Abstract
Rhizosphere diazotroph assemblages of salt marsh grasses are thought to be influenced by host plant species and by a number of porewater geochemical parameters. Several geochemical variables can adversely affect plant productivity and spatial distributions, resulting in strong zonation of plant species and growth forms. This geochemically induced stress may also influence the species compositions and distributions of rhizosphere diazotroph assemblages, but little is currently known about these organisms. The diversity and key physiological features of culturable, O2-tolerant rhizosphere diazotrophs associated with the tall and short growth forms of Spartina alterniflora and with Juncus roemerianus were examined. A total of 339 gram-negative strains were isolated by a root stab culture approach and morphologically and physiologically characterized by using API and BIOLOG tests. Eighty-six distinct groups composed of physiologically similar strains were identified. Of these groups, 72% were shown to be capable of N2 fixation through molecular analyses, and a representative strain was chosen from each diazotroph group for further characterization. Cluster and principal-components analysis of BIOLOG data allowed the designation of physiologically distinct strain groupings. Most of these groups were dominated by strains that were not identifiable to species on the basis of API or BIOLOG testing. Representatives of several families including the Enterobacteriaceae, Vibrionaceae, Azotobacteraceae, Spirillaceae, Pseudomonadaceae, and Rhizobiaceae were recovered, as well as strains with no clear taxonomic affiliations. This study identifies numerous potentially important physiological groups of the salt marsh diazotroph assemblage.
Intertidal salt marshes along the Atlantic coast of temperate North America are dominated by Spartina alterniflora (smooth cord grass), which grows in extensive and often monophyletic stands (46). High rates of macrophyte primary production and microbially mediated nutrient cycling are characteristic of these systems, resulting in substantial contributions to global carbon (16, 44) and nitrogen (13) budgets. Whole-system nutrient budgets indicate a net export of nitrogen from Spartina marshes (15), and the consensus of numerous studies is that primary productivity (11, 24, 48), as well as decomposition processes (28, 49), in Spartina marshes is nitrogen limited. Thus, nitrogen input through nitrogen fixation (diazotrophy) is potentially very important to maintaining high levels of macrophyte primary production in this ecosystem.
Spartina shows a range of growth morphologies that reflect nitrogen limitation and other environmental stress factors (8, 10). In the North Inlet, S.C., salt marsh and elsewhere, highly productive tall-form plants (≥1 m high) are found primarily near tidal creek banks while less productive short-form plants (≤0.5 m high) occur higher in the intertidal zone. Transitional zones of medium-height plants are also observed between the tall and short zones at many locations. These major differences in plant morphology and productivity result from several stress factors, whose impacts correspond to variations in elevation and sediment texture. The low-marsh growth zone of tall Spartina is characterized by low average porewater hydrogen sulfide levels and salinity relative to the high-marsh zone dominated by short Spartina. Active porewater exchange occurs along the creek banks, regulating dissolved solute levels and allowing some oxygen to penetrate these sediments (9). The high-marsh growth zone of short Spartina is more subject to interstitial porewater stagnation. Environmental stressors in the high-marsh sediments (high salinity and hydrogen sulfide concentrations, and low oxygen availability) reduce the efficiency of nitrogen uptake by Spartina (10), exacerbating nitrogen limitation in this zone. Porewater chemistry clearly has a strong impact on the growth and productivity of Spartina. These stressors may act as selective forces on the composition and diversity of rhizosphere microbial species including the diazotrophs. However, incomplete characterization of the rhizosphere microenvironment and its potential to shelter the resident microflora from such stresses limits our understanding of selection in the rhizosphere.
Most of the diazotrophic activity in Spartina marshes is closely associated with plant roots (30, 36, 51, 52), and this activity increases in response to treatments that stimulate plant primary production (21, 36, 51). Root exudates are thought to be the main source of carbon and energy for the microflora immediately surrounding active plant roots (20, 34), and diazotrophy in the Spartina rhizosphere is enhanced by amendment with extractable carbohydrates and carboxylic acids from Spartina tissues (7). In addition, Spartina transports significant quantities of oxygen into its rhizosphere, supporting aerobic respiration in sediments that would otherwise be anoxic and highly reduced (45). The rhizosphere of Spartina thus supports high levels of diazotrophic activity and fosters conditions (the “rhizosphere effect”) that may be conducive to maintenance of substantial diazotroph diversity. This diversity is poorly characterized at present.
Close associations between different species of diazotrophs and grasses are commonly observed. A variety of enterics (53), pseudomonads (5, 19, 25), and vibrios (39) have been isolated from the roots of assorted grasses. Clearly, numerous different diazotrophs can associate with the roots of grasses. Recent studies have highlighted the diversity of rhizosphere diazotrophs and illustrate the abundance of uncharacterized species associated with wetland grasses. Ueda et al. (47) constructed and screened a clonal library of PCR-amplified nifH sequences recovered from the rice rhizosphere and rhizoplane. Analysis of the nifH sequences revealed loosely defined clusters that contained sequences homologous to those of known diazotroph species (Clostridium pasteurianum, Klebsiella pneumoniae, and Azotobacter spp.). However, all of the nifH sequences recovered were unique, indicating a great diversity of uncharacterized diazotrophs in the rice rhizosphere. Resolution of PCR-amplified nifH sequences from tall and short Spartina rhizospheres by denaturing gradient gel electrophoresis has also revealed complex diazotroph assemblages (37). Denaturing gradient gel electrophoresis banding profiles show both overlapping and differentiating nifH sequences between the two zones. These molecular biological approaches demonstrate the occurrence of numerous diazotroph species in the rhizospheres of wetland grasses, including Spartina, but provide little and only inferential information on key ecophysiological characteristics of these organisms. To understand the relevant physiological features, distributions, and stress responses of these organisms, they must be isolated and characterized.
We have examined the diversity and key physiological characteristics of culturable oxygen-tolerant diazotrophs isolated from the rhizospheres of tall- and short-form Spartina. We also examined diazotrophs from Juncus roemerianus (black needlerush), which occurs near the terrestrial fringe and in small islands of slightly higher elevation in the short-Spartina zone. Comparison between culturable diazotrophs from Spartina and Juncus facilitates a preliminary assessment of the importance of plant host species versus environmental parameters in dictating diazotroph species distributions. These efforts have resulted in a detailed physiological examination of the culturable rhizosphere diazotroph assemblages in these salt marsh grasses and have revealed a large number of unidentified diazotroph species (3, 4).
