Abstract
Riboflavin production in the filamentous fungus Ashbya gossypii is limited by glycine, an early precursor required for purine synthesis. We report an improvement of riboflavin production in this fungus by overexpression of the glycine biosynthetic enzyme threonine aldolase. The GLY1 gene encoding the threonine aldolase of A. gossypii was isolated by heterologous complementation of the glycine-auxotrophic Saccharomyces cerevisiae strain YM13 with a genomic library from A. gossypii. The deduced amino acid sequence of GLY1 showed 88% similarity to threonine aldolase from S. cerevisiae. In the presence of the GLY1 gene, 25 mU of threonine aldolase specific activity mg−1 was detectable in crude extracts of S. cerevisiae YM13. Disruption of GLY1 led to a complete loss of threonine aldolase activity in A. gossypii crude extracts, but growth of and riboflavin production by the knockout mutant were not affected. This indicated a minor role of the enzyme in glycine biosynthesis of A. gossypii. However, overexpression of GLY1 under the control of the constitutive TEF promoter and terminator led to a 10-fold increase of threonine aldolase specific activity in crude extracts along with a 9-fold increase of riboflavin production when the medium was supplemented with threonine. This strong enhancement, which could not be achieved by supplementation with glycine alone, was attributed to an almost quantitative uptake of threonine and its intracellular conversion into glycine. This became evident by a subsequent partial efflux of the glycine formed.
The filamentous hemiascomycete Ashbya gossypii (Ashby and Novell) (10) is a biotechnologically important producer of vitamin B2 (riboflavin). Improved producer strains are used for commercial production of riboflavin (7) and give a yield of up to 15 g liter−1 (4). Since the first quantitative report (45), there have been numerous efforts to improve riboflavin production by A. gossypii by optimization of medium composition and fermentation conditions (20, 27) as well as by screening of antimetabolite-resistant mutants (38, 39). Since recombinant DNA techniques for A. gossypii have been established recently (42, 46), the way is now open for development of a targeted producer strain by molecular approaches.
Starting from GTP and ribulose-5-phosphate only six enzymatic reactions are specific for the biosynthetic pathway of riboflavin (3). However, metabolic flux to riboflavin is also determined by numerous nonspecific reactions providing sufficient amounts of the two starting metabolites. This is indicated by the observation that riboflavin production can be enhanced by supplementation of the culture medium with different riboflavin precursors, e.g., ribitol (24), purines (18), and glycine (7, 12, 18). The yield-enhancing effect of glycine, which is an important precursor during de novo purine biosynthesis, has also been described for riboflavin production by Candida flareri (13). Quantitative incorporation of glycine into the riboflavin molecule was already reported by Plaut in 1954 (29). It is mediated by glycine amide ribonucleotide synthetase, which catalyzes the formation of an amido linkage between glycine and 5-phosphoribosylamine, consuming one molecule of ATP. Against this background, the present study aimed at the improvement of fungal glycine biosynthesis, leading to a better precursor supply of purine and subsequent riboflavin formation.
Three main routes of glycine biosynthesis have been described so far. Among plants, animals, and microorganisms the most widespread glycine biosynthetic enzyme is serine hydroxymethyltransferase (SHMT; EC 2.1.2.1), which catalyzes the tetrahydrofolate-dependent cleavage of serine into glycine and 5,10-methylene-tetrahydrofolate. This reaction is the only one producing glycine in Escherichia coli (40). Accordingly, inactivation of the glyA gene, encoding SHMT, leads to glycine auxotrophy in E. coli. In Saccharomyces cerevisiae, this pathway of glycine biosynthesis is termed the glycolytic pathway, as it starts from the glycolytic intermediate 3-phosphoglycerate. It is contrasted with the gluconeogenic pathway, which starts from glyoxylate, a product of the anaplerotic glyoxylate cycle. Synthesis of alanine glyoxylate aminotransferase (EC 2.6.1.44), the key enzyme of the latter pathway, is subject to glucose repression, so that the gluconeogenic pathway is the major source of glycine only during growth of S. cerevisiae on nonfermentable carbon sources such as ethanol and acetate (43). Recently, this model of glycine biosynthesis in yeast has been expanded by a threonine aldolase (EC 4.1.2.5), which provides a significant amount of glycine during growth of S. cerevisiae on glucose (22, 25). Growth studies even suggested that threonine aldolase is the major source of glycine under these conditions, since disruption of the corresponding gene (GLY1) led to a strongly reduced growth rate in the absence of glycine. However, disruption of both SHM genes, encoding the two SHMT isoenzymes in yeast, did not significantly affect the growth rate (23).
The present work aimed at improving riboflavin production in A. gossypii by enhancing the biosynthesis of the riboflavin precursor glycine. Cloning, disruption, and overexpression of the A. gossypii GLY1 gene, encoding a threonine aldolase, are reported. Evidence is presented that overexpression of this glycine biosynthetic gene can lead to an increase in riboflavin production.
MATERIALS AND METHODS
Chemicals.
Lysing enzymes from Trichoderma harzianum were purchased from Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany. [3-14C]serine was supplied by Amersham Buchler GmbH and Co. KG, Germany.
Strains and growth conditions.
A. gossypii ATCC 10895 was used as an A. gossypii wild-type strain. S. cerevisiae YM13 (ATCC 201877; MATα ura3-1 trp1-1 ade2-1 his3-11,15 leu2-3,112 can1-100 shm1::HIS3 shm2::LEU2 gly1::URA3) (23) and its uracil auxotrophic derivative YM13F (see below) were used for complementation experiments with the A. gossypii genomic library. E. coli DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (11) was the recipient for plasmid amplification.
