were the most common in the species (100 %). The species were also frequently (93.75 %) associated with vanA which is responsible for resistance to vancomycin. A high prevalence of staphylococci harboured aac-(3)-1 (75 %), mecA (68.75 %) and ermA (62.50 %). These genes are related to resistance to gentamicin, methicillin and erythromycin, respectively.
Table 3.
Distribution of antibiotic resistance genes in staphylococci isolates from various food samples by PCR.
| Food type | Food sample | Number of antibiotic resistance gene |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aac-(3)-1 | acrA | ermA | ermB | mecA | strA | tetA | tetB | vanA | blaTEM | ||
| Cooked | Banku (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| Beans (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Fish (1) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | |
| Fufu (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Kenkey (1) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | |
| Pepper sauce (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Salad (1) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Soup (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | |
| Waakye (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Total (9) | 7 (77.78) | 9 (100) | 7 (77.78) | 5 (55.56) | 6 (66.67) | 9 (100) | 9 (100) | 5 (55.56) | 8 (88.89) | 4 (44.44) | |
| Vegetable | Cabbage (1) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| Carrot (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | |
| Tomato (1) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | |
| Total (3) | 2 (66.67) | 3 (100) | 1 (33.33) | 1 (33.33) | 3 (100) | 3 (100) | 3 (100) | 0 (0) | 3 (100) | 2 (66.67) | |
| Fruit | Apple (1) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| Banana (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | |
| Orange (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Pineapple (1) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | |
| Total (4) | 3 (75) | 4 (100) | 2 (50) | 2 (50) | 2 (50) | 4 (100) | 4 (100) | 1 (25) | 4 (100) | 3 (75) | |
| Total (16) | 12 (75) | 16 (100) | 10 (62.50) | 8 (50) | 11 (68.75) | 16 (100) | 16 (100) | 6 (37.50) | 15 (93.75) | 9 (56.25) | |
Cooked foods are local cuisines in Ghana, Food sample: Value in bracket represent the food samples with staphylococci isolates, Number of antibiotic resistance gene: Value in bracket represent the percentage of the number of antibiotic resistance gene present in each food sample.
The occurrence of beta-lactamase TEM (penicillins and cephalosporins resistance) was 56.25 %. The presence of erm(B) (50 %) and tet(B) (37.50 %) genes, conferring resistance to erythromycin and tetracycline, respectively, in the staphylococci were relatively low. There was no statistically significant difference (p > 0.05) in the prevalence of antibiotic resistance genes between the food samples and the food types.
3.3. Patterns of virulence factors of staphylococci based on PCR
A high number of staphylococci isolates harboured the exfoliative toxin genes eta (75 %) and etb (62.50 %), and toxic shock syndrome toxin 1 gene tst (62.50 %) based on PCR (Table 4). The prevalence of the intercellular adhesion gene icaA and the serine-aspartate repeat gene sdrE were similar in the staphylococci (31.25 %). The Staphylococcus species were also positive for hlg (25 %) and cna (12.50 %), which encode for γ-hemolysin and collagen-binding protein, respectively. The sec was the most common Staphylococcus enterotoxin (SE) gene in the isolates (87.5 %) followed by seb and see (62.50 %), sea (50 %) and sed (25 %) genes (Table 4). Statistically, a significant difference (p < 0.05) was seen in the prevalence of the virulence genes between the food samples. On the contrary, there was no significant difference ((p > 0.05) in the virulence gene profile between food types.
Table 4.
Distribution of virulence genes in staphylococci isolates from various food samples as revealed by PCR.
| Food type | Food sample | Number of virulence gene |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cna | eta | etb | hlg | icaA | sdrE | tst | sea | seb | sec | see | sed | ||
| Cooked | Banku (1) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) |
| Beans (1) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | |
| Fish (1) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Fufu (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | |
| Kenkey (1) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | |
| Pepper sauce (1) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | |
| Salad (1) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | |
| Soup (1) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | |
| Waakye (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | |
| Total (9) | 2 (22.22) | 9 (100) | 6 (66.67) | 2 (22.22) | 3 (33.33) | 3 (33.33) | 6 (66.67) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Vegetable | Cabbage (1) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) |
| Carrot (1) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Tomato (1) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | |
| Total (3) | 0 (0) | 2 (66.67) | 2 (66.67) | 1 (33.33) | 1 (33.33) | 1 (33.33) | 2 (66.67) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Fruit | Apple (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) |
| Banana (1) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | |
| Orange (1) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | |
| Pineapple (1) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | |
| Total (4) | 0 (0) | 1 (25) | 2 (50) | 1 (25) | 1 (25) | 1 (25) | 2 (50) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Total (16) | 2 (12.50) | 12 (75) | 10 (62.50) | 4 (25) | 5 (31.25) | 5 (31.25) | 10 (62.50) | 8 (50) | 10 (62.50) | 14 (87.50) | 10 (62.50) | 4 (25) | |
Cooked foods are local cuisines in Ghana, Food sample: Value in bracket represent all the staphylococci isolates from the food sample, Number of virulence gene: Value in bracket represent the percentage of the virulence gene present in the isolates from each food sample.
