Abstract
The detection of RAS mutations and co-mutations in liquid biopsy offers a novel paradigm for the dynamic management of metastatic colorectal cancer (mCRC) patients.
Expanding the results of the prospective OMITERC (OMIcs application from solid to liquid biopsy for a personalized ThERapy of Cancer) project, we collected blood samples at specific time points from patients who received a first-line chemotherapy (CT) for KRAS-mutated mCRC. CTC quantification was performed by CellSearch® system. Libraries from cfDNA were prepared using the Oncomine™ Colon cfDNA Assay to detect tumour-derived DNA in cfDNA. The analysis involved >240 hotspots in 14 genes.
Twenty patients with KRAS-mutated mCRC treated at the Medical Oncology Unit of Careggi University Hospital were prospectively enrolled. Nine patients had available data for longitudinal monitoring of cfDNA. After 6 weeks of first-line CT an increase of KRAS-mutated clone was reported in the only patient who did not obtain disease control, while all patients with decrease of KRAS clones obtained disease control. Overall, in patients with a short (<9 months) progression-free survival (PFS) we registered, at 6 weeks, an increase in cfDNA levels and in KRAS mutations or other co-mutations, i.e. PIK3CA, FBXW7, GNAS, and TP53. In selected cases, co-mutations were able to better anticipate radiological progressive disease (PD) than the increase of KRAS-mutated clones.
In conclusion, our study confirms plasma ctDNA as a crucial tool for anticipating PD at an early time point and highlights the value of a comprehensive assessment of clonal dynamics to improve the management of patients with mCRC.
1. Introduction
The management of patients with metastatic colorectal cancer (mCRC) is highly dependent on the molecular signature of the tumor at the diagnosis and throughout the continuum of care [1]. Although the one-size-fits-all approach is still widely used, an increasing number of patients receive tailored treatments [[2], [3], [4]]. Approximately half of mCRCs harbor mutations in the RAS genes and such mutations confer resistance to anti-EGFR monoclonal antibodies (moAbs); therefore, in microsatellite stable (MSS) RAS-mutated CRC, anti-angiogenic agents combined with standard chemotherapy (CT) are the backbone of the first-line of treatment [5].
Although the predictive role of RAS mutations has been well established, their prognostic role is still controversial [6,7]. Recently, the detection of RAS mutations in liquid biopsy, i.e. plasma, has offered a novel tool for diagnostic purposes, prognostication, and, last but not least, monitoring treatment response [8,9]. Beyond RAS, other mutations including PIK3CA, FBXW7 and SMAD4 can be longitudinally detected in plasma of mCRC patients; however, the clinical and prognostic implication of these mutations remains to be fully elucidated [10,11].
As advances in mCRC treatment move in the direction of precision medicine, tools for longitudinal molecular profiling and real-time monitoring of drug resistance are eagerly awaited [12]. We recently showed the results of the prospective OMITERC (OMIcs application from solid to liquid biopsy for a personalized ThERapy of Cancer) study, in which we confirmed the presence of circulating tumor cells (CTCs) at baseline as a negative prognostic factor for survival and cell-free DNA (cfDNA) as a promising biomarker for the longitudinal monitoring of treatment effect in patients with KRAS-mutated mCRC. Additionally, we highlighted the value of the dynamic changes of cfDNA over time in a higher number of patients [13].
Here, we report the results of the longitudinal analysis of RAS mutations and other co-mutations in cfDNA from patients with KRAS-mutated mCRC.
2. Patients and methods
2.1. Study population
We analyzed a subgroup of patients within the OMITERC study, for whom we examined a hotspot panel of cancer-associated gene mutations to identify tumor-specific variants in plasma at baseline and during treatment. OMITERC study was approved by Institutional review board of Azienda Ospedaliero-Universitaria Careggi (Comitato Etico Regionale for clinical experimentation of Toscana region – Italy - Area Vasta Centro). All patients gave written informed consent. Inclusion and exclusion criteria were previously described in the OMITERC study. .13
2.2. Study design and procedures
A complete description of the study design and procedures has been previously reported.13 Briefly, all procedures were adopted in accordance to international standards. Blood samples were collected from each patient in a cfDNA BCT (Streck) tube and in a CellSave Preservative tube (CellSearch; Menarini Silicon Biosystems) for molecular analysis and CTC assessment, respectively. Time points for collecting the blood samples were: baseline (T0), 6 weeks (T1), 3 months (T2), 6 months (T3) and 9 months (T4).