A. gossypii strains were maintained on solid complex medium (1% yeast extract, 1% glucose). For selection and selective growth of transformants, Geneticin was added to a final concentration of 400 μg/ml. For the determination of enzyme activities and riboflavin production, liquid cultures on the same medium (plus amino acid supplementation if required) were incubated at 28°C in 500-ml shake flasks, each containing 100 ml of medium, on a rotary shaker (Pilot-Shake system; Kühner AG) at 110 rpm. Growth experiments were performed on a minimal medium containing (per liter) 2.5 g of glucose, 1.5 g of NH4Cl, 0.5 g of asparagine, 0.2 g of NaCl, 0.4 g of MgSO4 · 7H2O, 50 mg of MnSO4 · H2O, 40 mg of CaCl2 · 2H2O, 0.1 g of myo-inositol, 0.25 g of nicotinic acid amide, and 0.2 g of yeast extract. After sterilization, 2 g of KH2PO4 per liter (pH 6.7) was added. Yeast strains were grown routinely on YPD medium (1% yeast extract, 2% peptone, 2% glucose) or on SD medium (0.67% Bacto Yeast Nitrogen Base and 2% glucose, with 20 mg each of uracil, adenine, histidine, and tryptophan per liter, 30 mg of leucine per liter, and 10 mM glycine or without selected supplements, depending on the selection required). E. coli strains containing plasmids were grown in 2× TY medium supplemented with 50 μg of ampicillin per ml (33).
5-FOA selection.
As described by Boeke et al. (5) 108 cells of S. cerevisiae YM13 precultivated on YPD medium were plated on 5-fluoroorotic acid (5-FOA) medium (5) containing 10 mM glycine. After 3 days of incubation at 30°C, about 350 spontaneous 5-FOA-resistant mutants were isolated. One of them was designated YM13F and subsequently used for screening the genomic library.
General recombinant DNA techniques.
Transformation of E. coli, plasmid preparation, restriction mapping, DNA ligations, Southern blotting, and other DNA manipulations were done by standard techniques (33). Plasmids were reisolated from S. cerevisiae by the method of Nasmyth and Reed (26). Transformation of S. cerevisiae was done by the method of Dohmen et al. (8).
PCR amplification.
PCR amplification was performed with Taq DNA polymerase or an enzyme mix containing Taq DNA polymerase and Pwo DNA polymerase (both obtained from Boehringer Mannheim GmbH, Mannheim, Germany) for fragments larger than 5 kb. Reactions were carried out as specified by the manufacturer. Each cycle included 1 min of denaturation (94°C), 1 min of annealing (65°C), and 2 min per 3 kb of fragment size of chain elongation (Taq polymerase, 72°C; Taq-Pwo polymerase, 68°C). After completion of the last cycle, the PCR products were purified by agarose gel electrophoresis and used for cloning.
Isolation of genomic DNA from A. gossypii.
High-molecular-weight genomic DNA was prepared from shake flask cultures grown for 16 h on liquid complex medium. About 4 to 10 g of the mycelium was harvested by filtration, washed twice with distilled water, suspended in 30 ml of SCE buffer (1 M sorbitol, 0.1 M sodium citrate, 60 mM EDTA) containing 30 μl of β-mercaptoethanol and 50 mg of lysing enzymes, and incubated at 37°C for 1 h. Subsequent addition of 3 ml each of 0.5 mM EDTA (pH 8.0) and 10% (wt/vol) sodium dodecyl sulfate (SDS) led to lysis of the protoplasts after about 5 min of incubation at room temperature. After proteinase K treatment (1 h at 50°C), 1 volume of 5 M ammonium acetate was added and the mixture was centrifuged (15 min at 10,000 rpm [Beckman JA-14 rotor]). The genomic DNA was precipitated from the supernatant with ethanol, treated with RNase A, extracted repeatedly with phenol-chloroform (1:1, vol/vol), and reprecipitated with ethanol before being resuspended in TE buffer.
Construction of the genomic library.
The A. gossypii GLY1 gene was isolated from a genomic library constructed in the S. cerevisiae-E. coli shuttle vector YEp352 (15). Chromosomal DNA isolated from A. gossypii ATCC 10895 was partly digested with Sau3A. Fragments of >8 kb were separated by sucrose density gradient centrifugation (33) and ligated into BamHI-restricted YEp352. The resulting plasmids were introduced into E. coli DH5α to construct the genomic library.
Transformation of A. gossypii.
Transformation of A. gossypii was done by an electroporation method developed by Revuelta (32a). Fungal mycelium was grown overnight in liquid complex medium, harvested by filtration, washed with 50 mM phosphate buffer containing 25 mM dithiothreitol (DTT), incubated in the same solution for 30 min at 30°C, and collected by filtration. The cells were subsequently washed with 10 mM Tris-HCl (pH 7.5) containing 270 mM sucrose and 1 mM MgCl2 and resuspended in 1 ml of the same solution. Aliquots of the suspension, together with the transforming DNA, were dispensed into 2-mm electrocuvettes and pulsed with a Gene Pulser (Bio-Rad, Munich, Germany) set at 1.5 kV/cm, 100 Ω, and 25 μF. After electroporation, the mycelium was plated on complex medium and incubated at 30°C for 6 h to allow regeneration of the cells. To apply selection pressure, 5 ml of top agar (complex medium plus 1.8 mg of Geneticin per ml) was subsequently added.
Clonal purification of A. gossypii transformants was carried out by the selection of Geneticin-resistant spores as described by Steiner et al. (42).
Sequence determination and alignment.
The GLY1 nucleotide sequence was determined on both strands by the dideoxynucleotide termination method of Sanger et al. (34). Homology searches were performed by using the sequence similarity search program BLAST (28). The CLUSTAL method (14) was used for multiple alignment of sequences.
Gene disruption.