3.4. Detection of Staphylococcus species based on WGS data
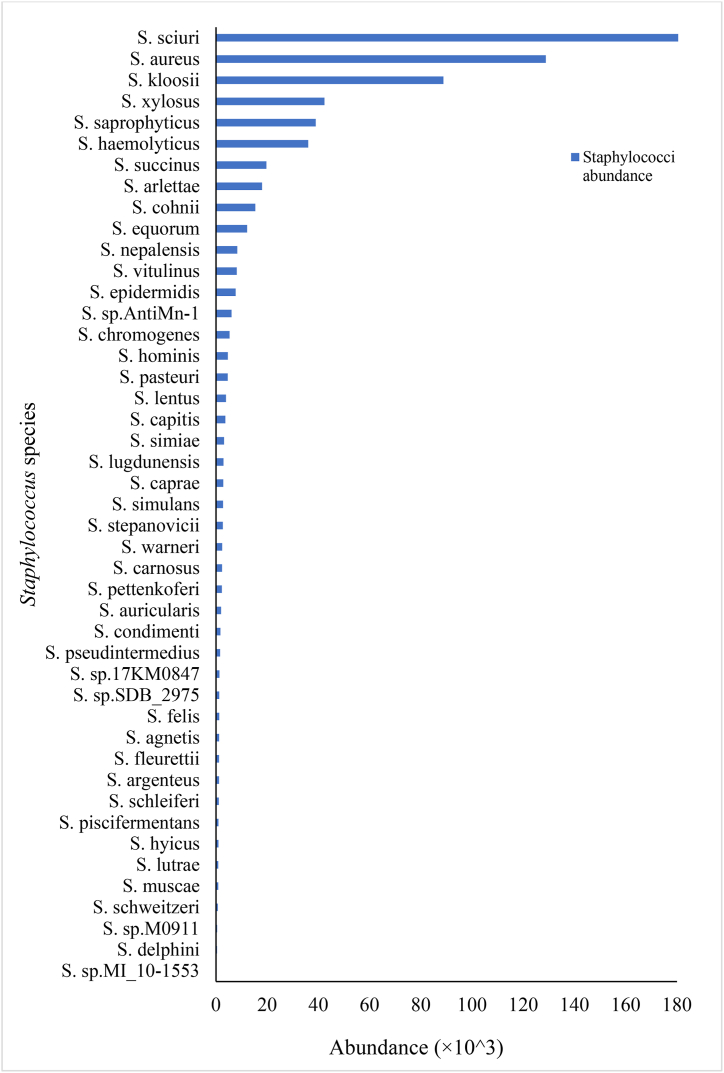
Genotypic identification based on the WGS data revealed 45 Staphylococcus species, comprising coagulase positive (CoP), coagulase-negative (CoN) and unspeciated staphylococci, and confirmed the phenotypic and PCR-based identification assay (Fig. 1). The species abundance ranged from 2.97 × 102 to 1630.13 × 103 and was evident for Staphylococcus sp MI 10–1553 and Staphylococcus sciuri (now Mammaliicoccus sciuri [36]), respectively. S. aureus showed a comparatively high abundance of 128.79 × 103. The presence and abundance of S. kloosii (88.85 × 103), S. xylosus (42.36 × 103), S. saprophyticus (38.95 × 103) and S. haemolyticus (36.04 × 103) were also detected. The food samples were also positive for S. succinus (19.73 × 103), S. arlettae (17.99 × 103), S. cohnii (15.39 × 103), S. equorum (12.19 × 103), S. nepalensis (8.36 × 103), S. vitulinus (8.12 × 103) and S. sp AntiMn-1 (6.07 × 103). S. epidermidis showed a higher abundance (7.72 × 103) compared with S. chromogenes (5.32 × 103), S. hominis (4.66 × 103), S. pasteuri (4.61 × 103), S. capitis (3.67 × 103), S. lugdunensis (2.96 × 103), S. caprae (2.86 × 103) and S. warneri (2.42 × 103).
Fig. 1.
Distribution of Staphylococcus species abundance based on WGS data.
3.5. Detection of genotypic determinants of resistance to antimicrobials based on CARD
A total of 125 antibiotic resistance determinants belonging to 82 groups of genes and proteins were detected from the staphylococci sequences based on the AMR gene family and drug class (Table S4). The majority of the determinants were associated with S. aureus (21.69 %), S. saprophyticus (18.78 %) and S. epidermidis (18.52 %). The presence of genes that encode antimicrobial resistance in S. haemolyticus, S. ludgunensis, S. warneri, S. hominis and S. capitis were 9.79, 7.94, 7.14, 5.56 and 4.50 %, respectively. The remaining staphylococcal species showed a low number of antibiotic-resistant determinants (less than 3 %).
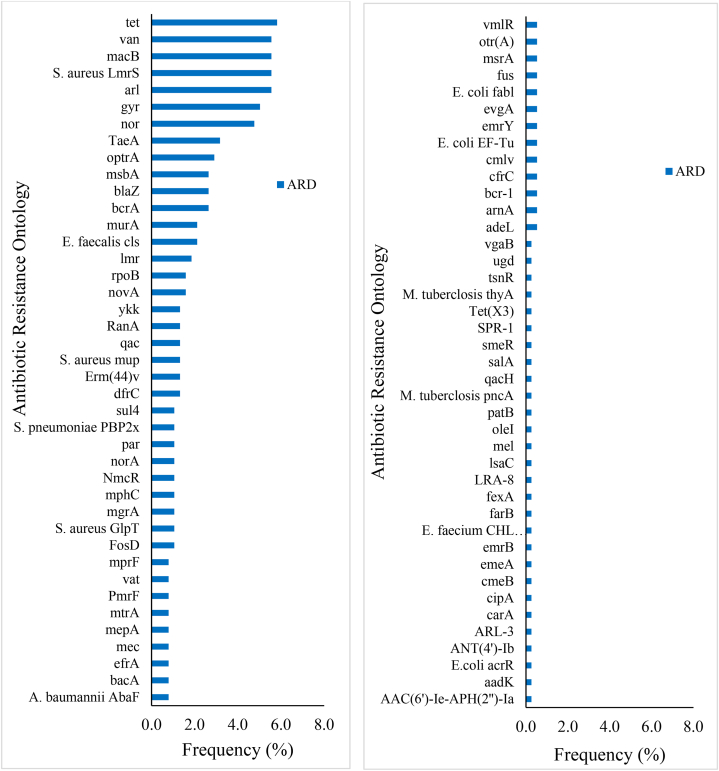
The tet (tetracycline resistance) was the most prevalent gene (5.82 %) and commonly comprised tetK (18.18 %), tetA (58) and tetT (13.64 %), and tet (35) and tetW (9.09 %) (Fig. 2, Table S4). The arl (multidrug resistance) gene also occurred frequently (5.56 %) and consisted mainly of arlS (52.38 %) and arlR (47.62 %). The remaining determinants included Staphylococcus aureus LmrS (5.56 %), macB (5.56 %), van (5.56 %) (vanHO, vanG, vanHF, vanKI, vanM and vanRM), gyr (5.03 %) (gyrA and gyrB) and nor (4.76 %) (S. aureus norA and norB) encoding vancomycin (van), fluoroquinolones (gyr) and multidrug (nor, macB, S. aureus LmrS) resistance. The genes taeA (3.17 %), blaZ (2.65 %), bcrA (2.65 %), msbA (2.65 %) and optrA (2.91 %) were detected to confer resistance to pleuromutilin, beta-lactams, bacitracin, nitroimidazoles and multidrug, respectively. The blaZ frequently comprised mecC-type and PC1 beta-lactamase. The Erm (44)v (multidrug resistance) and Streptococcus pneumoniae penicillin-binding protein 2× (PBP2x) (amoxicillin resistance) were 1.32 % and 1.06 %, respectively.