MATERIALS AND METHODS
Materials, reference cultures, and media.
API test strips were obtained from Fisher Scientific (Pittsburgh, Pa.). BIOLOG test plates were purchased from BIOLOG (Hayward, Calif.). Reference cultures of Azotobacter chroococcum (ATCC 9043), Acetobacter diazotrophicus (ATCC 49037), and Vibrio diazotrophicus (ATCC 33466) were purchased from the American Type Culture Collection (Rockville, Md.). Azospirillum brasilense Sp7, Azospirillum lipoferum Sp59b, Rhizobium leguminosarum bv. viceae (USDA 2370), and R. meliloti (USDA 1025) were provided by Peter van Berkum, U.S. Department of Agriculture. Azotobacter vinlandii UW was provided by Robert Robson, University of Reading, Reading, United Kingdom, and Klebsiella pneumoniae M5a1-UW was provided by Gary Roberts, University of Wisconsin. The cultivation medium and incubation conditions for the Azotobacter spp. have been described previously (14). Azospirillum spp. were grown on PYE (peptone yeast extract [33]) at 30°C. Rhizobium spp. (30°C) and K. pneumoniae (37°C) were grown on full-strength tryptic soy agar (Difco, Detroit, Mich.). Acetobacter diazotrophicus was grown at 30°C on mannitol agar (1), which contains the following (per liter of distilled water): Bacto Yeast Extract, 5.0 g; Bacto Peptone, 3.0 g; mannitol, 25.0 g; and Bacto Agar, 15.0 g. Vibrio diazotrophicus was grown in tryptic soy broth at 26°C.
The isolation media were based on the mineral salts recipe of Shieh et al. (40) and contained the following (per liter of distilled water): NaCl, 28 g; MgSO4, 0.5 g; CaCl2, 0.01 g; Na2MoO4, 0.01 g; and Trizma base, 6.06 g. Basal salts solutions were adjusted to pH 7.0 or 7.5, and 0.5 or 1.5% (wt/vol) Noble Agar (Difco) was added for semisolid tube and solid plating media, respectively. After being autoclaved, the media were supplemented with autoclaved FeCl3 (0.05 mM) and K2HPO4 (3 mM) and sterile-filtered citrate (60 mM [final concentration]), glucose (70 mM), malate (90 mM), or sucrose (30 mM).
Root culture inoculation and pure-culture isolation.
Short-form S. alterniflora plants were collected in January 1994, and tall-form S. alterniflora and J. roemerianus plants were collected in June 1995. All the plants were from our field study site on Goat Island in the North Inlet salt marsh, near Georgetown, S.C. (described previously [16, 31]). In the laboratory, roots were shaken and rinsed with deionized water to remove loosely associated sediment, stab inoculated into culture tubes of nitrogen-free semisolid media, and loosely capped. Root cultures were incubated in the dark at 30°C until clonal outgrowths from the roots were observed. Each clonal outgrowth was subcultured by stab inoculation into fresh nitrogen-free semisolid medium having the appropriate pH and carbon source and incubated at 30°C. Pure cultures were isolated by streak plating on nitrogen-free media and maintained on Bacto Marine Agar during characterization. Cultures were suspended in 5.0% dimethyl sulfoxide and 5.0% (final concentration) glycerol and stored frozen at −70°C.
Culture characterization.
Gram-staining characteristics and cell morphologies were determined by standard methods (18). Motility was observed in wet mounts. The oxygen requirements were determined with Bacto Marine Agar semisolid medium supplemented with sodium thioglycolate as specified for the thioglycolate medium in the USP recipe (2). Preliminary physiological characterization and grouping of strains were based on results of API 20E testing (bioMérieux Vitek, Inc.). Test results were obtained as specified by the manufacturer. API NFT strips were also used for organisms giving few positive results or inconclusive results in the API 20E tests. API 20E strip test descriptions are as follows: ONPG (β-galactosidase), ADH (arginine dihydrolase), LDC (lysine decarboxylase), ODC (ornithine decarboxylase), CIT (citrate utilization), H2S (sulfide production), URE (urease), TDA (tryptophane deaminase), IND (indole production), VP (Voges-Proskauer reaction), GEL (gelatin liquefaction), GLU (glucose fermentation), MAN (mannitol fermentation), INO (inositol fermetation), SOR (sorbitol fermentation), RHA (rhamnose fermentation), SAC (sucrose fermentation), MEL (melibiose fermentation), AMY (amygdalin fermentation), ARA (arabinose fermentation), and NO3 (nitrate reduction). API NFT strip test descriptions are as follows: NO3 (nitrate reduction), TRP (indole production from tryptophane), GLU1 (glucose fermentation), ADH (arginine dihydrolase), URE (urease), ESC (esculin hydrolysis), GEL (gelatinase), PNPG (β-galactosidase), GLU2 (d-glucose assimilation), ARA (arabinose assimilation), MNE (mannose assimilation), MAN (mannitol assimilation), NAG (N-acetyl-d-glucosamine assimilation), MAL (maltose assimilation), GNT (d-gluconic acid assimilation), CAP (capric acid assimilation), ADI (adipic acid assimilation), MLT (l-malic acid assimilation), CIT (citric acid assimilation), and PAC (phenylacetic acid assimilation). Strains that differed by three or fewer test results were grouped. Cytochrome oxidase tests (bioMérieux Vitek, Inc.) were conducted separately to supplement these API data.
Identification of nifH-containing strains.
