Abstract
Our ability to understand the function of the nervous system is dependent upon defining the connections of its constituent neurons. Development of methods to define connections within neural networks has always been a growth industry in the neurosciences. Transneuronal spread of neurotropic viruses currently represents the best means of defining synaptic connections within neural networks. The method exploits the ability of viruses to invade neurons, replicate, and spread through the intimate synaptic connections that enable communication among neurons. Since the method was first introduced in the 1970s, it has benefited from an increased understanding of the virus life cycle, the function of viral genome, and the ability to manipulate the viral genome in support of directional spread of virus and the expression of transgenes. In this unit, we review these advances in viral tracing technology and the way in which they may be applied for functional dissection of neural networks.
BASIC PROTOCOL 1:
RETROGRADE INFECTION OF CNS CIRCUITS BY PERIPHERAL INJECTION OF VIRUS
BASIC PROTOCOL 2:
TRANSNEURONAL ANALYSIS BY INTRACEREBRAL INJECTION
ALTERNATE PROTOCOL 1:
TRANSNEURONAL ANALYSIS WITH MULTIPLE RECOMBINANT STRAINS
ALTERNATE PROTOCOL 2:
CONDITIONAL REPLICATION AND SPREAD OF PRV
ALTERNATE PROTOCOL 3:
CONDITIONAL REPORTERS OF PRV INFECTION AND SPREAD
ALTERNATE PROTOCOL 4:
REPORTERS OF NEURAL ACTIVITY IN POLYSYNAPTIC CIRCUITS
SUPPORT PROTOCOL 1:
GROWING AND TITERING A PRV VIRAL STOCK
SUPPORT PROTOCOL 2:
IMMUNOHISTOCHEMICAL PROCESSING AND DETECTION
SUPPORT PROTOCOL 3:
DUAL-IMMUNOFLUORESCENCE LOCALIZATION
Keywords: herpesvirus, rabies, transneuronal, transgene expression
INTRODUCTION
Viral transneuronal tracing exploits the propensity of neurotropic viruses to invade neurons and produce infectious progeny that cross synapses to infect other neurons within a neural network. This approach was a relatively new technology when this unit was initially published in 1999. Although the method had been championed as a means for defining neural network organization in the early 1980s by Dolivo and colleagues (Dolivo, 1980) there remained considerable debate regarding the specificity of virus transport between neurons. Notable in this regard were concerns regarding lytic and non-synaptic release of virus into the extracellular environment and the potential of such release to undermine the interpretation of circuit-related transport. These concerns were addressed in a number of laboratories through detailed studies of the viral life cycle and the function of gene products in the viral genome. The latter proved to be particularly important in the engineering of strains of virus that were reduced in virulence, were transported selectively in the retrograde direction through neural circuitry, and expressed unique reporters of infection (Koyuncu et al., 2021; Pomeranz et al., 2005). Evidence regarding the specificity of viral transport through neural circuits has continued to accumulate as application of the method has become a staple of circuit studies and the tools available have become increasingly powerful. In this revision, we update protocols for the production and use of pseudorabies virus (PRV) for circuit analysis and introduce new approaches that have improved the ability to define the functional architecture of the nervous system. It is important to note that other viruses have proven to be powerful probes of circuit organization. Of particular importance in this regard is the increased use of rabies virus to define both polysynaptic and monosynaptic circuit organization (Kelly and Strick, 2000; Lavin et al., 2019). Our focus upon PRV emerges from the extensive use of this virus to define synaptology in a variety of systems and the substantial advances in the technology resulting from the construction of recombinant viruses. However, it is useful to note that methods relevant to the generation and use of Herpes Simplex Virus (HSV), the human pathogen, are in large part the same as those detailed for PRV. Additionally, the H129 strain of HSV is the only known alphaherpesvirus that is transported largely, but not exclusively, in the anterograde direction (Zemanick et al., 1991; Barnett et al., 1995; Archin and Atherton, 2002; Archin et al., 2003; Kelly and Strick, 2003; Rinaman and Schwartz, 2004; Song et al., 2009; Lo and Anderson, 2011; McGovern et al., 2012,a,b; Pisano et al., 2021; Vaughan and Bartness, 2012) through circuits, and we illustrate new recombinants of H129 that express unique reporters (Wojaczynski et al., 2015).
The protocols introduced in this unit emphasize aspects of experimental design that have the greatest import for successful use of PRV and its recombinants in circuit definition. Descriptions of aspects of the viral life cycle that are crucial to successful experimental design are highlighted and illustrated along with reference to published studies. We also emphasize the valuable insights regarding the temporal progression of infection that can be obtained through careful analysis of the distribution of virally encoded proteins within infected neurons. Finally, we detail new experimental approaches for dissecting complex circuitry in dual-infection paradigms, through conditional replication of virus, and through conditional reporter expression in targeted populations of neurons. These protocols are complemented by inclusion of support protocols detailing methods for growing and storage of high-titer viral stocks. It is important to note that references to published literature in this unit are used selectively to illustrate important issues related to viral tracing with PRV, and should not be viewed as comprehensive either with respect to the field or experimental studies of individual systems. Several reviews provide excellent perspectives on both the use of the method and the mechanisms that contribute to viral replication and spread through the nervous system (Jasmin, 1995; Loewy, 1998; Boldogkoi et al., 2002a, b, 2004, 2009; Mettenleiter, 2003; Song et al., 2005; Geerling et al., 2006; Ekstrand et al., 2008; Card and Enquist, 2012; Koyuncu et al., 2021; Hogue et al., 2015).
The use of viruses as transneuronal tracers of neuronal circuitry is now a commonly accepted means of defining the synaptic organization of neural networks. The method exploits the propensity of neurotropic viruses to invade neurons and produce infectious progeny that cross synapses to infect synaptically defined neural networks (Fig. 1). In essence, virus replication and spread provides a self-amplifying marker of neural connectivity. Importantly, no class of neurons has been shown to be refractile to infection by PRV. Further, the virus spreads through the entire somatodendritic compartment and therefore has access to all afferent axons synapsing upon infected neurons. Thus, well-designed temporal analyses of viral transport provide a comprehensive view of both the identity and organization of neurons that compose neural networks. In this unit we present the basic methodology necessary for effective use of this experimental approach with PRV. Since viral invasiveness is determined by properties of the virus and the host neurons, there are no “generic” approaches for applying the method. Consequently, knowledge of the life cycle of the virus and the basic organization of the system of interest should be carefully considered in the design and execution of experiments, as well as the interpretation of data.
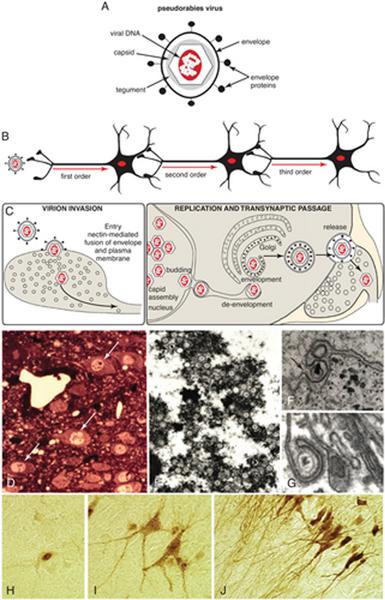
Figure 1.
(A) The structure of alpha herpesvirus virions and features characteristic of their neuroinvasiveness are illustrated. Viral DNA is sequestered within a capsid composed of virally encoded proteins. The capsid and a surrounding tegument of each virion are contained within a viral envelope acquired from the host cell. The envelope contains a second set of virally encoded proteins that are important for target cell recognition, attachment, and the receptor-mediated fusion event that leads to the release of the capsid into a permissive cell. (B) Recombinant strains of PRV-Bartha move selectively in the retrograde direction through neural circuits. (C) A model of virion assembly postulated for pseudorabies virus is illustrated. Assembly of virions is a multistep process that leads to assembly of mature virions in the cell soma. Virions traffic through the soma and dendrites of infected cells and are released in the vicinity of synaptic contacts. Adapted with permission from Card (1998). (D) Distinct nuclear inclusions (arrows) mark infected neurons and are clinically diagnostic for herpesvirus infection. (E) Transmission electron microscopy reveals that nuclear inclusions are sites of capsid assembly. (F) Capsids bud through the inner leaf of the nuclear envelope (arrow) in transit to the cytoplasm, where they acquire lipid bylayers from the trans-Golgi reticulum (G) or late endosomal compartment. (H-J) The distribution of viral antigens within infected cells provides an index of the stage of infection. At early stages (H), antigens are largely confined to the cell nucleus. As capsids migrate into the cell cytoplasm and acquire an envelope, viral antigens appear throughout the soma and proximal dendrites (I). Punctate staining on the cell soma and dendrites marks sites of transneuronal passage. At late stages of infection (J), viral antigens fill the entirety of the dendritic compartment.
PRV is a DNA virus from the same family, alpha herpesvirus, as the human pathogen Herpes Simplex Virus (HSV). The natural host of the virus is the pig, and it is the causal agent for Aujeszky’s disease (Kluge and Mare, 1974). PRV has a wide host range, infecting all mammals except higher primates (Fraser and Ramachandran, 1969; McCracken et al., 1973; Hagemoser et al., 1980; Hall et al., 1984, Laval and Enquist., 2020). PRV is not known to cause human infections in the laboratory. However, caution when using PRV is appropriate because there are reports of rare human infection in farm workers (Yang et al., 2019).
The use of PRV for viral tracing has benefited from mechanistic studies that have defined the role of virally encoded proteins in invasiveness, transynaptic passage, and virulence (Mettenleiter, 2000; Pomeranz et al., 2005; Mettenleiter et al., 2008; Koyuncu et al., 2021; Engel et al., 2015). These studies have identified attenuated strains useful for transneuronal analysis, and defined model systems that have proven to be of great value in defining the viral life cycle. This interdependent multidisciplinary approach has proven integral to establishing both the specificity and usefulness of PRV as a transneuronal tracer.
The complete genome sequences of virulent PRV (the Becker strain and the Kaplan strain), as well as the attenuated Bartha strain commonly used for circuit tracing have been published (Szpara et al., 2011). The structure of PRV particles (virions) and the life cycle that allows spread of virus through the nervous system are illustrated in Figure 1. There are four essential elements to the virion structure that contribute to its ability to (a) gain access to permissive cells, (b) be transported to the cell soma, (c) replicate to produce infectious progeny, and (d) spread through the parent cell to infect other neurons within a circuit via transneuronal spread. The ability to access permissive cells is directly dependent upon the interaction of virally encoded envelope proteins with extracellular matrix molecules and receptors on the surface of neurons. Binding of envelope proteins to heparin sulfate proteoglycans in the extracellular matrix restricts the spread of virions through the extracellular compartment and optimizes the ability of virions to find receptors on a permissive host. PRV uses the nectin receptor to invade neurons through receptor-mediated fusion of the virion envelope and the plasma membrane of the target cell (Campadelli-Fiume et al., 2000). Nectin is an adhesion molecule that is widely expressed in the nervous system, consistent with the ability of PRV to infect all classes of neurons (Mizoguchi et al., 2002; Takai et al., 2008). Fusion of the virion envelope and plasma membrane releases the capsid containing the viral genome within the host neuron. The viral capsid and associated tegument proteins are subsequently transported along microtubules via motor proteins to the cell soma, where the capsid disassembles to release the viral genome. The viral genome enters the cell nucleus through nuclear pores along with tegument proteins that initiate its expression. Expression of immediate early genes from the viral genome initiates a cascade of transcription that generates all of the proteins necessary for the assembly of new virions. Progeny capsids assembled in the cell nucleus acquire an envelope by budding through the inner leaf of the nuclear envelope. These particles gain access to the cell cytoplasm by a de-envelopement event involving fusion of the membrane acquired from the inner nuclear membrane with the outer nuclear membrane. The naked capsid then acquires two lipid bilayers from the trans-Golgi reticulum or a late endosomal compartment. This secondary envelopment process results in a mature enveloped virus residing within a transport vesicle. The outer membrane of the transport vesicle is essential for egress of virus particles from the parent neuron, and the viral envelope facilitates access to closely apposed synaptically connected neurons. Detailed descriptions of this process have been published (Card et al., 1993; Granzow et al., 1997; Enquist et al., 2002; Hogue et al., 2014).
The brain response to viral infection is integral to the specificity of viral transport through neural networks. This is due to the fact that, although the non-neuronal response to infection effectively isolates infected neurons, the ability of viral particles to pass through synapses defeats this effort to restrict spread. Consequently, naturally occurring wild-type strains of PRV spread bidirectionally through the nervous system via the intimate synaptic connections that compose neural circuits. It is important to note that the most effective PRV tracing strains are mutants that are markedly reduced in virulence but maintain their ability to pass retrogradely through circuits (Fig. 2). Reactive astrogliosis is fundamental to the ability of the non-neuronal response to isolate infected neurons. Astrocytes respond to infection by wrapping cells and their synaptic contacts with processes that separate them from adjacent cells and neuropil (Rinaman et al., 1993). Binding studies have demonstrated that astrocytic membrane also has a very high affinity for viral particles (Marchand and Schwab, 1987). Thus, viral particles that escape transneuronal passage are sequestered within surrounding astrocytes. Astrocytes have the capacity to replicate the PRV genome and package it within capsids. However, they are defective in secondary envelopment of capsids in the cell soma (Card et al., 1993). Without an envelope, capsids are incapable of leaving parent astrocytes and cannot contribute to the spread of progeny virus through the brain. The reactive response of glia cells to viral infection therefore supports circuit-related transport of virus instead of contributing to non-synaptic spread.
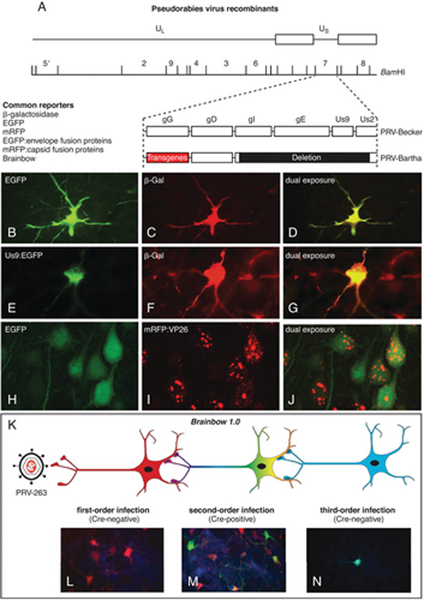
Figure 2.
(A) The organization of the PRV-Bartha genome and a common site of transgene insertion (gG) for recombinant viruses is illustrated. (B-J) Reporter expression from recombinant viruses useful in single- and dual-infection studies is illustrated. (C-D) In this case, either bacterial β-galactosidase (β-Gal) or jellyfish enhanced green fluorescent protein (EGFP) genes have been engineered into the gG locus, and the cell was infected by retrograde transport from two different projection targets from a collateralized axon. (E-G) Dual infection in the same paradigm resulting from replication of β-Gal and an EGFP Us9 fusion protein is illustrated. Note the differential concentration of the fusion protein in the Golgi complex of the cell cytoplasm. (H-I) Dual infection of cells with HSV-129 recombinant viruses expressing EGFP and a mRVP-capsid (VP26) fusion protein is illustrated. The fusion protein is differentially concentrated in the cell nucleus, whereas EGFP is a cytoplasmic marker, making dual-infected cells easily identified. (K) The experimental paradigm employing Cre-Lox technology for conditional expression of reporters from the Brainbow 1.0 cassette is illustrated. (L) The dTomato (red) reporter is the default maker of infected cells in the absence of Cre. (M) In the presence of Cre, the dTomato reporter is recombined from the viral genome to enable the expression of EYFP and mCerulean (cyan) reporters. (N) Neurons infected by transneuronal passage of recombined virus will only express the conditional reporters.
In the following sections, we present the viral transneuronal tracing method within the aspects of the life cycle of PRV that have influenced experimental design and interpretation of data. We first describe procedures for retrograde infection of CNS circuits by peripheral (Basic Protocol 1) and intracerebral injection (Basic Protocol 2) of PRV-Bartha and its recombinants. We supplement these basic protocols with the more sophisticated and powerful tracing approaches that have evolved from the construction of well-characterized recombinant viruses. These include the injection of multiple recombinant strains to define collateralization of neurons within a network (Alternate Protocol 2), the use of conditional replication of PRV to define neural networks synaptically connected to defined populations of neurons (Alternate Protocol 3), the use of conditional reporter expression to define the synaptic connections to targeted populations of neurons within complex circuits (Alternate Protocol 3), and the use of transgene expression from the viral genome to assess neuronal activity (Alternate Protocol 4). Support protocols are also included that detail methods for detecting virus, as well as details for the growth and titering of high tier viral stocks (neurostocks). Finally, we include a discussion of critical parameters that must be considered in experimental design and interpretation. We also provide a tabular listing of well characterized viral stocks that are available through the NIH-sponsored Center for Neuroanatomy with Neurotropic Viruses (CNNV; http://www.cnnv.pitt.edu; Table 1). The mission of this center (NIH Grant P40OD010996) is to develop well-characterized viral tracing reagents and make them available to investigators seeking to use the method. Reagents and resources available through the CNNV also include HSV and rabies virus. It is also important to note that other strains of PRV and other neurotropic viruses for circuit analysis are available through other investigators.
Table 1.