Fig. 2.
Distribution of antibiotic resistance determinants (ARD) of Staphylococcus species based on WGS data.
Resistance was also detected for daptomycin (cls), fosfomycin (murA, S. aureus GlpT, FosD), lincosamides (lmrB and lmrD), novobiocin (novA), rifampicin (rpoB), trimethoprim (dfrC), mupirocin (mupA and mupB), fluoroquinolones (qacA and qacB), aminoglycosides (RanA) and multidrug (ykkC, ykkD and mgrA). The frequencies of 21.4 % of the determinants were less frequent (less than 1 %). These included the Acinetobacter baumannii AbaF protein, vat (vatB and vatE), arnA, bcr-1, tsnR, fus (fusA and fusD), Mycobacterium tuberculosis thyA, ARL-3 and aminoglycoside acetyltransferase AAC (6′)-Ie-APH(2″)-Ia, aminoglycoside nucleotidyltransferase ANT (4′)-Ib and aadK which offer resistance to fosfomycin, streptogramins, polymyxin, bicyclomycin, thiostrepton, fusidic acid, para-aminosalicylic acid, beta-lactams and aminoglycosides, respectively. The mprF, E. coli fabl and NmcR determinants are associated with defensin (Bacillus subtilis mprF) and daptomycin (S. aureus mprF), isoniazid and triclosan and beta-lactam resistance, respectively. The mec (methicillin resistance) gene constituted 0.8 % and was attributed to mecD, mecl and mecR1.
3.6. Detection of genotypic drug resistance and mechanism
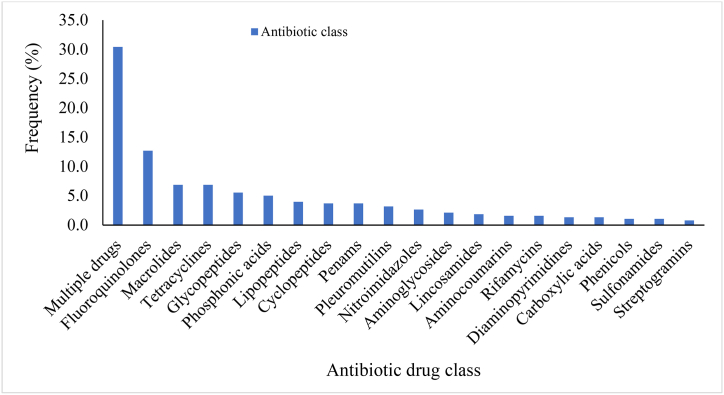
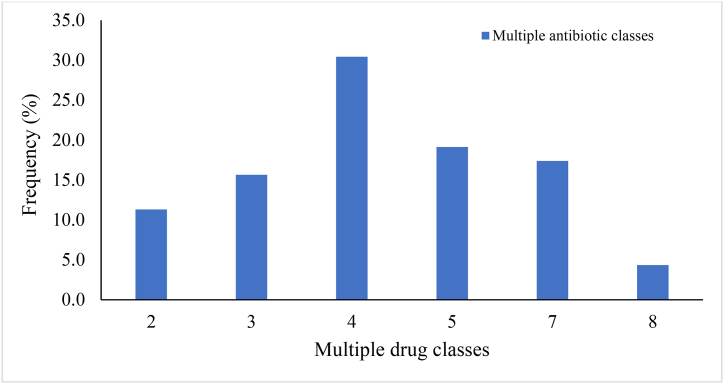
The resistance to multiple antimicrobials was predominant (30.42 %) and ranged from two drug classes (11.30 %) (tetracyclines and glycylcyclines, isoniazid (antitubercular agents) and triclosan) to eight drug classes (4.35 %) (fluoroquinolones, cephalosporins, glycylcyclines, penams, tetracyclines, rifamycins, phenicols and triclosan) (Fig. 3, Fig. 4). Multidrug resistance (resistance to antimicrobials from three or more classes) accounted for 86.96 % of the multiresistance. Resistance to four drug classes (carbapenems, cephalosporins, cephamycins and penams) was the most frequent (30.43 %), followed by five drug classes (19.13 %) (macrolides, aminoglycosides, oxazolidinones, diaminopyrimidines and phenicols), seven drug classes (17.39 %) (macrolides, lincosamides, streptogramins, tetracyclines, oxazolidinones, phenicols and pleuromutilins) and three drug classes (15.65 %) (aminoglycosides, tetracyclines and phenicols) (Fig. 4).