Dot blot hybridizations were next used to confirm the potential of each API strain group to fix N2. DNA was extracted from one member of each group by using a modified Marmur (29) procedure including a cetyltrimethylammonium bromide purification step (32). DNA quality (i.e., relative molecular weight) and quantity were assessed by gel electrophoresis and fluorometry, respectively. Dot blots were prepared with 1 μg of DNA/dot from each isolate by methods described previously (27) and with positively charged nylon membranes (Tropix, Inc., Bedford, Mass.). The blots were UV cross-linked, baked for 30 min at 80°C under vacuum, and stored at 4°C until used. Prehybridization and hybridization solutions consisted of 5× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate), 2× Denhardt’s solution (1× Denhardt’s solution is 0.02% bovine serum albumin, 0.02% Ficoll, and 0.02% polyvinylpyrrolidone), 0.1% N-lauroylsarcosine, and 0.02% sodium dodecyl sulfate (SDS) and contained 100 pmol of biotinylated nifH-specific probe, RL-43R (5′-ATGCCGATCCGCGAAAACAAAGCCCAGGAGATCTACATCGTC). Prehybridization (2 h) and hybridization (2 h) mixtures were incubated at 40°C. The blots were then washed twice in 2× SSC–0.1% SDS for 5 min each and twice in 0.5× SSC–0.1% SDS at 45°C for 15 min each. Probe detection used the Southern-Light chemiluminescence detection system (Tropix, Inc.). After a 72-h exposure, hybridization signal intensities were scored against positive and negative controls. Azospirillum lipoferum was used as the positive control, and Bacillus subtilis and Staphylococcus aureus served as the negative controls. These results were verified by PCR amplification with nifH-specific primers. PCR amplification was carried out in a 25-μl reaction volume. The amplification mixture included template DNA (25 ng), reaction buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 10 pmol of primers RL-25 (5′-CAGATCAG[C/A/G]CCGCC[C/G]AG[A/G]CG[A/C]AC) and RL-28 (5′-GGTAT[C/T]GG[C/T]AA[A/G]TC[G/C]AC[G/C]AC) per μl, and 0.1 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). DNA amplification was performed with the following temperature profile: initial denaturation (94°C for 30 s); 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a 2-min extension at 72°C. Amplimers were scored after electrophoresis in 1.5% agarose gels.
Diazotroph characterization.
Strains that hybridized with the nifH probe and showed a PCR amplimer of the appropriate size were examined further by using the BIOLOG system. Each group representative was grown overnight at 30°C on Bacto Marine Agar. Colonies were collected with sterile cotton swabs and resuspended in 25 ml of sterile saline (0.85% NaCl [pH 7.0]). Inoculum turbidity was measured spectrophotometrically (λ = 590 nm) and ranged from 53 to 59% transmittance. GN Microplate test panels were inoculated with these suspensions and incubated for 24 h at 30°C. The Microlog 2 automated system and accompanying software was used for data collection and to standardize optical density values.
Analysis of physiological data.
BIOLOG optical density values were analyzed in SYSTAT 7.0 (42) by cluster analysis with Euclidean distances, and a corresponding dendogram was constructed with average linkages. Principal-components analysis was conducted in SAS (38).
RESULTS AND DISCUSSION
Root inoculation into semisolid media was used for recovery of rhizosphere and rhizoplane diazotrophs. Clonal outgrowths from root surfaces were observed after incubation for 24 to 48 h at 30°C. These outgrowths either extended directly from the root surface or were observed as dense bands, most of which were located below the medium surface. A total of 67 strains were recovered from short Spartina plants, 103 were recovered from tall Spartina, and 169 were recovered from Juncus. Most of these organisms were facultative anaerobes with a preference for microaerophilic growth conditions, although a few obligate aerobes and microaerophiles were also isolated. Rhizosphere O2 supply reflects a balance between lacunal transport- (45) and mass transport (9)-derived inputs and consumption driven by the demands of root and microbial metabolism and abiotic chemically reduced species. Facultative anaerobes probably have a selective advantage in the rhizosphere, with regularly fluctuating oxic (during periods of active photosynthesis) and anoxic conditions. Obligate anaerobes were not examined in this study.
All 339 strains stained gram negative. The majority were motile rods, although eight Juncus strains and four tall-form and two short-form Spartina strains were nonmotile cocci. Pleomorphism was common in week-old cultures, and pigmentation was observed in some cases. Pigmented strains were isolated from all three plant types, with most being isolated from tall Spartina and Juncus. Fourteen red, yellow, or orange strains were isolated.
The carbon sources we used are important constituents of grass root exudates (carboxylic acids) and plant tissues (carbohydrates) (7, 43). Citrate- or malate-supplemented media yielded the majority of strains from all three plant types, with relatively few isolates being recovered from glucose- or sucrose-supplemented Spartina root cultures (Table 1). In contrast, sucrose-supplemented root cultures yielded a substantial number of strains from Juncus roots. Recovery of a large and diverse array of diazotrophs on the carboxylic acids (Table 1; also see Table 6) may reflect a true preference for these substrates by rhizosphere diazotrophs (26).
TABLE 1.
Strain yields from each plant source obtained by using the root stab culture technique and the four carbon sources listed
| Carbon source | No. of strains froma:
|
||
|---|---|---|---|
| SS | TS | J | |
| Citrate | 24 | 35 | 55 |
| Malate | 31 | 35 | 55 |
| Glucose | 6 | 19 | 4 |
| Sucrose | 6 | 14 | 55 |
| Total | 67 | 103 | 169 |
a SS and TS, short- and tall-form S. alterniflora, respectively; J, J. roemerianus.
TABLE 6.
BIOLOG substrate class (carbohydrate, carboxylic acid, amino acid) utilization by dendrogram clusters
| Substrate class | Substrate class utilizationa by cluster:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A (n = 3) | B (n = 7) | C (n = 3) | D (n = 3) | E (n = 6) | F (n = 3) | G (n = 3) | H (n = 3) | I (n = 3) | J (n = 3) | K (n =7) | |
| Carbohydrates | 27/28 | 19/28 | 21/28 | 28/28 | 22/28 | 23/28 | 20/28 | 23/28 | 14/28 | 14/28 | 10/28 |
| Carboxylic acids | 23/26 | 12/26 | 14/26 | 13/26 | 19/26 | 17/26 | 13/26 | 11/26 | 19/26 | 7/26 | 11/26 |
| Amino acids | 18/20 | 16/20 | 16/20 | 16/20 | 17/20 | 13/20 | 12/20 | 7/20 | 17/20 | 2/20 | 5/20 |
Number of class substrates used/total number of class substrates. Clusters A, B, and C fall into group 1; clusters D, E, F, and G fall into group 2; cluster H is group 3; cluster I is group 4; and clusters J and K fall into group 5.
