Abstract
The seasonal variations in community structure and cell morphology of pelagic procaryotes from a high mountain lake (Gossenköllesee, Austria) were studied by in situ hybridization with rRNA-targeted fluorescently labeled oligonucleotide probes (FISH) and image-analyzed microscopy. Compositional changes and biomass fluctuations within the assemblage were observed both in summer and beneath the winter ice cover and are discussed in the context of physicochemical and biotic parameters. Proteobacteria of the beta subclass (beta-proteobacteria) formed a dominant fraction of the bacterioplankton (annual mean, 24% of the total counts), whereas alpha-proteobacteria were of similar relative importance only during spring (mean, 11%). Bacteria of the Cytophaga-Flavobacterium cluster, although less abundant, constituted the largest fraction of the filamentous morphotypes during most of the year, thus contributing significantly to the total microbial biomass. Successive peaks of threadlike and rod-shaped archaea were observed during autumn thermal mixing and the period of ice cover formation, respectively. A set of oligonucleotide probes targeted to single phylotypes was constructed from 16S rRNA-encoding gene clone sequences. Three distinct populations of uncultivated microbes, affiliated with the alpha- and beta-proteobacteria, were subsequently monitored by FISH. About one-quarter of all of the beta-proteobacteria (range, 6 to 53%) could be assigned to only two phylotypes. The bacterial populations studied were annually recurrent, seasonally variable, and vertically stratified, except during the periods of lake overturn. Their variability clearly exceeded the fluctuations of the total microbial assemblage, suggesting that the apparent stability of total bacterioplankton abundances may mask highly dynamic community fluctuations.
Until recently, microbial ecologist studying aquatic bacteria faced a basic dilemma: they could either measure the abundance, biomass, growth rates, activity, etc. of the “average” bacterium under in situ conditions (e.g., see reference 13), ignoring the phylogenetic and physiological diversity of microbial communities, or they could isolate and ecophysiologically characterize individual bacterial strains (e.g., see reference 36) but were then not able to tell if these microorganisms were also common in the environment. Consequently, little knowledge has been gathered about the spatial and temporal abundance fluctuations of defined phylogenetic groups and of individual bacterial species in natural habitats. Molecular biological techniques used to identify microbes in environmental samples have recently provided new tools to study bacterioplankton biodiversity (e.g., see references 1, 9, 14, 15, and 19) and the in situ abundances of bacteria and archaea that could not be adequately distinguished before (2, 4, 5, 25). Microbiologists are now in a position to potentially elucidate the biogeography (24), population dynamics, and successions (28) not only of a few morphologically conspicuous microbes but of a large number of species, most of which might still be uncharacterized.
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes selectively visualizes bacterial cells with defined phylogenetic affiliations (3, 5). Based on a rapidly growing set of 16S (and, to a lesser extend, 23S) rRNA sequence data, it is probably the phylogenetically most sophisticated (22) approach for whole-cell in situ identification. On the other hand, FISH of plankton samples can be performed with minimal laboratory requirements (16), and evaluation relies on epifluorescence microscopy, which is a standard technique of aquatic microbial ecologists, e.g., for counting (30) and sizing (33) of picoplankton. In contrast to other identification approaches, FISH largely conserves the gestalt of the targeted microorganisms, i.e., their morphologies, cell sizes (26, 34), and cellular rRNA content (7, 32). So, despite the limitations of the method (as discussed in reference 5), its potential for the identification and cytometric analysis of planktonic microbes is just about to be recognized.
Recent investigations have reported that various freshwater microbial communities are dominated by bacteria which are phylogenetically affiliated with the alpha and beta subclasses of the class Proteobacteria (alpha- and beta-proteobacteria, respectively) and with members of the Cytophaga-Flavobacterium cluster (2, 6, 16, 19). These observations were based on single or short-term sampling schemes. The instantaneous community composition of the bacterioplankton, however, may not be representative for different seasons, and the typical ranges of annual community variability remain to be established.
