Abstract
In the absence of liquid suspension, dry biofilms can form upon hard surfaces within a hospital environment, representing a healthcare‐associated infection risk. Probiotic cleansers using generally recognized as safe organisms, such as those of the Bacillus genus, represent a potential strategy for the reduction of dry biofilm bioburden. The mechanisms of action and efficacy of these cleaners are, however, poorly understood. To address this, a preventative dry biofilm assay was developed using steel, melamine, and ceramic surfaces to assess the ability of a commercially available Bacillus spp. based probiotic cleanser to reduce the surface bioburden of Escherichia coli and Staphylococcus aureus. Via this assay, phosphate‐buffered saline controls were able to generate dry biofilms within 7 days of incubation, with the application of the probiotic cleanser able to prevent >97.7% of dry biofilm formation across both pathogen analogs and surface types. Further to this, surfaces treated with the probiotic mixture alone also showed a reduction in dry biofilm across both pathogen and surface types. Confocal laser scanning microscopy imaging indicated that the probiotic bacteria were able to germinate and colonize surfaces, likely forming a protective layer upon these hard surfaces.
Keywords: Bacillus, biofilm, CLSM‐FISH, probiotic, surface
We developed a test method for assessing the prevention of the adhesion of organisms to surfaces that could generate dry biofilm material. The study used a probiotic blend of Bacillus and a formulated product to assess their efficacy and visualize the presence of pathogen analogs on the surface of three material types.
1. INTRODUCTION
Within the clinic environment, dry biofilms have been described as any biofilm that is capable of forming upon a dry surface, while wet biofilms are those more closely associated with medical devices such as catheters (Ledwoch et al., 2018). Recent studies have indicated that organisms, including microbial pathogens, can survive on a multitude of surfaces, likely as dry biofilms, ranging from everyday items such as keyboards and folders (Ledwoch et al., 2018) to those of clinical relevance such as hard surfaces within the hospital environment (Caselli et al., 2016). The control of dry biofilms within a hospital setting is therefore important to manage surface pathogens and associated antimicrobial resistance gene loads. The dry biofilm mode of existence is thought to provide a protective niche that leads to reservoirs of clinically relevant pathogens. These protective niches, much like those found in wet biofilm, provide a resistance mechanism against environmental stressors and xenobiotic compounds. Conventional disinfection methods, such as the use of hypochlorite, are less effective against dry biofilms than against planktonic cells of Staphylococcus aureus (Almatroudi et al., 2016). Additional studies have also suggested the limitations of quaternary ammonium compounds and thermal treatment against dry biofilms (Almatroudi et al., 2018; Lineback et al., 2018). Dry biofilms, therefore, represent a viable target for the reduction of hospital‐acquired infections (HAIs) and the increased associated economic burden through surface pathogen control. Increasing resistance mechanisms to chemical treatments represent an increasing cost to healthcare services, with estimated treatment costs from healthcare‐associated infections reaching £1bn in the United Kingdom (Ebrahimi et al., 2017; Mackley, 2018).
In the search for alternative solutions to combat HAI and its causative agents, probiotic treatments of surfaces have been suggested by a number of studies (De Cesare et al., 2019; D'accolti et al., 2018, 2019). Here, the approach aims to use a nonpathogenic microorganism to colonize hard surfaces and effectively outcompete clinically relevant pathogens. The technologies available generally combine hard surface cleaning product formulations such as surfactants, chemical preservatives, and stabilizers to offer both an immediate biocidal challenge via the chemical formulation and a longer‐lasting impact via the biological agent. One of the major biological agents used for this approach is various species of the Bacillus genus. These are chosen due to their ability to be grown with relative ease at the industrial scale, coupled with their ability to form highly stable spores. These spores offer a prolonged product shelf life and increased flexibility with cleaning product formulations compared to preserved planktonic cells. Further to this, the approach of using Bacillus spp. aims to exploit the diverse antimicrobial secondary metabolite capabilities found within the different members of the genus to deliver a more natural and precise mode of pathogen control. Recently, this concept has been demonstrated in the clinical setting via multicenter hospital trials in Italy (Caselli et al., 2018, 2019). Within these studies, the use of Bacillus spp. based probiotic cleansers indicated a reduction in HAI from 4.8% to 2.3% over 18 months (Caselli et al., 2018), demonstrating the suitability of this approach for the prevention of pathogen accumulation upon hard surfaces.