Fig. 3.
Antibiotic classes of resistance determinants of Staphylococcus species based on WGS data.
Fig. 4.
Prevalence of determinants conferring resistance to multiple antimicrobials based on WGS data.
The encoding determinants were predominantly proteins of the major facilitator superfamily (MFS) (40 %), ATP-binding cassette F (ABC-F) (17.39 %), resistance-nodulation-cell division (RND) (6.96 %) and small multidrug resistance (SMR) (5.22 %) (Fig. S1). The Erm 23S ribosomal RNA methyltransferase (4.35 %), multidrug and toxic compound extrusion (MATE) (3.48 %), NmcA beta-lactamase (3.48 %) and ABC (2.61 %) were also present. Fluoroquinolones (moxifloxacin) alone was the most frequent drug class that the majority of the isolates were susceptible (12.70 %) (Fig. 3). This was followed by macrolides (mphC) and tetracyclines (emrY) (6.88 %), glycopeptides (vancomycin) (5.56 %) and phosphonic acids (fosfomycin) (5.03 %). Resistance to lipopeptides (daptomycin) (3.97 %), cyclopeptides (bacitracin) and penams (methicillin) (3.70 %), pleuromutilins (tiamulin) (3.17 %), nitroimidazoles (msbA) (2.65 %), aminoglycosides (aadK, ANT (4′)) (2.12 %) and lincosamides (lmrB) (1.85 %) were also detected. Resistance to aminocoumarins (novobiocin) and rifamycins (rifampicin) was at 1.59 % each.
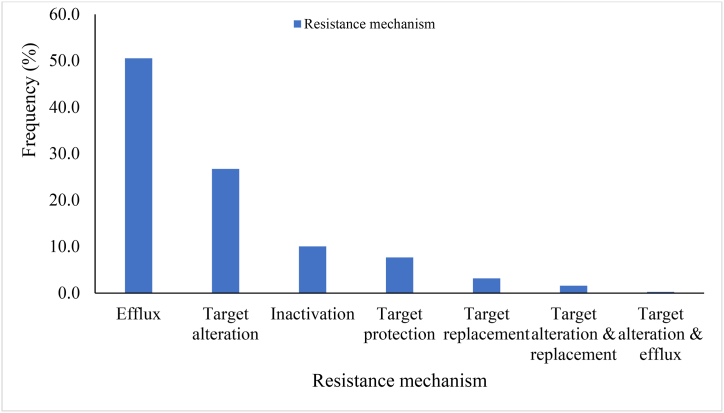
Low resistance (between 0.26 % and 1.32 %) was similarly reported for 11 antimicrobials. This included diaminopyrimidines (trimethoprim), carboxylic acids (mupirocin), phenicols (chloramphenicol), sulfonamides, streptogramins, bicyclomycin, elfamycins (kirromycin), fusidanes (fusidic acid) and antitubercular agents (pyrazinamide). Carbapenems and antimicrobial lipids were the least detected and were encoded by SPR beta-lactamase and antibacterial free fatty acids, respectively. The antibiotic resistance mechanisms of the determinants were mostly efflux pump (50.53 %) whilst a combination of target alteration and efflux pump (about 0.26 %) was the least resistance mechanism (Fig. 5). Target alteration, inactivation, target protection and target replacement were 26.72 %, 10.05 %, 7.67 % and 3.17 %, respectively. Target alteration plus replacement was 1.59 %.
Fig. 5.
Antibiotic resistance mechanisms of Staphylococcus species based on WGS data.
3.7. Detection of virulence factor determinants
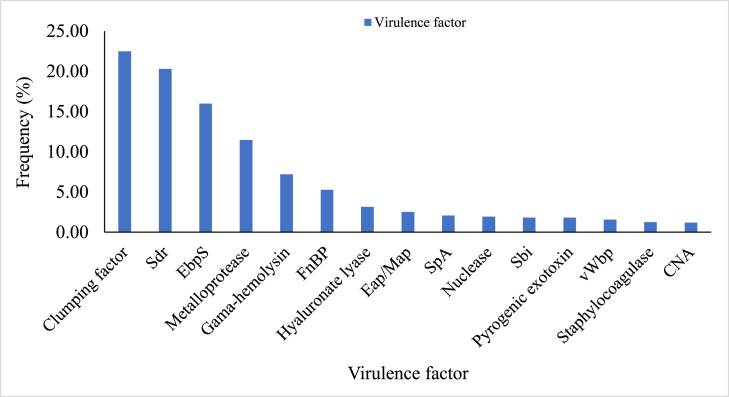
The major virulence factors of the Staphylococcus species were further identified from the sequences of the species (Fig. 6). A total of 15 virulence factors were recorded and commonly comprised clumping factor (clf) (22.49 %), serine-aspartate repeat, Sdr (sdr) (20.30 %), elastin binding protein, Ebps (ebp) (15.98 %) and metalloprotease (11.47 %). The sdrC and sdrD were the most frequent genes (61.11 % and 19.75 %, respectively) of Sdr. The major virulence factors similarly included γ-hemolysins (7.21 %), fibronectin-binding proteins, FnBPs (fnbA and fnbB) (5.26 %), hyaluronate lyase (3.13 %), extracellular adherence protein/MHC analogous protein of broad specificity (Eap/Map) (2.51 %), and staphylococcal protein A (SpA) (2.07 %). Nuclease, staphylococcal binder of immunoglobulins (Sbi), pyrogenic exotoxin and secreted von Willebrand factor binding protein (vWbp) were also present with frequencies ranging from 1.57 % to 1.94 %. The remaining major virulence factors recorded for the species were staphylocoagulase (1.25 %) and collagen-binding protein (CNA) (1.19 %) with related genes coa and cna, respectively.