A total of 243 of 339 strains gave adequate responses (at least three positive test results) on the API 20E test strips. However, 63 of 339 strains performed poorly on the API 20E test strips and were characterized with the API NFT strips. The remaining 33 strains were uncharacterized due to insufficient data from both test strips.
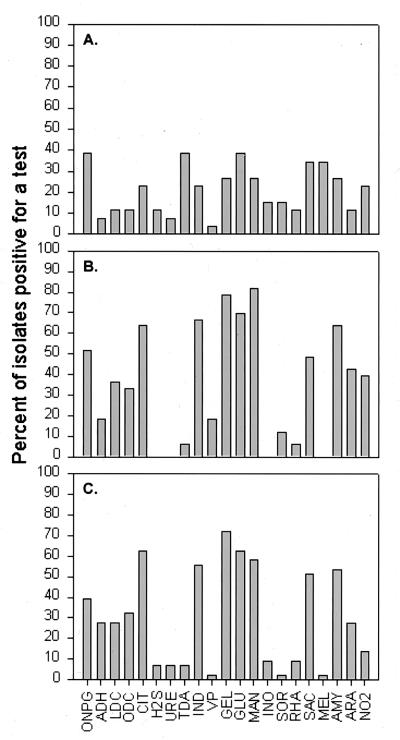
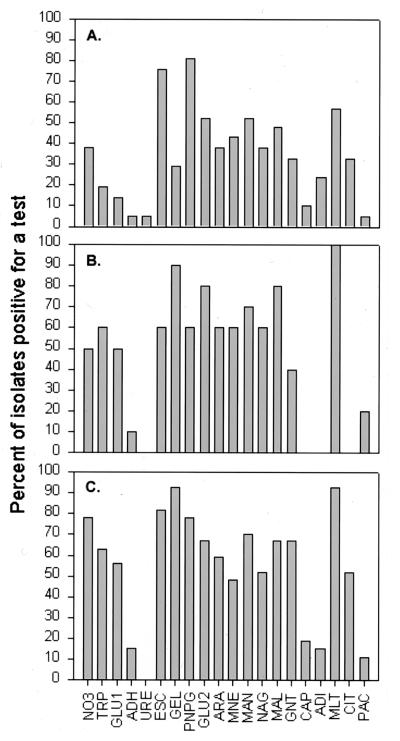
The results of the API 20E tests are summarized in Fig. 1. Overall, the test result profiles for the tall-Spartina and Juncus strain collections appear quite similar, differing substantially from these for the short Spartina strains. Strains from tall Spartina and Juncus frequently (≥50% of strains) showed positive results in the CIT, IND, GEL, GLU, MAN, and AMY API 20E tests. The result profiles of the API NFT tests for tall-Spartina and Juncus strains were also similar (Fig. 2). Strains from tall Spartina and Juncus were typically positive for the ESC, GEL, PNPG, GLU2, ARA, MAN, MAG, MAL, GNT, and MLT API NFT tests. These API results indicate striking physiological similarities between tall-Spartina and Juncus strain collections. However, the API test strips provided only a limited data set, and conclusions from such data should be drawn with caution. When strains were compared further by using the BIOLOG system, there was no strong similarity between tall-Spartina and Juncus strain collections. Key results from the three strain collections are summarized below.
FIG. 1.
Percentage of the total strain collection for each plant type that was positive for each API 20E test reaction. (A) Short-form S. alterniflora; (B) tall-form S. alterniflora; (C) J. roemerianus.
FIG. 2.
Percentage of the total strain collection for each plant type that was positive for each API NFT test reaction. (A) Short-form S. alterniflora; (B) tall-form S. alterniflora; (C) J. roemerianus.
At least some short-Spartina strains were positive for all tests on both API 20E and API NFT test strips, but no clear patterns of test results were evident. More strains positive for several API 20E tests were recovered from short Spartina with citrate- or malate-supplemented media than with glucose- or sucrose-containing media (Table 2). Only three strains from glucose- or sucrose-supplemented root cultures could be characterized with the API 20E test strips, and very few positive results were obtained. Citrate medium strains (hereafter referred to as citrate strains) were more frequently positive for fermentation of several carbohydrates (MAN, INO, SOR, RHA, MEL, AMY, and ARA) than were the malate strains. Malate strains were more frequently positive than citrate strains for GEL and IND.
TABLE 2.
API 20E test summary for short-form S. alterniflora strains
| Test(s) positive for strains froma:
| |||||||
|---|---|---|---|---|---|---|---|
| C (n = 18)
|
M (n = 18)
|
S (n = 2)
|
G (n = 1)
|
||||
| ≥50% | 0% | ≥50% | 0% | ≥50% | 0% | ≥50% | 0% |
| MEL | H2S | GEL | URE | H2S | All other tests | TDA | All other tests |
| URE | ARA | GEL | IND | ||||
| VP | GLU | ||||||
| SAC | |||||||
a Tests that were positive for ≥50% of the strain collection or for no strains (0%) are listed for each carbon source (C, citrate; M, malate; S, sucrose; G, glucose) used in strain isolation. Sample sizes (n) are given in parentheses.
A total of 22 short-Spartina strains were characterized by the API NFT tests; the majority of these (13 strains) were from the malate-supplemented medium. This limited data set prevented detailed comparisons across carbon sources. Broadly, all API NFT tests except URE were positive for at least some strains, but very few tests could be considered diagnostic (positive for ≥50% of isolates) for these short-Spartina strains. The most useful tests were PNPG and MLT.
Tall-Spartina strains grew well on all four carbon sources, although more strains were recovered from citrate- or malate-supplemented media (Table 3). None of these strains gave positive results for the H2S, URE, INO, or MEL tests on the API 20E strips, and very few gave positive results for ADH (n = 8), TDA (n = 4), VP (n = 8), or SOR (n = 4). Overall, citrate and malate strains had similar result profiles. These strains were often positive for LDC, ODC, and GLU, while the sucrose and glucose strains yielded very few positive results for these tests (two, two, and eight, respectively). Sucrose and glucose strains were differentiated by their results in the ONPG and ARA tests. AMY and GEL were frequently positive for strains recovered on all carbon sources.