Observations from multicenter studies (Caselli et al., 2018, 2019) offer a good initial insight as to the efficacy of using probiotic‐based cleansers toward the control of HAI; however, the underpinning microbiology as to the exact mechanisms of action, especially in relation to the dry biofilm mode of life, is still somewhat lacking, especially regarding the ability of Bacillus spp. spores to germinate and colonize hard surfaces. A reproducible model system is therefore needed to both improve scientific understanding and provide a tool to better engineer probiotic technologies toward hard surface pathogen control. There have been methods proposed for both the generation of dry biofilms on surfaces using a variety of static (Adator et al., 2018) and mixed reactor approaches (Almatroudi et al., 2015) with varying levels of exposure to liquid media requiring up to 12 days to generate and using specialist equipment such as a CDC biofilm reactor (Almatroudi et al., 2015, 2016, 2018). However, the current methodologies are typically considered for a reactive approach to testing such that a biofilm is developed on the surface before being treated through chemical interventions. Within the present study, we have developed a simple and reproducible model for assessing the ability of probiotic treatment of surfaces in a preventative study design for dry surface biofilms.
2. MATERIALS AND METHODS
2.1. Probiotic cleanser
The probiotic cleanser used was a commercially available general all‐purpose cleaner provided by Genesis Biosciences. The product consists of a blend of B. amyloliquefaciens, B. licheniformis, B. subtilis, B. pumilus, and B. megaterium spores at a combined concentration of 6.7 × 107 CFU/mL suspended in a liquid cleaning formulation that contained nonionic surfactants, benzothiazolinone (BIT) preservatives, and organic based chelation agents. A cleanser formulation‐free Bacillus blend was prepared in the same manner, with the liquid cleaning formulation replaced with phosphate‐buffered saline (PBS).
2.2. Dry biofilm formation
Dry biofilms were formed on the test surfaces using the modified method of Adator et al. (2018). Briefly, test microorganisms (Staphylococcus aureus NCIMB 9518 or Escherichia coli NCIMB 8879) were prepared to a concentration of 1.5–5.0 × 108 CFU/mL from 48‐h stock plates on tryptone soya agar (TSA; Neogen) in maximum recovery diluent (MRD; Neogen) and 50 µL deposited onto the test surface. Three test surfaces were selected, namely, stainless steel (3.1 cm2; SYSPAL), melamine (4.0 cm2; Wickes), and ceramic (4.0 cm2; Wickes), that were sterilized through autoclaving before use. The treated surfaces were then left at room temperature for 7 days in a sterile environment at room temperature and humidity. To assess the viability of the dry biofilm, surviving organisms were enumerated following an initial rinse of the test piece with 1 mL of sterile PBS to remove loosely attached cells; test pieces were face down into a sterile 5 cm Ø pot containing 10 mL of Dey–Engley neutralizing broth (Neogen) and 5 g glass beads (3 mm Ø). Following orbital shaking at 120 rpm for 30 min at room temperature, a range of dilutions of the resulting mixture was prepared in PBS and plated out onto TSA. Surviving organisms were then determined following incubation at 37°C. A set of test pieces was also treated with uninoculated MRD as a negative control.
2.3. Spore status following application
To understand the extent to which germination was occurring through the applied Bacillus spores, the spore blend or formulated product (100 µL) was dispensed on each test surface type (n = 6) and left for 1 h at room temperature. After this time, the organisms were recovered from the surface using the method described in 2.2; following orbital shaking, three of the replicates were enumerated as per 2.2 to give a total number of cells, while the remaining three replicates were heat shocked at 80°C for 20 min before TSA enumeration to give a spore count, with the difference between these two being the estimate of vegetative cells.
2.4. Antidry biofilm assay
To determine the extent of dry biofilm prevention by the probiotic blend of Bacillus, surfaces were pretreated with the following: (1) the probiotic cleanser (Bacillus + formulation), (2) the Bacillus blend prepared in PBS, and (3) PBS as a control. In treatments containing Bacillus, a concentration of 1.0–3.0 × 107 CFU/mL was confirmed by dilution of the product in PBS and plating out onto TSA, followed by incubation at 37°C for 24 h. Each test surface was treated in triplicate with 100 µL cleanser, Bacillus blend, or control and left at room temperature until visibly dry, ∼1 h. The treated surfaces were then challenged with 50 µL of the 1.5–5.0 ×108 CFU/mL suspension of microorganisms and incubated at room temperature for 7 days. The surviving organisms were then recovered as per the section above. To differentiate between Bacillus spp. and surviving challenge organism, S. aureus was recovered on Baird Parker Agar (Neogen) and E. coli was recovered on MacConkey agar (Neogen) supplemented with 0.5% sodium deoxycholate. Before testing, both media types were validated to ensure that Bacillus spp. were inhibited.