Fig. 6.
Virulence factors of Staphylococcus species based on WGS data.
4. Discussion
The high number of 45 staphylococcal species identified supports the idea of significant species diversity of staphylococci in foods and reinforces the notion of diverse Staphylococcus species in Africa [16]. The most abundant staphylococcal species, S. sciuri (M. sciuri), has a wide host range and adapts to different habitats [37,38]. The staphylococci identified in this study are clinically relevant and known to cause a variety of community-acquired, farm-acquired and nosocomial-acquired infections such as pneumonia and meningitis in humans [16,[39], [40], [41]]. The observation made in the abundance of these pathogens underscores the need for pragmatic measures including the creation and enforcement of awareness and personal hygiene to mitigate the occurrence of Staphylococcus infections.
Staphylococci are increasingly associated with antimicrobial multiresistance mechanisms [42]. The staphylococci identified in this study showed resistance to tetracycline, levofloxacin, ciprofloxacin, norfloxacin, gentamicin and amikacin. This confirms the findings of earlier studies that reported a high antibiotic resistance profile in staphylococci [40,43,44]. The susceptibility of all the staphylococci isolates to nitrofurantoin may denote its potency in treating Staphylococcus infections. Nitrofurantoin was similarly detected as the most active antibiotic against strains of Staphylococcus isolates from cheese as well as patients with urogenital infections [45,46]. The PCR and WGS assays confirmed the phenotypic responses of the species to the antibiotics and affirmed a critical therapeutic implication to human health in the face of the ever-increasing shortage of effective antibiotics against staphylococci [47,48]. The pattern of antibiotic resistance observed in S. aureus, S. saprophyticus and S. epidermidis in this study was consistent with previously reported findings in clinical isolates of these species from the oral cavity [40,44,49]. The resistance determinants identified such as blaZ, tetL, tetK, mecA, ermA, ermB, rpoB and gyrA have been reported in staphylococci [50].
The optrA and cfrC confer MDR and commonly coexist in MRSA with a high potential for dissemination among gram-positive bacteria [[51], [52], [53]]. Tigecycline is the last line of defence against severe infection and is threatened by the increasing occurrence of tet (X3) genes in animals, meat for consumption and humans [[54], [55], [56]]. The production of PBP2a in staphylococci is regulated by the blaZ-blaI-blaR1 and mecA-mecI-mecRI systems and confers resistance to methicillin and all beta-lactams [[57], [58], [59]]. The resistance of the staphylococci isolates to tetracyclines, penicillins, sulfonamides, trimethoprim, macrolides, lincosamides, fluoroquinolones, aminoglycosides, pleuromutulins and polymyxins could be associated with the frequent use of these antibiotic classes in food-producing animals and plant [60]. The multidrug resistance pattern ranged from three to eight drug classes. The high prevalence of staphylococci resistance to fluoroquinolones is influenced by the high occurrence of efflux pumps and target alteration resistance mechanisms in the species [61]. The efflux pump functions to maintain low-intracellular concentrations of antibiotics by exporting the antibiotic agents from bacteria cells before reaching their targets [62,63]. The MFS, ABC, RND, SMR and MATE proteins detected are the main families of efflux pumps in bacteria in terms of energy source and structure [64]. The findings suggest a strong tendency for XDR in Staphylococcus species and affirm the rapid increase in the prevalence of XDR in bacterial species [65,66].
The study revealed a high prevalence of major virulence genes in the staphylococcal species which were commonly associated with adherence (clf, ebpS), exoenzyme (coa, vWbp), immune evasion (Sbi, spA, nuc) and toxin production (hlg, eta). The staphylococcal isolates also had the toxinogenic ability to cause food poisoning (sea to see), stimulate the epithelial secretion of proinflammatory cytokines to induce toxic shock syndrome with high mortality (tst) and destroy desmosomal cell attachments to cause scalded skin syndrome (eta, etb) [41,67]. The high frequency of the clumping factor signifies the abundance of the gene in the species. The presence of the gene, as well as the genes for fibronectin-binding proteins, elastin-binding proteins and collagen-binding proteins, is crucial for the staphylococcal species to bind, colonise and invade host cells [68]. The presence of the icaA gene together with sec and hlg confers enhanced pathogenicity in staphylococci [16]. The putative roles of these virulence factors in the antimicrobial resistance of staphylococci have been documented and explored as targets for novel therapeutic agents against staphylococcal infections [[69], [70], [71]].
5. Conclusions
Food samples had a high level of staphylococcal contamination, which varied in composition. The identified species comprised CoNs and CoPs staphylococcal species and are commonly implicated in various animal and human infections. The staphylococcal species were multidrug-resistant to three or more drug classes. Staphylococci were frequently associated with the tet genes, efflux pump proteins and susceptibility to fluoroquinolones. The presence of adhesins, exoproteins and pyrogenic toxin superantigens was commonly detected. Interestingly, these isolates were susceptible to nitrofurantoin. The next-generation sequencing technique revealed better insight into the diversity of staphylococci and their antimicrobial resistance and virulence genes. The prevalence, phenotypic and genetic multiresistance of Staphylococcus species to antimicrobials remains a critical health concern and there is a need for routine monitoring of staphylococcal levels in foods to ensure food and human safety. This study was limited to the assessment of antibiotic resistance and virulence patterns in staphylococcal isolates. A few primers were used for PCR analysis. A tracer study is required to identify the possible sources of antimicrobial-resistant staphylococci in food.