TABLE 3.
API 20E test summary for tall-form S. alterniflora strains
| Tests positive for strains froma:
| |||||||
|---|---|---|---|---|---|---|---|
| C (n = 31)
|
M (n = 25)
|
S (n = 11)
|
G (n = 17)
|
||||
| ≥50% | 0% | ≥50% | 0% | ≥50% | 0% | ≥50% | 0% |
| LDC | H2S | ONPG | H2S | ONPG | H2S | ONPG | ADH |
| ODC | URE | LDC | URE | CIT | URE | IND | H2S |
| CIT | INO | ODC | TDA | MAN | VP | GEL | URE |
| IND | SOR | CIT | INO | AMY | INO | TDA | |
| GEL | RHA | IND | MEL | ARA | SOR | INO | |
| GLU | MEL | GEL | MEL | RHA | |||
| MAN | GLU | MEL | |||||
| SAC | MAN | ||||||
| AMY | AMY | ||||||
a Tests that were positive for ≥50% of the strain collection or for no strains (0%) are listed for each carbon source (C, citrate; M, malate; S, sucrose; G, glucose) used in strain isolation. Sample sizes (n) are given in parentheses.
Fourteen strains from tall Spartina were examined with the API NFT strips. None of these strains were positive for CIT or CAP tests. Not surprisingly, glucose (n = 4) and sucrose (n = 5) strains showed overall similar test result profiles. Interestingly, all glucose strains were positive for TRP but none of the sucrose strains were. Four of the five sucrose strains were positive for ARA, MNE, and NAG, while only a single glucose strain was positive for MNE. Malate strains (n = 4) were positive for all tests except ADH, URE, ADI, and PAL.
Many strains were recovered from Juncus roots for all carbon sources except glucose (Table 4). Citrate strains were differentiated from the others by giving positive results in the H2S, TDA, GLU, SAC, AMY, and ARA tests. Malate strains were frequently positive for GEL (96%), ONPG (65%), IND (50%), and MAN (50%). Less than 10% of the citrate, malate, and sucrose strain collections gave positive results in the VP, INO, SOR, RHA, and MEL tests.
TABLE 4.
API 20E test summary for Juncus strains
| Tests positive for strains froma:
| |||||||
|---|---|---|---|---|---|---|---|
| C (n = 36)
|
M (n = 26)
|
S (n = 55)
|
G (n = 3)
|
||||
| ≥50% | 0% | ≥50% | 0% | ≥50% | 0% | ≥50% | 0% |
| CIT | VP | ONPG | H2S | None | H2S | CIT | ADH |
| GEL | SOR | IND | URE | INO | GEL | LDC | |
| GLU | GEL | TDA | SOR | ODC | |||
| AMY | MAN | VP | MEL | H2S | |||
| RHA | URE | ||||||
| MEL | IND | ||||||
| VP | |||||||
| GLU | |||||||
| MAN | |||||||
| INO | |||||||
| SOR | |||||||
| RHA | |||||||
| MEL | |||||||
| NO3 | |||||||
a Tests that were positive for ≥50% of the strain collection or for no strains (0%) are listed for each carbon source (C = citrate, M = malate, S = sucrose, and G = glucose) used in strain isolation. Sample sizes (n) are given in parentheses.
Twenty-seven Juncus strains were characterized with the API NFT strips. Only a single glucose strain gave some positive results on these strips. Strains from sucrose, malate, and citrate were positive for most tests, with URE being the notable exception. Citrate (n = 13) and malate (n = 7) strains showed very similar utilization profiles and differed markedly only in the frequencies of positive results for a few key tests (MNE, ARA, NAG, and CIT). In addition, neither citrate or malate strains assimilated ADI, although sucrose strains were frequently (67%) positive for this test.
The two pH values used did not appear to impose any significant selective pressure in these isolations. We routinely record rhizosphere porewater pH values between 7.0 and 7.5 at our study site (37), and this difference is not likely to be significant to rhizosphere microbiota in the salt marsh.
API 20E and API NFT profiles were useful for eliminating redundancies in the culture collection, both within and across plant types. Strains differing by no more than three test results were grouped, yielding a total of 86 groups. The groups ranged in size from 30 strains to 1 strain. Four API groups were arbitrarily chosen, and the strains within them were tested by the BIOLOG system. Each of these groups had effectively homogeneous BIOLOG substrate utilization profiles, differing by no more than three test results among member strains. This validates the API grouping approach.
All nifH hybridization results, positive or negative, were verified by PCR amplification. Of the 86 API strain groups, 54 (63%) had nifH sequences and were presumably capable of nitrogen fixation. The fixed-nitrogen-free media efficiently selected for diazotrophs, approximately 72% of the total strains recovered. However, nitrogen-efficient but nondiazotrophic microbes were also recovered. Persistence of these organisms may have been due to nitrogen inputs from the root tissues during the original enrichments and to trace nitrogen levels in the media during isolation. Presumably these organisms are adapted to scavenging low levels of nitrogen from the environment (23, 53).