2.5. CLSM imaging
Initially, surfaces (steel, melamine, and ceramic) were inoculated with 100 µL of Bacillus blend (1.5 × 107 CFU/mL) or PBS, then left to air dry for 1 h (or until visibly dry). Once dried, 50 µL of the 1.5–5.0 × 108 CFU/mL microorganism was added to the surface and then left at room temperature for 7 days. After this, surfaces were rinsed with 1 mL PBS to remove transient organisms. Fluorescence in situ hybridization (FISH) of the inoculated surfaces was performed using previously described methods (Ainsworth et al., 2006). The buffer used for the hybridization was composed of 0.9 M NaCl, 0.01% sodium dodecyl sulfate (SDS), 0.01 M Tris‐HCl (pH 7.2), and 35% formamide, with all probes (Table 1) used at a final concentration of 5 ng/µL. The hybridization was conducted at 46°C for 1.5 h, followed by a 10‐min wash in a buffer (0.08 M NaCl, 0.01% SDS, and 0.01 M Tris‐HCl (pH 7.2)) at 46°C. FISH confocal laser scanning microscopy (FISH‐CLSM) of samples was performed at the Bioimaging Facility at the University of Huddersfield, UK, using a Zeiss LSM880 inverted confocal microscope, and images were processed using Zen 2.1 software (Zeiss Microscopy).
Table 1.
Fluorescent probes were used in the study.
3. RESULTS
3.1. Dry biofilm development on surfaces
The dry biofilm model system was able to generate biofilms on the surface of all 3 coupon types (stainless steel, ceramic, and melamine) after 7 days of incubation at room temperature (Figure 1). Surfaces inoculated with E. coli showed the greatest degree of variation by surface, with 3.7–3.8 Log organisms recovered per cm2 on stainless steel and ceramic, respectively, while 4.7 Log organisms were recovered from melamine. Recovery of S. aureus was more consistent, ranging between 4.9 and 5.1 Log organisms across the three test surfaces.
Figure 1.
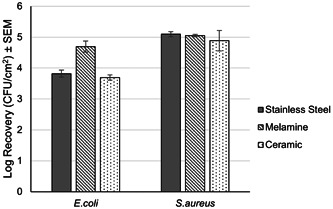
Log recovery of surviving organisms across the three surface types when inoculated with Escherichia coli and Staphylococcus aureus after 7 days of incubation at room temperature and RH, n = 3. CFU, colony‐forming unit; RH, relative humidity.
3.2. Status of Bacillus spp. on inoculated surfaces
Recovery of the Bacillus spp. inoculated on each of the surfaces to determine the number of vegetative cells and spores (Figure 2) indicated that both vegetative cells and spores were present following an hour of incubation at room temperature. On surfaces that were inoculated solely with the blend of Bacillus strains, vegetative cells on the surfaces ranged from 43% to 78%, being highest on the steel surfaces and least on melamine. The range of vegetative cells on the surface was narrower upon surfaces treated with the formulated Bacillus blend, ranging from 43% to 65%, with ceramic having the lowest proportion of vegetative cells. The total recovery of Bacillus from each test surface can be seen in Table A1.
Figure 2.
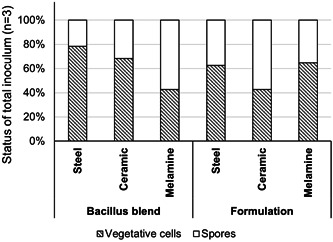
Status of Bacillus spp. recovered from steel, ceramic, and melamine surfaces indicated the presence of both vegetative cells and spores present on all surface types.
3.3. Anti‐dry biofilm properties of the Bacillus blend
The application of the Bacillus blend, without additional cleaning chemistry, was able to decrease the surface colonization of both E. coli and S. aureus when compared to the PBS control, as determined via enumeration techniques (Figure 3). In comparison to the PBS control, the presence of E. coli on both melamine and ceramic was reduced by >97%, while a reduction of 67.7% ± 15.7% was observed on the steel surfaces. For S. aureus, reductions in microbial load after Bacillus blend treatment were much lower than that of E. coli, with ceramic yielding a reduction of 71.3%.