Strains of PRV Available from CNNV
| Virus | Attributes |
|---|---|
|
| |
| PRV-Becker | Wild-type laboratory strain of PRV. Anterograde and retrograde, but highly virulent. |
| PRV-Bartha | Attenuated vaccine strain of PRV most widely used for retrograde transneuronal tracing. “Attenuated” means that animals live several days longer after infection without significant symptoms compared to PRV-Becker. While much less virulent than PRV-Becker, PRV-Bartha does kill animals after infection. |
| PRV-NIA3 | Wild-type, highly virulent PRV isolate. It is transported bidirectionally through circuits and has been used in tracing experiments in animal species (e.g., cats) that are refractory to infection with other PRV strains. |
| PRV-BaBlu | PRV-Bartha containing a b-galactosidase reporter gene (lacZ gene) inserted into gG locus. The gG promoter drives expression of the lacZ gene, and the virus is selectively transported retrogradely. |
| PRV-91 | PRV-Becker with a deletion of the gE gene. The virus is attenuated compared to PRV-Becker and is only transported retrogradely through neural circuits |
| PRV-98 | PRV-Becker with a deletion of the gI gene. The virus is attenuated compared to PRV-Becker and is only transported retrogradely through neural circuits. |
| PRV-151 | PRV-Becker containing the CMV-EGFP reporter gene cassette inserted into the gG locus of the viral genome. The CMV promoter drives expression of the EGFP gene, and virus is transported bidirectionally. |
| PRV-152 | PRV-Bartha containing the CMV-EGFP reporter gene cassette inserted into the gG locus of the viral genome. The CMV promoter drives expression of the EGFP gene and the virus is selectively transported retrogradely. |
| PRV-180 | PRV-Becker expressing mRFP-VP26 (capsid) fusion protein. The fusion protein is incorporated into capsids; strongly expressed in nuclei of infected cells; transported both in the anterograde and retrograde directions. |
| PRV-GS443 | PRV-Becker expressing EGFP-VP26 (capsid) fusion protein. The fusion protein is incorporated into capsids; strongly expressed in nuclei of infected cells; transported both in the anterograde and retrograde directions. |
| PRV-181 | PRV-Becker recombinant expressing mRFP-VP26 (capsid) and GFP-VP22 (tegument) fusion proteins. Both fusion proteins are incorporated into dual-colored viral particles. Transported both in the anterograde and retrograde directions. |
| PRV-614 | PRV-Bartha containing the CMV-mRFP reporter gene cassette inserted into the gG locus of the viral genome. Isogenic with PRV-152. Often used with PRV-152 in dual-infection studies. |
| PRV-813 | PRV-Becker lacking the Us3 gene. This virus is modestly less virulent than PRV Becker, but not as attenuated as PRV Bartha. Anterograde and retrograde tracer. |
| PRV-823 | PRV-Becker lacking the Us3 gene expressing mRFP-VP26 (capsid) fusion protein. This virus is less virulent than PRV-813 but not as attenuated at PRV Bartha. Anterograde and retrograde tracer. |
| PRV-833 | PRV-Becker lacking the Us3 gene expressing mRFP-VP26 (capsid) fusion protein and eGFP-VP22 (tegument) fusion protein. This virus is modestly less virulent than PRV-823 and is an anterograde and retrograde tracer. |
| PRV-Ba2001 | PRV-Bartha whose replication and expression of tau-EGFP is conditional upon Cre recombinase mediated recombination of the viral genome. A retrograde only tracer (This recombinant is replaced with an updated version called PRV-Introvert). |
| PRV-263 | PRV-Bartha containing the Brainbow 1.0 cassette in the gG locus. Default expression of dTomato reporter unless in the presence of cre-recombinase (Cre). Cre excises the dTomato gene and liberates expression of EYFP or mCerulean. |
| PRV-267 | PRV-Bartha containing the cre-recombinase gene in the gG locus. Biologically active Cre is expressed in a circuit related fashion with viral transneuronal passage and replication. |
| PRV-290 | PRV-Bartha containing the mTurquoise2 reporter gene cassette inserted into the gG locus of the viral genome. Isogenic with PRV-152 and PRV-614. Can be used with PRV-152 and/or PRV-614 for dual- and triple-infection studies. |
| PRV-IE180 null | PRV-Becker null mutant with deletions in IE180 and Us9 genes, with hSyn-Cre reporter gene cassette replacing the Us9 locus. It produces long-term constitutive Cre expression, but there is no viral replication or spread unless trans-complemented with IE180. |
| PRV-Introvert-GFP | PRV-Bartha whose replication and expression of GFP is conditional upon Cre recombinase-mediated recombination of the viral genome. A retrograde-only tracer. |
| PRV-Introvert-mCherry | PRV-Bartha whose replication and expression of mCherry is conditional upon Cre recombinase-mediated recombination of the viral genome. A retrograde-only tracer. |
NOTE: All protocols involving PRV must conform to the rules and regulations mandated for the use of class II infectious pathogens. Experiments using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals. BSL II regulations also mandate that experiments must be conducted in facilities approved for the use of BSL II infectious agents. The requirements of these facilities are detailed later in the unit.
BASIC PROTOCOL 1
RETROGRADE INFECTION OF CNS CIRCUITS BY PERIPHERAL INJECTION OF VIRUS
As noted earlier, the pig is the natural host of PRV, and the virus has a wide host range, infecting all mammals except higher primates. Human herpesviruses and rabies virus have been successfully employed to study the organization of circuits in higher species. Experiments using PRV-Bartha and its recombinants capitalize upon the clearly documented restricted phenotype of viral replication and spread established in rodents. Upon infecting neurons, PRV-Bartha produces infectious progeny that traffic into the somatodendritic compartment to move across synapses and spread from post-synaptic to pre-synaptic neurons (i.e., retrogradely) through neurons comprising a circuit. The molecular basis of the restricted retrograde transport phenotype is well documented and derives from a deletion in the unique short region of the viral genome that eliminates the capacity of particles in the cell body to be sorted and transported in axons (Fig. 1). As a result, spread from pre-synaptic axons to post-synaptic cells is completely abrogated (i.e., an anterograde spread defect). The deletion responsible for the retrograde-only spread phenotype also reduces the virulence (e.g., lengthens the mean time to death with markedly reduced peripheral neuropathy) and cytopathology of viral infection. Also, compared to wild-type infections, PRV-Bartha spread slower in neuronal circuits. This phenotype has been mapped to point mutations in a gene (UL21) whose function is not well understood (Curanovic et al., 2009; Szpara et al., 2011). Collectively, these features of viral replication and spread of PRV-Bartha based viruses have made these viruses popular tools for circuit analysis.
Studies of PRV and HSV have documented differential affinities of virus for different compartments of neurons, as well as glial cells (Vahlne et al., 1978, 1980; Marchand and Schwab, 1987). These viruses have a high affinity for axon terminals and glial cells that contributes to the restricted zone of uptake following injection of peripheral organs or the brain. Consequently, variations in the density of innervation or the cytoarchitecture of a target organ will have profound effects upon the onset of replication and the progression of infection within a circuit. In this protocol, retrograde infection of CNS circuitry is achieved through inoculation of the peripheral targets of autonomic or somatic motor neurons. Emphasis is placed upon critical steps of the method that must be followed to ensure reproducible and reliable patterns of infection. Where appropriate, reference is made to sections of the Commentary that provide a more comprehensive treatment of a particular topic.
Materials
Experimental animals, e.g., rodents
Betadine
Sterile physiological saline (0.9% w/v NaCl)
High-titer PRV (see Support Protocol 1; Table 1)
Phosphate-buffered saline (PBS; see recipe)
PLP fixative for perfusion (see recipe)
Animal clippers
Surgical instruments (will vary depending on required surgery)
10-μl Hamilton syringe equipped with a 26-G needle that has a sharpened beveled tip (sterilize by autoclaving; do not use cold sterilization as the solution will compromise the titer of the inoculum)
Sutures and/or wound clips
Appropriate syringes and needles for injection of analgesics into experimental animal
Heating pad or heat lamp
Additional reagents and equipment for animal anesthesia (Davis, 2008), injection of animals (Donovan & Brown, 2005), and perfusion fixation (unit 1.1) [*Copy Editor: Here and throughout, use the following Lit Citations in place of unit references. Please also add to Lit Cited section. Davis, J.A. (2008), Mouse and Rat Anesthesia and Analgesia. Current Protocols in Neuroscience, 42: A.4B.1- A.4B.21. https://doi.org/10.1002/0471142301.nsa04bs42; Donovan, J. and Brown, P. (2005), Parenteral Injections. Current Protocols in Neuroscience, 33: A.4F.1-A.4F.9. https://doi.org/10.1002/0471142301.nsa04fs33; Paletzki, R. F., & Gerfen, C. R. (2019). Basic neuroanatomical methods. Current Protocols in Neuroscience, 90, e84. doi: 10.1002/cpns.84]
CAUTION: PRV and other neurotropic viruses are class 2 infectious agents that require a laboratory which meets Biosafety Level 2 (BSL-2) regulations as defined in Health and Human Services Publication 88–8395 (see Critical Parameters). Among these regulations is the requirement that infected animals be confined to the BSL-2 laboratory throughout the experiment and that the laboratory be dedicated to the viral studies. These regulations may vary among institutions, and it is the responsibility of the investigator to determine the regulations that may be peculiar to their institution. It is also important to ensure that the laboratory be equipped with all of the reagents and equipment necessary for the experiment. Aseptic surgical procedures should be used, but it is also important to include precautions that protect against compromising the concentration of the inoculum (see Critical Parameters). PRV is not known to cause human infections in the laboratory. However, caution when using PRV is appropriate because there are reports of rare human infection in farm workers (Yang et al., 2019).
Perform surgery
-
1
Deeply anesthetize animal with anesthetic (Davis, 2008).
Animals should be deeply anesthetized so that they are unresponsive to sensory stimuli (e.g., tail pinch).
-
2
Perform the surgical procedure appropriate for the target organ.
-
3
Prepare the site of the incision as a sterile surgical field. Shave fur from the region and sterilize the field with betadine. Make the incision through the skin and underlying musculature to expose the area/organ to be injected.
Inject virus and allow post-inoculation recovery
The principal features that will determine the method of injection are the density of the axonal innervation and cytoarchitecture of the tissue to be injected (Fig. 2). The following sections detail the general procedures common to injecting virus into a peripheral tissue. However, it is important to review the discussion in this unit on details of experimental design of the injection strategy that may vary according to the injected tissue (see Critical Parameters).
-
4Draw up and express sterile physiological saline prior to loading the 10-μl Hamilton syringe with virus. Repeat this process two or three times to eliminate contaminants and ensure that the syringe is delivering accurate aliquots. Perform the same draw-and-express procedure with virus; this prevents dilution of the virus with saline that may be in the bore of the needle.The saline draw ensures a tight seal of the plunger with the glass barrel of the syringe and increases the likelihood of accurately delivering the desired amount of virus to the injection target. However, it is imperative that residual saline be removed from the needle to prevent dilution of the inoculum. The importance of purging the syringe of saline cannot be overemphasized, since the microliter syringes are loaded with small volumes of virus and residual saline will dilute the inoculum. Reductions in titer resulting from dilution will introduce variability in the extent of infection between animals and, under the worst circumstances, may prevent productive replication of virus altogether (see Understanding Results).
-
5Inject the inoculum into the target tissue (the amount and number of injection sites will depend upon the density of innervation and tissue architecture). See the Critical Parameters section for a more detailed discussion of the factors that should be considered.IMPORTANT NOTE: Do not remove or disrupt connective tissue or fascial sheaths associated with the injection target prior to injection. Envelope glycoproteins of PRV and other alpha herpesviruses have high affinity for extracellular matrix proteins, and these sheaths serve to limit diffusion of the virus from the injection site.
-
6Close incisions with sutures and/or wound clips, provide a subcutaneous injection of analgesic, and allow the animal to recover on a heating pad or under a heat lamp. When the animal is conscious, return it to its home cage and ensure that it is ambulating normally. House animals individually after virus injection. Provide food and water ad libitum and standardize the photoperiod to 12-hr light per 24-hr cycle. Weigh animals daily, as precipitous weight loss provides an important indication of viral pathogenesis.Use sutures to muscle layers of body cavities (such as the abdomen) or use wound clips to close incisions through the skin.BSL-2 regulations stipulate that infected animals must be confined to the BSL-2 laboratory throughout the experiment. Thus, the laboratory should contain a HEPA-filtered unit for housing the animals.Injected animals should be carefully monitored post-inoculation to ensure that the viral infection is not compromising their ability to attend to their bodily needs. Most institutions require that the animals be monitored twice a day, typically first thing in the morning and late in the afternoon.When attenuated strains of virus are used, essentially all elements of a circuit can be characterized within a post-inoculation interval during which animals are free of symptoms. Nevertheless, it should be emphasized that even attenuated strains of virus will ultimately cause death at long survival intervals, and animals usually exhibit signs of infection at these late stages of infection. Monitoring the weight of an animal provides a reliable means of predicting the onset of debilitating consequences of infection. In unpublished observations, we noted that animals exhibit a precipitous drop of ~20% body weight during the day preceding imminent death. Thus, it is probable that any animal that becomes lethargic and experiences a large weight loss is at risk. These animals should be anesthetized and perfused.
Perform perfusion fixation of tissues
-
7
At the appropriate post-inoculation interval, anesthetize animals (Davis, 2008) and sacrifice animals by transcardiac perfusion fixation. Perfuse the entire animal with PBS followed by PLP fixative to inactivate virus throughout the animal (see unit 1.1 for perfusion fixation protocols). Remove the brain and any other tissues to be included in the analysis. Post-fix this tissue in PLP fixative for 1 to 2 hr at 4°C.
In our experience, tissue can be post-fixed for a minimum of 2 hr or as long as 3 days, prior to cryoprotection.
As noted above, the entire animal should be perfused. Studies of brain circuitry classically clamp the descending aorta and perfuse only the head, since preservation of brain structure and antigenicity is the preeminent goal. Since aldehydes inactivate virus, transcardiac perfusion inactivates virus throughout the animal, a particularly important goal after peripheral inoculation.
Dispose of waste properly
-
8
Treat everything that comes in contact with an infected animal during the course of an experiment as contaminated biohazardous material, including the perfused carcass, instruments, gauze, and syringes used for injections, as well as animal bedding, food, water, and caging. Place disposable materials, including animal tissues, in a biohazard bag and incinerate according to institutional regulations. Decontaminate instruments and equipment with ethanol, then wash them with standard detergents, and autoclave.
Although cold sterilization procedures are generally adequate, sterilization through autoclaving is preferential. Perfused carcasses are considered as contaminated waste, even though the perfusion fixation has presumably inactivated the virus.
Analyze tissues
-
9
Perform immunohistochemical analysis of tissues (see Support Protocols 2 and 3).
BASIC PROTOCOL 2
TRANSNEURONAL ANALYSIS BY INTRACEREBRAL INJECTION
Strick and colleagues championed intracerebral injection of HSV as a means of defining multisynaptic circuits in primate brain (Zemanick et al., 1991; Hoover and Strick, 1993, 1999; Middleton and Strick, 2001, 2002). This method has subsequently been widely applied using PRV in a wide variety of circuits, and is now routine in rodents. Studies of HSV have identified strains that move preferentially in either the anterograde or retrograde direction through a circuit (e.g., Zemanick et al., 1991). Studies of PRV have shown that the virulent wild-type virus will spread in both directions through a circuit after injection into the brain, and that the attenuated PRV-Bartha strain and its recombinants only spread retrogradely through a circuit following intracerebral injection (Card et al., 1998, 1999). Given these facts, it is of preeminent importance to select a strain of virus in which the direction of transport has been rigorously defined. Table 1 provides a list of such viruses currently available through the CNNV (CNNV; http://www.cnnv.pitt.edu).
Many of the procedures and issues detailed in Basic Protocol 1 are relevant to experiments involving intracerebral injection. Since the fundamental methods outlined in this protocol are quite similar to those applied in classical tract tracing, it is advisable to consult other investigators and publications to obtain detailed guidance for procedures that have been developed for accurate intracerebral injection with classical tracers. The following protocol provides guidance for the use of PRV-Bartha and Bartha recombinants in studies involving retrograde trans-synaptic passage following intracerebral inoculation.
Materials
Experimental animals, e.g., rodents
High-titer PRV (see Support Protocol 1; Table 1)
Phosphate-buffered saline (PBS; see recipe)
PLP fixative (see recipe for 4% paraformaldehyde-lysine-periodate)
Stereotaxic apparatus (David Kopf Instruments)
Stereotaxic atlas (e.g., Paxinos and Watson, 2013; Swanson, 1998)
Surgical instruments
Syringe with needle (see note below)
Sterile bone wax and gel foam
Wound clips
Heating pad or heat lamp
Additional materials for animal anesthesia (Davis, 2008), injection of animals (Donovan & Brown, 2005), monitoring of post-surgical condition of animals (Basic Protocol 1), perfusion fixation (unit 1.1), and immunohistochemical localization of neurochemicals (see Support Protocol 2 and units 1.1 & 1.2) [*Copy Editor – here are citations for Units 1.1 and 1.2: Paletzki, R. F., & Gerfen, C. R. (2019). Basic neuroanatomical methods. Current Protocols in Neuroscience, 90, e84. doi: 10.1002/cpns.84; Volpicelli-Daley, L.A. and Levey, A. (2003), Immunohistochemical Localization of Proteins in the Nervous System. Current Protocols in Neuroscience, 25: 1.2.1–1.2.17. https://doi.org/10.1002/0471142301.ns0102s25]
IMPORTANT NOTE: The needle, cannula, or pulled-glass pipet used for virus injection will influence the zone of viral diffusion. Hamilton microliter syringes equipped with fixed needles of 26- to 32-G are adequate for injection of large cell groups. The needles affixed to these syringes are either blunt or have a sharpened tip with the opening on the beveled surface. If a beveled needle is used, the opening should be directed towards the cell group of interest. This is quite important, since the affinities of virions for extracellular matrix molecules restrict virus diffusion to the immediate vicinity of the injection. More restricted injections can be made with glass pipets. The pipets are pulled using standard procedures (see Chapter 6), and the tip broken back so that the internal diameter is 15 to 20 μm. The shaft of the pipet is placed over the needle of a Hamilton syringe preloaded with virus, and the interface is sealed with beeswax. It is important to keep the length of the glass sleeve as short as possible, since the space between the glass and the needle becomes a reservoir for virus that must be filled before virus can be ejected from the pipet tip.
Perform surgery
-
1Deeply anesthetize animal (Davis, 2008).Animals should be deeply anesthetized so that they are unresponsive to sensory stimuli (e.g., tail pinch).