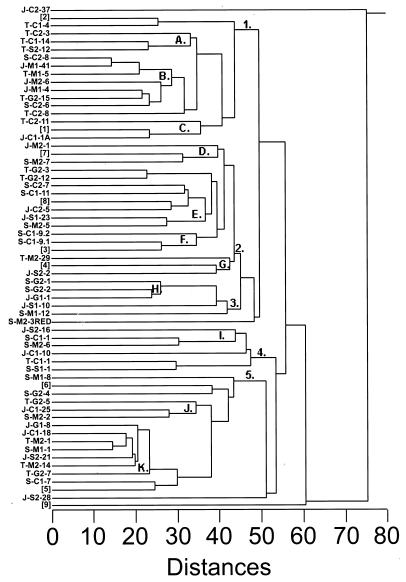
Standardized BIOLOG OD values were used to construct a dendrogram in order to facilitate identification of physiological groups within the total culture collection (Fig. 3). Major branch points were examined by principal-components analysis to determine the key substrate classes (i.e., carbohydrates, carboxylic acids, and amino acids) responsible for major strain subdivisions (Table 5). Carbohydrate, carboxylic acid, and amino acid utilization accounted for 100% of the variability in the diazotroph strain data sets. Groups 1 and 5 preferentially utilized carboxylic acids, and group 4 preferentially used carbohydrates. Groups 2 and 3 preferred both carbohydrates and amino acids to carboxylic acids. Organisms falling outside these five groups utilized either very few of the BIOLOG substrates or all of them. A more detailed examination was conducted on clusters containing three or more API groups to again identify specific substrate class preferences (Table 6). Clusters A, B, C, and D preferentially utilized amino acids, and clusters E, F, and H preferentially utilized carbohydrates. Clusters G, I, J, and K showed no clear substrate class preferences. Overall, clear differences do exist among clusters based on substrate utilization profiles, although some overlap in these profiles is apparent. Signature substrates were identified for several clusters and are as follows: cluster A, glucuronamide; cluster C, alaninamide and glycyl-l-aspartic acid; cluster F, α-lactose, α-d-lactose lactulose, and 2-aminoethanol; cluster H, l-rhamnose; and cluster I, γ-aminobutyric acid.
FIG. 3.
Dendrogram constructed from Euclidean distances of BIOLOG test results for unknown diazotroph strains and known diazotroph species. Strains are designated by zone (S, short-form S. alterniflora; T, tall-form S. alterniflora; J, J. roemerianus), carbon source (C, citrate; M, malate; G, glucose; S, sucrose), pH (1, pH 7.0; 2, pH 7.5), and strain number. Known diazotrophs are as follows: [1] Vibrio diazotrophicus, [2] Azospirillum brasilense, [3] Azotobacter chroococcum, [4] Klebsiella pneumoniae, [5] Azospirillum lipoferum, [6] Azotobacter diazotrophicus, [7] Rhizobium meliloti, [8] Rhizobium leguminosarum, and [9] Azotobacter vinlandii. Strain groupings and clusters are illustrated numerically and by letter at the appropriate node, respectively.
TABLE 5.
Principal-components analysis of normalized BIOLOG optical density values for major dendrogram branches, represented by major strain groupsa
| Substrate class | Result for strain group:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
5
|
|||||||||||
| PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | |
| Carbohydrates | −0.68 | 0.33 | 0.65 | 0.72 | 0.17 | −0.67 | 0.64 | 0.18 | 0.75 | 0.71 | 0.00 | 0.71 | −0.68 | 0.33 | 0.65 |
| Carboxylic acids | 0.72 | 0.13 | 0.68 | 0.11 | 0.93 | 0.35 | −0.49 | 0.84 | 0.22 | −0.71 | 0.00 | 0.71 | 0.72 | 0.13 | 0.68 |
| Amino acids | 0.14 | 0.93 | −0.33 | 0.69 | −0.33 | 0.65 | 0.59 | 0.51 | −0.63 | 0.00 | 1.00 | 0.00 | 0.14 | 0.93 | −0.33 |
a BIOLOG tests results for three major substrate groups, the carbohydrates, carboxylic acids, and amino acids, accounted for 100% of the variability in the data sets.
These data also permitted us to perform a preliminary evaluation of the taxonomy of some diazotroph clusters (Fig. 3). The majority of the clusters contained unidentified organisms that probably belong to the families Vibrionaceae and Enterobacteriaceae. However, these families are distinguished primarily on the basis of a small set of phenotypic traits, i.e., negative Gram reaction, bacillus cell shape, facultative oxygen requirement, and cytochrome oxidase production. Substantially more data are needed to extend these preliminary identifications. Fermentation and substrate utilization results for other clusters are consistent with their placement in the families Rhizobiaceae, Spirillaceae, Azotobacteraceae, and Pseudomonadaceae. Based on the results obtained in this study, clusters C, D, and F were the only taxonomically coherent clusters. Other clusters contained organisms apparently differing in taxonomic affiliations. Identification to species level was possible for several strains. Cluster B contained three strains that were identified as Vibrio parahaemolyticus, Vibrio anguillarum, and Vibrio alginolyticus. One strain in cluster K was identified as Pseudomonas syringae, and another from the same cluster was apparently a Flavobacterium species but did not correspond to any documented species in this genus. Similar major taxonomic groupings have been identified in other nonlegume diazotroph-plant systems (5, 50, 53). Overall, the clusters identified were diverse and contained representatives from several taxonomic groupings. The API and BIOLOG tests provided convenient tools for characterization, although additional physiological and molecular biological testing will be required for complete classification.
Known diazotrophs were also examined to evaluate the utility of BIOLOG results for identification of rhizosphere diazotrophs (data not shown). Most cultures of known diazotrophs showed fewer than three discrepancies between the BIOLOG test results and the known characteristics of the organisms (22). However, others (Azotobacter chroococcum, Vibrio diazotrophicus, and Rhizobium meliloti) produced BIOLOG test results differing substantially from their known physiological traits. All of our known and unknown cultures were grown on the same rich medium, Bacto Marine Agar, before inoculation of the BIOLOG test plates. This was done to promote consistency in test results, allowing comparisons among the strains to be made. However, several of our type strains are routinely cultivated on defined media containing low levels of NaCl, consistent with their terrestrial origins. Bacto Marine Agar, while supporting growth, was stressful for some of these organisms, perhaps resulting in atypical test results. Similar stress-driven atypical results could occur for some of the unknown strains, but in the interests of consistency and expediency, results from Bacto Marine Agar cultures were used throughout.
Primary productivity in the salt marsh requires substantial new nitrogen inputs from diazotrophy to offset the export of fixed nitrogen (6, 23). Clearly, diazotrophs and diazotrophic interactions are very important in this environment. It has been suggested for many ecosystems that taxonomically uncharacterized species are ubiquitous, and consequently our understanding of microbial diversity is presently quite poor (12, 17, 35, 41). It is clear that even well-studied functional groups, such as the diazotrophs, contain many unknown species. This study provides a substantial amount of new information on this important group of organisms and has identified several key features of diazotrophs from the S. alterniflora and J. roemerianus rhizospheres and rhizoplanes. Isolation on laboratory medium is known to bias microorganism recovery, with the potential for loss of important species. However, this technique does yield the pure cultures required for ecophysiological studies and permits some characterization of the physiological diversity of diazotrophs in the rhizospheres of salt marsh grasses. Future studies will quantitatively monitor the physiological groups of diazotrophs identified here to address diazotroph assemblage stability, interactions with host plants, and responses to geochemical stressors.