Figure 3.
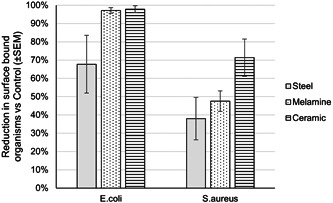
The Bacillus blend, with no additional cleaning chemistry, was capable of decreasing the microbial load on all three test surfaces when challenged with both Escherichia coli and Staphylococcus aureus when compared to the phosphate‐buffered saline control.
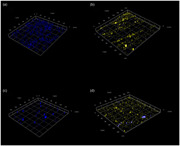
FISH‐CLSM imaging of the steel test surfaces indicated that upon coupons treated with PBS only, there was an attachment and proliferation of either E. coli or S. aureus that was not removed with washing (Figure 4). In contrast, when steel coupons were pretreated with the Bacillus blend, there was a reduction in the visible accumulation of either E. coli or S. aureus, with the majority of the surface instead showing fluorescence related to that of Bacillus spp. Similar observations were made on ceramic and melamine test pieces (Figures A1 and A2).
Figure 4.
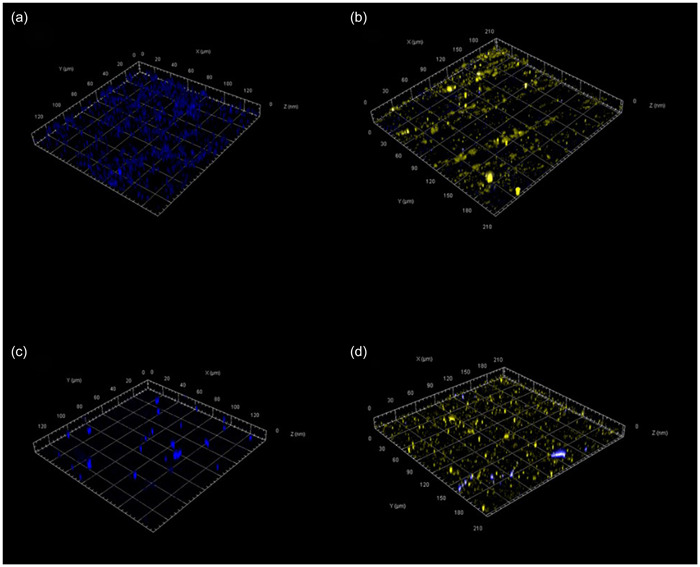
Example confocal laser scanning microscopy images showing dry biofilm formation of Escherichia coli (a) and Staphylococcus aureus (c) on stainless steel surfaces fluorescing blue. Stainless steel surfaces pretreated with the Bacillus blend before the addition of E. coli (b) and S. aureus (d) are shown with Bacillus fluorescing yellow.
3.4. Antidry biofilm properties of the probiotic cleanser
When surfaces were pretreated with the probiotic cleanser formulation (Bacillus spores and cleaning formulation), there was no evidence of any recoverable organisms from steel, melamine, or ceramic when challenged with E. coli, representing a 100% reduction in dry biofilm compared to the control surfaces (Figure 5). There was a greater degree of variability observed across the S. aureus‐challenged test surfaces, where microbial loads were decreased by a minimum of 97.7% compared to the control surfaces. In the case of ceramic and steel surfaces, the reduction in viable biofilm‐associated cells was 99.6% ± 0.1% and 99.8% ± 0.0%, respectively.
Figure 5.
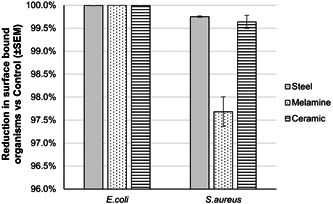
The probiotic cleanser (Bacillus spores and cleaning formulation) was capable of decreasing the microbial load on all three test surfaces when challenged with both Escherichia coli and Staphylococcus aureus compared to the phosphate‐buffered saline control.