For CTCs, the Cell Search system was used for enrichment and counting, followed by CTC isolation by dielectrophoresis (DEParray) in samples with a number of CTCs>5. Isolated single CTCs have been submitted to WGA before NGS sequencing.
CfDNA was extracted from 2 mL plasma using the QIAsymphony Circulating DNA Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. CfDNA quantification was performed using the Qubit 3.0 fluorometer (Thermo Fisher Scientific). Libraries from cfDNA were prepared using the Oncomine™ Colon cfDNA Assay to detect colon (or other related gastrointestinal) tumour-derived DNA in cfDNA. The panel allows the analysis of single nucleotide variants and short indels that are frequently detected in colon/gastrointestinal cancers, involving >240 hotspots in 14 genes (AKT1, APC, BRAF, CTNNB1, EGFR, ERBB2, FBXW7, GNAS, KRAS, MAD4, MAP2K1, NRAS, PIK3CA, TP53). Ten nanograms of cfDNA were used to prepare barcoded libraries using the Ion AmpliSeq™ Library kit 2.0 and Ion Xpress™ barcode adapters (Thermofisher Scientific). Library purification was performed using the Agentcourt AMPure XP reagent (Beckman Coulter). Libraries were checked by capillary electrophoresis on the Agilent Bioanalyzer using the Agilent High Sensitivity DNA kit and quantified by the Ion Library Quantitation Kit (Thermofisher Scientific) on the StepOne Plus system (Thermofisher Scientific). Ion 520™ & Ion 530™ Kit-OT2 (Thermofisher Scientific) on the Ion OneTouch™ 2 System and Ion One Touch ES were used for template preparation. Sequencing of both cfDNA and DNA obtained from GWA of single CTCs was performed on the Ion S5 using Ion 520 Chips (Thermofisher Scientific). The run was set in order to obtain at least 1000 × coverage for each sample. For CTC obtained DNA, data analysis was performed using the Ion Reporter Software 5.10 and the workflow AmpliSeq CHPv2 single sample with a careful exclusion of usual single white blood cells and/or gDNA variants. Instead, for cfDNA data analysis was carried out using the Ion Reporter Software 5.10 and the workflow Oncomine Colon Liquid Biopsy-w1.4-DNA-Single Sample. The uniformity of coverage (a quality parameter of sequencing) was 100 % in all the analyzed samples.
2.3. Clinical data and outcomes
Clinical data including all available demographic information, medical history, diagnosis, sites of metastatic disease, type of first-line and subsequent lines of CT, surgery, pathological features, molecular analysis, clinical outcomes, laboratory tests were collected from patients’ medical records and analyzed using descriptive statistics. Radiological response was carefully assessed according to RECIST, version 1.1 [14]. For target lesions, partial response (PR) was defined as a decrease in the sum of the longest diameter of at least 30 %, taking as reference the baseline sum of the longest diameter. PD was defined as the onset of new lesions and/or 20 % or greater increase in the sum of the longest diameter of measured lesions, taking as reference the nadir. Complete response (CR) was defined as the disappearance of all target lesions for at least one month. Stable disease (SD) was defined as neither a sufficient tumor reduction to qualify for PR nor a sufficient increase to qualify for PD. Overall survival (OS) was defined as the time from the start of treatment to death from any cause. Progression-free survival (PFS) was defined as the time from the start of treatment to PD, or death from any cause, whichever occurred first.
3. Results
Twenty patients with KRAS-mutated mCRC treated at the Medical Oncology Unit of Careggi University Hospital, Florence, Italy, from April 2016 to October 2018, were enrolled in the OMITERC study. 13 Nine patients had available data for longitudinal monitoring of cfDNA at baseline and during treatment. All patients were dead at the data cutoff analysis (October 2022). Patient characteristics are shown in Table 1. The median age was 68 years (range 53–84). Five patients were male and 4 females. All except one had an Eastern Cooperative Oncology Group (ECOG) performance status of 0. Primary tumor site was right colon in 3, left colon or rectum in 5, and transverse colon in 1. Four patients had single organ metastases, while 5 patients had a multi-organ involvement.