Funding
This work was financially supported by the Directorate of Research, Innovation and Consultancy of the University of Cape Coast under the grant code RSG/GRP/CANS/2020/107.
Ethics approval
This study was approved by the University of Cape Coast Institutional Review Board (UCCIRB/EXT/2020/46).
Availability of data and materials
The datasets used during the current study are available from the corresponding author upon request. The WGS data are publicly available in the MG-RAST database with accession numbers 4963321.3 and 4963322.3.
CRediT authorship contribution statement
Daniel Sakyi Agyirifo: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Theophilus Abonyi Mensah: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Andrews Senyenam Yao Senya: Writing – review & editing, Writing – original draft, Methodology, Investigation. Alphonse Hounkpe: Writing – review & editing, Writing – original draft, Methodology, Investigation. Cindy Deladem Dornyoh: Writing – review & editing, Writing – original draft, Methodology, Investigation. Emmanuel Plas Otwe: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Directorate of Research, Innovation and Consultancy of the University of Cape Coast for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21584.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Eromo T., Tassew H., Daka D., Kibru G. Bacteriological quality of street foods and antimicrobial resistance of isolates in Hawassa, Ethiopia. Ethiop J Health Sci. 2016;26(6):533–542. doi: 10.4314/ejhs.v26i6.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feglo P., Sakyi K. Bacterial contamination of street vending food in Kumasi, Ghana. J. Med. Biomed. Sci. 2012;1(1):1–8. [Google Scholar]
- 3.Karikari A.B., Kpordze S.W., Yamik D.Y., Saba C.S. Ready-to-Eat foods as sources of extended spectrum β-lactamase producing Salmonella and E. coli in tamale, Ghana. Front Trop Dis. 2022;3 [Google Scholar]
- 4.Wang Y.-T., Lin Y.-T., Wan T.-W., Wang D.-Y., Lin H.-Y., Lin C.-Y., et al. Distribution of antibiotic resistance genes among Staphylococcus species isolated from ready-to-eat foods. J. Food Drug Anal. 2019;27(4):841–848. doi: 10.1016/j.jfda.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayeni F.A. In: Staphylococcus aureus. Hemeg H., Ozbak H., Afrin F., editors. IntechOpen Limited; UK: 2018. Prevalence, diagnosis and local susceptibility of staphylococci infections; pp. 75–90. [Google Scholar]
- 6.Fungwithaya P., Boonchuay K., Narinthorn R., Sontigun N., Sansamur C., Petcharat Y., et al. First study on diversity and antimicrobial-resistant profile of staphylococci in sports animals of Southern Thailand. Vet. World. 2022;15(3):765–774. doi: 10.14202/vetworld.2022.765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otarigho B., Falade M.O. Analysis of antibiotics resistant genes in different strains of Staphylococcus aureus. Bioinformation. 2018;14(3):113–122. doi: 10.6026/97320630014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenul C., Horswill A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2019;7(2):1–21. doi: 10.1128/microbiolspec.gpp3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K-i, Oguchi A., et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359(9320):1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 10.Malachowa N., DeLeo F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010;67(18):3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi C.C., Pereira M.F., Giambiagi-deMarval M. Underrated Staphylococcus species and their role in antimicrobial resistance spreading. Genet. Mol. Biol. 2020;43(1 Suppl 2) doi: 10.1590/1678-4685-GMB-2019-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saber H., Jasni A.S., Jamaluddin T.Z.M.T., Ibrahim R. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) species. Malys J Med Sci. 2017;24(5):7. doi: 10.21315/mjms2017.24.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bissong M.E.A., Tahnteng B.F., Ateba C.N., Akoachere J.-F.T.K. Pathogenic potential and antimicrobial resistance profile of Staphylococcus aureus in milk and beef from the Northwest and Southwest Regions of Cameroon. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/6015283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z., Shah H.N., Misra R., Chen J., Zhang W., Liu Y., et al. The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob. Resist. Infect. Control. 2018;7(1):1–10. doi: 10.1186/s13756-018-0367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunes R.S.C., Del Aguila E.M., Paschoalin V.M.F. Safety evaluation of the coagulase-negative staphylococci microbiota of salami: superantigenic toxin production and antimicrobial resistance. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/483548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asante J., Amoako D.G., Abia A.L., Somboro A.M., Govinden U., Bester L.A., et al. Review of clinically and epidemiologically relevant coagulase-negative Staphylococci in Africa. Microb. Drug Resist. 2020;26(8):951–970. doi: 10.1089/mdr.2019.0381. [DOI] [PubMed] [Google Scholar]
- 17.Becker K., Both A., Weißelberg S., Heilmann C., Rohde H. Emergence of coagulase-negative staphylococci. Expert Rev. Anti Infect. Ther. 2020;18(4):349–366. doi: 10.1080/14787210.2020.1730813. [DOI] [PubMed] [Google Scholar]
- 18.Gherardi G., Di Bonaventura G., Savini V. Elsevier; 2018. Staphylococcal Taxonomy. Pet-To-Man Travelling Staphylococci: A World in Progress; pp. 1–10. [Google Scholar]