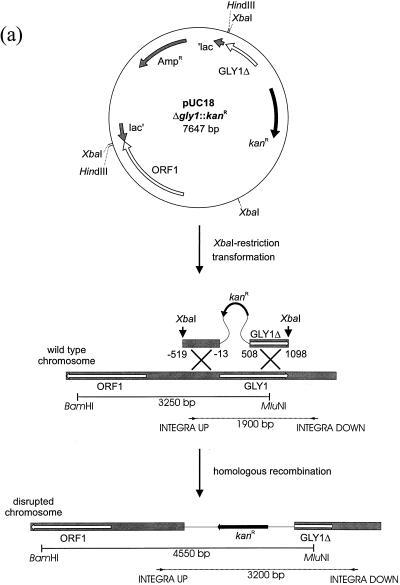
GLY1 knockout mutants were constructed by replacing the 5′-terminal 500 bp of the gene by a Geneticin resistance cassette. The 3.7-kb HindIII fragment from the subclone YEp352 GB 26-9-6, which carried the GLY1 gene (except the 3′-terminal 48 bp) and 2.7 kb upstream from the gene, was inserted into pUC18 (44) at the HindIII site. The resulting plasmid, pUC18GLY1, was used as a template in a PCR with the primers GLY1UP (5′-CCTGGGCTCGAGACTTGTAGTCAACTGTAGCAG-3′) and GLY1DOWN (5′-CTTGTTCTCGAGAAGGCTTGGTGTATGGAGAAC-3′), resulting in amplification of the whole plasmid except the 5′-terminal 500 bp of the GLY1 gene. At the underlined position of each primer, an XhoI site was introduced so that the PCR product could be religated after XhoI cleavage (pUC18GLY1Δ500). A 1.8-kb SalI fragment of plasmid pAG231 (42), which carried the E. coli kanamycin resistance gene under the control of the A. gossypii TEF promoter and terminator (TEF is translation elongation factor 1α), was subsequently introduced into the XhoI site of pUC18GLY1Δ500. The kanamycin resistance gene can be used as a dominant marker conferring resistance to Geneticin in eukaryotes (16). The resulting plasmid pUC18Δgly1::kanr (see Fig. 3a) was digested with XbaI and used to transform A. gossypii to Geneticin resistance. Since pUC18 does not contain an autonomous replicating sequence for A. gossypii and homologous recombination is described as the main mechanism for DNA integration in A. gossypii (12), disruption of GLY1 was expected in all Geneticin-resistant transformants.
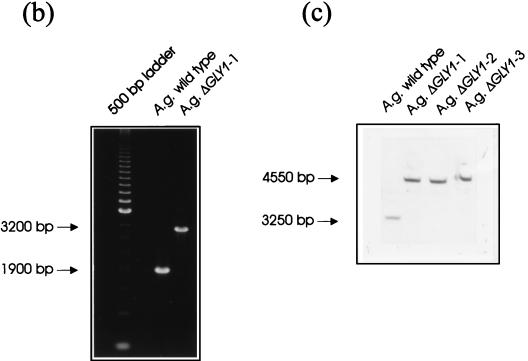
FIG. 3.
Disruption of the GLY1 gene. (a) Schematic representation of the disruption construct pUC18Δgly1::kanr and the single-step gene disruption procedure. The intact genomic GLY1 gene was replaced by an incomplete copy together with the Kanr gene by means of a double crossover. On the XbaI fragment, DNA regions identical to the chromosome are shaded. The numbers refer to the start of the GLY1 gene. (b and c) Disruption of the gene was verified by PCR (b) and Southern hybridization (c) in the knockout mutants ΔGLY1-1, ΔGLY1-2, and ΔGLY1-3. For Southern analysis, genomic DNA was digested simultaneously with BamHI and MluNI, as indicated, and a 941-bp SacI-MluNI fragment was used as a probe (Fig. 1). For PCR, the primers INTEGRA-UP (5′-AGGAGCGTTACGTCCAACGGTCGTTCTGTG-3′) and INTEGRA-DOWN (AATGGTAGAGCTACTAGCCCTCCGCAATA-3′), which were derived from sequences upstream and downstream, respectively, of the GLY1 gene, were used.
Construction of the GLY1 overexpression plasmid pAG203GLY1.
PCR amplification of the GLY1 gene was carried out to permit cloning into the expression plasmid pAG203 (19). It uses the TEF promoter for a strong and constitutive expression. The primers GLY1-START (5′-AAACCCAGCATGCAACAGGATATGGAACTACCAGAG-3′) and GLY1-STOP (5′-CATCGAGTTAACTTAATACTTGTAGGTCTTGAT-3′) were designed on the basis of the nucleotide sequence of the GLY1 gene. To facilitate cloning, additional restriction sites (SphI and HpaI, respectively; exchanged nucleotides are underlined) were introduced into each primer. Introduction of the SphI site into primer GLY1-START was necessary to enable the cloning in frame to the ATG initiation codon available on the plasmid pAG203 and led to a modification of the second codon from AAT to CAA. This corresponds to the conserved amino acid exchange Asn→Gln. The amplified PCR product was digested with SphI and HpaI, purified by agarose gel electrophoresis, and inserted into SphI-ScaI-linearized pAG203. The resulting plasmid was designated pAG203GLY1.
Determination of total riboflavin and mycelial dry weight.
For the determination of total riboflavin, cells were disrupted by the addition of 100 μl of lysing enzymes to 1 ml of sample taken from liquid cultures. After incubation at 30°C for 1 h, 900 μl of distilled water was added. The resulting homogenate was centrifuged (5 min at 13,000 rpm [Eppendorf F45-18-11 standard rotor]), filtered (pore size, 0.45 μm; Millipore, Eschborn, Germany) and analyzed for riboflavin by high-pressure liquid chromatography as described by Schmidt et al. (39).
For the determination of mycelial dry weight, samples were taken from liquid cultures and filtered through paper filters (diameter, 5 cm). The collected mycelium was dried overnight at 110°C and weighed.