Table 1.
Patient characteristics.
| ID | Age | Sex | PS ECOG | Primary tumor location | Metastatic sites | KRAS | I Line CT | Best response | PFS (months) |
|---|---|---|---|---|---|---|---|---|---|
| COL001 | 67 | M | 0 | Transverse colon | Liver, peritoneum | pG12D | FOLFOX bevacizumab | SD | 10.0 |
| COL003 | 76 | F | 0 | Left colon/rectum | Liver, peritoneum | pG12A | FOLFOX bevacizumab | PR | 8.0 |
| COL004 | 84 | M | 1 | Left colon/rectum | Liver | pG12D | 5-FU | SD | 8.2 |
| COL005 | 61 | M | 0 | Left colon/rectum | Liver, lung, lymph nodes | pG13D | FOLFOX bevacizumab | PD | 3.8 |
| COL006 | 75 | M | 0 | Left colon/rectum | Lung, local recurrence | pG12C | FOLFOX bevacizumab | PR | 10.0 |
| COL007 | 53 | F | 0 | Left colon/rectum | Liver, lung, lymph nodes | pG12S | FOLFOXIRI bevacizumab | PR | 12.2 |
| COL012 | 60 | F | 0 | Rigt colon | Liver | pG12A | FOLFOX bevacizumab | PR | 7.0 |
| COL015 | 62 | F | 0 | Rigt colon | Liver | pG13V | FOLFOX | CR | 15.6 |
| COL017 | 78 | M | 0 | Rigt colon | Liver | pA146K | FOLFOX bevacizumab | PR | 9.1 |
Abbreviations: CR: complete response; CT: chemotherapy; FOLFOX: 5-fluorouracil, oxaliplatin; FOLFOXIRI: 5-fluorouracil, oxaliplatin, irinotecan; PD: progressive disease; PFS: progression-free survival; PR: partial response; SD: stable disease; 5-FU: 5-fluorouracil.
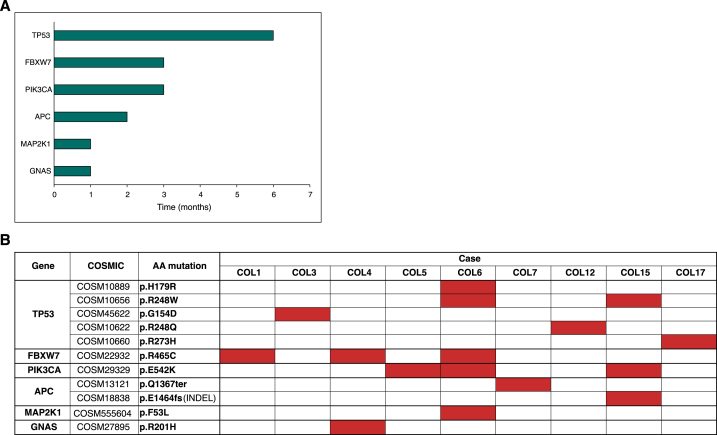
Liver metastases were present at the beginning of treatment in the vast majority of patients (8 out of 9). All patients received a first-line treatment: 5-fluorouracil (5FU), oxaliplatin (FOLFOX) in 1 patient, FOLFOX plus bevacizumab in 6, FOLFOXIRI (5FU, oxaliplatin, and irinotecan) plus bevacizumab in 1, and 5FU as single agent in 1. The distribution of KRAS hotspot variants from formalin fixed paraffin embedded tissue samples was as follows: 2 pts G12D, 2 pts G12A, 1 pt G12C, 1 pt G12S, 1 pt G13D, 1 pt G13V, and 1 pt A146K. 17 COSMIC mutations were identified, at baseline or on-treatment, in the 9 selected patients of KRAS-mutated mCRC. These 17 mutations included 6 mutations of TP53, 3 of PIK3CA, 3 of FBXW712, 2 of APC, and one each of MAPK21 and GNAS. Detailed data for each gene are shown in Fig. 1 (A, B). The median number of significant mutations for each patient was two (2.8 range; 2–7). Of note, Oncomine™ Colon cfDNA Assay used varying cfDNA input concentrations and subsequently determined varying limits of detection (LODs). The median input was 16,8 ng cfDNA (range 6,1–20 ng) and the LODs varied from 0,16 % to 0,69 %.