4. DISCUSSION
There are clear challenges in producing methodologies for the testing of antimicrobial products against biofilms. While the testing of disinfectants within the BS:EN test methods mainly focus on surface tests such as BS EN 13697:2015 + A1:2019 (Anonymous, 2020) in which organisms are dried onto a surface over a short period (>1 h), those produced by ASTM have focused on the growth of biofilms using the CDC reactor (Biosurface Technologies, US [Anonymous, 2019]) but maintained in solution. In both cases, these test methods have a strong focus on the removal of biomass from a surface rather than the prevention of its accumulation. The data generated within this study suggest that with modifications, simple methodologies for the production of dry biofilms, such as those proposed by Adator et al. (2018), provide a simple but reproducible test model for assessing the ability of antimicrobials to prevent or reduce the formation of dry biofilms. The test method has also identified the potential for Bacillus‐based probiotics to act as a preventative measure to reduce the bioburden on surfaces associated with dry biofilms.
Through the use of the model system, it was demonstrated that Bacillus spp. spores contained within a probiotic cleanser were able to both germinate and persist upon a variety of hard surfaces. These observations tie in with those of Caselli et al. (2016, 2019), where similar levels and persistence were documented. The use of CLSM microscopy combined with FISH probes further reinforced microbial count data, showing the ability of Bacillus spp. to actively colonize steel, ceramic, and melamine surfaces. The ability of Bacillus spp. to actively colonize hard surfaces suggests that potential competitive exclusion mechanisms are at play between the added probiotic bacteria and pathogenic microorganisms. The use of three test surfaces (steel, ceramic, and melamine) demonstrated that the test can be easily modified to suit specific surface testing requirements. It is well documented that there are multiple properties concerning both the surfaces and microorganisms that could influence adhesion (Zheng et al., 2021), and variation in adhesion of both pathogen analogs and Bacillus sp. was also observed here. Further understanding of the metabolic activities of surface‐bound Bacillus spp. when exposed to clinically relevant pathogens would be of benefit to better understand the microbial mechanisms behind this. This data would be a valuable tool in refining bespoke consortia of different Bacillus species to enhance the efficacy of future probiotics products.
The impact of combining a hard surface cleaning formulation with a biological agent, in this case, Bacillus spp., was further understood in this work. The use of agents such as surfactants and chemical preservatives such as the common biocide BIT was able to provide an initial challenge to the pathogenic microorganisms but not the probiotic spores. This is due to the fact that the probiotic cleanser formulation was designed in a manner to provide a biocidal but not sporicidal challenge to preserve its integrity during storage. It is likely that as organic fouling and natural attrition diluted the chemical challenge, the concurrent germination of the probiotic spores allowed the Bacillus spp. to gain an immediate foothold in niche areas that could support microbial life.
AUTHOR CONTRIBUTIONS
Richard Wormald: Data curation (equal); investigation (equal); methodology (equal); software (equal); visualization (equal); writing—review and editing (equal). Paul N. Humphreys: Conceptualization (equal); supervision (equal); writing—review and editing (equal). Christopher J. Charles: Conceptualization (equal); supervision (equal); writing—review and editing (equal). Simon P. Rout: Conceptualization (equal); funding acquisition (lead); methodology (equal); supervision (equal); visualization (equal); writing—original draft (lead).
CONFLICT OF INTEREST STATEMENT
Christopher J. Charles is a current employee of Genesis Biosciences and was involved in the conceptualization of the project and manuscript review but had no role in the collection of data, analysis, or interpretation of the data. The remaining authors declare no conflict of interest.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
This research was funded by the National Biofilm Innovation Centre, Grant Number BB/R012415.
1.
See Table A1 and Figures A1 and A2.
Table A1.
Total recovery of Bacillus from surfaces.
| Log10 recovery | ||
|---|---|---|
| Application | Bacillus only | Bacillus cleanser |
| Initial | 6.82 | 6.92 |
| Steel | 6.78 ± 0.009 | 6.67 ± 0.09 |
| Ceramic | 6.55 ± 0.05 | 6.52 ± 0.03 |
| Melamine | 5.92 ± 0.2 | 6.02 ± 0.1 |
Figure A1.

Confocal laser scanning microscopy comparison of ceramic surfaces inoculated with Staphylococcus aureus with no pretreatment (a) and inoculated following treatment with the Bacillus sp. blend (b), and nonpretreated surfaces challenged with Escherichia coli (c) and following treatment with the Bacillus sp. blend (d). S. aureus and E. coli are stained blue, with the Bacillus stained yellow.
Figure A2.