Cell extraction.
A. gossypii mycelium was harvested by filtration, rinsed with distilled water, and resuspended in 50 mM HEPES–NaOH buffer (pH 7.0)–1 mM DTT–20 μM pyridoxal phosphate at a ratio of 2 to 5 ml/g (wet weight) of mycelium. The cells were disrupted in a French press (Aminco, Silver Spring, Md.) at 20,000 lb/in2, and the resulting homogenate was centrifuged at 20,000 × g for 20 min. The supernatant was desalted on a Sephadex G-25 column (NAP-10; Pharmacia Biotech, Uppsala, Sweden) for use in enzyme assays and is subsequently referred to as the crude extract. All procedures were carried out at 4°C.
S. cerevisiae cell extracts were prepared as described previously (25).
Enzyme assays.
Threonine aldolase activity was determined by quantification of the glycine formed in a high-pressure liquid chromatography system as previously described (25). In a final volume of 250 μl, a typical assay mixture contained 80 mM threonine, 100 mM HEPES–NaOH (pH 7.0), 30 μM pyridoxal phosphate, and 75 μl of crude extract. SHMT activity was measured by a modification of the method of Geller and Kotb (9). [3-14C]serine incorporation into tetrahydrofolate-bound products was measured. A typical assay mixture contained 50 mM Tris-HCl (pH 8.0), 0.2 mM serine, 0.024 mM [3-14C]serine, 2 mM tetrahydrofolate, 0.25 mM pyridoxal phosphate, 2.5 mM EDTA (pH 8.0), 3 mM DTT, and 0.25 mg of protein (from crude extracts). The labeled 5,10-methylene tetrahydrofolate was separated from unreacted serine by streaking 20-μl aliquots of the assay mixture onto DEAE-cellulose filter circles (diameter, 2.3 cm; Whatman DE 81) and washing the filters in distilled water to remove unreacted labeled serine.
Protein concentrations were determined spectrophotometrically by the method of Bradford (6) at 595 nm with the Serva Blue G dye binding reagent. Bovine serum albumin was used as a standard.
Nucleotide sequence accession number.
Sequence data have been submitted to the EMBL database and are listed under accession no. AJ005442.
RESULTS
Cloning and sequencing of the A. gossypii GLY1 gene.
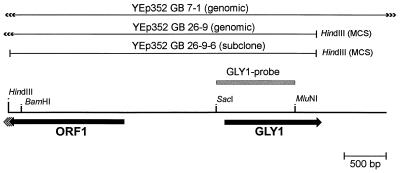
To isolate the GLY1 gene from A. gossypii, heterologous complementation of the glycine auxotrophic yeast strain YM13 (shm1::HIS3 shm2::LEU2 gly1::URA3) with a plasmid library of genomic A. gossypii DNA in YEp352 (Ampr URA3) was performed. However, no transformants could be isolated by direct selection for glycine prototrophy. Therefore, uracil auxotrophic mutants of the yeast strain YM13 were isolated by selection for 5-FOA resistance (for details, see Materials and Methods) to allow preselection of plasmid-containing clones. One of the spontaneous 5-FOA-resistant mutants (YM13F) was subsequently used for a second transformation with the genomic library. Preselection for uracil prototrophy led to the isolation of 70,000 transformants, which were replica plated on minimal medium without glycine. From this medium, 25 glycine-prototrophic clones were isolated. Curing from the plasmid by two subsequent cultivations on complex medium as well as retransformation with the isolated plasmids showed that complementation was linked to uracil prototrophy, which indicated that it was due to a cloned fragment. Restriction analysis of the isolated plasmids showed that they carried inserts of 10 to 12 kb, all of them containing the same gene. Subcloning proceeding from the genomic clone YEp352 GB 26-9 revealed complementation of a 3.7-kb HindIII fragment (YEp352 GB 26-9-6), which was subsequently sequenced. It carried two incomplete open reading frames (ORFs), 1,326 bp of ORF1 and 1,098 bp of ORF2 (Fig. 1). The deduced amino acid sequence of ORF2 showed 88% similarity to threonine aldolase (GLY1) from S. cerevisiae. The predicted amino acid sequence of ORF1 displayed 54% similarity to YEL043w, a hypothetical 106.1-kDa protein of unknown function encoded in the GLY1-GDA1 intergenic region of S. cerevisiae. The striking similarity of ORF2 to the S. cerevisiae GLY1 gene, together with the detection of 25 mU of threonine aldolase specific activity mg of protein−1 in the yeast transformants, in contrast to <0.1 mU mg of protein−1 in the control (25), led to the conclusion that a GLY1 homologue had been isolated from A. gossypii. The full sequence of the A. gossypii GLY1 gene was subsequently determined by partial sequencing of the genomic clone YEp352 GB 7-1 (Fig. 1). The A. gossypii GLY1 gene has an ORF of 1,146 bp encoding a predicted protein of 382 amino acids.
FIG. 1.
Schematic representation of the sequenced genomic DNA insert. The positions of the two ORFs GLY1 and ORF1 are indicated by black arrows. The solid bars above the map represent the sequences present in the relevant genomic clones YEp352 GB 26-9 and YEp352 GB 7-1 and in the subclone YEp352 GB 26-9-6. The 3′-terminal HindIII sites in YEp352 GB 26-9 and YEp352 GB 26-9-6 originate from the multiple-cloning site (MCS) of the cloning vector YEp352. A 941-bp SacI-MluNI fragment that was used as a probe for Southern hybridization is indicated by the shaded bar.
Alignment of A. gossypii threonine aldolase with other proteins.