ACKNOWLEDGMENTS
We acknowledge Peter Noble and Holmes Finch for their help with the statistical analyses, and we acknowledge the Belle W. Baruch Institute for Marine Biology and Coastal Research for access to field sites and for use of the BIOLOG Automated System.
This research was supported by NSF award DEB-9407596 to C.R.L. C.E.B. and A.L.A. acknowledge partial support from the Howard Hughes Undergraduate Research Program of the Department of Biological Sciences, University of South Carolina.
REFERENCES
- 1.American Type Culture Collection. Catalogue of bacteria and bacteriophages. Rockville, Md: American Type Culture Collection; 1992. [Google Scholar]
- 2.Atlas R M. Handbook of microbiological media. Boca Raton, Fla: CRC Press, Inc.; 1993. [Google Scholar]
- 3.Bagwell C E, Lovell C R. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Physiological diversity within the rhizosphere diazotroph assemblages of salt marsh grasses, abstr. N-41; p. 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagwell C E, Lovell C R, Piceno Y M, Ashburne A L. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Differences between diazotrophic assemblages from the rhizosphere of salt marsh grasses, abstr. N-208; p. 415. [Google Scholar]
- 5.Bally R, Thomas-Bauzon D, Heulin T, Balandreau J, Richard C, De Ley J. Determination of the most frequent N2-fixing bacteria in a rice rhizosphere. Can J Microbiol. 1983;29:881–887. [Google Scholar]
- 6.Biondini M, Klein D A, Redente E F. Carbon and nitrogen losses through root exudation by Agropyron cristatum, A. smithii, and Bouteloua gracilis. Soil Biol Biochem. 1988;20:477–482. [Google Scholar]
- 7.Boyle C D, Patriquin D G. Carbon metabolism of Spartina alterniflora Loisel in relation to that of associated nitrogen-fixing bacteria. New Phytol. 1981;89:275–288. [Google Scholar]
- 8.Bradley P M, Dunn E L. Effect of sulfide on the growth of three salt marsh halophytes of the Southeastern US. Am J Bot. 1989;76:1707–1713. [Google Scholar]
- 9.Bradley P M, Morris J T. Physical characteristics of salt marsh sediments: ecological implications. Mar Ecol Prog Ser. 1990;61:245–252. [Google Scholar]
- 10.Bradley P M, Morris J T. Influence of oxygen and sulfide concentration on nitrogen uptake kinetics in Spartina alterniflora. Ecology. 1990;71:282–287. [Google Scholar]
- 11.Bradley P M, Morris J T. The influence of salinity on the kinetics of NH4+ uptake in Spartina alterniflora. Oecologia. 1991;85:375–380. doi: 10.1007/BF00320613. [DOI] [PubMed] [Google Scholar]
- 12.Brussard L, Behan-Pelletier V M, Bignell D, Brown V K, Didden W, Folgarait P, Fragoso C, Freckman D W, Gupta V, Hattori T, Hawksworth D L, Klopatek C, Lavelle P, Malloch D W, Rusek J, Soderstrom B, Tiedje J M, Virginia R A. Biodiversity and ecosystem functioning in soil. Ambio. 1997;26:563–570. [Google Scholar]
- 13.Capone D G, Carpenter E J. Nitrogen fixation in the marine environment. Science. 1982;217:1140–1142. doi: 10.1126/science.217.4565.1140. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y P, Lopez-de-Victoria G, Lovell C R. Utilization of aromatic compounds as carbon and energy sources during growth and N2-fixation by free-living nitrogen fixing bacteria. Arch Microbiol. 1993;159:207–212. [Google Scholar]
- 15.Dame R, Chrzanowski T, Bildstein K, Kjerfve B, McKellar H, Nelson D, Spurrier J, Stancyk S, Stevenson H, Vernberg J, Zingmark R. The outwelling hypothesis and North Inlet, South Carolina. Mar Ecol Prog Ser. 1986;33:217–229. [Google Scholar]
- 16.Dame R F, Kenny P D. Variability of Spartina alterniflora primary production in the euhaline North Inlet estuary. Mar Ecol Prog Ser. 1986;32:71–80. [Google Scholar]
- 17.Freckman D W, Blackburn T H, Brussard L, Hutchings P, Palmer M A, Snelgrove P V R. Linking biodiversity and ecosystem functioning of soils and sediments. Ambio. 1997;26:556–562. [Google Scholar]
- 18.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. [Google Scholar]
- 19.Haahtela K, Wartiovaara T, Sundman V, Skujins J. Root-associated N2 fixation (acetylene reduction) by Enterobacteriaceae and Azospirillum strains in cold-climate spodosols. Appl Environ Microbiol. 1981;41:203–206. doi: 10.1128/aem.41.1.203-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale M G, Moore L D, Griffin G J. Root exudates and exudation. In: Dommergues Y R, Krupa S V, editors. Interactions between non-pathogenic soil microorganisms and plants. New York, N.Y: Elsevier Scientific Publishing Co.; 1978. pp. 163–203. [Google Scholar]
- 21.Hanson R B. Nitrogen fixation (acetylene reduction) in a salt marsh amended with sewage sludge and organic carbon and nitrogen compounds. Appl Environ Microbiol. 1977;33:846–852. doi: 10.1128/aem.33.4.846-852.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensyl W R, editor. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 23.Herman R P, Provencio K, Torrez R, Seager G W. Seasonal and spatial population dynamics of the nitrogen efficient guild in a desert bajada grassland. Appl Environ Microbiol. 1994;60:1160–1165. doi: 10.1128/aem.60.4.1160-1165.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkinson C S, Schubauer J P. Static and dynamic aspects of nitrogen cycling in the salt marsh graminoid Spartina alterniflora. Ecology. 1984;65:961–969. [Google Scholar]