-
2
Secure the head in a stereotaxic frame. Make an incision to expose the cranium and use coordinates determined using a stereotaxic to position the syringe over the skull over the desired area of injection. Drill a hole in the cranium at this location and pierce the dura mater in preparation for lowering the cannula/needle/glass pipet into the brain parenchyma.
Inject virus and allow post-inoculation recovery
-
3Affix the syringe to the stereotaxic arm and lower the tip of the needle or glass pipet to the desired dorsoventral coordinate through the hole in the cranium—the needle should be lowered into the tissue slowly and left in place for approximately 5 min to allow the tissue to relax around the tip of the needle. Inject the virus at 10 nl/min and leave the needle/pipet in situ for a minimum of 5 min after completion of the injection, to reduce reflux of virus along the injection tract. Slowly withdraw the needle/pipet from the brain, fill the hole in the cranium with bone wax or gel foam, and close the scalp with wound clips.Typically, injection of 50 to 100 nl of virus will produce a restricted zone of viral uptake (see Critical Parameters for more detail). However, the cytoarchitecture of the area of injection will also be influential. Detailed information regarding the extent of viral diffusion following injection of virus alone or in combination with classical tracers can be found in Jasmin et al. (1997), O’Donnell et al. (1997), and Card et al. (1999).
-
4
Allow the animal to recover on a heating pad or under a heat lamp and then return it to its home cage in the animal housing unit. Monitor the animal according to the procedures detailed in Basic Protocol 1.
Perform perfusion fixation of tissues
- At the desired post-inoculation interval, anesthetize and perfuse the entire animal with PBS followed by 4% paraformaldehyde-lysine-periodate PLP fixative (see unit 1.1 and Table 2 for perfusion fixation protocols). Prepare the tissue for immunohistochemical localization of infected neurons (see Support Protocol 2 and units 1.1 & 1.2).For guidance in determining appropriate survival intervals, see Critical Parameters.
Table 2.
Components of 4% Paraformaldehyde-Lysine-Periodate PLP Fixative Solution
| 100 ml | 250 ml | 500 ml | 1000 ml | |
|---|---|---|---|---|
|
| ||||
| Paraformaldehyde | 4.00 g | 10.00 g | 20.00 g | 40.00 g |
| Sodium metaperiodate | 0.22 g | 0.54 g | 1.10 g | 2.14 g |
| Lysine | 1.37 g | 3.43 g | 6.85 g | 13.70 g |
ALTERNATE PROTOCOLS
The complexity inherent within CNS circuitry often makes it difficult to define connections specific to particular populations of neurons within a neural network at long post-inoculation survival intervals. Microdissection of connections specific to neurons within these neural networks has been enabled through the construction of viruses that express unique reporters either constitutively or conditionally, replicate conditionally, and express transgenes from the viral genome (Fig. 3). Dual-infection paradigms allow an assessment of the collateralization of axons within neural networks (Billig et al., 2000; Cano et al., 2001; McEwan et al., 2021). Combining viral infection with Cre-lox technology (Sauer, 1987) has allowed definition of connections specific to targeted populations of neurons. One such method developed by Friedman and colleagues in 2001 and upgraded in 2017, renders replication of virus conditional upon the presence of cre recombinase (Cre) within neurons (DeFalco et al., 2001; Pomeranz et al., 2017). Another approach combining the use of the Brainbow cassette developed by Lichtman, Sanes, and colleagues (Livet et al., 2007) with viral tracing allows conditional reporter expression in Cre-expressing neurons and their synaptic partners in infected neural networks (Card et al., 2011a,b). The ability of PRV-Bartha recombinants to spread retrogradely through neural circuits also provides a powerful means of delivering biologically active Cre in a circuit-related fashion (Card et al., 2011a) and to assess the activity of neural networks through the expression of calcium indicator proteins (Boldogkoi et al., 2009; Granstedt et al., 2009, 2010). The following four alternate protocols provide guidelines for applying each of these experimental approaches.
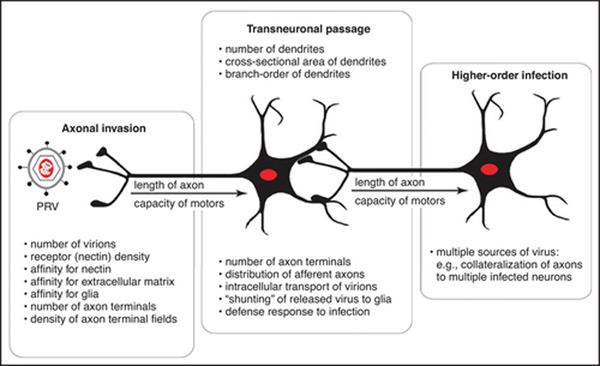
Figure 3.
The factors that influence replication and spread of PRV-Bartha recombinants through neural circuits are illustrated.
ALTERNATE PROTOCOL 1
Transneuronal Analysis with Multiple Recombinant Strains
The use of recombinant strains of PRV constitutively expressing different reporter proteins provides a powerful means of defining collateralization of axons within complex neural circuits (Fig. 3). The unique reporters expressed by these recombinants can be detected by their native fluorescence (e.g., EGFP, mRFP) or by immunocytochemical localization using monospecific antibodies (e.g., b-galactosidase and other nonfluorescent reporters). There are a variety of factors that should be considered in the design of these studies and the interpretation of data. Thus, the issues discussed in Critical Parameters and illustrated in Figure 2 should be carefully considered before embarking upon this type of experiment. Collection of data in fluorescence studies can be difficult without a clear understanding of the extent of infection within a circuit. We therefore recommend processing one bin of tissue for immunoperoxidase localization of viral proteins to obtain a permanent record of the distribution and density of infected neurons. The data derived from the immunoperoxidase localizations can then be used to direct the analysis of putative dual-infected neurons using immunofluorescence methods (unit 2.1). [*Copy Editor: Combs, C. A., & Shroff, H. (2017). Fluorescence microscopy: A concise guide to current imaging methods. Current Protocols in Neuroscience, 79, 2.1.1– 2.1.25. doi: 10.1002/cpns.29] The following protocol is based upon the use of isogenic PRV-Bartha recombinants that express unique reporters. This method is compatible with either peripheral Basic Protocol 1) or intracerebral (Basic Protocol 2) injection paradigms.
The goal of experiments of this type is to determine if a single population of neurons provides a common influence upon neurons in two different areas through collateralized axonal projections. To address this issue, two recombinant viruses are injected into separate targets and a temporal analysis is conducted to determine if the retrograde trans-synaptic passage of the two viruses ultimately leads to infection of a single population of neurons. However, the number of neurons that contributes to each circuit often differs in the circuits under study (see Figs. 3 and 4). Thus, it is likely that different post-inoculation intervals would be required for each virus to reach the parent collateralized neurons, and injection of the two viruses would have to be temporally separated to accommodate this difference. The importance of designing the experiment so that the two viruses reach the putative site of the collateralized neurons cannot be overstated. This is due to the fact that infection of a neuron can render it refractory to infection by a second virus that arrives after the initial infection is established (Kim et al., 1999; Banfield et al., 2003). This phenomenon is called superinfection exclusion or homologous interference. It has been shown that 6 hours after infection with a PRV recombinant, cells are completely refractory to superinfection with a second PRV recombinant. Furthermore, neurons are only capable of replicating a small number of herpesviral genomes (Kobiler et al., 2010; Taylor et al., 2012). These considerations raise the possibility of false negatives, e.g., the inability of a second virus to infect neurons previously infected by another virus. Consequently, the single most important aspect of the experimental design is to determine the temporal kinetics of invasion of each recombinant virus in single-injection paradigms. Data from these single-injection studies will allow an informed determination of the timing of the injection of the two viruses (discussed in greater detail in Critical Parameters). Importantly, there is one replication-incompetent PRV recombinant called PRV-IE180 null that is not susceptible to superinfection exclusion (Wu et al., 2014). This Becker strain mutant contain deletions in IE180 and Us9 genes and expresses Cre-recombinase in a constitutive fashion from the human synapsin promoter, allowing long-term retrograde labelling of neurons (Oyibo et al., 2013).
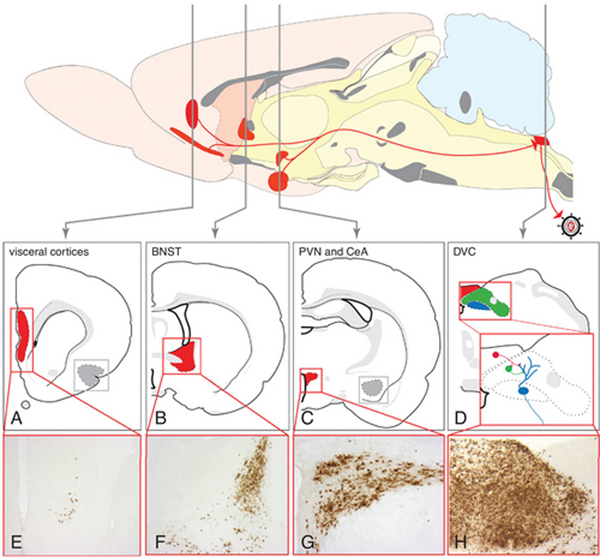
Figure 4.
The sequential retrograde trans-synaptic passage of PRV-Bartha from the stomach wall of the rat is illustrated. Retrograde transport of virus from the stomach wall results in infection of parasympathetic neurons in the dorsal motor vagal nucleus (blue in D) and transneuronal spread of virus to neurons in the adjacent NTS (green in D) and area postrema (red in D). Replication and spread of infection to higher-order neurons occurs with advancing survival. Included in this extended network is the paraventricular hypothalamic nucleus (C and G; PVN), central nucleus of the amygdala (CeA), bed nucleus of the stria terminalis (B, F; BNST), and visceral cortices (A, E). This model has been used extensively to define the synaptic organization of preautonomic neural networks and to define approaches for statistical evaluation of the progression of infection. See Card et al. (2005a) for details.
Selection of recombinant viruses
As noted above, the preeminent concern in this type of experiment is that one virus will interfere with the replication of the second strain. One way of reducing this possibility is to use recombinant viruses that are isogenic and have the same titer. Other factors that are important to the success of the experiment are (1) selection of viruses that are attenuated for virulence, but maintain their invasiveness phenotype; (2) demonstration that the reporter proteins of each virus are efficiently expressed in quantities that can be detected on the basis of their native fluorescence or with immunofluorescence; and (3) availability of specific antisera produced in different species that will produce reliable localizations of each reporter protein. The native fluorescence of reporters can be used without immunofluorescence amplification, but it is important to first define the kinetics of reporter-gene expression to determine the post-inoculation interval when fluorescence is most intense within the area of interest. A large body of literature employing this approach has accumulated since the initial publication of this unit in 1999. It is evident from those studies that isogenic recombinants of PRV-Bartha that express EGFP [PRV-152; (Billig et al., 2000; Smith et al., 2000; Schneeberger et el., 2019; Pisano et al., 2021)], mRFP (PRV-614; Banfield et al., 2003), or mTurquoise2 (PRV-290; Hogue et al., 2018), work extremely well in this experimental paradigm; these are available through the CNNV (http://www.cnnv.pitt.edu; Table 1).
Injection of the recombinant viruses
The single most important aspect of injection in this paradigm is to ensure that syringes are not contaminated with both viruses. The best way to ensure this is to dedicate separate syringes to the different viruses. Otherwise, the procedures detailed in the earlier protocols should be used for injection of each recombinant.
Post-inoculation survival and tissue processing
The timing of the two injections should be adjusted to optimize the possibility that both viruses will reach neurons with collateralized axons within the same approximate post-inoculation interval. Nevertheless, temporal analyses should still be conducted to ensure that both of the recombinant viruses are moving through the circuitry according to the temporal profiles established in the single-injection analyses. Tissue sections should be processed using the dual-labeling immunofluorescence protocol detailed in Support Protocol 3.
ALTERNATE PROTOCOL 2
Conditional Replication and Spread of PRV
The conditional replication approach championed by Friedman and colleagues (DeFalco et al., 2001) relies upon Cre-mediated recombination of the PRV genome to generate infectious progeny. The recombinant virus engineered for this approach, PRV-2001, only replicates in the presence of Cre and expresses an EGFP reporter. The recombinant carries a floxed stop cassette in front of the thymidine kinase (TK) gene, whose expression is essential for viral DNA synthesis and production of infectious progeny in nonmitotic cells. In the presence of Cre, the loxP stop cassette is removed from the viral genome, promoting the expression of thymidine kinase and a tau-EGFP reporter. As a result, viral DNA synthesis occurs, and infectious progeny are produced within an EGFP-filled neuron. The resulting progeny are replication competent, leave the parent neuron by retrograde transport across synapses, and are capable of replicating in, and being transported through, non-Cre-expressing cells. Thus, PRV-2001 spreads efficiently through neural networks presynaptic to the first infected neuron that expressed Cre.
The obvious limitation to this experiment is the expression of Cre. In the proof-of-principle study establishing the approach, PRV-2001 was injected into the tuberal hypothalamus of transgenic mice engineered to express Cre in neuropeptide Y (NPY) neurons. Injection of PRV-2001 into the tuberal hypothalamus of these animals resulted in viral replication only within arcuate NPY neurons and their presynaptic partners. The same approach was employed by Yoon et al. (2005) and Campbell and Herbison (2007a, b) to define neural networks presynaptic to Cre-expressing LHRH neurons in the rostral hypothalamus. An important consideration in studies using transgenic Cre-expressing animals is to ensure that the Cre-expressing neurons are sequestered within an area where it is possible to make a focal injection of PRV-2001. In many cases, phenotypically restricted expression of Cre in neurons is not functionally defined; e.g., NPY is widely expressed in areas outside of the arcuate hypothalamic nucleus. Consequently, the success of these experiments is dependent upon isolating injections of PRV-2001 to localized populations of Cre-expressing neurons. Lentivirus or adeno-associated virus (AAV) vectors represent another means of obtaining targeted Cre expression (Schneeberger et al, 2019).
The methods for using this experimental approach are the same as those detailed in the prior sections. Those procedures that use PRV-2001 in combination with lentivirus vectors to induce Cre expression are detailed in the next section. The experiment can be designed such that the injection of PRV-2001 is made into the area containing the Cre-expressing neurons (for direct infection) or into a projection target containing their axons (to infect the Cre neurons by retrograde transport). The latter approach provides a further means of dissecting complex networks.
More recently, Pomeranz et al., (2017) developed an improved version of PRV-2001, called PRV-Introvert. Similarly, activation of TK and GFP or mCherry is dependent on the expression of Cre recombinase. However, PRV-Introvert drives fluorescent reporter expression to higher levels by using the potent, inmediate-early CMV promoter. Furthermore, the risk of leaky viral replication is eliminated with the insertion of a synthetic intron into the PRV TK gene, and inversion of part of the TK exon between two lox sites. Thus, PRV-Introvert should be the primary choice over PRV-2001.
ALTERNATE PROTOCOL 3
Conditional Reporters of PRV Infection and Spread
As noted above, the primary limitation of PRV-2001 methodology is that the virus must have access to Cre-expressing neurons to replicate. This requirement eliminates the ability to conduct studies in which the Cre-expressing neurons are a synapse or more away from a projection target. Combining the development of conditional Brainbow reporter cassettes (Livet et al., 2007) with viral transneuronal tracing has provided a powerful means of addressing this limitation (Fig. 3). The Brainbow cassette places genes encoding fluorescent proteins in sequence, separated by loxP stop cassettes. In the absence of Cre, the cassette only expresses the first gene in the sequence. In the presence of Cre, the first gene is recombined from the cassette either alone or in combination with the second gene. Thus, the presence of Cre provides a means of conditional reporter expression.
The PRV-263 strain of PRV contains the Brainbow 1.0L cassette in the gG locus of PRV-Bartha (Fig. 2), and is therefore isogenic with PRV-152 (EGFP reporter) and PRV-614 (mRFP reporter). The virus is replication competent in all neurons, is transported retrogradely through neural circuits, and expresses dTomato fluorescence unless in the presence of Cre. When Cre is present, the dTomato gene is recombined from the viral genome alone or in combination with the mCerulean gene. Cre-mediated recombination therefore produces conditional expression of either the mCerulean or EYFP gene products. Recombination of the viral genome by Cre is permanent, so subsequent retrograde spread of virus from the Cre-expressing neurons will also mark neurons with the conditional reporters of infection.
Similar to studies employing PRV-2001, the utility of PRV-263 is directly related to the ability to obtain targeted expression of Cre. Transgenic animals are certainly compatible with the use of PRV-263. However, the use of viral vectors allows delivery of Cre to neurons, either regionally or phenotypically defined. In the proof-of-principle experiments demonstrating the utility of PRV-263, targeted expression of Cre was achieved using lentivirus vectors (Card et al., 2011b). Two vectors used in the study differed only in the promoter that controlled expression of Cre. Phenotypically restricted expression of Cre in catecholamine neurons was achieved using a vector in which expression of Cre was controlled by a synthetic dopamine-b-hydroxylase (DbH) promoter (Hwang et al., 2001). The activity of this promoter is controlled by the Phox2a or Phox2b transcription factor, thereby restricting expression of Cre to noradrenaline- or adrenaline-containing neurons (Card et al., 2006, 210). Regionally restricted expression of Cre was achieved by placing Cre under the control of the elongation factor 1a (EF1-a) promoter, which is ubiquitous and active in all classes of neuron.
Experimental design
Successful use of PRV-263 depends upon the presence of Cre within the cell group of interest. If viral vectors are used to generate Cre expression, the vector injection must precede injection of PRV-263 to ensure stable levels of Cre expression. Parametric studies of Cre expression from the viral vector are therefore a necessary prerequisite to informed experimental design. Analysis of Cre expression following central injection of the aforementioned lentivirus vectors demonstrated that robust expression of Cre was achieved within 7 days of injection and that expression was stable through 64 days, the longest post-injection interval examined. The temporal kinetics and magnitude of Cre expression will vary between vectors and in response to the use of different promoters, so it is essential that parametric studies be conducted for each vector.