Fig. 1.
A, B. Distribution and frequency of co-mutations at baseline or on-treatment in the patient population.
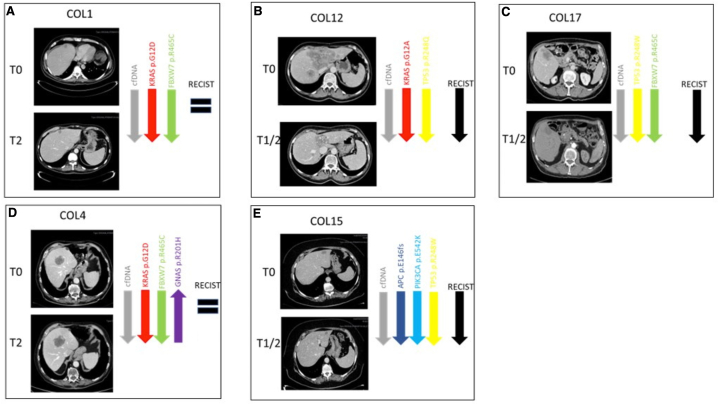
All 9 patients had at least one measurable lesion. Disease reassessment was performed every 2–3 months according to clinical practice. The evaluation of treatment response, according to RECIST 1.1 criteria, revealed the following best responses: CR in 1 patient (11.1 %), PR in 5 patients (55.6 %%), SD in 2 patients (22.2 %), and PD in 1 patient (11.1 %) (Table 1). Then, we calculated the depth of response selecting maximum 2 target lesions per organ. Four patients (i.e. #ptCOL6, #ptCOL12 and #ptCOL15, #ptCOL17) were deep responders achieving a reduction in tumor size of 36 %, 40 %, 54 % and 100 %, respectively (radiological assessment for #ptCOL12, #ptCOL15, and #ptCOL17 was reported in Fig. 2 B, E, C).
Fig. 2.
A - E. A summary of longitudinal assessment with detailed computed tomography scans and molecular clone trends in five patients (A #ptCOL1, B #ptCOL12, C #ptCOL17, D #ptCOL4, and E #ptCOL15) for whom radiographic data were available. Radiological evaluation was performed according to RECIST criteria, version 1.1. #ptCOL12 (B), #ptCOL15 (E), and #ptCOL17 (C) obtained a PR; #ptCOL1 (A), #ptCOL4 (D) obtained a SD.
3.1. Longitudinal analysis
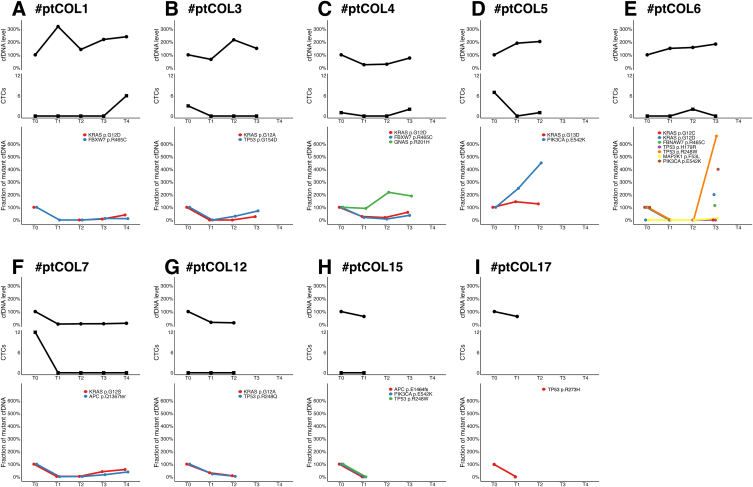
After 6 weeks of first-line CT (T1) a decrease in cfDNA levels has been observed in 67 % patients (6 out of 9; #ptCOL3, #ptCOL4, #ptCOL7, #ptCOL12, #ptCOL15, #ptCOL17), and at the same time we registered a decrease in KRAS mutations (Fig. 3 B, C, F–I). The level of cfDNA seems not to be closely associated with treatment response (X-squared test p-value = 0.3173).