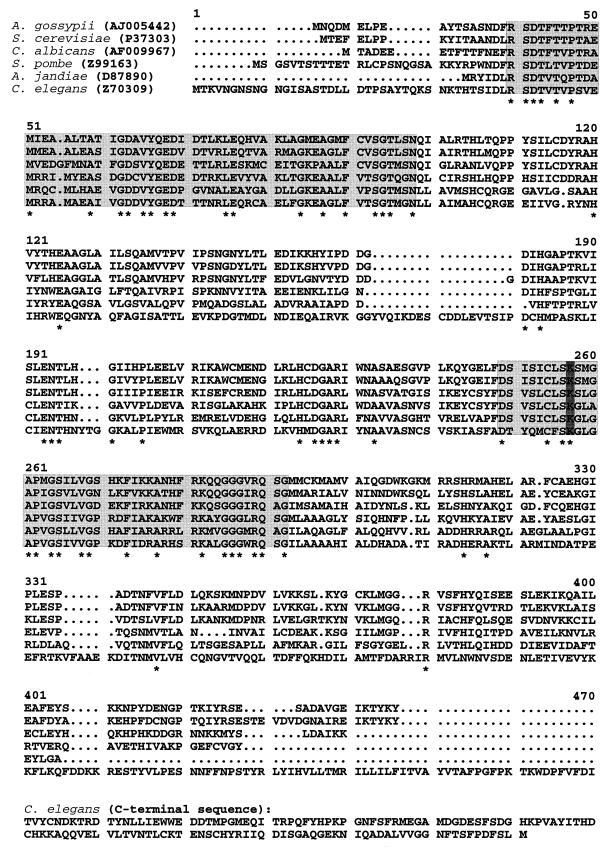
A multiple amino acid sequence alignment that includes threonine aldolase from A. gossypii and other proteins whose sequences have previously been reported is shown in Fig. 2. Apart from yeast threonine aldolase, with 88% similarity, the putative A. gossypii protein showed the highest similarity to two other fungal proteins, namely, threonine aldolase from Candida albicans (76%) and a hypothetical protein from Schizosaccharomyces pombe (62%). We also detected high similarity to a protein from the bacterium Aeromonas jandiae (62%), which was recently identified as an l-allo-threonine aldolase (21), as well as to a hypothetical protein from the nematode Caenorhabditis elegans (54%). Apart from the C. elegans protein, all six proteins are of similar length. Whereas the C and N termini of the proteins are only weakly conserved, the central part contains two highly conserved domains. One of them contains Lys199 of the l-allo-threonine aldolase of Aeromonas jandiae, which was recently identified as the pyridoxal 5′-phosphate binding lysine residue essential for the aldol cleavage reaction by site-directed mutagenesis (21). This residue is conserved among all six proteins. Threonine aldolase from Aeromonas jandiae is by far the smallest of the six proteins, lacking the weakly conserved C- and N-terminal regions. This indicates that these residues might not be important for the enzymatic activity of the protein and explains why the formation of a truncated protein from plasmids YEp352 GB 26-9 and YEp352 GB 26-9-6 led to complementation of glycine auxotrophy in S. cerevisiae YM13.
FIG. 2.
Multiple alignment of putative proteins with high similarity to threonine aldolase from A. gossypii: threonine aldolase from S. cerevisiae, GLY1 from Candida albicans, hypothetical protein from Schizosaccharomyces pombe, l-allo-threonine aldolase from Aeromonas jandiae, and GLY1-like protein from Caenorhabditis elegans. EMBL database accession numbers are shown in parentheses. Residues conserved among all sequences are denoted by asterisks. Dots represent gaps introduced into a sequence for alignment. The numbers above the aligned sequences refer to the alignment positions. K199 of the l-allo-threonine aldolase from A. jandiae, which was recently identified as the pyridoxal 5′-phosphate binding lysine residue (21), is indicated by a dark grey box. It is located within one of two highly conserved regions, highlighted in light grey. A C-terminal extension of the C. elegans protein with no homology to the other five proteins is shown below.
Characterization of GLY1 knockout mutants.
The chromosomal GLY1 gene was inactivated by gene disruption. A linearized DNA fragment containing the Geneticin resistance cassette flanked by the 3′ end of the GLY1 gene on one side and by the upstream region of GLY1 on the other side was used to transform A. gossypii ATCC 10895 to Geneticin resistance (Fig. 3a). Gene disruption was confirmed by PCR (Fig. 3b). As expected, a 1.3-kb shift of the signal was observed. Beyond that, Southern hybridization (Fig. 3c) demonstrated that replacement with the resistance marker had occurred at the GLY1 locus.
Inactivation of GLY1 resulted in a complete loss of detectable threonine aldolase activity (<0.1 mU mg of protein−1). This indicated that GLY1 is the only gene encoding a threonine aldolase in A. gossypii. However, SHMT specific activity still reached the wild-type level (2.5 to 3 mU mg of protein−1), which demonstrated that this reaction is catalyzed by a different enzyme in A. gossypii. Disruption of GLY1 did not lead to a requirement for glycine. Growth and riboflavin production on minimal medium without supplementary glycine turned out to be unchanged in the GLY1 disruption mutants.
Overexpression of GLY1.