The power of PRV-263 over PRV-2001 derives from the ability to inject virus at sites synaptically distant from the Cre-expressing neurons. This allows an analysis of “microcircuits” within the context of larger labeled networks defined by a projection target. In our proof-of-principle studies we combined injection of PRV-263 into the kidney with lentivirus vector injection into a region of brainstem (RVLM) that exerts a regulatory influence over arterial blood pressure through neural control over peripheral vascular beds and the kidneys. Cre expression from the lentivirus vector was under the control of a synthetic DbH promoter, which restricted expression of Cre to catecholamine neurons within RVLM expressing the Phox2a transcription factor. Injection of PRV-263 into the kidney produced retrograde transneuronal spread of virus through the preautonomic network synaptically linked to the kidney (Schramm et al., 1993; Huang and Weiss, 1999; Cano et al., 2004). Infected neurons without access to Cre were marked by the default dTomato reporter of infection. In contrast, Cre-expressing catecholamine neurons in RVLM expressed the conditional reporters, as did neurons in the network synaptically linked to the Cre-expressing catecholamine neurons. This experimental approach is compatible with both peripheral (Basic Protocol 1) and central (Basic Protocol 2) injection of virus.
The collection of data from studies employing PRV-263 is labor intensive and requires an informed perspective on both the distribution of infected neurons within the neuraxis as well as the magnitude of infection within infected cell groups. Thus, similar to the approach described in Protocol 1, the fluorescence analysis should be preceded by an immunoperoxidase localization of infected neurons. The supplemental information published in our proof-of-principle study (Card et al., 2011b) contains a detailed description of the sampling strategy that we developed to optimize collection of the fluorescence data.
ALTERNATE PROTOCOL 4
REPORTERS OF NEURAL ACTIVITY IN POLYSYNAPTIC CIRCUITS
The transneuronal tracing capabilities of viruses provides a powerful means of defining circuit organization but, until recently, the method has been refractory to providing insight into the functional activity of the labeled networks. Since viral infection induces the expression of Fos (a proto-oncogene that is expressed in neurons following depolarization), this commonly used assay of neural activity is not compatible with viral tracing technology (Cano et al., 2003). However, due to the construction of viral strains that express genetically-encoded calcium indicators (GECI), it is now possible to characterize the activity in neural networks defined by viral transport (Boldogkoi et al., 2009; Granstedt et al., 2009, 2010; Granstedt et al., 2013; Sun et al., 2013). PRV-Bartha and PRV-Kaplan recombinants engineered to express GECIs were successfully used to characterize activity in viral-labeled circuits of retinal explants (Boldogkoi et al., 2009). Similarly, Granstedt and colleagues constructed a recombinant of PRV-614 (PRV-369) in which the mRFP gene in the gG locus was replaced with the G-CaMP2 open reading frame under the control of the cytomegalovirus immediate-early promoter (Granstedt et al., 2009, 2010). Retrograde infection of neurons in the submandibular gland by injection of PRV-369 into the salivary gland allowed real-time, two-photon microscopy imaging of neuronal activity in living animals. Calcium transients due to spontaneous activity as well as stimulation were documented in the infected neurons.
Virus-induced alteration of neuronal activity is an important consideration in evaluating the calcium transients observed in studies using PRV-369 and other recombinants expressing FCIPs. Granstedt and colleagues demonstrated changes in both the frequency and duration of neuronal firing at longer survival intervals and suggested that these changes may be due to virus-induced toxicity and/or the host defense responses to viral infections. These data are consistent with previous slice recording studies in which neurons infected by retrograde transneuronal transport of PRV-152 (EGFP reporter) were shown to exhibit normal physiological signatures at an early point following viral infection (Smith et al., 2000; Irnaten et al., 2001). They also make the important observation that evaluating activity within the network with FCIPs should be rigorously investigated within the temporal progression of infection to clarify activity reflective of network function compared to that resulting from virus-induced alterations of neural activity.
Analysis of neuronal activity using expression of GECIs from the viral genome is among the newest viral transneuronal tracing applications. Accordingly, there is very little literature to draw from in recommending specific protocols. The reader is referred to those studies that have applied the technology for guidance regarding its use, strengths and limitations.
SUPPORT PROTOCOL 1
GROWING AND TITERING A PRV VIRAL STOCK
Stocks of PRV are grown in vitro in monolayers of pig kidney (PK15) cells using relatively straightforward tissue culture procedures. Some of the methods for growing the virus differ among strains (e.g., multiplicity of infection and incubation time). The following protocol produces high-titer (108 to 109 pfu/ml) stocks of the attenuated vaccine strain PRV-Bartha and its derivatives.
Materials
PK15 cells grown in 100-mm dishes containing DMEM/10% FBS/pen-strep (see recipe). PK15 cells can be obtained from investigators who routinely work with PRV (e.g., CNNV) or from the American Type Tissue Culture Collection (ATCC).
PRV-Bartha—and other strains of PRV—are not commercially available but, like PK15 cells, can be readily obtained from investigators who study PRV or through the CNNV (http://www.cnnv.pitt.edu).
Trypsin-EDTA (see recipe)
Unsupplemented DMEM containing 2% FBS
DMEM/2% FBS/pen-strep (see recipe)
Phosphate-buffered saline (PBS; see recipe), 37°C
DMEM/1% methocel/sodium bicarbonate/2% FBS/pen-strep (see recipe)
0.5% methylene blue in 70% methanol
100-mm sterile plastic dishes
Plastic cell scraper
Sterile 50-ml screw-cap plastic tubes
Sterile 1.7-ml microcentrifuge tubes, snap cap
Inverted microscope
Cup sonicator
Screw-cap cryovials
Centrifuge with Fotodyne 24-place rotor
Cryoguard M-40 thermal exposure indicators (Controlled Chemicals)
6-well tissue culture plates
Rocking platform
Additional reagents and equipment for tissue culture (Phelan, 2007) [*Copy Editor: Phelan, M.C. (2007), Techniques for Mammalian Cell Tissue Culture. Current Protocols in Neuroscience, 38: A.3B.1-A.3B.19. https://doi.org/10.1002/0471142727.nsa03bs38 ]
Prepare crude viral stocks
-
1
Prepare monolayers of PK15 cells the day before infection. Grow the cells in 100-mm sterile plastic dishes containing DMEM/10% FBS/pen-strep (typically three plates for each viral stock). Split the cells using trypsin-EDTA so that the monolayers reach 90% confluence the following day (see Phelan, 2007 for tissue culture techniques).
-
2
Resuspend the PRV-Bartha virus in DMEM containing 2% FBS (unsupplemented) so that 1.0 ml will provide a multiplicity of infection (MOI) of 0.01. Aspirate medium from each dish of PK15 cells, wash once with PBS and aspirate, and add the virus in 1.0 ml of medium at that MOI.
For example, for an MOI of 0.01, infect 3 × 106 PK15 cells with 3 × 104 plaque forming units (pfu) of PRV-Bartha.
-
3
Adsorb virus stock to cells for 1 hr in a humidified 37°C, 5% CO2 incubator. Gently tilt the plates every 15 min to ensure that the medium is uniformly distributed over the surface of the monolayer.
-
4
Remove unadsorbed virus particles by aspirating the medium and replacing it with 10 ml of fresh 37°C DMEM/2% FBS/pen-strep.
-
5Incubate monolayers for 2 to 3 days in a humidified 37°C, 5% CO2 incubator. When the monolayer shows extensive cytopathic effects, harvest the cells and medium with a plastic scraper. Combine the contents of all infected plates in one 50-ml, screw cap conical plastic tube. Mix the contents carefully. From this tube, transfer 1 ml to each of ten microcentrifuge tubes. Store the remaining crude stock in the 50-ml screw cap tube to be used as backup. Freeze all tubes at –70°C.The length of time to incubate infected monolayers will depend upon the rate at which virus spreads through the monolayer. This time is monitored by the appearance of cytopathic changes in infected cells that can be determined by periodically examining the plate of cells with an inverted microscope. Visible cytopathic effects are the distinct rounding of cells and margination of the chromatin.These crude stocks can be stored frozen at –70°C for long periods of time or can be used immediately for the preparation of purified “neurostocks” according to steps 6 to 10.
Prepare neurostocks
-
6If the crude stocks from step 5 were stored frozen, rapidly thaw the required amount in a 37°C water bath and gently mix the samples every 5 min until all ice has just disappeared. Place the individual samples on wet ice and then sonicate each sealed tube using a cup sonicator (10 pulses for a total of 10 sec at an amplitude of 80%) to disperse aggregated virus and separate virions from cellular debris. Invert the samples to mix contents and store on ice in preparation for division into aliquots.Keep the tubes sealed during the sonication to avoid creation of aerosols.
-
7Divide the viral stock into 1-ml aliquots in sterile 1.7-ml microcentrifuge tubes or screw-cap cryovials. Either store the vials at –70°C or continue with the separation of cellular debris to prepare the neurostock.Use one of these vials to determine the titer of the stock (see steps 11 to 18).
-
8
If the virus stock has been stored frozen, thaw and sonicate each sample as described in step 6 before proceeding with the rest of the purification. Remove the cellular debris from each sample by centrifuging for 5 min at 2000 × g (5000 rpm in the Fotodyne 24 place rotor) at room temperature.
-
9
Pipet off the supernatant, being careful to exclude the cell debris in the loose pellet at the bottom of each tube.
-
10Divide the supernatant containing purified virus into 100- to 200-μl aliquots in screw-cap cryovials, snap-freeze on dry ice, and store at –70°C.Do not aliquot in volumes less than 100 μl, since smaller volumes may produce freezing artifacts that reduce the titer. Do not freeze in snap-cap tubes because the ultra-cold, very dry environment in the freezer will “freeze-dry” the sample and inactivate the virus particles.Virus can be stored frozen in aliquots at –70°C for extended periods of time without consequence as long as the temperature remains ≤–70°C. Under no circumstances should these samples be thawed and refrozen, as this will reduce the titer of the sample. To help detect freezer failures or unexpected increases in temperature, the samples should be stored with Cryoguard M-40 thermal exposure indicators (Controlled Chemicals). These indicators remain green at temperatures below –50°C but turn irreversibly red when the temperature rises above –40°C. Samples in a box containing red thermal indicators should be discarded.
Determine virus concentration or titer in plaque-forming units (pfu)
-
11Prepare monolayers of PK15 cells the day before the assay is to be performed by splitting a 100-mm dish of cells that has reached confluence and plating the cells in three 6-well tissue culture plates.Cells should reach about 90% confluence at the time of use in step 14.
-
12Rapidly thaw and sonicate a 1-ml tube of crude stock (from step 7) using the procedures in step 6.Sonication of a neurostock will aid in dispersing viral aggregates.
-
13Prepare a series of 10-fold dilutions of virus as follows.
- Place 0.9 ml of DMEM/2% FBS/pen-strep in each of seven sterile microcentrifuge tubes.
- Add 0.1 ml of the viral stock to the first tube, mix well, and add 0.1 ml of that mixture to the second tube.
- Repeat this procedure using new tubes for the desired number of dilutions.
-
14
Aspirate the medium from each well of the 6-well plates and wash the cells with 37°C PBS. Add 0.2 ml each of the 10–5, 10–6, and 10–7 virus dilution to duplicate wells and return the plates to the incubator.
-
15
Adsorb the virus to the monolayers in each well for 1 hr in a humidified 37°C, 5% CO2 incubator, taking care to rock the plate every 15 min to ensure that there is an even distribution of virus over the monolayer.
-
16Remove the plates from the incubator and aspirate the inoculum from each well. Next, add 3 ml of DMEM/1% methocel/sodium bicarbonate/2% FBS/pen-strep. Return the plates to the incubator and leave them undisturbed for 2 to 3 days.This viscous methocel solution facilitates the formation of infected cell foci (plaque formation) by restricting the diffusion of virus particles from infected cells.It is essential that the plates be undisturbed during incubation to avoid interfering with plaque formation on the monolayer, thereby ensuring accurate determination of the titer of the viral stock. PRV-Bartha titers can be reliably determined after 2 days of incubation, but other strains that spread less efficiently may require a longer incubation period.
-
17Remove the plates from the incubator and aspirate the methocel solution from each plate. Wash each well once with 37°C PBS and then add 1 ml of 0.5% methylene blue in 70% methanol. Place the plates on a rocking platform for 10 min at room temperature to ensure even staining of the monolayer. Lastly, rinse off the excess stain under a gentle stream of tap water to visualize the viral plaques.Plaques result from virus-induced killing of cells in the monolayer, which lift off the plastic. Plaques appear as clear holes in the monolayer surrounded by a halo of rounded cells. The staining procedure kills both virus and cells.
-
18To calculate the titer in plaque-forming units per ml (pfu/ml), count the total number of plaques on all countable plates, divide by the total volume plated on these plates based on the lowest dilution giving countable numbers of plaques, and multiply by the reciprocal of the lowest dilution that gave countable plates.As an example, if there are 12 plaques in the well containing 0.2 ml of the 10–7 dilution and 175 plaques in the well containing 0.2 ml of the 10–6 dilution, the total number of plaques would be 187 plaques. The volume plated would be 0.22 ml of the 10–6 dilution. Thus, 187 plaques divided by 0.22 ml of the 106 gives you a titer of 8.5 × 108 pfu/ml. This method is more accurate than determining the titer based on 12 plaques, or a titer based on the average of titers determined for each dilution.
SUPPORT PROTOCOL 2
IMMUNOHISTOCHEMICAL PROCESSING AND DETECTION
Localization of viral antigens with the immunoperoxidase method provides a reliable first step in defining and documenting the extent of viral transport through neuronal circuitry in each case (Fig. 4). One can then further dissect the organization and phenotype of the circuit using dual-labeling methods. The avidin-biotin modification (Hsu et al., 1981) of the peroxidase-antiperoxidase method is presented for this purpose. It produces excellent signal against a low background with high dilutions of primary antibody.
These experiments are labor intensive, and it is obviously beneficial to obtain the maximum amount of information from each case. For example, as noted above, it may subsequently become desirable to establish the phenotype of neurons initially demonstrated by viral transport to contribute to a polysynaptic circuit. Since one cannot predict all possible components of a circuit that may be defined by viral transport, there is a necessary delay in the ability to perform the phenotypic characterizations. This delay could be problematic if measures were not taken to protect the antigenicity of the tissue. The following protocol for tissue processing and storage is designed to protect antigenicity and thereby maximize the amount of information that can be obtained from an experiment. Central to this approach is the storage of sectioned tissue in a glycol-based cryoprotectant that preserves tissue antigenicity for extended periods. The cryoprotectant developed by Watson et al. (1986) has these properties. Our parametric studies have demonstrated that tissue from the same animal stored in the cryoprotectant at –20oC, but processed for immunocytochemical localization of viral antigens 10 years apart, showed no discernable differences in either the pattern or density of labeling.
Materials
Perfused tissue from experimental animal (see Basic Protocol 1 or 2)
4% paraformaldehyde-lysine-periodate (PLP) fixative (see recipe)
0.1 M and 10 mM sodium phosphate buffer, pH 7.4 (Crawley et al., 1997) [*Copy Editor: (1997), Common Stock Solutions, Buffers, and Media. Current Protocols in Neuroscience, 00: A.2A.1-A.2A.8. https://doi.org/10.1002/0471142301.nsa02as00 ]
20% to 30% (w/v) sucrose in 0.1 M sodium phosphate buffer, pH 7.4
Glycol-based cryoprotectant (see recipe)
0.5% (w/v) sodium borohydride in PBS (prepare fresh; optional)
0.5% (v/v) H2O2/30% (v/v) methanol in PBS (prepare fresh; optional)
Primary antibody solution (see recipe and Table 3) [*Copy Editor, here and elsewhere, the authors refer to the primary antibody solution but then cite Table 1. I changed it to Table 3, but please ask them to confirm. It’s Table 1 in the 2014 version.]
Table 3.
Preparation of a Dilution Series for Primary Antibody Incubation
| Final dilution | Stock (μl) | Serum (μl) | TX-100 (μl)a | PBS (μl) |
|---|---|---|---|---|
|
| ||||
| Using a 1:10 stock of primary antibody | ||||
| 1:1,000 | 10 | 10 | 30 | 950 |
| 1:2,000 | 5 | 10 | 30 | 955 |
| 1:5,000 | 2 | 10 | 30 | 958 |
| Using a 1:100 stock of primary antibody | ||||
| 1:1,000 | 100 | 10 | 30 | 860 |
| 1:10,000 | 10 | 10 | 30 | 950 |
| 1:20,000 | 5 | 10 | 30 | 955 |
| Using a 1:500 stock of primary antibody | ||||
| 1:10,000 | 50 | 10 | 30 | 910 |
| 1:20,000 | 25 | 10 | 30 | 935 |
| 1:50,000 | 10 | 10 | 30 | 950 |
The volumes of Triton X-100 are for a 10% solution. This diluted solution can be more reliably pipetted than the viscous concentrated solution sold by the manufacturer.
Biotinylated, affinity-purified secondary antibody against IgG of species used to raise primary antibody
Normal serum generated in same species as secondary antibody
10% (v/v) Triton X-100
Vectastain Elite kit (Vector Laboratories)
Diaminobenzidine (DAB)
Tris-buffered saline (TBS), pH 7.6 (Crawley et al., 2007)
30% H2O2
Bleach
50%, 70%, 95%, and 100% ethanol
Xylene
Resin for mounting coverslips (Cytoseal 60; Stephens Scientific)
Rocking platform
Freezing microtome, chucks, and freezing medium (Shandon M-1 Embedding Matrix; Thermo Scientific)
Plexiglas compartments with porous nets on the bottom (Brain Research Laboratories)
Paintbrush with fine bristles
Gelatin-coated (subbed) microscope slides (unit 1.1)
Coverslips
Prepare tissue
-
1Remove perfused tissue and post-fix it in PLP fixative for 2 to 4 hr at 4°C.This is sufficient to preserve tissue structure without compromising antigenicity. However, the tissue can be stored in the fixative at 4°C for as long as 3 days without consequence. Longer periods of post fixation should be avoided, as aldehydes may hide antibody epitopes by cross-linking proteins, and thereby reduce antigenicity.