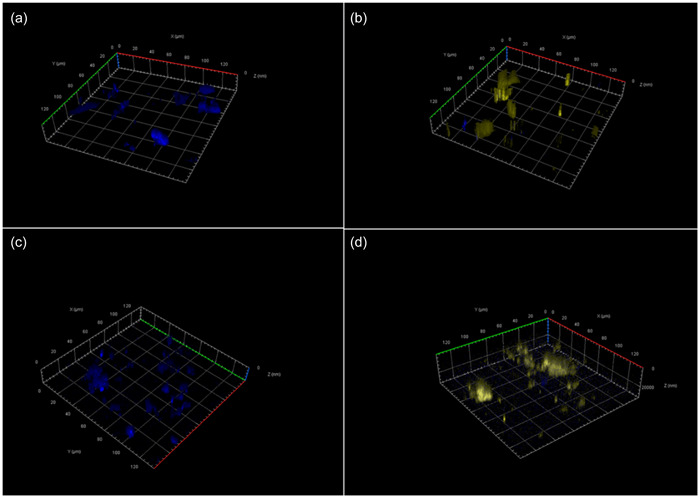
Confocal laser scanning microscopy comparison of melamine surfaces inoculated with Staphylococcus aureus with no pretreatment (a) and inoculated following treatment with the Bacillus sp. blend (b), and nonpretreated surfaces challenged with Escherichia coli (c) and following treatment with the Bacillus sp. blend (d). S. aureus and E. coli are stained blue, with the Bacillus stained yellow.
Wormald, R. , Humphreys, P. N. , Charles, C. J. , & Rout, S. P. (2023). Bacillus‐based probiotic cleansers reduce the formation of dry biofilms on common hospital surfaces. MicrobiologyOpen, 12, e1391. 10.1002/mbo3.1391
DATA AVAILABILITY STATEMENT
All data are presented in this published article.
REFERENCES
- Adator, E. H. , Cheng, M. , Holley, R. , Mcallister, T. , & Narvaez‐Bravo, C. (2018). Ability of Shiga toxigenic Escherichia coli to survive within dry‐surface biofilms and transfer to fresh lettuce. International Journal of Food Microbiology, 269, 52–59. [DOI] [PubMed] [Google Scholar]
- Ainsworth, T. D. , Fine, M. , Blackall, L. L. , & Hoegh‐Guldberg, O. (2006). Fluorescence in situ hybridization and spectral imaging of coral‐associated bacterial communities. Applied and Environmental Microbiology, 72, 3016–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almatroudi, A. , Gosbell, I. B. , Hu, H. , Jensen, S. O. , Espedido, B. A. , Tahir, S. , Glasbey, T. O. , Legge, P. , Whiteley, G. , Deva, A. , & Vickery, K. (2016). Staphylococcus aureus dry‐surface biofilms are not killed by sodium hypochlorite: Implications for infection control. Journal of Hospital Infection, 93, 263–270. [DOI] [PubMed] [Google Scholar]
- Almatroudi, A. , Hu, H. , Deva, A. , Gosbell, I. B. , Jacombs, A. , Jensen, S. O. , Whiteley, G. , Glasbey, T. , & Vickery, K. (2015). A new dry‐surface biofilm model: An essential tool for efficacy testing of hospital surface decontamination procedures. Journal of Microbiological Methods, 117, 171–176. [DOI] [PubMed] [Google Scholar]
- Almatroudi, A. , Tahir, S. , Hu, H. , Chowdhury, D. , Gosbell, I. B. , Jensen, S. O. , Whiteley, G. S. , Deva, A. K. , Glasbey, T. , & Vickery, K. (2018). Staphylococcus aureus dry‐surface biofilms are more resistant to heat treatment than traditional hydrated biofilms. Journal of Hospital Infection, 98, 161–167. [DOI] [PubMed] [Google Scholar]
- Anonymous . (2019). Standard test method for determining disinfectant efficacy against biofilm grown in the CDC biofilm reactor using the single tube method (ASTM E2871‐19). ASTM International.
- Anonymous . (2020). Chemical disinfectants and antiseptics, quantitative non‐porous surface test for the evaluation of bactericidal and/or fungicidal activity of chemical disinfectants used in food, industrial, domestic and institutional areas. Test method and requirements without mechanical action (phase 2, step 2) (BS EN 13697:2015 + A1:2019‐TC). British Standards Institute.