The size distribution of planktonic bacteria, and particularly the appearance of filamentous cells, has come into the focus of aquatic microbial ecology in the context of studies of predator-prey interactions. It has been shown both in the laboratory (18, 37) and in field experiments (20) that the filamentous morphotype is a phenotypic adaptation of some microbes to protistan grazing, but there are probably numerous other causes for bacteria to elongate far beyond their typical sizes (e.g., see reference 23). Threadlike bacteria have been observed throughout the year in the plankton of a hypertrophic lake (41) but were also found in midwinter in an oligotropic alpine lake (31).
In earlier studies, we demonstrated FISH to be an appropriate tool for the monitoring of spatial (2) and short-term temporal (26) dynamics of different phylogenetic groups of the planktonic microbial community in a high mountain lake. Here we report on the seasonal and vertical abundance distributions of pelagic members of Bacteria and Archaea in Gossenköllesee and analysis of the community structure at different levels of taxonomic resolution. We applied published domain- and group-specific oligonucleotide probes (5) but also used the sequence information from a 16S rRNA-encoding gene (rDNA) library obtained from Gossenköllesee bacterioplankton 1 year earlier to construct specific probes targeted at individual bacterial populations. Particular attention was paid to the changes in abundance and taxonomic composition of the filamentous bacterial morphotypes which were recognized as a permanently important fraction of the planktonic procaryotes in Gossenköllesee. Additionally, we monitored the seasonal changes in the biomass size distributions of the nonfilamentous fraction of the pelagic microbial community.
MATERIALS AND METHODS
Sampling site and procedure.
The study site, Gossenköllesee, is a small, oligotrophic high-mountain lake in the Central Alps (Tyrol, Austria), situated above the timberline at an altitude of 2,417 m (11). The catchment area of Gossenköllesee is listed as a United Nations Educational, Scientific, and Cultural Organization Man and Biosphere Reserve. Annual water temperatures typically range between 0 and 15°C, and thermal mixing occurs from late October to mid-November and from late June to early July (44). During the summer months, the lake is exposed to high levels of solar UV radiation (40). The lake was ice covered from mid-November 1996 until the end of June 1997, and during this period, dissolved oxygen decreased to well below saturation in the lower water layers (44). Table 1 depicts the development of temperature, dissolved oxygen, conductivity, and the chlorophyll a concentration during the study period (from reference 44).
TABLE 1.
Temperature, dissolved oxygen, alkalinity, and chlorophyll a concentrations in Gossenköllesee during the study perioda
| Date | Temp (°C)
|
O2 (% saturation)
|
Alkalinity (μeq liter−1)
|
Chlorophyll a concn (μg liter−1)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Surface | 4 m | 8 m | Surface | 4 m | 8 m | Surface | 4 m | 8 m | Surface | 4 m | 8 m | |
| July 1996 | 9.4 | 8.6 | 7.4 | 116 | 105 | 0.7 | 1.2 | 1.9 | ||||
| August 1996 | 15.4 | 12.4 | 7.4 | 0.8 | 1.0 | 5.0 | ||||||
| September 1996 | 9.3 | 8.7 | 6.2 | 100 | 101 | 111 | 1.3 | 1.8 | 2.2 | |||
| October 1996 | 4.5 | 4.3 | 4.5 | 95 | 116 | 120 | 94 | 93 | 93 | 1.4 | 1.7 | 2.5 |
| November 1996 | 0.7 | 3.9 | 4.1 | 105 | 115 | 97 | 91 | 91 | 98 | 0.8 | 3.0 | 3.2 |
| December 1996 | 0.6 | 4 | 4.2 | 95 | 120 | 85 | 89 | 90 | 105 | 1.8 | 6.9 | 3.2 |
| January 1997 | 0.4 | 3.9 | 4.1 | 96 | 116 | 64 | 84 | 106 | 97 | 1.0 | 6.0 | 3.1 |
| February 1997 | 0 | 3.6 | 4.1 | 99 | 114 | 40 | 102 | 93 | 108 | 0.9 | 7.5 | 2.4 |
| March 1997 | 0.6 | 3.6 | 4.2 | 92 | 117 | 34 | 101 | 89 | 111 | 0.8 | 6.1 | 2.1 |
| April 1997 | 0.3 | 3.4 | 4.2 | 92 | 110 | 29 | 84 | 86 | 110 | 0.2 | 3.3 | 1.7 |
| May 1997 | 0.6 | 2.9 | 4 | 89 | 101 | 45 | 58 | 93 | 108 | 0.4 | 1.3 | 0.6 |
| June 1997 | 0.3 | 3 | 4 | 88 | 95 | 30 | 8 | 92 | 101 | 0.3 | 0.8 | 1.0 |
| July 1997 | 7.6 | 5.9 | 5.8 | 106 | 105 | 109 | 90 | 88 | 92 | 0.8 | 1.5 | 1.5 |
a Adapted from reference 44.