-
2Rinse the tissue in 0.1 M sodium phosphate buffer, pH 7.4, and place it in a vial containing 20% to 30% sucrose in 0.1 M sodium phosphate buffer at 4°C, until the tissue sinks to the bottom of the vial, which indicates that satisfactory cryoprotection has been achieved.Adequate cryoprotection generally occurs within 12 to 24 hr at 4°C.
Section tissue
-
3Prepare the tissue for sectioning in the preferred plane of section. Given that virus is often transported extensively through the CNS, particularly at long survival intervals, we typically section the entire brain in the coronal plane. In that case we have obtained excellent reproducible results using the following procedure:
- Bisect the brain at the level of the midbrain and place the two halves on the chuck of a freezing microtome (also see unit 1.1 for sectioning procedures).
- Freeze the tissue to –50°C and section it at 35 μm per section throughout the rostrocaudal extent of the forebrain and brainstem. Place sections sequentially into six 1 ml vials of glycol-based cryoprotectant (Watson et al., 1986).
- Repeat this sequential collection of sections until all of the tissue has been sectioned.Each vial with then contain a representative sample of sections through the brain at a frequency of 210 μm. This frequency provides an accurate sampling of all regions of the neuraxis.
-
4Store the tissue in the glycol-based cryoprotectant at –20°C.As noted above, our experience with the cryoprotectant created by Watson et al. (1986) has demonstrated that this solution preserves antigenicity of tissue for periods of at least 10 years. This has the obvious benefit of permitting a systematic analysis of the synaptology and phenotype of neurons in a polysynaptic circuit defined by viral transport.
Remove cryoprotectant from tissue
-
5Place the tissue in Plexiglas compartments with porous nets on the bottom and remove the cryoprotectant using four 15-min washes with 0.1 M sodium phosphate buffer, pH 7.4 (100 ml per wash), with agitation on a rocker platform.These chambers are available from Brain Research Laboratories in a number of different sizes and with different numbers of compartments. They allow easy movement of tissue through multiple buffer changes.The cryoprotectant is fairly viscous and has a tendency to adhere to tissue. As a result, if it is not completely removed, it will prevent antibody penetration and produce patchy, unreliable staining. If patchy staining is routinely encountered, increase the number and duration of washes.
-
6Optional: Reduce problems with high background by pretreating the tissue at this point as follows:
- Wash for 15 min in 0.5% sodium borohydride in PBS.This solution bubbles, so it is important to monitor throughout the treatment and continually re-immerse the tissue in the solution rather than allowing it to accumulate on surface bubbles.
- Wash four times as described in step 5, each time for 15 min in 0.1 M sodium phosphate buffer.
- Wash for 15 min in 0.5% H2O2/30% methanol in PBS.
- Wash four times, each time for 15 min in 0.1 M sodium phosphate buffer.This pretreatment eliminates background without compromising antigenicity.
-
7Transfer the tissue to the primary antibody solution (Table 3) for localization of infected neurons. Dilute the primary with PBS containing Triton X-100 and normal serum according to Table 3. Incubate tissue 24 to 48 hr in the primary antiserum at 4°C, and then allow it to warm to room temperature over 30 min.If necessary, the length of this step can be extended to 3 days without compromising viral antigenicity. However, longer periods of incubation should be accompanied by longer post-incubation buffer washes, to keep background staining low.
-
8
Wash the tissue four times with agitation at room temperature, each time for 15 min in 10 mM sodium phosphate buffer, pH 7.4.
-
9
Dilute the biotinylated affinity-purified secondary antibody 1:200 by mixing the following:
5 μl of biotinylated secondary antibody
20 μl of normal serum generated in same species as secondary antibody
30 μl of 10% (v/v) Triton X-100
945 μl PBS.
-
10
Place the tissue in the diluted secondary antibody solution and incubate 60 to 90 min at room temperature.
-
11
Wash tissue as in step 8.
-
12Combine 5 μl each of Vectastain Elite kit components A and B 90 min prior to use. Just prior to use, bring to a final volume of 1 ml with 960 μl PBS and 30 μl of 10% Triton X-100. Incubate tissue in this solution for 90 to 120 min at room temperature with agitation.This solution will generate the avidin-biotin complex, which in this kit is conjugated to horseradish peroxidase, and may be detected with DAB and hydrogen peroxide as in the following steps.
-
13
Wash with buffer as in step 8.
-
14Prepare a “saturated” solution of diaminobenzidine (DAB) in TBS, pH 7.6. Filter the solution prior to use.Saturated in this context is defined as the amount of DAB that will go into a solution during 5 min of vigorous stirring at room temperature (do this in a hood). This eliminates the necessity of weighing out this carcinogen.
-
15
Preincubate the tissue in the saturated DAB solution for 10 min at room temperature with occasional agitation.
-
16Add 35 μl of 30% H2O2 per 100 ml of DAB solution and monitor the reaction visually by occasionally examining a section under the microscope. To accomplish this, use a paintbrush with fine bristles to transfer a section to a bath containing TBS, pH 6, and then use the brush to manipulate the section onto the portion of a microscope slide that has been submerged in the buffer. (Make sure that the slide is not gelatin coated, since the tissue section will be returned to the DAB solution following examination.) Briefly examine the wet section under the microscope to determine the density of the immunoperoxidase reaction product relative to background staining.The best staining is achieved when the specific signal is prominent, and the background is low. Of course, this can only be determined in tissue sections that contain infected neurons, so it is important to make an informed selection of the section that will be analyzed.The goal of this procedure is to optimize specific staining (i.e., produce the most intense signal) while keeping the background staining low. This is typically achieved within 3 min when the reagents are used at the recommended dilutions.
-
17
Terminate the reaction by washing the tissue in multiple changes of fresh sodium phosphate buffer. Inactivate the DAB solution, and all washes, with bleach. Dispose of the solutions according to the biohazard regulations of your institution.
-
18
Organize the sections from rostral to caudal in a dish of buffer and then mount the sections on gelatin-coated slides and let them dry on the slides overnight at room temperature.
-
19
Dehydrate the slides using an ethanol series (ethanol solutions of 50%, 70%, 95%, 95% again, 100%, and then 100% again, 10 min each). Clear the slides in three changes of xylene (15 min each), and coverslip with resin for microscopic observation.
SUPPORT PROTOCOL 3
DUAL-IMMUNOFLUORESCENCE LOCALIZATION
Localization of unique reporters expressed by PRV recombinants provides a powerful method for further functional dissection of a polysynaptic circuit. Different-color fluorophores conjugated to species-specific secondary antibodies can be used to determine the phenotype of neurons that contribute to a multisynaptic projection, or for simultaneous localization of recombinant viruses that express unique reporters (Fig. 3). This approach has been greatly aided by the recent development of fluorophores that are resistant to fading so that tissues can be dehydrated, cleared, and coverslipped with minimal loss of signal.
Materials
Sectioned tissue in cryoprotectant (Support Protocol 2)
Primary antibodies generated against PRV or reporter proteins (see note below)
Primary antibodies generated against phenotypic markers of neurons that contribute to the circuit of interest. These antibodies should be generated in a species different from those raised against PRV (see recipe).
Secondary antibodies generated against the IgG of the two species used for the primary antibodies, conjugated, respectively to Cy2 and Cy3 (Jackson ImmunoResearch Laboratories)
Light-proof vials
Fluorescence microscope (unit 2.1)
Additional reagents and equipment for immunohistochemical processing and detection of tissues (see Support Protocol 2) and fluorescence microscopy (unit 2.1) [*Copy Editor: Combs, C. A., & Shroff, H. (2017). Fluorescence microscopy: A concise guide to current imaging methods. Current Protocols in Neuroscience, 79, 2.1.1– 2.1.25. doi: 10.1002/cpns.29]
NOTE: Antibodies against reporter proteins should be generated in different species. Reporters commonly used in the construction of recombinant strains of PRV are β-galactosidase and the enhanced green fluorescent protein (EGFP) (Fig. 3). Antibodies generated against both of these reporters are commercially available from a variety of vendors.
Wash cryoprotectant from tissue using the same method described in the immunoperoxidase procedure (see Support Protocol 2, step 5).
- Place the tissue in a mixture of the two primary antibodies diluted with PBS containing Triton X-100 and normal serum according to Table 3. Incubate 24 to 48 hr at 4°C.The antibodies must be generated in different species to prevent cross-reactivity and maintain the specificity of immunolabeling.The ideal dilution for immunofluorescence labeling using the following procedures is 1/10th the dilution that produces ideal labeling with the immunoperoxidase method described in Support Protocol 2. Thus, if an antibody is used at a 1:20,000 dilution for immunoperoxidase localizations, one can assume that it will work well at 1:2000 for immunofluorescence. However, this should be determined directly in separate parametric studies with each antiserum before initiating the dual-labeling investigations.
Wash the tissue with multiple changes of 10 mM sodium phosphate buffer, pH 7.4, over 30 min at room temperature with agitation.
- In light-proof vials, dilute the Cy2- and Cy3-conjugated, species-specific secondary antibodies to a final concentration of 1:500 with PBS and incubate the tissue for 2 hr in this solution at room temperature with agitation.These fluorophores produce green (Cy2) or red (Cy3) fluorescence and, unlike FITC and rhodamine, are resistant to fading. Cy2 and Cy3 are not as sensitive to light as other fluorophores; nevertheless, the incubations should be performed in light-proof vials and the processed slides stored in light-tight boxes to produce and preserve the optimal signal.
Wash the tissue as in step 3.
Mount the sections on gelatin-coated slides and air dry overnight. Dehydrate and coverslip the sections as described in step 19 of Support Protocol 2.
Examine slides using a fluorescence microscope (unit 2.1).
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see Crawley et al. (1997); for suppliers, see suppliers appendix. [*Copy Editor: (1997), Common Stock Solutions, Buffers, and Media. Current Protocols in Neuroscience, 00: A.2A.1-A.2A.8. https://doi.org/10.1002/0471142301.nsa02as00]
DMEM/1% methocel/sodium bicarbonate/2% FBS/pen-strep
Weigh out a 2% (w/v) solution of methocel (methylcellulose) in distilled water. Add a stir bar and shake to disperse. Autoclave 30 min using a liquid cycle; be sure to keep the stir bar in the solution during autoclaving. Resuspend the autoclaved methocel by stirring overnight at 4°C. Prepare the final solution in a sterile bottle by mixing equal volumes of 2× DMEM and the methocel suspension. Add 2.5 ml of 100× pen-strep (Life Technologies) to 500 ml of the final solution. Add 5 ml of 7.5% (w/v) sodium bicarbonate per 500 ml medium to provide optimal buffering capacity. Lastly, add heat-inactivated fetal bovine serum (FBS; Crawley et al., 2007) for a final concentration of 2%. Store up to 3 months at 4°C.
DMEM/2% (or 10%) FBS/pen-strep
Modify Dulbecco’s modified Eagles medium (DMEM; available commercially from a variety of vendors; store 100-ml and 500-ml aliquots in sterile screw-cap bottles) as follows.
Just prior to use, add:
100 U/ml penicillin G
100 μg/ml streptomycin
Sodium bicarbonate to 7.5% (w/v) final
Heat-inactivated fetal bovine serum (FBS; Crawley et al., 2007)) to 2% or 10% (v/v)
Store prepared medium up to 3 months at 4°C
Use only qualified, performance-tested, mycoplasma-, virus-, and endotoxin-tested FBS for tissue culture. It is best to use the same lot number as much as possible.
Glycol-based cryoprotectant
300 g sucrose
10 g polyvinyl-pyrrolidone (PVP-40)
500 ml 0.1 M sodium phosphate buffer, pH 7.2 (Crawley et al., 2007)
300 ml ethylene glycol
Mix 200 g of sucrose and 10 g of PVP-40 in a 1500-ml beaker. Add 500 ml 0.1 M phosphate buffer slowly and stir until the sucrose and PVP-40 dissolve. Add the remaining (100 g) sucrose with stirring. Stir in the ethylene glycol. Add distilled water to bring the final volume to 1000 ml. Store indefinitely at –20°C.
From Watson et al. (1986).
Phosphate-buffered saline
0.1 M sodium phosphate buffer, pH 7.4 (Crawley et al., 2007)
2.7 mM KCl
1.5 mM KH2PO4
137 mM NaCl
Store up to 6 months at room temperature
PLP fixative
To prepare 4% paraformaldehyde-lysine-periodate (PLP), heat the desired volume of 0.1 M sodium phosphate buffer, pH 7.4 (Crawley et al., 2007), to 70°C in a hood on a hot plate with stirring. Add the appropriate amount of paraformaldehyde and stir vigorously until the solution clears. Chill the solution to room temperature on ice and then add the appropriate amounts of periodate and lysine (see Table 2) while stirring. Filter the solution and store at 4°C until it is used for perfusion.
It is best to use this solution the same day it is made.
From McLean and Nakane (1974).
Primary antibody solution
The best signal is achieved with a polyclonal antiserum generated against inactivated virus. The advantage of this approach is that the immunoreactivity will identify all virally encoded proteins and thereby permit identification of neurons at all stages of productive infection (Fig. 1 and 4). Although not commercially available, numerous laboratories have prepared antisera specific for PRV and are willing to provide aliquots to those interested in conducting tracing studies with PRV. The authors have raised rabbit polyclonal antibodies against acetone-inactivated wild-type virus. These antibodies provide very reliable staining at dilutions of 1:10,000. Limited aliquots of these rabbit polyclonal antisera are available through the CNNV (http://www.cnnv.pitt.edu). A rabbit polyclonal antiserum against PRV is also available commercially from Thermo Scientific (PA1–081). In direct comparisons, we demonstrated that this antiserum produces staining equivalent to our rabbit polyclonals.
Dilute antibodies with 10 mM sodium phosphate buffer, pH 7.4 (Crawley et al., 2007) containing 0.3% (v/v) Triton X-100 to increase antibody penetration and 1% normal serum to reduce background. The normal serum should be of the same species used to generate the secondary antibodies. A final volume of 1 ml is adequate for incubation of one vial of tissue. Table 3 provides a convenient means of preparing various dilutions of antiserum.
Trypsin-EDTA
Obtain the stock solution containing 0.5% trypsin and 5.3 mM EDTA (Life Technologies) and store this stock at –20°C in 2-ml aliquots. To prepare the final working concentrations for tissue culture, dilute in PBS (see recipe) to 0.1% trypsin and 1.06 mM EDTA.
COMMENTARY
Background Information
Viral transneuronal analysis shares many of the methodological considerations important for the successful use of classical anterograde and retrograde monosynaptic tracers. Nevertheless, the viral transneuronal method differs considerably from classical methods in that it is dependent upon the tropism, invasiveness, and life cycle of live pathogens. The fact that these pathogens exploit the polarized architecture and synaptic associations of neurons provides the fundamental basis for the method and emphasizes the importance of understanding the biological mechanisms through which these pathogens parasitize the nervous system. This mechanistic foundation has provided much of the support for the specificity of transynaptic passage of virus and is central to successful application of the method. The following sections review issues fundamental to the successful use of PRV for transneuronal analysis. Although this protocol focuses exclusively on transsynaptic tracing with PRV, it is important to note that rabies rabies virus (RV) has been widely used as a purely retrograde tracer in animal models including rodents and non-human primates. RV is an enveloped RNA virus, member of the Rhabdoviridae family with a bullet-shaped virion of 180×75 nm in size. RV is retrogradely transported trenassynaptically [*Copy Editor: Please ask the author to confirm the spelling of this word.] in a time-dependent fashion (Kelly and Strick., 2000). PRV and RV are sometimes confused due to naming similarities. However, they are completely different viruses with PRV containing a large 120 kB, enveloped double-stranded DNA genome, and RV containing a small 12 kB single-stranded (negative sense) RNA genome. Wild-type PRV can spread to both presynaptic and postsynaptic connected neurons, whereas RV can only spread to presynaptic neurons (purely retrograde). Although multiple RV strains have been used for tracing studies, the most common ones are CVS-N2c and SAD-B19 (Callaway., 208; Reardon et al., 2016; Wickersham et al., 2007). Replication deficient, pseudotyped RV recombinants have been developed more recently to further control tracing properties, rendering these mutants to be monosynaptically-restricted and to only infect a defined subset of input cells. The Center for Neuroanatomy with Neurotropic Viruses (CNNV) not only shares with investigators a large collection of PRV mutants for tracing studies, but also RV and HSV-1 recombinants (http://www.cnnv.pitt.edu). Currently, the CNNV has a repository of 16 RV recombinants from the CVS-N2c strain including both standard and pseudotyped mutants expressing a plethora of reporter genes including mCherry, GFP, Chr2, YFP, dsRed, tdTomato, and GCaMP6.
Critical Parameters
Safe handling of viruses
PRV and other alpha herpesviruses are designated Class 2 infectious agents on the basis of CDC/NIH criteria. PRV is not known to cause human infections in the laboratory. However, caution when using PRV is appropriate because there are reports of rare human infection in farm workers (Yang et al., 2019). Therefore, the biosafety level 2 (BSL-2) classification mandates that all experiments be conducted in a laboratory that meets the criteria defined in Biosafety in Microbiological and Biomedical Laboratories (BMBL) 6th Editiion from the CDC and the NIH. The regulations in that publication stipulate that BSL-2 laboratories (1) be dedicated to the viral studies and have restricted access, (2) contain a biosafety cabinet for handling of virus, (3) contain a HEPA-filtered housing unit for infected animals, and (4) have containers for disposal of infectious waste. Procedures mandated for experimental procedures in BSL-2 laboratories and for disposal of waste are also detailed. Some institutions interpret these regulations differently, so investigators should check with their respective institutional biosafety officers regarding the regulations, to obtain approval for conducting the studies. Importantly, action plans for dealing with accidents should be approved by the institutional biosafety official and discussed with all personnel participating in the study.