- Caselli, E. , Arnoldo, L. , Rognoni, C. , D'accolti, M. , Soffritti, I. , Lanzoni, L. , Bisi, M. , Volta, A. , Tarricone, R. , & Brusaferro, S. (2019). Impact of a probiotic‐based hospital sanitation on antimicrobial resistance and HAI‐associated antimicrobial consumption and costs: A multicenter study. Infection Drug Resistance, 12, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli, E. , Brusaferro, S. , Coccagna, M. , Arnoldo, L. , Berloco, F. , Antonioli, P. , Tarricone, R. , Pelissero, G. , Nola, S. , Conte, A. , Tognon, L. , Villone, G. , Trua, N. , Mazzacane, S. , & La Fauci, V. (2018). Reducing healthcare‐associated infections incidence by a probiotic‐based sanitation system: A multicentre, prospective, intervention study. PLoS ONE, 13, e0199616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli, E. , D'accolti, M. , Vandini, A. , Lanzoni, L. , Camerada, M. T. , Coccagna, M. , Branchini, A. , Antonioli, P. , Balboni, P. G. , Di Luca, D. , & Mazzacane, S. (2016). Impact of a probiotic‐based cleaning intervention on the microbiota ecosystem of the hospital surfaces: Focus on the resistome remodulation. PLoS ONE, 11, e0148857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cesare, A. , Caselli, E. , Lucchi, A. , Sala, C. , Parisi, A. , Manfreda, G. , & Mazzacane, S. (2019). Impact of a probiotic‐based cleaning product on the microbiological profile of broiler litters and chicken caeca microbiota. Poultry Science, 98, 3602–3610. [DOI] [PubMed] [Google Scholar]
- D'accolti, M. , Soffritti, I. , Lanzoni, L. , Bisi, M. , Volta, A. , Mazzacane, S. , & Caselli, E. (2019). Effective elimination of Staphylococcal contamination from hospital surfaces by a bacteriophage–probiotic sanitation strategy: A monocentric study. Microbial Biotechnology, 12, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'accolti, M. , Soffritti, I. , Piffanelli, M. , Bisi, M. , Mazzacane, S. , & Caselli, E. (2018). Efficient removal of hospital pathogens from hard surfaces by a combined use of bacteriophages and probiotics: Potential as sanitizing agents. Infection drug resistance, 11, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi, A. , Arvaneh, Z. , Mahzounieh, M. , & Lotfalian, S. (2017). Antibiotic resistance induction by benzalkonium chloride exposure in nosocomial pathogens. International Journal of Infection, 4, e40296. [Google Scholar]
- Kudoh, J. , & Ikeuchi, T. (1985). Nucleotide sequences of the sporulation gene spo0A and its mutant genes of Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America, 82, 2665–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledwoch, K. , Dancer, S. J. , Otter, J. A. , Kerr, K. , Roposte, D. , Rushton, L. , Weiser, R. , Mahenthiralingam, E. , Muir, D. D. , & Maillard, J. Y. (2018). Beware biofilm! Dry biofilms containing bacterial pathogens on multiple healthcare surfaces; a multi‐centre study. Journal of Hospital Infection, 100, e47–e56. [DOI] [PubMed] [Google Scholar]
- Lineback, C. B. , Nkemngong, C. A. , Wu, S. T. , Li, X. , Teska, P. J. , & Oliver, H. F. (2018). Hydrogen peroxide and sodium hypochlorite disinfectants are more effective against Staphylococcus aureus and Pseudomonas aeruginosa biofilms than quaternary ammonium compounds. Antimicrobial Resistance & Infection Control, 7, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackley, A. (2018). Raising standards of infection prevention and control in the NHS. House of Commons Library, Number CDP‐2018‐0116.
- Tang, Y. Z. , Gin, K. Y. H. , & Lim, T. H. (2005). High‐temperature fluorescent in situ hybridization for detecting Escherichia coli in seawater samples, using rRNA‐targeted oligonucleotide probes and flow cytometry. Applied and Environmental Microbiology, 71, 8157–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. (2010). Simultaneous detection and differentiation of Staphylococcus species in blood cultures using fluorescence in situ hybridization. Medical Principles and Practice, 19, 218–221. [DOI] [PubMed] [Google Scholar]
- Zheng, S. , Bawazir, M. , Dhall, A. , Kim, H. E. , He, L. , Heo, J. , & Hwang, G. (2021). Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Frontiers in Bioengineering and Biotechnology, 9, 643722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are presented in this published article.