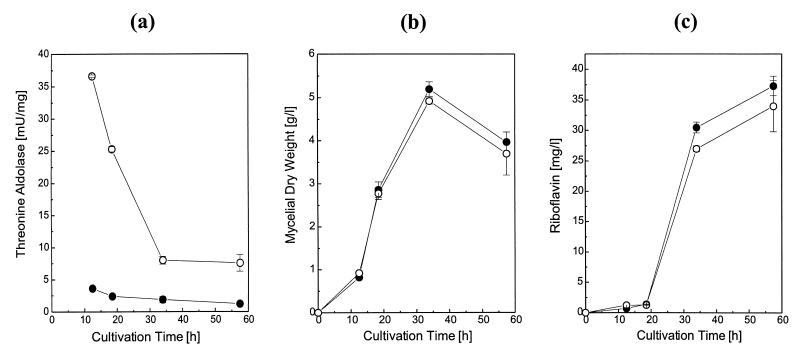
Although disruption of GLY1 had demonstrated that threonine aldolase is probably not essential for glycine biosynthesis in the A. gossypii wild-type strain, overexpression of the gene seemed to be promising with regard to an improvement of glycine biosynthesis. Overexpression of the GLY1 gene was achieved by introducing the expression plasmid pAG203GLY1 (see Materials and Methods) into A. gossypii ATCC 10895. Transformants showed a ca. 10-fold increase in threonine aldolase specific activity over the whole time course of cultivation (Fig. 4a). The growth remained unchanged (Fig. 4b). However, it was not possible to obtain a constant level of threonine aldolase specific activity throughout cultivation. As in the control strain, which was transformed only with the control plasmid pAG203, enzyme activity decreased drastically from 12 to 34 h of cultivation. To ensure that this decrease was not due to a decrease of promoter activity, β-galactosidase was expressed under the control of the TEF promoter by using the plasmid pAG110 (41). In this case, 70% of the starting activity was still detectable at the end of the cultivation time (data not shown).
FIG. 4.
Time courses of threonine aldolase specific activity (a), growth (b), and riboflavin production (c) in A. gossypii pAG203 (•) and A. gossypii pAG203GLY1 (○). Experimental details are described in Materials and Methods.
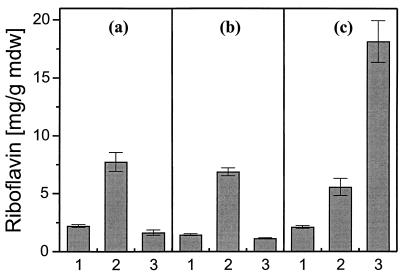
As shown in Fig. 4c, overexpression of threonine aldolase alone did not lead to an improved riboflavin production in this experiment. Therefore, riboflavin production was subsequently investigated on complex medium supplemented with glycine or threonine. Figure 5 demonstrates that supplementation with 80 mM glycine led to an about threefold increase in riboflavin production in the wild-type strain as well as in the two plasmid-containing strains A. gossypii pAG203 and A. gossypii pAG203GLY1. However, if the culture medium was supplemented with 50 mM threonine, an eightfold increase in riboflavin production was detectable in A. gossypii pAG203GLY1 whereas threonine did not affect riboflavin production in the two control strains. When the amino acid concentrations of the culture medium were determined after cultivation, it became apparent that in strain A. gossypii pAG203GLY1, the threonine concentration had dropped from 50 ± 5 to 6 ± 0.5 mM and simultaneously the glycine concentration had increased from 2 ± 0.2 to 41 ± 4 mM. In the control strain A. gossypii pAG203, the decrease in threonine concentration (from 50 ± 5 to 32 ± 3 mM) and the increase in glycine concentration (from 2 ± 0.2 to 6 ± 0.5 mM) were much smaller. Thus, the yield-enhancing effect of threonine aldolase overexpression plus threonine supplementation must be attributed to an increased uptake of extracellular threonine and its intracellular conversion into glycine. The weaker effect of glycine supplementation could be due to a lower uptake: this amino acid concentration decreased only slightly from 82 ± 2 to 79 ± 1 mM.
FIG. 5.
Riboflavin production per mycelial dry weight (mdw) of A. gossypii wild type (a), pAG203 (b), and pAG203GLY1 (c) on liquid complex medium without amino acid supplementation (lanes 1), with 80 mM glycine (lanes 2), and with 50 mM threonine (lanes 3). Riboflavin was determined after 3 days of cultivation. All values are means of three independent experiments.
DISCUSSION
Threonine aldolase is thought to be the major source of glycine in the yeast S. cerevisiae (23, 25). Therefore, it was a promising candidate for the improvement of glycine biosynthesis and subsequent riboflavin production in the closely related filamentous fungus A. gossypii.
The ORF isolated by heterologous complementation of the glycine-auxotrophic S. cerevisiae strain YM13 was identified as encoding threonine aldolase of A. gossypii by detection of the enzymatic activity of the corresponding protein in crude extracts as well as by an amino acid alignment with threonine aldolases from S. cerevisiae, C. albicans, and Aeromonas jandiae. The remarkably high homology (88%) between the deduced amino acid sequences of the two threonine aldolases from A. gossypii and S. cerevisiae underlines the close relationship between the two fungi, which was concluded from an alignment of ITS1 and ITS2 sequences by Prillinger et al. (31) and led to a new definition of the family Saccharomycetaceae that included both unicellular saprophytic yeasts and dimorphic or filamentous parasitic fungi. Furthermore, colocation of the GLY1 gene and a YEL043w homologue in A. gossypii is another good example of conservation of gene order in A. gossypii and S. cerevisiae. Only recently was the A. gossypii THR4 gene located in a four-gene cluster that is conserved between A. gossypii and S. cerevisiae (1).
Surprisingly, heterologous complementation of S. cerevisiae YM13 did not lead to the isolation of a gene encoding SHMT. This is probably due to the particular importance of threonine aldolase for glycine biosynthesis in yeast. We had already reported in a previous paper (25) that growth of S. cerevisiae YM13 can be completely restored by transformation with a plasmid containing the GLY1 gene. On the other hand, McNeil et al. (23) demonstrated that both gly1 shm1 and gly1 shm2 double mutants are severely impaired in growth. Taking into consideration that heterologous expression of an SHM gene from A. gossypii in yeast is probably even weaker than that of the homologous gene, it becomes conceivable that this might not be sufficient to permit growth of the glycine-auxotrophic yeast strain.