Water samples from three depth layers (the surface, 4 m, and 8 m) were collected monthly between 4 July 1996 and 25 June 1997 with a 5-liter Schindler-Patalas sampler. Portions of 100 ml were filtered with polycarbonate membrane filters (pore size, 0.2 μm; diameter, 47 mm, Osmonics, Livermore, Calif.) for later in situ hybridization and fixed as described previously (16). Filters were stored in small petri dishes at −20°C until further processing. Additionally, 100-ml subsamples were fixed with formalin (final concentration, 2% [vol/vol]) for bacterial abundance and cell size determination.
Abundance and biomass size distribution of the microbial assemblage.
Formalin-fixed subsamples of 5 to 20 ml were filtered with black membrane filters (pore size, 0.22 μm; diameter, 25 mm; Osmonics) and stained with 4′,6′-diamidino-2-phenylindole (DAPI), and bacterial abundances were determined by epifluorescence microscopy (30). At least 200 filaments per sample were counted at a low magnification (×400). The cell sizes of more than 400 DAPI-stained bacteria per sample were measured by using the semiautomated image analysis system described by Posch et al. (31). The analysis was limited to nonfilamentous bacterial morphotypes (<10-μm cell length). Cell volumes were estimated from measured area and perimeter (c.f. reference 31) and converted into cell dry mass by using the allometric conversion factor of Loferer-Krößbacher et al. (21). Biomass allocation into 0.4-μm cell length classes was calculated as described by Pernthaler et al. (27).
Design and characterization of oligonucleotide probes.
Water samples for DNA extraction were taken at Gossenköllesee on 11 December 1995 from a depth of 3 m. Around 900 ml of 50-μm-prescreened lake water was filtered with hydrophilic filters (Durapore [pore size, 0.2 μm; diameter, 47 mm]; Millipore Corp., Bedford, Mass.) until the filters were completely clogged with plankton biomass. During filtration, the samples were kept at ambient water temperatures (2 to 3°C). The filters were cut into smaller sections and stored at −20°C until further processing. DNA extraction from the filters was performed by following the protocol of Fuhrman et al. (14). Almost full-length bacterial 16S rDNA fragments were amplified by PCR from the extracted DNA by using two general bacterial 16S rDNA primers (38) on a Hybaid OmniGene thermocycler (MWG-Biotech, Ebersberg, Germany). Amplification conditions were 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min for 30 cycles after an initial preheating step of 94°C for 3 min. PCR amplification products were purified and cloned as described previously (38). Fifty 16S rDNA clones were partially sequenced (300 to 500 nucleotides) by using a LICOR DNA 4000 automated sequencer (MWG-Biotech) (38). These partial sequences were added to the 16S rRNA sequence database of the Technical University of Munich with the program package ARB (43). Two clones each affiliated with the alpha (GKS59 and GKS69)- and beta (GKS16 and GKS98)-proteobacteria were selected, and their 16S rDNA inserts were fully sequenced. The clone identifications, EMBL accession numbers, and nearest relatives in the database are presented in Table 2. By using the PROBE_DESIGN tool of ARB (43), specific oligonucleotide probes were constructed. In order to minimize the risk of accidentally detecting more than one population by FISH, we designed two oligonucleotide probes per clone that were targeted at different positions on the 16S rRNA and each of which showed at least one mismatch with all other known sequences. All probes were purchased from Interactiva (Ulm, Germany) and labeled with the indocarbocyanine dye Cy3. The sequences and target positions of the clone-specific probes are given in Table 2.