Fig. 3.
A - I. Longitudinal assessment of CTC, ctDNA levels, KRAS-mutated clones and co-mutations. An increase of KRAS-mutated clone at T1 was observed in #ptCOL5 (D) who was the only patient experiencing PD as best response. In selected cases (#ptCOL3 [B], #ptCOL4 [C] and #ptCOL6 [E]), PD was anticipated by the increase/emergence of co-mutations before the increase of KRAS-mutated clones.
Additionally, we observed an increase of KRAS-mutated clone at T1 in the only patient (#ptCOL5) who did not obtain disease control; in contrast, all patients with decrease of KRAS clones, at this early time point, obtained disease control. These preliminary results may suggest that an early radiological tumor assessment may be carefully evaluated in mCRC patients who experience an early increase of the driver mutation.
Median PFS was 9.1 months (range 3.8–15.6). In patients with short PFS (<9 months) we registered, at an early time point, an increase in cfDNA levels and in KRAS mutations or other co-mutations, i.e. PIK3CA (#ptCOL5, #ptCOL6), FBXW7 and GNAS (#ptCOL4), TP53 (#ptCOL3, #ptCOL6) (Fig. 3 B - E). Overall, DNA clonality elevation was observed at an earlier time point than radiological progression in 6 out of 9 patients. On average, the time from DNA clonality elevation and disease progression was 3 months.
Interestingly, the increase of co-mutations was able to anticipate the increase of RAS-mutated clones and, consequently, radiologic PD in #ptCOL3, #ptCOL4 and #ptCOL6. In particular, in #ptCOL3 a restart of value of the TP53 p.G154D was registered at T2, while the increase of KRAS p.G12A was evident later (at T3); in #ptCOL4 an increase of GNAS p.R201H was registered at T2, while the increase of KRAS p.G12D at T3; in #ptCOL6 the emergence p.G12D and the increase of TP53 p.R248W and PIK3CA p.E542K anticipated PD, while the baseline driver clone KRAS p.G12C remained undetectable.
Regarding CTCs, they were evaluated at baseline and during treatment simultaneously with cfDNA level and emerged that they were present at baseline in 50 % of patients (4 out of 9; #ptCOL3, #ptCOL4, #ptCOL5 and #ptCOL7) and in all cases they decreased to zero at T1.
In #ptCOL1 and #ptCOL6 CTCs were not detectable at baseline while emerged for the first-time during treatment (at T4 and T2, respectively) along with an increase in cfDNA levels; in #ptCOL12 and #ptCOL15 CTCs were never detected. Details of this trial have been previously reported in OMITERC study, where we showed that overall, the presence of CTCs at baseline was associated with poor PFS, OS and treatment response. .13
4. Discussion
The assessment of RAS mutational status at baseline is not sufficient to fully exploit the possibilities that this biomarker could offer. Some efforts have been made to elucidate the prognostic effect of mutant allele frequency (MAF) at baseline. In a retrospective and prospective multicentric study, Elez et al. showed that plasma RAS MAF was not associated with primary tumor location or metastatic tumor burden. In contrast, patients with peritoneal metastases had lower levels of MAF than those with liver (p = 0.0003), lung (p = 0.044) or lymph node (p = 0.025) metastases. In addition, reduced OS and PFS were observed in patients with high MAF (p = 0.005 and p = 0.009, respectively) [15].
To date, given the recent advances in liquid biopsy, understanding tumor temporal heterogeneity has become challenging for clinicians [16,17]. In particular, the longitudinal assessment of RAS mutational status in mCRC has found potential application in selecting RAS and BRAF wild-type (WT) patients who may benefit from the rechallenge with anti-EGFR moAbs, since the emergence of RAS mutations has been described as one of the main mechanisms of acquired resistance to anti-EGFR-based CT [18]. In addition, in RAS and BRAF WT patients, the detection of oncogenic fusions or other oncogenic drivers offers the opportunity to select targeted therapies [19]. However, even in RAS-mutated CRC patients, quantification of RAS at various time points facilitates the real-time treatment monitoring [20].