- 19.Gashaw M., Berhane M., Bekele S., Kibru G., Teshager L., Yilma Y., et al. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: a cross sectional study. Antimicrob. Resist. Infect. Control. 2018;7(1):1–8. doi: 10.1186/s13756-018-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphreys H., Becker K., Dohmen P., Petrosillo N., Spencer M., Van Rijen M., et al. Staphylococcus aureus and surgical site infections: benefits of screening and decolonization before surgery. J. Hosp. Infect. 2016;94(3):295–304. doi: 10.1016/j.jhin.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Kosecka-Strojek M., Sabat A.J., Akkerboom V., Becker K., Van Zanten E., Wisselink G., et al. Development and validation of a reference data set for assigning Staphylococcus species based on next-generation Sequencing of the 16S-23S rRNA Region. Front. Cell. Infect. Microbiol. 2019;9:278. doi: 10.3389/fcimb.2019.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghebremedhin B., Layer F., Konig W., Konig B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J. Clin. Microbiol. 2008;46(3):1019–1025. doi: 10.1128/JCM.02058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Land M., Hauser L., Jun S.-R., Nookaew I., Leuze M.R., Ahn T.-H., et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics. 2015;15(2):141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fijałkowski K., Peitler D., Karakulska J. Staphylococci isolated from ready-to-eat meat–identification, antibiotic resistance and toxin gene profile. Int. J. Food Microbiol. 2016;238:113–120. doi: 10.1016/j.ijfoodmicro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Kim E., Kim H.-J., Yang S.-M., Kim C.-G., Choo D.-W., Kim H.-Y. Rapid identification of Staphylococcus species isolated from food samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Biotechnol. 2019;29(4):548–557. doi: 10.4014/jmb.1901.01046. [DOI] [PubMed] [Google Scholar]
- 26.Agyirifo D.S., Mensah T.A., Senya A.S.Y., Assan M., Otwe E.P. Antimicrobial susceptibility and resistance genes profiles in gram negative isolates from automated teller machines in Cape Coast, Ghana. Afr J Biol Sci. 2022;4(2):92–102. [Google Scholar]
- 27.Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. Performance Standards for Antimicrobial Susceptibility Testing: M100. [Google Scholar]
- 28.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Version 12.0 Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2022. pp. 33–38.http://www.eucast.org Available from: [Google Scholar]
- 29.Dong Q., Wang Q., Zhang Y., Chen Y., Wang H., Ding H. Prevalence, antimicrobial resistance, and staphylococcal toxin genes of blaTEM‐1a‐producing Staphylococcus aureus isolated from animals in Chongqing, China. Veterinary Medicine and Science. 2023;9(1):513–522. doi: 10.1002/vms3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J., Shi C., Song M., Xu X., Yang P., Paoli G., et al. Phenotypic and genotypic antimicrobial resistance traits of foodborne Staphylococcus aureus isolates from Shanghai. J. Food Sci. 2014;79(4):M635–M642. doi: 10.1111/1750-3841.12405. [DOI] [PubMed] [Google Scholar]
- 31.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(suppl_1):D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapierre L., Cornejo J., Zavala S., Galarce N., Sánchez F., Benavides M.B., et al. Phenotypic and genotypic characterization of virulence factors and susceptibility to antibiotics in Salmonella infantis strains isolated from chicken meat: first findings in Chile. Animal. 2020;10(6):1049. doi: 10.3390/ani10061049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhaiyan M., Wirth J.S., Saravanan V.S. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020;70(11):5926–5936. doi: 10.1099/ijsem.0.004498. [DOI] [PubMed] [Google Scholar]
- 37.Boamah V.E., Agyare C., Odoi H., Adu F., Gbedema S.Y., Dalsgaard A. Prevalence and antibiotic resistance of coagulase-negative Staphylococci isolated from poultry farms in three regions of Ghana. Infect. Drug Resist. 2017;10:175–183. doi: 10.2147/IDR.S136349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Ripa L., Gómez P., Alonso C.A., Camacho M.C., Ramiro Y., de la Puente J., et al. Frequency and characterization of antimicrobial resistance and virulence genes of coagulase-negative staphylococci from wild birds in Spain. detection of tst-carrying S. sciuri isolates. Microorganisms. 2020;8(9):1317. doi: 10.3390/microorganisms8091317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akinkunmi E., Lamikanra A. Species distribution and antibiotic resistance in coagulase-negative staphylococci colonizing the gastrointestinal tract of children in Ile-Ife, Nigeria. Trop. J. Pharmaceut. Res. 2010;9(1):35–43. [Google Scholar]
- 40.Garbacz K., Wierzbowska M., Kwapisz E., Kosecka-Strojek M., Bronk M., Saki M., et al. Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J. Oral Microbiol. 2021;13(1) doi: 10.1080/20002297.2021.1983322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilczyszyn W.M., Kosecka-Strojek M., Międzobrodzki J. Pet-To-Man Travelling Staphylococci: A World in Progress. Elsevier; 2018. Phage-associated virulence determinants of Staphylococcus aureus; pp. 173–183. [Google Scholar]
- 42.Singh N.H., Singh R., Chongtham U. Speciation and antibiotic susceptibility pattern of coagulase negative staphylococci in a tertiary care hospital of Manipur, India. J. Clin. Diagn. Res. 2022;16(3):DC20–D24. [Google Scholar]
- 43.Cui J., Liang Z., Mo Z., Zhang J. The species distribution, antimicrobial resistance and risk factors for poor outcome of coagulase-negative staphylococci bacteraemia in China. Antimicrob. Resist. Infect. Control. 2019;8(1):65. doi: 10.1186/s13756-019-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szczuka E., Jabłońska L., Kaznowski A. Coagulase negative staphylococci: pathogenesis, occurrence of antibiotic resistance genes and in vitro effects of antimicrobial agents on biofilm-growing bacteria. J. Med. Microbiol. 2006;65(12):1405–1413. doi: 10.1099/jmm.0.000372. [DOI] [PubMed] [Google Scholar]
- 45.Kayili E., Sanlibaba P. Prevalence, characterization and antibiotic resistance of Staphylococcus aureus isolated from traditional cheeses in Turkey. Int. J. Food Prop. 2020;23(1):1441–1451. [Google Scholar]
- 46.Sina H., Semassa J.A., Dougnon V.T., Adjile A.A., Baba-Moussa F., Bankolé H.S., et al. Antibiotics resistance profile of Staphylococci isolated from urogenital infections and toxins production of Staphylococcus aureus strains. Ann. Med. Health Sci. Res. 2018;2018(8):29–34. [Google Scholar]