Disruption of GLY1 in A. gossypii led to a complete loss of detectable threonine aldolase activity, whereas SHMT still reached the wild-type level. This indicated that the aldol cleavage reaction of serine into glycine and 5,10-methylene tetrahydrofolate is catalyzed by a separate enzyme in A. gossypii. This is in agreement with the situation in yeast (23) but contrasts with that in E. coli (36) and rat liver (35), where threonine aldolase activity was demonstrated for purified SHMT. In S. cerevisiae, disruption of the GLY1 gene leads to a strongly reduced growth rate in the absence of glycine whereas the disruption of both SHM genes does not (23). These studies suggested that threonine aldolase is the major source of glycine in yeast. In contrast, disruption of GLY1 did not lead to a requirement for glycine in A. gossypii. Additionally, a decrease in riboflavin production, indicating a reduced glycine supply, was not detectable. That means that threonine aldolase plays only a minor role during glycine biosynthesis of A. gossypii or that its function can be fully compensated by other glycine biosynthetic pathways.
A ca. 10-fold increase in threonine aldolase specific activity was reached by overexpression of GLY1 under the control of the TEF promoter and terminator. However, there was still a drastic decrease in activity during the course of cultivation. Although TEF promoters from different sources have been shown to be comparatively strong (2, 37), activity was demonstrated to depend on the age of the tissue for the TEF promoter from Lycopersicon esculentum (30). Such a promoter-dependent kind of regulation can be excluded in our case, because β-galactosidase, expressed under the control of the TEF promoter, did not show a similar time course. Consequently, the decrease in threonine aldolase specific activity must be attributed to a promoter-independent type of regulation on the mRNA or protein level. Apart from these in vitro data, overexpression of threonine aldolase was also demonstrated in vivo by the almost quantitative conversion of extracellular threonine into glycine in the overexpression strain A. gossypii pAG203GLY1.
Overexpression of GLY1, together with threonine supplementation of the culture medium, led to a strong enhancement of riboflavin production, which could not be achieved by glycine supplementation alone. From the almost quantitative conversion of the extracellular threonine into glycine, we conclude that this enhancement must be due to an increased uptake of extracellular threonine and its subsequent intracellular conversion into glycine. Although most of the glycine produced was subsequently excreted into the medium, this obviously improved the intracellular availability of glycine and led to an enhanced riboflavin production. The finding that improvement of riboflavin production requires both overexpression of threonine aldolase and threonine supplementation leads to the conclusion that threonine biosynthesis must be the limiting factor under these conditions. We have evidence that feedback inhibition of aspartokinase, an important regulatory enzyme during threonine biosynthesis in S. cerevisiae (32) and Corynebacterium glutamicum (17), is responsible for this limitation. Elimination of this feedback inhibition could therefore be the key to further improvement of riboflavin production without any amino acid supplementation of the culture medium.
ACKNOWLEDGMENTS
This work was supported in part by the “Fonds der chemischen Industrie.”
We thank Andrew L. Bognar for kindly providing us with S. cerevisiae YM13. Thanks are also due to H. Seulberger, BASF AG (Ludwigshafen, Germany), for managing the sequencing of the A. gossypii GLY1 gene and to P. Philippsen for supplying plasmids pAG231, pAG203, and pAG110. We are also grateful to J. Heinisch for helpful suggestions about the molecular biology of yeast.
REFERENCES
- 1.Altmann-Jöhl R, Philippsen P. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol Gen Genet. 1996;250:69–80. doi: 10.1007/BF02191826. [DOI] [PubMed] [Google Scholar]
- 2.Axelos M, Bardet C, Liboz T, Van Thai A L, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana elongation factor EF-1α: molecular cloning, characterization and expression. Mol Gen Genet. 1989;219:106–112. doi: 10.1007/BF00261164. [DOI] [PubMed] [Google Scholar]
- 3.Bacher A. Biosynthesis of flavins. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 215–249. [Google Scholar]
- 4.Bigelis R. Industrial products of biotechnology: application of gene technology. In: Rehm H J, Reed G, editors. Biotechnology. 7b. Weinheim, Germany: VCH; 1989. p. 243. [Google Scholar]
- 5.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Demain A L. Riboflavin oversynthesis. Annu Rev Microbiol. 1972;26:369–388. doi: 10.1146/annurev.mi.26.100172.002101. [DOI] [PubMed] [Google Scholar]
- 8.Dohmen R J, Strasser A W M, Höner C B, Hollenberg C P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 9.Geller A M, Kotb M Y. A binding assay for serine hydroxymethyltransferase. Anal Biochem. 1989;180:120–125. doi: 10.1016/0003-2697(89)90098-5. [DOI] [PubMed] [Google Scholar]
- 10.Guillermond P. Recherches sur quelques Ascomycetes inferieurs isoles de la stigmatomycose des graines de cotonnier. Essai sur la phylogenie des Ascomycetes. Rev Gen Bot. 1928;40:328–342. , 397–414, 474–485, 555–574, 606–624, 690–704. [Google Scholar]
- 11.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning—a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 12.Hanson A M. Microbial production of pigments and vitamins. In: Peppler H J, editor. Microbial technology. New York, N.Y: Reinhold; 1967. pp. 222–250. [Google Scholar]
- 13.Heefner D L, Weaver C A, Yarus M J, Burdzinski L A, Gyure D C, Foster E W. Riboflavin producing strains of microorganisms, method for selecting, and method for fermentation. 1988. Patent cooperation treaty WO 88/09822. [Google Scholar]
- 14.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 15.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 16.Jiminez A, Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces. Nature. 1980;287:869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- 17.Kalinowski J, Cremer J, Bachmann B, Eggeling L, Sahm H, Pühler A. Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol Microbiol. 1991;5:1197–1204. doi: 10.1111/j.1365-2958.1991.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan C, Demain A L. Nutritional studies on riboflavin overproduction by Ashbya gossypii. In: Aheaon D G, editor. Recent trends in yeast research. Atlanta: Georgia State University; 1970. pp. 137–159. [Google Scholar]
- 19.Kurth, R., P. Philippsen, S. Steiner, and M. Wright. 1992. New promoter region. Patent cooperation treaty WO 92/00379.