A prospective study conducted by Tie et al. identified the early decrease in circulating tumor DNA (ctDNA) as a reliable predictor of therapeutic response. ctDNA levels before cycle 2 anticipated radiological response assessed at 8–10 weeks [21]. Similarly, Osumi et al. found an association between change in ctDNA levels at early time points during CT (2 weeks and 8 weeks after treatment start) and clinical outcomes in patients with mCRC, suggesting the possibility of predicting tumor response by analyzing longitudinal change in ctDNA [22].
Consistent with previous observations, in our study, the assessment of RAS cfDNA mutations at an early time point (day 40) was associated with treatment response at the radiological assessment performed, on average, two months later. A prospective study conducted by Klein-Scory et al. combined BEAMing and ddPCR to detect RAS mutations in liquid biopsy in patients with mCRC treated with first-line CT. Results were in line with our study, confirming the longitudinal assessment of RAS mutational status in cfDNA as a feasible biomarker for treatment monitoring. In this case series, all patients with a rapid disappearance of RAS mutations in cfDNA obtained disease control, while no decrease of RAS in cfDNA was observed in the patient who experienced a rapid PD [23].
Given the observational nature, our study has several limitations, including the lack of longitudinal data in 11 patients, because blood samples had not been obtained due to clinical issues, the heterogeneous assessment of disease response (every 2–3 months according to clinical practice) and the lack of blinded independent radiologic evaluation. Although limited in sample size, our study suggests broadening the horizon of longitudinal monitoring from the analysis of RAS to the comprehensive analysis of all the other clones that may emerge or change during the therapy. As consequence, the opportunity to monitor other concomitant or acquired mutations (i.e. PIK3CA, FBXW7, GNAS, TP53) aims at predicting PD and short PFS anticipating radiological assessment. In selected cases of our cohort, PD was not only due to KRAS-mutation clones but also to concomitant or acquired mutations in other genes. However, the real impact of mutations beyond RAS in treatment resistance for KRAS-mutated CRC patients is little explored. Less than 10 % of CRCs harbor mutations in the tumor suppressor FBXW7 gene. To date, the prognostic role of these mutations is not fully understood, although several retrospective studies suggested a negative impact on survival in metastatic patients. 11 PIK3CA mutations occur in approximately 20 % of CRCs. Although evidence is still limited, PIK3CA mutations have been reported to confer resistance to anti-EGFR moAbs, but further studies are needed to fully elucidate their prognostic and predictive role. 10, [24] Since the limited sample size of this study precluded an extended statistical analysis, larger studies based on liquid biopsy are required to provide a precise estimate of the contribution of non-KRAS clones in first-line CT failure. These results, if validated in prospective cohorts, could raise awareness among clinicians to modify the timing of radiologic reassessment in certain clinical contexts. However, the exact timing of longitudinal sampling still remains to be defined in order to appreciate clinically relevant changes. In our study, an early timepoint suitable for predicting response was 6 weeks.
In conclusion, the results obtained are extremely relevant since they may enable early detection of PD, which may affect the planning for combination treatments or treatment switching, thereby providing the potential to hasten disease management decisions.
Our study reinforces the potential application of plasma cfDNA for efficiently anticipating PD in patients with mCRC and highlights the value of NGS in revealing clonal dynamics of tumors in response to therapy.
Ethics approval and consent to participate
The study was approved by the Local Ethical Committee (Comitato Etico di Area Vasta Centro AOU Careggi, Firenze, Italy), reference number CEAVC BIO 16.028_10033_bio. Written informed consent to participate was obtained from all patients. The study was performed in accordance with the Declaration of Helsinki.
Data availability
The data presented in this study are available on request to the corresponding author.