- 47.Cheung G.Y.C., Bae J.S., Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomsen I.P., Liu G.Y. Targeting fundamental pathways to disrupt Staphylococcus aureus survival: clinical implications of recent discoveries. JCI Insight. 2018;3(5) doi: 10.1172/jci.insight.98216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michels R., Last K., Becker S.L., Papan C. Update on coagulase-negative staphylococci-what the clinician should know. Microorganisms. 2021;9(4):830. doi: 10.3390/microorganisms9040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watkins R R., Holubar M., David M.Z. Antimicrobial resistance in methicillin-resistant Staphylococcus aureus to newer antimicrobial agents. Antimicrob. Agents Chemother. 2019;63(12) doi: 10.1128/AAC.01216-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendes R.E., Deshpande L., Streit J.M., Sader H.S., Castanheira M., Hogan P.A., et al. ZAAPS programme results for 2016: an activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J. Antimicrob. Chemother. 2018;73:1880–1887. doi: 10.1093/jac/dky099. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y., Lv Y., Cai J., Schwarz S., Cui L., Hu Z., et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015;70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 53.Li S.M., Zhou Y.F., Li L., Fang L.X., Duan J.H., Liu F.R., et al. Characterization of the multi-drug resistance gene cfr in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front. Microbiol. 2018;9:2925. doi: 10.3389/fmicb.2018.02925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruce S.A., Smith J.T., Mydosh J.L., Ball J., Needle D.B., Gibson R., et al. Shared antibiotic resistance and virulence genes in Staphylococcus aureus from diverse animal hosts. Sci. Rep. 2022;12:4413. doi: 10.1038/s41598-022-08230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Y.-Y., Liu Y., Chen Y., Huang F.-M., Chen R.-C., Xiao Y.-H., et al. Sporadic dissemination of tet (X3) and tet (X6) mediated by highly diverse plasmidomes among livestock-associated acinetobacter. Microbiol. Spectr. 2021;9(3) doi: 10.1128/Spectrum.01141-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manoharan M., Sistla S., Ray P. Prevalence and molecular determinants of antimicrobial resistance in clinical isolates of Staphylococcus haemolyticus from India. Microb. Drug Resist. 2021;27(4):501–508. doi: 10.1089/mdr.2019.0395. [DOI] [PubMed] [Google Scholar]
- 57.Chambers H.F. Solving staphylococcal resistance to beta-lactams. Trends Microbiol. 2003;11:145–148. doi: 10.1016/s0966-842x(03)00046-5. [DOI] [PubMed] [Google Scholar]
- 58.Kakoullis L., Papachristodoulou E., Chra P., Panos G. Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics. 2021;10:415. doi: 10.3390/antibiotics10040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venter H., Henningsen M.L., Begg S.L. Antimicrobial resistance in healthcare, agriculture and the environment: the biochemistry behind the headlines. Essays Biochem. 2017;61:1–10. doi: 10.1042/EBC20160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zalewska M., Błażejewska A., Czapko A., Popowska M. Antibiotics and antibiotic resistance genes in animal manure–consequences of its application in agriculture. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.610656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashem R.A., Yassin A.S., Zedan H.H., Amin M.A. Fluoroquinolone resistant mechanisms in methicillin-resistant Staphylococcus aureus clinical isolates in Cairo, Egypt. J Infect Dev Ctries. 2013;7(11):796–803. doi: 10.3855/jidc.3105. [DOI] [PubMed] [Google Scholar]
- 62.Amaral L., Martins A., Spengler G., Molnar J. Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front. Pharmacol. 2014;4(168):1–11. doi: 10.3389/fphar.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapoor G., Saigal S., Elongavan A. Action and resistance mechanisms of antibiotics: a guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017;33(3):300–305. doi: 10.4103/joacp.JOACP_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reygaert W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501. doi: 10.3934/microbiol.2018.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faires M.C., Traverse M., Tater K.C., Pearl D.L., Weese J.S. Methicillin-resistant and -susceptible Staphylococcus aureus infections in dogs. Emerg. Infect. Dis. 2010;16:69–75. doi: 10.3201/eid1601.081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fitzgerald J.R. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Bzdil J., Zouharova M., Nedbalcova K., Sladecek V., Senk D., Holy O. Oxacillin (methicillin) resistant staphylococci in domestic animals in the Czech republic. Pathogens. 2021;10:1585. doi: 10.3390/pathogens10121585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Downer R., Roche F., Park P.W., Mecham R.P., Foster T.J. The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 2002;277:243–250. doi: 10.1074/jbc.M107621200. [DOI] [PubMed] [Google Scholar]
- 69.Machuca M.A., Sosa L.M., Gonzalez C.I. Molecular typing and virulence characteristic of methicillin-resistant Staphylococcus aureus isolates from pediatric patients in Bucaramanga, Colombia. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0073434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Risser F., López-Morales J., Nash M.A. Adhesive virulence factors of Staphylococcus aureus resist digestion by coagulation proteases thrombin and plasmin. ACS Bio Med Chem Au. 2022;XXXX(XXX) doi: 10.1021/acsbiomedchemau.2c00042. XXX-XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rungelrath V., DeLeo F.R. Staphylococcus aureus, antibiotic resistance, and the interaction with human neutrophils. Antioxidants Redox Signal. 2021;34(6):452–470. doi: 10.1089/ars.2020.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are available from the corresponding author upon request. The WGS data are publicly available in the MG-RAST database with accession numbers 4963321.3 and 4963322.3.