- 20.Lago B D, Kaplan L. Vitamin fermentations: B2 and B12. Adv Biotechnol. 1981;3:241–246. [Google Scholar]
- 21.Liu J-Q, Dairi T, Kataoka M, Shimizu S, Yamada H. l-allo-Threonine aldolase from Aeromonas jandiae DK-39: gene cloning, nucleotide sequencing, and identification of the pyridoxal 5′-phosphate-binding lysine residue by site-directed mutagenesis. J Bacteriol. 1997;179:3555–3560. doi: 10.1128/jb.179.11.3555-3560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J-Q, Nagata S, Dairi T, Misono H, Shimizu S, Yamada H. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific l-threonine aldolase that catalyzes cleavage of l-allo-threonine and l-threonine to glycine—expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 23.McNeil J B, McIntosh E M, Taylor B V, Zhang F, Tang S, Bognar A L. Cloning and molecular characterization of three genes, including two genes encoding serine hydroxymethyltransferase, whose inactivation is required to render yeast auxotrophic for glycine. J Biol Chem. 1994;269:9155–9165. [PubMed] [Google Scholar]
- 24.Mehta S M, Mattoo A K, Modi V V. Ribitol and flavinogenesis in Eremothecium ashbyi. Biochem J. 1972;130:159–166. doi: 10.1042/bj1300159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monschau N, Stahmann K-P, Sahm H, McNeil J B, Bognar A L. Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol Lett. 1997;150:55–60. doi: 10.1111/j.1574-6968.1997.tb10349.x. [DOI] [PubMed] [Google Scholar]
- 26.Nasmyth K A, Reed S I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Özbas T, Kutsal T. Comparative study of riboflavin production from two microorganisms: Eremothecium ashbyi and Ashbya gossypii. Enzyme Microb Technol. 1986;8:593–596. [Google Scholar]
- 28.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaut G W E. Biosynthesis of riboflavin. II. Incorporation of C14-labelled compounds into ring A. J Biol Chem. 1954;211:111–116. [PubMed] [Google Scholar]
- 30.Pokalsky A R, Hiatt W R, Ridge N, Rasmussen R, Houck C M, Shewmaker C K. Structure and expression of elongation factor 1α in tomato. Nucleic Acids Res. 1989;17:4661–4673. doi: 10.1093/nar/17.12.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prillinger H, Schweigkofler W, Breitenbach M, Briza M, Staudacher F, Lopandic K, Molnar O, Weigang F, Ibi M, Ellinger A. Phytopathogenic filamentous (Ashbya, Eremothecium) and dimorphic fungi (Holleya, Nematospora) with needle-shaped ascospores as new members within the Saccharomycetaceae. Yeast. 1997;13:945–960. doi: 10.1002/(SICI)1097-0061(199708)13:10<945::AID-YEA150>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 32.Ramos C, Calderon I L. Overproduction of threonine by Saccharomyces cerevisiae mutants resistant to hydroxynorvaline. Appl Environ Microbiol. 1992;58:1677–1682. doi: 10.1128/aem.58.5.1677-1682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Revuelta, J. L. Personal communication.
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schirch L, Gross T. Serine transhydroxymethylase: identification as the threonine and allothreonine aldolase. J Biol Chem. 1968;243:5651–5655. [PubMed] [Google Scholar]
- 36.Schirch L, Hopkins S, Villar E, Angelaccio S. Serine hydroxymethyltransferase from E. coli: purification and properties. J Bacteriol. 1985;163:1–7. doi: 10.1128/jb.163.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirmaier F, Philippsen P. Identification of two genes coding for the translation elongation factor EF-1α of S. cerevisiae. EMBO J. 1984;3:3311–3315. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt G, Stahmann K-P, Sahm H. Inhibition of purified isocitrate lyase identified itaconate and oxalate as potential antimetabolites for the riboflavin overproducer Ashbya gossypii. Microbiology. 1996;142:411–417. doi: 10.1099/13500872-142-2-411. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt G, Stahmann K-P, Kaesler B, Sahm H. Correlation of isocitrate lyase activity and riboflavin formation in the riboflavin overproducer Ashbya gossypii. Microbiology. 1996;142:419–426. doi: 10.1099/13500872-142-2-419. [DOI] [PubMed] [Google Scholar]
- 40.Stauffer G V. Biosynthesis of serine, glycine, and one-carbon units. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 506–513. [Google Scholar]
- 41.Steiner S. Expressionsstudien im filamentösen Pilz Ashbya gossypii unter Verwendung von Signalsequenzen des TEF-Gens. Ph.D. thesis. Giessen, Germany: Justus Liebig University; 1991. [Google Scholar]
- 42.Steiner S, Wendland J, Wright M C, Philippsen P. Homologous recombination as the main mechanism for DNA integration and cause of rearrangements in the filamentous ascomycete Ashbya gossypii. Genetics. 1995;140:973–987. doi: 10.1093/genetics/140.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulane R, Ogur M. Genetic and physiological control of serine and glycine biosynthesis in Saccharomyces. J Bacteriol. 1972;109:34–43. doi: 10.1128/jb.109.1.34-43.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 45.Wickerham L J, Flickinger M H, Johnston R M. The production of riboflavin by Ashbya gossypii. Arch Biochem. 1946;9:95–98. [PubMed] [Google Scholar]
- 46.Wright M C, Philippsen P. Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene. 1991;109:99–105. doi: 10.1016/0378-1119(91)90593-z. [DOI] [PubMed] [Google Scholar]