CRediT authorship contribution statement
Daniele Lavacchi: Writing – review & editing, Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation. Stefania Gelmini: Writing – review & editing, Methodology, Formal analysis, Data curation. Adele Calabri: Writing – review & editing, Methodology, Formal analysis, Data curation. Gemma Rossi: Writing – review & editing, Investigation, Formal analysis, Data curation. Lisa Simi: Methodology, Formal analysis. Enrico Caliman: Writing – review & editing, Investigation, Data curation. Irene Mancini: Methodology, Formal analysis, Data curation. Francesca Salvianti: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Giulia Petroni: Investigation, Formal analysis, Data curation. Alessia Guidolin: Investigation, Data curation. Federico Scolari: Software, Methodology, Formal analysis, Data curation. Luca Messerini: Validation, Supervision, Investigation, Formal analysis, Data curation. Serena Pillozzi: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Formal analysis, Data curation. Pamela Pinzani: Writing – review & editing, Writing – original draft, Validation, Supervision, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Lorenzo Antonuzzo: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Lorenzo Antonuzzo reports financial support was provided by Tuscany Region. Lorenzo Anronuzzo reports a relationship with Tuscany Region that includes: funding grants. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We wish to thank the OMITERC consortium and in particular Prof. Mario Pazzagli and Dr. Francesco Di Costanzo for contributing in the conceptualization and funding of the project.
Abbreviations
- cfDNA
cell-free DNA
- CRC
colorectal cancer
- CR
complete response
- ctDNA
circulating tumor DNA
- CT
chemotherapy
- CTC
circulating tumor cell
- ECOG
Eastern Cooperative Oncology Group;
- FOLFOX
5-fluorouracil, oxaliplatin
- FOLFOXIRI
5-fluorouracil, oxaliplatin, irinotecan
- LOD
limits of detection
- mCRC
metastatic colorectal cancer patients
- MAF
mutant allele frequency
- moAb
monoclonal antibody
- MSS
microsatellite stable
- OMITERC
OMIcs application from solid to liquid biopsy for a personalized ThERapy of Cancer
- OS
overall survival
- PD
progressive disease
- PFS
progression-free survival
- PR
partial response
- SD
stable disease
- WT
wild-type
- 5-FU
5-fluorouracil
References
- 1.National Comprehensive Cancer Network. Colon cancer (Version 2.2021). https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf.
- 2.Lavacchi D., Roviello G., D'Angelo A. Tumor-agnostic treatment for cancer: when how is better than where. Clin. Drug Invest. 2020 Jun;40(6):519–527. doi: 10.1007/s40261-020-00915-5. PMID: 32307639. [DOI] [PubMed] [Google Scholar]
- 3.Loupakis F., Antonuzzo L., Bachet J.B., et al. Practical considerations in the use of regorafenib in metastatic colorectal cancer. Ther Adv Med Oncol. 2020;12 doi: 10.1177/1758835920956862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danesi R., Fogli S., Indraccolo S., et al. Druggable targets meet oncogenic drivers: opportunities and limitations of target-based classification of tumors and the role of Molecular Tumor Boards. ESMO Open. 2021 Apr;6(2) doi: 10.1016/j.esmoop.2020.100040. Epub 2021 Feb 2. PMID: 33540286; PMCID: PMC7859305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-Alfonso P., Grande E., Polo E., et al. The role of antiangiogenic agents in the treatment of patients with advanced colorectal cancer according to K-RAS status. Angiogenesis. 2014 Oct;17(4):805–821. doi: 10.1007/s10456-014-9433-6. Epub 2014 May 3. PMID: 24793846. [DOI] [PubMed] [Google Scholar]
- 6.Lavacchi D., Fancelli S., Roviello G., et al. Mutations matter: an observational study of the prognostic and predictive value of KRAS mutations in metastatic colorectal cancer. Front. Oncol. 2022 Nov 29;12 doi: 10.3389/fonc.2022.1055019. PMID: 36523988; PMCID: PMC9745189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Haar J., Ma X., Ooft S.N., et al. Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat. Med. 2023 Mar;29(3):605–614. doi: 10.1038/s41591-023-02240-8. Epub 2023 Mar 2. PMID: 36864254; PMCID: PMC10033412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lastraioli E., Antonuzzo L., Fantechi B., et al. KRAS and NRAS mutation detection in circulating DNA from patients with metastatic colorectal cancer using BEAMing assay: concordance with standard biopsy and clinical evaluation. Oncol. Lett. 2021 Jan;21(1):15. doi: 10.3892/ol.2020.12276. Epub 2020 Nov 6. PMID: 33240421; PMCID: PMC7681220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lastraioli E., Lavacchi D., Palmieri V.E., et al. Evaluation of RAS mutational status through BEAMing assay to monitor disease progression of metastatic colorectal cancer: a case report. Anti Cancer Drugs. 2020 Oct;31(9):979–982. doi: 10.1097/CAD.0000000000000923. PMID: 32889896. [DOI] [PubMed] [Google Scholar]