- 25.Lindberg T, Granhall U. Isolation and characterization of dinitrogen-fixing bacteria from the rhizosphere of temperate cereals and forage grasses. Appl Environ Microbiol. 1984;48:683–689. doi: 10.1128/aem.48.4.683-689.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livingstone D C, Patriquin D G. Nitrogenase activity in relation to season, carbohydrates and organic acids in a temperate zone root association. Soil Biol Biochem. 1980;12:543–546. [Google Scholar]
- 27.Lovell C R, Hui Y. Design and testing of a functional group-specific DNA probe for the study of natural populations of acetogenic bacteria. Appl Environ Microbiol. 1991;57:2602–2609. doi: 10.1128/aem.57.9.2602-2609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marinucci A C, Hobbie J E, Helfrich J V K. Effect of litter nitrogen on decomposition and microbial biomass in Spartina alterniflora. Microb Ecol. 1983;9:27–40. doi: 10.1007/BF02011578. [DOI] [PubMed] [Google Scholar]
- 29.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 30.McClung C R, van Berkum P, Davis R E, Sloger C. Enumeration and localization of N2-fixing bacteria associated with roots of Spartina alterniflora Loisel. Appl Environ Microbiol. 1983;45:1914–1920. doi: 10.1128/aem.45.6.1914-1920.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris J T, Haskin B. A 5-yr record of aerial primary production and stand characteristics of Spartina alterniflora. Ecology. 1990;71:2209–2217. [Google Scholar]
- 32.Murry H G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.New P B, Kennedy I R. Regional distribution and pH sensitivity of Azospirillum associated with wheat roots in eastern Australia. Microb Ecol. 1989;17:299–309. doi: 10.1007/BF02012842. [DOI] [PubMed] [Google Scholar]
- 34.Newman E I. The rhizosphere: carbon sources and microbial populations. In: Fitter A H, editor. Ecological interactions in soil, plants, microbes, and animals. British Ecological Society special publication 4. Oxford, England: Blackwell Scientific Publications Ltd.; 1985. pp. 107–121. [Google Scholar]
- 35.Palmer M, Covich A P, Finlay B J, Gibert J, Hyde K D, Johnson R K, Kairesalo T, Lake S, Lovell C R, Naiman R J, Ricci C, Sabater F, Strayer D. Biodiversity and ecosystem processes in freshwater sediments. Ambio. 1997;26:571–577. [Google Scholar]
- 36.Patriquin D G. Factors affecting nitrogenase activity (acetylene reducing activity) associated with excised roots of the emergent halophyte Spartina alterniflora Loisel. Aquat Bot. 1978;4:193–210. [Google Scholar]
- 37.Piceno Y M, Lovell C R. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. A comparison of diazotroph assemblages in Spartina alterniflora rhizospheres in two marsh zones using denaturing gradient gel electrophoresis (DGGE) analysis, abstr. N-133; p. 338. [Google Scholar]
- 38.SAS Institute Inc. SAS/STAT software: changes and enhancements through release 6.12. Cary, N.C: SAS Institute Inc.; 1997. [Google Scholar]
- 39.Shieh W Y, Simidu U, Maruyama Y. Isolation of a nitrogen-fixing Vibrio species from the roots of eelgrass (Zostera marina) J Gen Appl Microbiol. 1987;33:321–330. [Google Scholar]
- 40.Shieh W Y, Simidu U, Maruyama Y. New marine nitrogen-fixing bacteria isolated from an eelgrass (Zostera marina) bed. Can J Microbiol. 1988;34:886–890. doi: 10.1007/BF02075812. [DOI] [PubMed] [Google Scholar]
- 41.Snelgrove P, Blackburn T H, Hutchings P A, Alongi D M, Grassle J F, Hummel H, King G, Koike I, Lambshead P J D, Ramsing N B, Solis-Weiss V. The importance of marine sediment biodiversity in ecosystem processes. Ambio. 1997;26:578–583. [Google Scholar]
- 42.SPSS Inc. SYSTAT 7.0. Chicago, Ill: SPSS Inc.; 1997. [Google Scholar]
- 43.Street H E, Elliott M C, Fowler M W. The physiology of roots. In: Dommergues Y R, Krupa S V, editors. Interactions between non-pathogenic soil microorganisms and plants. New York, N.Y: Elsevier Scientific Publishing Co.; 1978. pp. 69–130. [Google Scholar]
- 44.Teal J M, Howes B L. Interannual variability of a salt-marsh ecosystem. Limnol Oceanogr. 1996;41:802–809. [Google Scholar]
- 45.Teal J M, Kanwisher J W. Gas transport in the marsh grass, Spartina alterniflora. J Exp Bot. 1966;17:355–361. [Google Scholar]
- 46.Turner R E. Geographic variations in salt marsh macrophyte production: a review. Contrib Mar Sci. 1976;20:47–68. [Google Scholar]
- 47.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valiela I, Teal J M. Nutrient limitation in salt marsh vegetation. In: Reimold R J, Queen W H, editors. Ecology of halophytes. New York, N.Y: Academic Press, Inc.; 1974. pp. 547–563. [Google Scholar]
- 49.Valiela I, Teal J M, Allen S D, van Etten R, Goehringer D, Volkmann S. Decomposition in salt marsh ecosystems: the phases and major factors affecting disappearance of above-ground organic matter. J Exp Mar Biol Ecol. 1985;89:29–54. [Google Scholar]
- 50.Vose P B. Developments in nonlegume N2-fixing systems. Can J Microbiol. 1983;29:837–850. [Google Scholar]
- 51.Whiting G J, Gandy E L, Yoch D C. Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass Spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl Environ Microbiol. 1986;52:108–113. doi: 10.1128/aem.52.1.108-113.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiting G J, Morris J T. Nitrogen fixation (C2H2 reduction) in a salt marsh: its relationship to temperature and an evaluation of an in situ chamber technique. Soil Biol Biochem. 1986;18:515–521. [Google Scholar]
- 53.Wright S F, Weaver R W. Enumeration and identification of nitrogen-fixing bacteria from forage grass roots. Appl Environ Microbiol. 1981;42:97–101. doi: 10.1128/aem.42.1.97-101.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]