Maintaining viable virus stocks
The production and maintenance of high-titer viral stocks is essential to the transneuronal tracing method. Infection of an animal requires delivery of a minimum concentration of biologically active virus. Thus, it is absolutely essential to maintain high-titer stocks of virus that will provide useful data. In parametric analyses using rats, a minimum titer of a total of 105 pfu is necessary to achieve productive replication of PRV in 100% of injected animals (Card et al., 1995). When viral titer is reduced by ten-fold (104 total pfu), only 20% of animals become infected. Viral particles injected into tissue may bind unproductively and never reach a permissive cell. The site of injection and its tissue architecture dictates the amount of virus particles required for efficient and reproducible infection. Additionally, studies involving intracerebral injection have shown that the onset and progression of viral infection within a circuit is dependent, at least in part, upon the concentration of the inoculum (Card et al., 1999). Thus, a general rule of thumb is that the higher the concentration of virus, the earlier the onset of viral replication within a multisynaptic circuit. Of course, the number of cells susceptible to and available for infection at the site of virus injection is also a critical parameter.
The proteins in the lipid envelope of alpha herpesviruses are essential for infection of permissive cells, so anything that damages this envelope and its constituent proteins will reduce the titer of the stock, often dramatically. Ultraviolet light, bleach, detergents, alcohols, aldehydes, and organic solvents all reduce viral titer in this fashion. Ice-crystal formation resulting from freeze-thaw cycles also reduces titer through damage to the viral envelope. As a general rule, the small volumes of virus (50 to 100 μl) that result from the division of the neurostocks into aliquots should not be thawed and refrozen more than once. Rather, they should be thawed immediately prior to use and the unused portion should be inactivated and discarded. Similarly, neurostocks should be transferred immediately to a low-temperature freezer and stored at less than or equal to –70°C until use. Storing these aliquots at temperatures higher than –70°C will reduce their titer. Stocks should never be frozen at –20°C under any circumstances, since titer will drop four to five log units due to ice formation, with resulting virion damage. Also, it is important to avoid creation of large surface areas exposed to air during frozen storage (e.g., placing small volumes in tubes with a wide internal diameter), as this practice invariably results in reduced titers due to freeze-drying in the cold, dry freezer.
It is often desirable to use a single aliquot of virus to inject more than one animal during the same sitting. If this is the case, the virus should be kept cool by storing vials on wet ice during the period throughout the multiple surgeries. In unpublished studies, the authors have determined that the titer of PRV-Bartha or its derivatives will not exhibit a detectable reduction over a period of 3 to 6 hr when stored in this manner.
Selecting a strain of virus
Although directionally selective strains of HSV have been identified (e.g., Zemanick et al., 1991; Wojaczynski et al., 2015), there is no known strain of PRV that is transported only in the anterograde direction through CNS circuitry. PRV-Bartha and related strains are well known for their selective retrograde spread through the brain following inoculation of peripheral targets innervated by autonomic or somatic motor neurons (e.g., Fig. 4). These strains also move selectively in the retrograde direction after intracerebral injection. Although early studies suggested that Bartha had the capacity to infect by anterograde spread through visual circuitry (Card et al., 1991), it is now clear that the virus only spreads by retrograde transneuronal passage (Pickard et al., 2002; Smeraski et al., 2004).
The use of recombinant viruses expressing reporter proteins, such as β-galactosidase or EGFP, in dual-injection paradigms imposes additional demands upon selecting and characterizing the strains of virus that will be used in the analysis. Ideally, the two recombinants will only differ in the transgenes that they contain. In other words, the genome of the two strains should be isogenic, differing only in the transgenes that they express. The goal is to use two strains that are attenuated for virulence, equivalent in the temporal aspects of their invasive characteristics, and that have the ability to simultaneously replicate in the same neurons. Prior studies have shown that it is possible to coinfect neurons with strains of virus that are not isogenic (e.g., Jansen et al., 1995; Levatte et al., 1998; Kim et al., 1999). However, the use of nonisogenic viruses that differ in invasiveness and virulence can produce false negatives due to superinfection exclusion as explained above. This interference is more prevalent when the virulence of one strain exceeds that of the other. In addition, recent studies demonstrate that cells only replicate a small number (approximately 7) of viral genomes (Kobiler et al., 2010, Taylor et al., 2012). Thus, it is imperative to conduct single-infection studies to determine the temporal progression of infection in each component of the circuit and to design dual-infection experiments so that virus reaches putative collateralized neurons within the same time frame. Construction of isogenic strains is designed to reduce the variables that can lead to this type of interference.
Recombinant viruses must be evaluated for the efficiency of transgene expression in the paradigm under investigation, especially when transgene expression is the sole indicator of viral invasiveness. It is therefore important to define both the magnitude and temporal expression of transgenes in single-injection studies, examining the invasiveness of the recombinants in the same circuitry that will be the subject of the dual-injection experiments. Dual-immunofluorescence localizations of transgenes and viral proteins provide the most rigorous determination of the effectiveness of the transgene as an identifier of the invasiveness of recombinants. The two fluorophores identifying the location of the transgenes and viral immunoreactivity should be entirely colocalized in these single-injection experiments. If neurons exhibit viral antigen, but no transgene immunoreactivity, then the transgene is not being expressed by all infected neurons. If this is the case, one should either seek out another recombinant strain or determine if the measures outlined below (see Troubleshooting) can solve the problem. The large number of recombinant viruses currently available has dramatically improved the efficiency of labeling of neurons in dual-infection studies and has also improved the ability to discriminate dual-infected neurons. Of particular note in this regard is the construction of PRV-Bartha and HSV recombinants that express fluorophores as capsid fusion proteins (Kobiler et al., 2011; Kramer et al., 2012; Taylor et al., 2012a,b; Granstedt et al., 2013b; Kratchmarov et al., 2013, Wojaczynski et al., 2015; Hogue et al., 2015). Since capsids are assembled within the cell nucleus at an initial stage of viral replication, the fluorescent fusion proteins provide an efficient means of labeling the nucleus of infected neurons. When these recombinants are combined with recombinants that express diffusible cytoplasmic reporters (e.g., PRV-152 and PRV-614), the compartmentalized reporters provide easily discriminated markers of dual- and single-infected neurons (Fig. 3).
The cellular staining resulting from viral replication is another important feature that should be evaluated when selecting a strain of virus and in interpreting the temporal kinetics of viral infection within a circuit. Immunocytochemical localization of infected neurons with polyclonal antibodies that react with viral structural proteins (proteins present in mature virus particles) will identify more than 40 proteins expressed from the viral genome in different cellular compartments. When combined with the stepwise progression of virion replication and assembly, the different stages of the viral life cycle are reflected in the intracellular distribution of viral antigens detected by immunohistochemistry (Fig. 1. For neurons in early stages of viral replication, viral structural proteins will be concentrated within cell nuclei due to the import and assembly of capsid proteins within the cell nucleus. As immature virus particles enter the cytoplasm for envelopment, viral immunoreactivity is not only concentrated in the cell nucleus, but also appears in the soma and proximal dendrites. As mature virions participate in secondary envelopment, they traffic throughout the somatodendritic compartment (late stage of infection), and viral protein immunoreactivity provides an increasing dense stain of the dendritic tree, including its most distal processes. The differential distribution of viral antigens characteristic of each of these phases provides a powerful means of deciphering connectivity of neurons in complex networks that contain a high degree of reciprocity of connections among neurons contributing to the circuits. Recent studies have performed transneuronal circuit analysis with PRV in whole intact brains, providing a more holistic connectivity map of the CNS. This approach is based on two innovative methods: tissue clearing, and light-sheet microscopy to image whole organs, animal embryos, flies and worms without the need for laborious tissue sectioninig (Maturana et al., 2020; Pisano et al., 2021 & 2022; Schneeberger et el., 2019).
Designing an experiment.
Aside from selection of the particular virus strain that will be used, four primary considerations should be evaluated when designing a transneuronal tracing study. These include (1) the amount of virus stock that will be injected, (2) the cytoarchitecture of the area of injection, (3) the distance that virus particles must travel in axons and the estimated number of neurons that may contribute to the circuit, and (4) the number of time points after infection that should be examined to document the sequential movement of virus through the polysynaptic circuit (Fig. 2). Collectively, these factors influence the outcome and interpretation of an experiment and will dictate the number of experimental groups included in the experimental design and the post-inoculation time periods sampled.
Concentration of injected virus and cytoarchitecture of injection site
The concentration of virus particles in pfu (determined on the PK15 cell line) per injected volume is heavily dependent upon the cytoarchitecture of the area of inoculation. This fact is in large part due to the variable affinities of alpha herpesviruses for different cellular receptors found on cells of the tissue under study (Vahlne et al., 1978, 1980; Marchand and Schwab, 1987; Sams et al., 1995), and the demonstration that infectivity and the onset of viral replication in a circuit is concentration dependent (Card et al., 1995, 1999). PRV and HSV have very high affinities for axon terminals, astroglia, and extracellular matrix proteins. Therefore, diffusion of virus from the injection site is markedly restricted in areas where these elements are present in high concentration. In contrast, when they are present in low density, or when the area of injection has a high extracellular fluid volume, viral particle diffusion will be greater and the concentration of virus accumulated by permissive cells within the region of injection will be lower. As a result, larger amounts of virus stock are necessary to elicit a productive replication of virus. The importance of considering these factors in planning and executing an experiment is illustrated by the following examples.
A comparison of the invasiveness of PRV following injection of the kidney and spleen effectively illustrates the dramatic influences that tissue architecture exerts upon the outcome of infection (Cano et al., 2001, 2004). The spleen is a highly vascular organ in which an “open” circulation substantially increases the fluid content of its parenchyma relative to other organs. In contrast, the kidney contains a “closed” circulation network in which capillary beds shunt blood directly from the arterioles to venioles. This difference in architecture dramatically influences the amount of virus that must be injected to elicit a productive replication of virus in the neurons innervating each of these organs. The increased fluid content of the spleen parenchyma causes an immediate dilution of virus injected into this organ that is not observed after injection of the same concentration of virus into the kidney. As a result, 1 μl of virus (1 × 108 pfu/ml) produces a robust infection of neurons innervating the kidney, while 6 μl of the same viral stock must be injected into the spleen to elicit an infection of the neurons giving rise to the splenic innervation. Of similar importance is the density of innervation of solid tissues; e.g., tissues with a high density of innervation require injection of a smaller amount of virus than tissues with a sparse innervation. This point is illustrated by studies that have injected peripheral fad pads, which have a sparse neural innervation (Bamshad et al., 1998, 1999; Cano et al., 2003; Bartness and Song, 2005). Similar to the spleen, injection of large amount of virus stock is necessary to reproducibly infect circuits innervating adipose tissue, in this case because of the combination of the sparse neural innervation and the high affinity of virus for extracellular matrix molecules that restrict spread of virions from the site of injection. Injection of larger volumes of virus into multiple sites circumvents these problems. However, the amount of virus and the number of injections necessary to obtain reproducible results must be determined empirically in parametric studies.
Intracerebral injection of PRV requires the same consideration of tissue architecture in experimental design. The high affinity of PRV for axon terminals has an important influence on the outcome of infection. The availability of virus particles that can invade neurons is directly related to the concentration of the injected virus, terminal field density, and the concentration of astroglia in the area of viral injection (Fig. 2). These factors combine to provide an advantage for neurons that densely innervate the region of injection over those that provide a moderate or sparse innervation of the same region. This advantage was demonstrated directly in an analysis of the uptake of different concentrations of PRV in the striatum (Card et al., 1999). Decreasing concentrations of PRV did not compromise the onset of viral replication in the dense nigrostriatal dopaminergic projection system, but caused a temporal delay or entirely blocked the onset of viral replication in other neurons giving rise to striatal afferents of moderate or sparse density (Card et al., 1999). Furthermore, the magnitude of the delay correlated with the density of innervation. When one considers the demonstrated affinity of alpha herpesviruses for astroglia and extracellular matrix molecules, it becomes apparent that several aspects of tissue architecture will influence the amount of virus available to neuronal elements in the region. Thus, knowledge of the tissue architecture of a region is essential for planning intracerebral studies, particularly when afferents of moderate or sparse density are central to the circuit of interest.
Length of transport and number of neurons in a circuit
The distance that virus particles must travel through the brain and the number of neurons that must replicate the virus as it moves through a polysynaptic circuit have an important influence upon experimental design (Fig. 2). It is probable that other variables (e.g., neuronal activity and cellular metabolism) also influence the progression of infection through a circuit, but these influences are more difficult to measure. Since the ultimate goal of a transneuronal study is to define the sequential passage of virus through a circuit, a temporal analysis including multiple post-inoculation intervals should be an integral component of a transneuronal tracing study. The timing and frequency of the post-inoculation intervals must to be determined empirically for each circuit, but a good strategy is to define the longest post-inoculation time point that contains infected neurons in all of the cell groups of interest, and then to work backwards toward the first-order neuron. Since ~6 hr on average is the midpoint of maximal production of infectious progeny by an infected cell, one can estimate this time point on the basis of the number of neurons that are thought to contribute to a circuit and the distance that separates them. Nevertheless, the unexpected inclusion of interneurons within a circuit, and other factors, may render these initial estimates inaccurate. Therefore, one should not be surprised if the initial results to not correspond with these estimates.
Verifying the route of viral transport
With advancing times after initial infection, it becomes increasingly possible that a cell group was infected through a route other than that initially hypothesized. Similarly, it is often difficult to distinguish projection neurons, whose cell bodies are infected by retrograde transport of virus particles in axons from a distant site, from interneurons in the same area that were infected by trans-synaptic passage of virus from the infected projection neurons. The most direct way of addressing these issues is to verify the postulated route of viral transport by incorporating lesions or knife cuts in the experimental design and to use classical monosynaptic tracers in combination with the viral transneuronal analysis.
Lesions have proven to be crucial in eliminating alternative routes of viral transport in studies involving intracerebral injection of virus. This approach is illustrated by the results of two early studies involving the intracerebral injection of PRV-Bartha. O’Donnell et al. (1997) used electrolytic lesions of the ventral pallidum or globus pallidus to confirm that these areas were involved in disynaptic circuits connecting the nucleus accumbens with the mediodorsal thalamic nucleus. Similarly, Jasmin et al. (1997) used ibotenic acid lesions to demonstrate that a polysynaptic nociceptive pathway included the parabrachial nucleus in the midbrain. In both cases, these control studies were essential to establishing the postulated routes of viral transport, since the post-inoculation intervals were long, and the temporal analysis could not exclude alternative routes of viral transport. Other circumstances where the post-inoculation interval is shorter may be less dependent upon such verification, but in most cases, it will be desirable to use lesions or knife cuts to validate the contribution of one or more cell groups to the postulated circuit. In these cases, the lesions should be made ~1 week prior to the injection of the virus, and all other aspects of the analysis should reproduce those employed in the initial viral-tracing experiments. Separation of the timing of the lesion and viral injection appear to be important, since there are suggestions that the inflammatory response generated by the lesion may negatively influence viral replication and transport (Denes et al., 2006). Additionally, the post-inoculation survival should be well in excess of the time necessary to infect all elements of the circuit in cases not involving lesions.
The use of viral tracing in combination with conventional monosynaptic tracers can be quite effectively applied to discriminate first-order projection neurons from local circuit neurons that synapse upon those neurons and reside in the same vicinity. This is effectively illustrated by experiments in which PRV-Bartha and the β subunit of cholera toxin (βCT) were simultaneously injected into the wall of the stomach. Both tracers invaded the peripherally projecting process of preganglionic parasympathetic neurons and were then retrogradely transported to accumulate in the parent neurons in the dorsal motor vagal nucleus (DMN) of the caudal brainstem. However, PRV replicated and passed trans-synaptically to infect neurons in the immediately adjacent nucleus of the solitary tract, while βCT remained trapped within the projection neurons of the DMN. Similar approaches could be used to discriminate interneurons from projection neurons in motor nuclei that innervate peripheral targets, or in intracerebral-injection paradigms. However, it is important to recognize that PRV is not compatible with all classical tracers. LaVail et al. (1993) have shown that FluoroGold interferes with the ability of HSV to invade and replicate within trigeminal neurons when the two are simultaneously applied to scarified cornea. Thus, it is important to conduct parametric studies in well-characterized systems to ensure that the classical tracer does not interfere with viral replication.
Troubleshooting
Low rates of infection
If animals show no signs of infection, or if labeling of a new neuronal circuit gives unpredictable results, one must immediately suspect that the concentration of biologically active virus is too low. The best strategy in these cases is to determine the infectivity of the same stock in another circuit where viral transport parameters are well known. If variable results are observed in that analysis, it is probable that the titer of the virus has dropped during storage and handling. This loss of infectivity can occur after prolonged storage of viral aliquots, even in the absence of a rise in temperature if freeze-drying occurs. Thus, the titer of the stock should be re-examined, and, if the titer is low, the stock should be discarded.
Variability in the progression of infection
Many of the reasons for variability in the progression of infection were discussed in the protocols. Variations in the temporal course of viral transport through the CNS after peripheral injection may be related to variability in the diffusion of virus particles from the injection site. This is not unusual in peripheral-injection paradigms, since the innervating axons often arborize diffusely and the cytoarchitecture of the injection target makes it difficult to precisely determine the diffusion of the virus from the injection site. Virus particles are about 200 nm in diameter, and diffusion rates will be slow and limited in most tissues. The best approach under these circumstances is to measure the progression of viral infection from the first-order neuron in the CNS. For example, the first CNS infection after injection of an autonomic target will occur in preganglionic neurons. In the case of a stomach-muscle injection, these neurons will be found in the intermediolateral cell column of thoracic spinal cord and the dorsal motor vagal nucleus of caudal brainstem (Fig. 4). Once these neurons become infected, the progression of infection through the balance of the polysynaptic CNS circuit synapsing upon them should occur in a predictable manner. If the number of neurons in the sites of first-order CNS infection vary dramatically from one animal to another, and one is sure that equivalent amounts of virus are being injected in a consistent manner, it should be determined if the titer of the viral stock has become compromised.