- 10.Sartore-Bianchi A., Martini M., Molinari F., et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009 Mar 1;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. Epub 2009 Feb 17. PMID: 19223544. [DOI] [PubMed] [Google Scholar]
- 11.Korphaisarn K., Morris V.K., Overman M.J., et al. FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget. 2017 Jun 13;8(24):39268–39279. doi: 10.18632/oncotarget.16848. PMID: 28424412; PMCID: PMC5503612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazouji O., Ouhajjou A., Incitti R., et al. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front. Cell Dev. Biol. 2021 May 24;9 doi: 10.3389/fcell.2021.660924. PMID: 34150757; PMCID: PMC8213391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvianti F., Gelmini S., Mancini I., et al. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: the OMITERC prospective study. Br. J. Cancer. 2021 Jul;125(1):94–100. doi: 10.1038/s41416-021-01399-6. Epub 2021 May 5. PMID: 33953347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Elez E., Chianese C., Sanz-García E., et al. Impact of circulating tumor DNA mutant allele fraction on prognosis in RAS-mutant metastatic colorectal cancer. Mol. Oncol. 2019;13(9):1827–1835. doi: 10.1002/1878-0261.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roviello G., Lavacchi D., Antonuzzo L., et al. Liquid biopsy in colorectal cancer: No longer young, but not yet old. World J. Gastroenterol. 2022 Apr 21;28(15):1503–1507. doi: 10.3748/wjg.v28.i15.1503. PMID: 35582130; PMCID: PMC9048462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osumi H., Shinozaki E., Yamaguchi K., Zembutsu H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019 Apr;110(4):1148–1155. doi: 10.1111/cas.13972. Epub 2019 Mar 4. PMID: 30742729; PMCID: PMC6447957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cremolini C., Rossini D., Dell'Aquila E., et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol. 2019 Mar 1;5(3):343–350. doi: 10.1001/jamaoncol.2018.5080. PMID: 30476968; PMCID: PMC6439839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Li R., Li J., et al. Comprehensive analysis of oncogenic fusions in mismatch repair deficient colorectal carcinomas by sequential DNA and RNA next generation sequencing. J. Transl. Med. 2021 Oct 17;19(1):433. doi: 10.1186/s12967-021-03108-6. PMID: 34657620; PMCID: PMC8522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavacchi D., Fancelli S., Roviello G., et al. Mutations matter: an observational study of the prognostic and predictive value of KRAS mutations in metastatic colorectal cancer. Front. Oncol. 2022 Nov 29;12 doi: 10.3389/fonc.2022.1055019. PMID: 36523988; PMCID: PMC9745189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tie J., Kinde I., Wang Y., et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015 Aug;26(8):1715–1722. doi: 10.1093/annonc/mdv177. Epub 2015 Apr 7. PMID: 25851626; PMCID: PMC4511218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osumi H., Shinozaki E., Yamaguchi K., et al. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci. Rep. 2019 Nov 22;9(1) doi: 10.1038/s41598-019-53711-3. PMID: 31758080; PMCID: PMC6874682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein-Scory S., Wahner I., Maslova M., et al. Evolution of RAS mutational status in liquid biopsies during first-line chemotherapy for metastatic colorectal cancer. Front. Oncol. 2020;10:1115. doi: 10.3389/fonc.2020.01115. Published 2020 Jul 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Shi Yl, Zhou K., et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9:739. doi: 10.1038/s41419-018-0776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request to the corresponding author.