TABLE 2.
Probes designed from bacterial 16S rDNA sequences that were retrieved from Gossenköllesee in December 1995
| Clone | EMBL accession no. | Nearest relative in ARB database (% similarity) | Probe sequence | rRNA positiona | % FAb |
|---|---|---|---|---|---|
| GKS16 | AJ224987 | Rhodoferax fermentans (93.9) | 5′-TGCTACTCTACCGTTCG-3′ | 63–80 | 20 |
| 5′-GAACCGTTTCGTTCCGTA-3′c | 442–460 | 20 | |||
| GKS59 | AJ224988 | Rhodobacter sphaeroides (95.5) | 5′-GGGCCAATCCCTTCCC-3′ | 220–236 | 30 |
| 5′-CAGGTTGGCGCACCACCT-3′ | 1434–1452 | 30 | |||
| GKS69 | AJ224989 | Sphingomonas subarctica (95.5) | 5′-GGGCTCATCCTTCGGCGA-3′ | 218–236 | 35 |
| 5′-GGTCAGCTGCCTCCTTTG-3′c | 1451–1469 | 20–35 | |||
| GKS98 | AJ224990 | Alcaligenes xylosoxidans (96.2) | 5′-GACATACTCTAGCTCGG-3′ | 646–663 | 35 |
| 5′-CCCCACCGTGGTAATCGC-3′c | 1459–1477 | 20–35 |
a Escherichia coli numbering (8).
b FA, formamide concentration in the hybridization buffer.
c Used to monitor seasonal population dynamics.
dequate hybridization conditions were established by slot blot hybridization of in vitro RNA transcripts from the four 16S rDNA inserts with oligonucleotide probes at increasing concentrations of formamide (cf. reference 29). Plasmids containing inserts GKS16, GKS59, GKS69, and GKS98 were extracted from overnight cultures of the respective clones (QIAprep Spin Plasmid kit; Qiagen Inc., Chatsworth, Calif.), linearized with restriction enzyme NotI (GKS16 and GKS59) or ApaI (GKS69 and GKS98) (Boehringer, Mannheim, Germany), and purified (QIAquick PCR purification kit; Qiagen Inc.). rRNA was transcribed with the SP6/T7 rRNA transcription kit (Boehringer), and products were examined by polyacrylamide gel electrophoresis (4%) and ethidium bromide staining. Transcripts were denatured with 2% buffered glutaraldehyde and blotted in triplicate onto nylon membranes (Magna Charge; Micron Separations, Westboro, Mass.) in a Minifold II slot blot apparatus (Schleicher & Schuell, Dassel, Germany). The membranes were hybridized with radioactively labeled (42) probes GKS16-442, GKS59-1434, GKS69-1451, and GKS98-1459 at 46°C for 1.5 h and subsequently washed at 48°C for 30 min. The hybridization and washing buffers were identical to those used for FISH (see, e.g., reference 38). Probe-conferred radioactivity was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
FISH.