One has greater control in experiments involving intracerebral injection, and the progression of infection is, therefore, more predictable than in experiments involving peripheral injection. If variability does occur in intracerebral paradigms, one should investigate the following potential confounding variables. First, one should make sure that the inoculum is not being drawn up the cannula tract as the cannula/pipet is being withdrawn from the brain parenchyma. This artifact is sometimes difficult to determine, since virions have a preferential affinity for axon terminals and are transported to a distant site following uptake. The best approach to resolving problems of this nature is to extend the post-injection interval following completion of injection and removal of the pipet/cannula. Second, if one is injecting a cell group in close proximity to the ventricles, it is quite probable that some of the inoculum may escape into the cerebrospinal fluid (CSF). If this happens, the effective concentration of virus at the injection site will be reduced, and will produce substantial variability in the uptake of virus from one animal to another. It is generally quite easy to identify this problem, since leakage of virus into the CSF will lead to viral replication in ependymal cells lining the ventricles (Chen et al., 1999) as well as infection of a distinct population of midbrain serotoninergic neurons that project into the ventricles (Larsen et al., 1996). To address this problem: (1) pulled glass pipets should be used, since they cause less tissue damage; (2) the virus stock should be injected at a slower rate, so that it is more easily accommodated by the parenchyma; and (3) the pipet should be kept in place for a longer time after injection so that the inoculum invades cellular elements at the injection site efficiently. Each of these parameters will have to be determined empirically according to the cell group that is being injected.
False negatives
Two circumstances promote the possibility of false negatives. The first is the failure to detect viral immunoreactivity in a cell group known to project to the area of injection. One should always be aware of the possibility that not every infected neuron will produce infectious virus or express the reporter gene. Such a phenomenon was directly demonstrated in the work of Rotto-Percelay et al. (1992), who demonstrated that dorsal root ganglion neurons showed no signs of viral replication after injection of the gastrocnemius muscle, but contained viral DNA. This repression of viral genomes is a difficult issue to deal with experimentally and emphasizes the importance of being conservative in interpreting viral tracing data. In essence, the presence of viral proteins can be confidently interpreted as evidence that a neuron is part of a circuit, but negative data should not be used to exclude a neuron from a circuit.
False negatives related to inefficient expression or lack of expression of transgenes by recombinant viruses can also confound the interpretation of dual-injection experiments. The potential interference of one strain of virus with the replication of a second strain was discussed earlier (see Critical Parameters). Those studies emphasize the importance of selecting isogenic strains that are attenuated for virulence. However, even attenuated strains of PRV are replicating and do ultimately evoke an immune response (Denes et al., 2006, Laval et al., 2018). Mabon et al. (1999) have demonstrated that treatment with the immunosuppressant cyclosporin A improves the efficiency of transgene expression in sympathetic preganglionic neurons infected with replication-deficient strains of HSV type 1. This finding suggests that a similar treatment may be effective in dual-injection paradigms involving recombinant viruses, but this concept remains to be tested.
Understanding Results
The sequential passage of virus through synaptically linked neurons follows a well scripted and identifiable path. Thus, viral proteins will appear sequentially within neurons according to the hierarchy of synaptic connections (e.g., Fig. 3). When the neurons contributing to a circuit are spatially separated, it is a relatively straightforward process to follow progressive movement of virus through the circuit by conducting a temporal analysis in which animals are sacrificed at multiple post-inoculation intervals. However, there will be instances when projection neurons and interneurons are intermixed, making it difficult to determine first- versus second-order infection. Under those circumstances, insights into temporal progression can be derived from examination of the intracellular distribution of viral antigens (Fig. 1).
The length of time necessary to complete an experiment is directly dependent upon the route of injection, the extent of the circuit that one is investigating, and the virulence of the virus used. Most PRV strains used for tracing are replicating, polysynaptic tracers that will ultimately kill the animal under study. It is useful to determine the average time to the appearance of terminal symptoms (one particularly reliable determinant is a precipitous weight loss) for each virus in each paradigm. For most experiments using peripheral injection of replicating PRV-Bartha strains, infected animals can be expected to live no longer than 5 or 6 days. Experiments involving peripheral inoculation of CNS targets (e.g., muscles, organs, or ganglia) require the longest periods of time, since the virus must be transported from the periphery into the CNS. In general, 4 days are adequate to obtain extensive transneuronal labeling of the neuraxis in most experiments involving peripheral inoculation, but it is necessary to determine this directly in each paradigm. Adequate transport in studies involving intracerebral injection of virus can generally be achieved within 72 hr. For rodent experiments involving central injection of PRV-Bartha, animals can be expected to live no longer than 4 days.
Quantitative studies asserting plasticity within a circuit or reorganization of synaptic connections within circuitry in response to injury or disease are difficult to perform due to the inherent variability involved in the initial injection of virus. However, incorporation of internal quantitative measures that standardize comparisons among groups of animals can be successfully employed to demonstrate plastic changes in circuit organization. For example, statistically significant differences in the developmental assembly of preautonomic circuitry in response to experience were shown to occur using retrograde transneuronal transport of PRV from the viscera (Card et al., 2005a). This study was accomplished by standardizing comparisons of retrograde spread of infection of forebrain cell groups to numbers of neurons in the dorsal motor vagal complex of the caudal brainstem (Fig. 4). Similarly, quantitative measures were used to demonstrate plastic reorganization of neural circuits following traumatic brain injury (Card et al., 2005b). In all such studies it is essential to incorporate internal quantitative measures that control for variability that may occur in viral transport and replication.
To obtain reproducible results, all tissue must be processed quickly following transcardiac perfusion fixation. The tissue should be post-fixed, cryoprotected in sucrose, sectioned, and placed into cryopreservative for storage at –20°C, without delay. This approach standardizes the preservation of all tissue and reduces the possibility that tissue antigenicity will be compromised by storage at lower temperature. It also permits a systematic characterization of the organization and phenotype of neurons labeled by viral transport over a more protracted period of time.
Acknowledgements
We would like to acknowledge NIH funding P40OD010996 in support of the Center for Neuroanatomy of Neurotropic Viruses (CNNV).
Footnotes
Conflict of Interest Statement
Esteban A. Engel is an employee and equity holder of Sparks Therapeutics, a Roche company located in Philadelphia, PA 19104, USA.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- Archin NC, van den Boom L, Perelygina L, Hilliard JM, and Atherton SS 2003. Delayed spread and reduction in virus titer after anterior chamber inoculation of a recombinant of HSV-1 expressing IL-16. Invest. Ophthalmol. Vis. Sci 44: 3066–3076. [DOI] [PubMed] [Google Scholar]
- Archin NM and Atherton SS 2002. Rapid spread of a neurovirulent strain of HSV-1 through the CNS of BALB/c mice following anterior chamber inoculation. J. Neurovirol. 8: 122–135. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Aoki VT, Adkison MG, Warren WS, and Bartness TJ 1998. Central nervous system origins of the sympathetic nervous system outflow to white adipose tissue. Am. J. Physiol 275: R291–R299. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Song CK, and Bartness TJ 1999. CNS origins of the sympathetic nervous system outflow to brown adipose tissue. Am. J. Physiol 45: R1569–R1578. [DOI] [PubMed] [Google Scholar]
- Banfield BW, Kaufman JD, Randall JA, and Pickard GE 2003. Development of pseudorabies virus strains expressing red fluorescent proteins: New tools for multisynaptic labeling applications. J. Virol 77: 10106–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett EM, Evans GD, Sun N, Perlman S, and Cassel MD 1995. Anterograde tracing of trigeminal afferent pathways from the murine tooth pulp to cortex using herpes simplex virus type 1. J. Neurosci 15: 2972–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartness TJ and Song CK 2005. Innervation of brown adipose tissue and its role in thermogenesis. Can. J. Diabetes 29: 420–428. [Google Scholar]
- Billig I, Foris JM, Enquist LW, Card JP, and Yates BJ 2000. Definition of neuronal circuitry controlling the activity of phrenic and abdominal motoneurons in the ferret using recombinant strains of pseudorabies virus. J. Neurosci 20: 7446–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkoi Z, Reichart A, Toth IE, Sik A, Erdelyi F, Medveczky I, Llorens-Cortes C, Palkovits M, and Lenkei Z. 2002a. Construction of recombinant pseudorabies viruses optimized for labeling and neurochemical characterization of neural circuitry. Mol. Brain Res 109: 105–118. [DOI] [PubMed] [Google Scholar]
- Boldogkoi Z, Szabo A, Vrbova G, and Nogradi A. 2002b. Pseudorabies virus-based gene delivery to rat embryonic spinal grafts. Human Gene Ther. 13: 719–729. [DOI] [PubMed] [Google Scholar]
- Boldogkoi Z, Sik A, Denes A, Reichart A, Toldi J, Gerendai I, Kovacs KJ, and Palkovits M. 2004. Novel tracing paradigms—genetically engineered herpesviruses as tools for mapping functional circuits within the CNS: present status and future prospects. Progr. Neurobiol 72: 417–445. [DOI] [PubMed] [Google Scholar]
- Boldogkoi Z, Balint K, Awatramani GB, Balya D, Busskamp V, Viney TJ, Lagali PS, Duebel J, Pasti E, Tombacz D, Toth JS, Takacs IF, and Scherf BG 2009. Genetically timed, activity-sensor and rainbow transsynaptic viral tools. Nat. Methods6: 127–130. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008. Dec;18(6):617–23. doi: 10.1016/j.conb.2009.03.007. Epub 2009 Apr 6. PMID: 19349161; PMCID: PMC2698966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campadelli-Fiume G, Cocchi F, Menotti L, and Lopez M. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol 10: 305–309. [DOI] [PubMed] [Google Scholar]
- Campbell RE and Herbison AE 2007a. Defining the gonadotrophin-releasing hormone neuronal network: Transgenic approaches to understanding neurocircuitry. J. Neuroendocrinol 19: 61–573. [DOI] [PubMed] [Google Scholar]
- Campbell RE and Herbison AE 2007b. Definition of brainstem afferents to gonadotropin-releasing hormone neurons in the mouse using conditional viral tract tracing. Endocrinology 148: 5884–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano G, Sved AF, Rinaman L, Rabin BS, and Card JP 2001. Characterization of the central nervous system innervation of the rat spleen using viral transneuronal tracing. J. Comp. Neurol 439: 1–18. [DOI] [PubMed] [Google Scholar]
- Cano G, Passerin AM, Schiltz JC, Card JP, Morrison SF, and Sved AF 2003. Anatomical substrates for the central control of sympathetic outflow to intercapsular adipose tissue during cold exposure. J. Comp. Neurol 460: 303–326. [DOI] [PubMed] [Google Scholar]
- Cano G, Card JP, and Sved AF 2004. Dual viral transneuronal tracing of central autonomic circuits involved in the innervation of the two kidneys in the rat. J. Comp. Neurol 471: 462–481. [DOI] [PubMed] [Google Scholar]
- Card JP 1998. Exploring brain circuitry with neurotropic viruses: New horizons in neuroanatomy. Anat. Rec 253: 176–185. [DOI] [PubMed] [Google Scholar]
- Card JP and Enquist LW 2012. Use and visualization of neuroanatomical viral transneuronal tracers. In Visualization Techniques From Immunohistochemistry to Magnetic Resonance Imaging, Vol. 70 (Badoer E, ed.) pp. 225–268. Humana Press, London. [Google Scholar]
- Card JP, Whealy ME, Robbins AK, Moore RY, and Enquist LW 1991. Two alpha-herpesvirus strains are transported differentially in the rodent visual system. Neuron 6: 957–969. [DOI] [PubMed] [Google Scholar]
- Card JP, Rinaman L, Lynn RB, Lee B-H, Meade RP, Miselis RR, and Enquist LW 1993. Pseudorabies virus infection of the rat central nervous system: Ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci 13: 2515–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Dubin JR, Whealy ME, and Enquist LW 1995. Influence of infectious dose upon productive replication and transynaptic passage of pseudorabies virus in rat central nervous system. J. NeuroVirol 1: 349–358. [DOI] [PubMed] [Google Scholar]
- Card JP, Levitt P, and Enquist LW 1998. Different patterns of neuronal infection after intracerebral injection of two strains of pseudorabies virus. J. Virol 72: 4434–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Enquist LW, and Moore RY 1999. Neuroinvasiveness of pseudorabies virus injected intracerebrally is dependent on viral concentration and terminal field density. J. Comp. Neurol 407: 438–452. [DOI] [PubMed] [Google Scholar]
- Card JP, Levitt P, Gluhovsky MY, and Rinaman L. 2005a. Early experience modifies the postnatal assembly of autonomic emotional motor circuits in rats. J. Neurosci 25: 9102–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Santone DJ, Gluhovsky MY, and Adelson PD 2005b. Plastic reorganization of hippocampal and neocortical circuitry in experimental traumatic brain injury in the immature rat. J. Neurotrauma 22: 989–1002. [DOI] [PubMed] [Google Scholar]
- Card JP, Sved JC, Craig B, Raizada M, Vazquez J, and Sved AF 2006. Efferent projections of rat rostroventrolateral medulla C1 catecholamine neurons: Implications for the central control of cardiovascular regulation. J. Comp. Neurol 499: 840–859. [DOI] [PubMed] [Google Scholar]
- Card JP, Lois J, and Sved AF 2010. Distribution and phenotype of Phox2a-containing neurons in the adult Sprague-Dawley rat. J. Comp. Neurol 518: 2202–2220. [DOI] [PubMed] [Google Scholar]
- Card JP, Kobiler O, Ludmir EB, Desai V, Sved AF, and Enquist LW 2011a. A dual infection pseudorabies virus conditional reporter approach to identify projections to collateralized neurons in complex neural circuits. PLos One 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Kobiler O, McCambridge J, Ebdlahad S, Shan Z, Raizada MK, Sved AF, and Enquist LW 2011b. Microdissection of neural networks by conditional reporter expression from a Brainbow herpesvirus. Proc. Natl. Acad. Sci. U.S.A. 108: 3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yang M, Miselis RR, and Aston-Jones G. 1999. Characterization of transsynaptic tracing with central application of pseudorabies virus. Brain Res. 838: 171–183. [DOI] [PubMed] [Google Scholar]
- Curanovic D, Lyman MG, Bou-Abboud C, Card JP, and Enquist LW 2009. Repair of the UL21 locus in pseudorabies virus Bartha enhances the kinetics of retrograde, transneuronal infection in vitro and in vivo. J. Virol 83: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco J, Tomishima MJ, Liu H, Zhao C, Cai X, Marth JD, Enquist LW, and Friedman JM 2001. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291: 2608–2613. [DOI] [PubMed] [Google Scholar]
- Denes A, Boldogkoi Z, Hornyak A, Palkovits M, and Kovacs KJ 2006. Attenuated pseudorabies virus-evoked rapid innate immune response in the rat brain. J. Neuroimmunol 180: 88–103. [DOI] [PubMed] [Google Scholar]
- Dolivo M. 1980. A neurobiological approach to neurotropic viruses. Trends Neurosci. 3: 149–152. [Google Scholar]
- Ekstrand MI, Enquist LW, and Pomerantz RJ 2008. The alpha-herpesviruses: Molecular pathfinders in nervous system circuits. Trends Mol. Med 14: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel EA, Song R, Koyuncu OO and Enquist LW (2015), Investigating the biology of alpha herpesviruses with MS-based proteomics. Proteomics, 15: 1943–1956. 10.1002/pmic.201400604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW, Tomishima MJ, Gross MJ, and Smith GA 2002. Directional spread of alpha-herpesvirus in the nervous system. Vet. Microbiol 86: 5–16. [DOI] [PubMed] [Google Scholar]
- Fraser G. and Ramachandran SP 1969. Studies of the virus of Aujeszky’s disease. I. Pathogenicity for rats and mice. J. Comp. Pathol 79: 435–444. [DOI] [PubMed] [Google Scholar]
- Geerling JC, Mettenleiter TC, and Loewy AD 2006. Viral tracers for the analysis of neural circuits. In Neuroanatomical Tract-Tracing 3: Molecules, Neurons, Systems (Zaborszky L. Wouterloud FG, and Lanciego JL, eds.) pp. 263–303. Springer, New York. [Google Scholar]
- Granstedt AE, Bosse JB, Thiberge SY, and Enquist LW 2013a. In vivo imaging of alphaherpesvirus infection reveals synchronized activity dependent on axonal sorting of viral proteins. Proc. Natl. Acad. Sci. U.S.A. 110: E3516–E3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granstedt AE, Szpara ML, Kuhn B, Wang S-H, and Enquist LW 2009. Fluorescence-based monitoring of in vivo neural activity using a circuit-tracing pseudorabies virus. PLos One 4: e6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granstedt AE, Kuhn B, Wang S-H, and Enquist LW 2010. Calcium imaging of neuronal circuits in vivo using a circuit-tracing pseudorabies virus. Cold Spring Harb. Protoc 2010. (4): pdb.prot5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granstedt AE, Brunton BW, and Enquist LW 2013b. Imaging the transport dynamics of single alphaherpesvirus particles in intact peripheral nervous system explants from infected mice. mBio 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Weiland F, Jons A, Klupp BG, Karger A, and Mettenleiter TC 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: A reassessment. J. Virol 71: 2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemoser WA, Kluge JP, and Hill HT 1980. Studies on the pathogenesis of pseudorabies in domestic cats following oral inoculation. Can. J. Comp. Med 44: 192–197. [PMC free article] [PubMed] [Google Scholar]
- Hall LB Jr., Kluge JP, Evans LE, and Hill HT 1984. The effect of pseudorabies (Aujeszky’s) virus infection on young mature boars and boar fertility. Can. J. Comp. Med 48: 192–197. [PMC free article] [PubMed] [Google Scholar]
- Hogue IB, Bosse JB, Hu JR, Thiberge SY, Enquist LW. Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog. 2014. Dec 4;10(12):e1004535. doi: 10.1371/journal.ppat.1004535. PMID: 25474634; PMCID: PMC4256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue IB; Bosse JB; Engel EA; Scherer J; Hu J-R; Del Rio T; Enquist LW Fluorescent Protein Approaches in Alpha Herpesvirus Research. Viruses 2015, 7, 5933–5961. 10.3390/v7112915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue IB, Card JP, Rinaman L, Staniszewska Goraczniak H, Enquist LW. Characterization of the neuroinvasive profile of a pseudorabies virus recombinant expressing the mTurquoise2 reporter in single and multiple injection experiments. J Neurosci Methods. 2018. Oct 1;308:228–239. doi: 10.1016/j.jneumeth.2018.08.004. Epub 2018 Aug 8. PMID: 30098326; PMCID: PMC6294127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover JE and Strick PL 1993. Multiple output channels in the basal ganglia. Science 259: 819–821. [DOI] [PubMed] [Google Scholar]
- Hoover JE and Strick PL 1999. The organization of cerebellar and basal ganglia outputs to primary motor cortex as revealed by retrograde transneuronal transport of herpes simplex virus type 1. J. Neurosci 15: 1446–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SM, Raine L, and Fanger H. 1981. Use of avidin-biotin-peroxidase techniques: A comparison between ABC and unlabeled antibody (PAP) procedures. J. Histochem. Cytochem 29: 577–580. [DOI] [PubMed] [Google Scholar]
- Huang JH and Weiss ML 1999. Characterization of the central cell groups regulating the kidney in the rat. Brain Res. 845: 77–91. [DOI] [PubMed] [Google Scholar]
- Hwang D-Y, Carlezon WA, Isacson O, and Kim K-S 2001. A high-efficiency synthetic promoter that drives transgene expression selectively in noradrenergic neurons. Human Gene Ther. 12: 1731–1740. [DOI] [PubMed] [Google Scholar]
- Irnaten M, Neff RA, Wang J, Loewy AD, Mettenleiter TC, and Mendelowitz D. 2001. Activity of cardiorespiratory networks revealed by transsynaptic virus expressing GFP. J. Neurophysiol 85: 435–438. [DOI] [PubMed] [Google Scholar]
- Jansen ASP, Van Nguyen X, Karpitskiy V, Mettenleiter TC, and Loewy AD 1995. Central command neurons of the sympathetic nervous system: Basis of the fight-or-flight response. Science 270: 253–260. [DOI] [PubMed] [Google Scholar]
- Jasmin L. 1995. Pseudorabies virus as a neuroanatomical tracer. J. NeuroVirol 1: 326–327. [DOI] [PubMed] [Google Scholar]
- Jasmin L, Burkey AR, Card JP, and Basbaum AI 1997. Transneuronal labeling of a nociceptive pathway, the spino-(trigemino-)parabrachio-amygdaloid, in the rat. J. Neurosci 17: 3751–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RM and Strick PL 2000. Rabies as a transneuronal tracer of circuits in the central nervous system. J. Neurosci. Methods 103: 63–71. [DOI] [PubMed] [Google Scholar]
- Kelly RM and Strick PL 2003. Cerebellar loops with motor cortex and prefrontal cortex of a nonhuman primate. J. Neurosci 23: 8432–8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Moore RY, Enquist LW, and Card JP 1999. Circuit-specific co-infection of neurons in the rat central nervous system with two pseudorabies virus recombinants. J. Virol 75: 9521–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge JP and Mare CJ 1974. Swine pseudorabies: abortion, clinical disease, and lesions in pregnant gilts infected with pseudorabies virus (Aujeszky’s disease). Am. J. Vet. Res 35: 991–995. [PubMed] [Google Scholar]
- Kobiler O, Lipman Y, Therkelsen K, Daubechies I, and Enquist LW 2010. Herpesviruses carrying a Brainbow cassette revel replication and expression of limited numbers of incoming genomes. Nat. Comm 1: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O, Brodersen P, Taylor MP, Ludmir EB, and Enquist LW 2011. Herpesvirus replication compartments originate with single incoming viral genomes. mBio 2: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu OO, Enquist LW, Engel EA. Invasion of the Nervous System. Curr Issues Mol Biol. 2021;41:1–62. doi: 10.21775/cimb.041.001. Epub 2020 Jul 29. PMID: 32723924. [DOI] [PubMed] [Google Scholar]
- Kramer T, Greco MA, Taylor MP, Ambrosini AE, Cristea IM, and Enquist LW 2012. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe 12: 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch’ng TH, Cristea IM, and Enquist LW 2013. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J. Virol 87: 9431–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PJ, Hay-Schmidt A, Vrang N, and Mikkelsen JD 1996. Origin of projections from the midbrain raphe nuclei to the hypothalamic paraventricular nucleus in the rat: A combined retrograde and anterograde tracing study. Neuroscience 70: 963–988. [DOI] [PubMed] [Google Scholar]
- Laval K; Enquist LW The Neuropathic Itch Caused by Pseudorabies Virus. Pathogens 2020, 9, 254. 10.3390/pathogens9040254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval K, Enquist LW. The Neuropathic Itch Caused by Pseudorabies Virus. Pathogens. 2020. Mar 31;9(4):254. doi: 10.3390/pathogens9040254. PMID: 32244386; PMCID: PMC7238046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail JH, Carter SR, and Topp KS 1993. The retrograde tracer FluoroGold interferes with the infectivity of herpes simplex virus. Brain Res. 625: 57–62. [DOI] [PubMed] [Google Scholar]
- Lavin TK, Jin L, Wickersham IR. Monosynaptic tracing: a step-by-step protocol. J Chem Neuroanat. 2019. Dec;102:101661. doi: 10.1016/j.jchemneu.2019.101661. Epub 2019 Aug 10. PMID: 31408693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levatte MA, Mabon PJ, Weaver LC, and Dekaban GA 1998. Simultaneous identification of two populations of sympathetic preganglionic neurons using recombinant herpes simplex virus type 1 expressing different reporter genes. Neuroscience 82: 1253–1267. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, and Lichtman JW 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–63. [DOI] [PubMed] [Google Scholar]
- Lo L. and Anderson DJ 2011. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy AD 1998. Viruses as transneuronal tracers for defining neural circuits. Neurosci. Biobehav. Rev 22: 679–684. [DOI] [PubMed] [Google Scholar]
- Mabon PJ, Weaver LC, and Dekaban GA 1999. Cyclosporin A reduces the inflammatory response to a multi-mutant herpes simplex virus type-1 leading to improved transgene expression in sympathetic neurons in hamsters. J. NeuroVirol 5: 268–279. [DOI] [PubMed] [Google Scholar]
- Marchand CF and Schwab ME 1987. Binding, uptake and retrograde axonal transport of herpes virus suis in sympathetic neurons. Brain Res. 383: 262–270. [DOI] [PubMed] [Google Scholar]
- Maturana CJ, Verpeut JL, Pisano TJ, Dhanerawala ZM, Esteves A, Enquist LW, Engel EA. Small Alphaherpesvirus Latency-Associated Promoters Drive Efficient and Long-Term Transgene Expression in the CNS. Mol Ther Methods Clin Dev. 2020. Apr 14;17:843–857. doi: 10.1016/j.omtm.2020.04.004. PMID: 32368565; PMCID: PMC7191541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken RM, McFerran JB, and Dow C. 1973. The neural spread of pseudorabies virus in calves. J. Gen. Virol 20: 17–28. [DOI] [PubMed] [Google Scholar]
- McEwan S, Kwon H, Tahiri A, Shanmugarajah N, Cai W, Ke J, Huang T, Belton A, Singh B, Wang L, Pang ZP, Dirice E, Engel EA, El Ouaamari A. Deconstructing the origins of sexual dimorphism in sensory modulation of pancreatic β cells. Mol Metab. 2021. Nov;53:101260. doi: 10.1016/j.molmet.2021.101260. Epub 2021 May 21. PMID: 34023484; PMCID: PMC8258979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern AE, Davis-Poynter N, Farrell MJ, and Mazzone SB 2012a. Transneuronal tracing of airways-related sensory circuitry using herpes simplex virus 1, strain H129. Neuroscience 207: 148–166. [DOI] [PubMed] [Google Scholar]
- McGovern AE, Davis-Poynter N, Rakoczy J, Phipps S, Simmons DG, and Mazzone SB 2012b. Anterograde neuronal circuit tracing using a genetically modified herpes simplex virus expressing EGFP. J. Neurosci. Methods 209: 158–167. [DOI] [PubMed] [Google Scholar]
- McLean IW and Nakane PK 1974. Distribution of retinal axons within the lateral hypothalamic area. J. Histochem. Cytochem 22: 1077–1083. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC 2000. Aujeszky’s disease (pseudorabies) virus: The virus and molecular pathogenesis—state of the art. Vet. Res 31: 99–115. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC 2003. Pathogenesis of neurotropic herpesviruses: Role of viral glycoproteins in neuroinvasion and transneuronal spread. Virus Res. 92: 197–206. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC, Keil GM, and Fuchs W. 2008. Molecular biology of animal herpesviruses. In Animal Viruses: Molecular Biology (Mettenleiter TC and Sobrino F, eds.). Caister Academic Press, Norfolk, U.K. [Google Scholar]
- Middleton FA and Strick PL 2001. Cerebellar projections to the prefrontal cortex of the primate. J. Neurosci 21: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA and Strick PL 2002. Basal-ganglia “projections” to the prefrontal cortex of the primate. Cerebral Cortex 12: 926–935. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Nakanishi H, Kimura K, Matsubara K, Ozaki-Kuroda K, Katata T, Honda T, Kiyohara Y, Heo K, Higashi M, Tsutsumi T, Sonoda S, Ide C, and Takai Y. 2002. Nectin: An adhesion molecule involved in formation of synapses. J. Cell Biol. 156: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell P, Lavin A, Enquist LW, Grace AA, and Card JP 1997. Interconnected parallel circuits between rat nucleus accumbens and thalamus revealed by retrograde transynaptic transport of pseudorabies virus. J. Neurosci 17: 2143–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyibo HK, Znamenskiy P, Oviedo HV, Enquist LW, Zador AM. Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front Neuroanat. 2014. Sep 3;8:86. doi: 10.3389/fnana.2014.00086. PMID: 25232307; PMCID: PMC4153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G. and Watson C. 2013. The Rat Brain in Stereotaxic Coordinates. Elsevier Science & Technology Press. [Google Scholar]
- Pickard GE, Smeraski CA, Tomlinson CC, Banfield BW, Kaufman J, Wilcox CL, Enquist LW, and Sollars PJ 2002. Intravitreal injection of the attenuated pseudorabies virus PRV Bartha results in infection of the hamster suprachiasmatic nucleus only by retrograde transsynaptic transport via autonomic circuits. J. Neurosci 22: 2701–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano TJ, Dhanerawala ZM, Kislin M, Bakshinskaya D, Engel EA, Hansen EJ, Hoag AT, Lee J, de Oude NL, Venkataraju KU, Verpeut JL, Hoebeek FE, Richardson BD, Boele HJ, Wang SS. Homologous organization of cerebellar pathways to sensory, motor, and associative forebrain. Cell Rep. 2021. Sep 21;36(12):109721. doi: 10.1016/j.celrep.2021.109721. PMID: 34551311; PMCID: PMC8506234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano TJ, Hoag AT, Dhanerawala ZM, Guariglia SR, Jung C, Boele HJ, Seagraves KM, Verpeut JL, Wang SS. Automated high-throughput mouse transsynaptic viral tracing using iDISCO+ tissue clearing, light-sheet microscopy, and BrainPipe. STAR Protoc. 2022. Apr 14;3(2):101289. doi: 10.1016/j.xpro.2022.101289. PMID: 35496792; PMCID: PMC9038781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Reynolds AE, and Hengartner CJ 2005. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev 69: 462–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Ekstrand MI, Latcha KN, Smith GA, Enquist LW, Friedman JM. Gene Expression Profiling with Cre-Conditional Pseudorabies Virus Reveals a Subset of Midbrain Neurons That Participate in Reward Circuitry. J Neurosci. 2017. Apr 12;37(15):4128–4144. doi: 10.1523/JNEUROSCI.3193-16.2017. Epub 2017 Mar 10. PMID: 28283558; PMCID: PMC5391685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A. Rabies Virus CVS-N2c(ΔG) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron. 2016. Feb 17;89(4):711–24. doi: 10.1016/j.neuron.2016.01.004. Epub 2016 Jan 21. PMID: 26804990; PMCID: PMC4760870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaman L. and Schwartz GJ 2004. Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J. Neurosci 24: 2782–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaman L, Card JP, and Enquist LW 1993. Spatiotemporal responses of astrocytes, ramified microglia, and brain macrophages to central neuronal infection with pseudorabies virus. J. Neurosci 13: 685–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotto-Perceley DM, Wheeler JG, Osorio FA, Platt KB, and Loewy AD 1992. Transneuronal labeling of spinal interneurons and sympathetic preganglionic neurons after pseudorabies virus injections in the rat medial gastrocnemius muscle. Brain Res. 574: 291–306. [DOI] [PubMed] [Google Scholar]
- Sams JM, Jansen AS, Mettenleiter TC, and Loewy AD 1995. Pseudorabies virus mutants as transneuronal tracers. Brain Res. 687: 182–190. [DOI] [PubMed] [Google Scholar]
- Sauer B. 1987. Functional expression of the Cre-Lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol 7: 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm LP, Strack AM, Platt KB, and Loewy AD 1993. Peripheral and central pathways regulating the kidney: A study using pseudorabies virus. Brain Res. 616: 251–262. [DOI] [PubMed] [Google Scholar]
- Schneeberger M, Parolari L, Das Banerjee T, Bhave V, Wang P, Patel B, Topilko T, Wu Z, Choi CHJ, Yu X, Pellegrino K, Engel EA, Cohen P, Renier N, Friedman JM, Nectow AR. Regulation of Energy Expenditure by Brainstem GABA Neurons. Cell. 2019. Jul 25;178(3):672–685.e12. doi: 10.1016/j.cell.2019.05.048. Epub 2019 Jun 27. PMID: 31257028; PMCID: PMC7481042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeraski CA, Sollars PJ, Ogilvie MD, Enquist LW, and Pickard GE 2004. Suprachiasmatic nucleus input to autonomic circuits identified by retrograde transsynaptic transport of pseudorabies virus from the eye. J. Comp. Neurol 471: 298–313. [DOI] [PubMed] [Google Scholar]
- Smith BN, Banfield BW, Smeraski CA, Wilcox CL, Dudek FE, Enquist LW, and Pickard GE 2000. Pseudorabies virus expressing enhanced green fluorescent protein: A tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. U.S.A 97: 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CK, Enquist LW, and Bartness TJ 2005. New developments in tracing neural circuits with herpesviruses. Virus Res. 111: 235–249. [DOI] [PubMed] [Google Scholar]
- Song CK, Schwartz GJ, and Bartness TJ 2009. Anterograde transneuronal viral tract tracing reveals central sensory circuits from white adipose tissue. Am. J. Physiol 3: 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011. Oct;7(10):e1002282. doi: 10.1371/journal.ppat.1002282. Epub 2011 Oct 13. PMID: 22022263; PMCID: PMC3192842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XR, Badura A, Pacheco DA, Lynch LA, Schneider ER, Taylor MP, Hogue IB, Enquist LW, Murthy M, and Wang SS 2013. Fast GCaMPs for improved tracking of neuronal activity. Nat. Comm 4: 2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW 1998. Brain Maps: Structure of the Rat Brain. Elsevier, Amsterdam. [Google Scholar]
- Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, and Enquist LW 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of Alphaherpesviruses. PLoS Pathogens 7: e1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, and Ogita H. 2008. Nectins and nectin-like molecules: Roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 9: 603–615. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Kobiler O, and Enquist LW 2012a. Alphaherpesvirus axon-to-cell spread involves limited virion transmission. Proc. Natl. Acad. Sci. U.S.A 109: 17046–17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Kramer T, Lyman MG, Kratchmarov R, and Enquist LW 2012b. Visualization of an alphaherpesvirus membrane protein that is essential for anterograde axonal spread of infection in neurons. mBio 3: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahlne A, Nystrom B, Sandberg M, Hamberger A, and Lycke E. 1978. Attachment of herpes simplex virus to neurons and glial cells. J. Gen. Virol 40: 359–371. [DOI] [PubMed] [Google Scholar]
- Vahlne A, Svennerholm B, Sandberg M, Hamberger A, and Lycke E. 1980. Differences in attachment between herpes simplex type 1 and type 2 viruses to neurons and glial cells. Infect. Immun 28: 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CH and Bartness TJ 2012. Anterograde transneuronal viral tract tracing reveals central sensory circuits from brown fat and sensory denervation alters its thermogenic responses. Am. J. Physiol 302: R1049–R1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RE, Wiegand ST, Clough RW, and Hoffman GE 1986. Use of cryoprotectant to maintain long-term peptide immunoreactivity and tissue morphology. Peptides 7: 155–159. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann K-K, Young JAT, and Callaway EM 2007. Monosynaptic restriction of transsynaptic tracing from single genetically targeted neurons. Neuron 53: 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007. Jan;4(1):47–9. doi: 10.1038/nmeth999. Epub 2006 Dec 10. PMID: 17179932; PMCID: PMC2755236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojaczynski GJ, Engel EA, Steren KE, Enquist LW, Patrick Card J. The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct Funct. 2015;220(3):1395–420. doi: 10.1007/s00429-014-0733-9. Epub 2014 Mar 2. PMID: 24585022; PMCID: PMC4152387. [DOI] [PMC free article] [PubMed] [Google Scholar]; O BW, Engel EA, Enquist LW. Characterization of a replication-incompetent pseudorabies virus mutant lacking the sole immediate early gene IE180. mBio. 2014. Nov 11;5(6):e01850. doi: 10.1128/mBio.01850-14. PMID: 25389174; PMCID: PMC4235210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guan H, Li C, Li Y, Wang S, Zhao X, Zhao Y, Liu Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int J Infect Dis. 2019. Oct;87:92–99. doi: 10.1016/j.ijid.2019.08.007. Epub 2019 Aug 10. PMID: 31408708. [DOI] [PubMed] [Google Scholar]
- Yoon H, Enquist LW, and Dulac C. 2005. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123: 669–682. [DOI] [PubMed] [Google Scholar]
- Zemanick MC, Strick PL, and Dix RD 1991. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc. Natl. Acad. Sci. U.S.A 88: 8048–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.