FISH of filter sections with oligonucleotide probes, counterstaining with DAPI, and mounting for microscopic evaluation were performed as described previously (16). The group-specific probes were ARCH915 (domain Archaea), EUB338 (domain Bacteria), ALF1b (alpha-proteobacteria), BET42a (beta-proteobacteria), GAM42a (gamma-proteobacteria), and CF319a (members of the Cytophaga-Flavobacterium cluster). Probe sequences and respective formamide concentrations in the hybridization buffers were given by Snaidr et al. (38). Epifluorescence microscopy was carried out as described before (2, 16) on a Zeiss Axioplan or a Zeiss Axioplan II, each equipped with an HBO 100-W mercury lamp and a 100× Plan Neofluar objective. At low relative abundances of hybridized cells (≤5% DAPI staining) at least 1,000 DAPI-stained objects were evaluated per sample; otherwise, between 500 and 1,000 cells were inspected. The lower boundaries depicted in Fig. 2, 4, and 5 represent the empirical limits of reliable quantification by our counting protocol (usually 1% of the DAPI-stained cell counts). Absolute densities of hybridized bacteria were calculated as the product of their relative abundances on filter sections (percentage of DAPI-stained objects) and the DAPI-stained direct cell counts. Both the absolute numbers and the fractions of hybridized cells were evaluated separately for filamentous bacteria. Therefore, the lower limits of these counts differ from those of all of the cells. Two counting protocols were tested. For bacteria hybridizing with EUB338 and CF319a, the abundances of hybridized cells were quantified as fractions of DAPI-stained filaments, in analogy with the standard counting technique. For ARCH915, first the fraction of all hybridized cells (filaments and nonfilaments) was determined as a percentage of the total DAPI-stained cell counts. Then, the relative abundance of filaments in at least 500 hybridized cells was evaluated. This second protocol was much less time consuming but turned out to be too imprecise for the calculation of absolute filament abundances. Accordingly, only the relative fractions of filamentous archaea are depicted in the results (see Fig. 2).
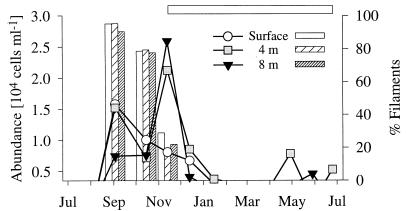
FIG. 2.
Seasonal abundances of pelagic archaea (lines) and percentages of filamentous morphotypes (bars). The horizontal bar outlines the period of ice cover.
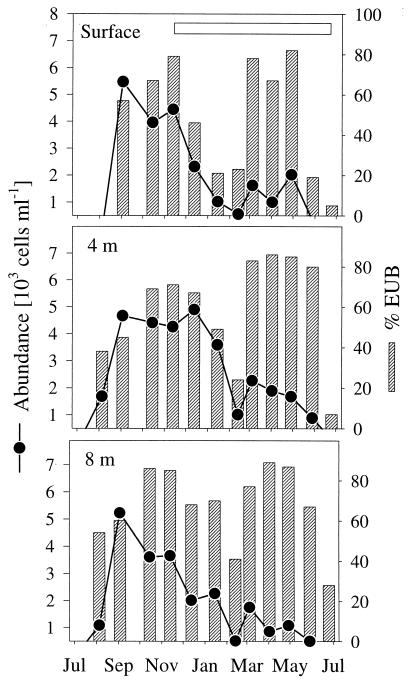
FIG. 4.
Seasonal dynamics of filamentous bacteria of the Cytophaga-Flavobacterium cluster and their fraction of bacteria (% EUB). The horizontal bar indicates the period of ice cover.
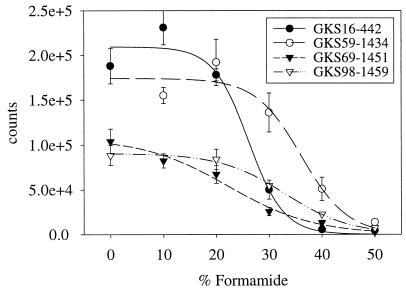
FIG. 5.
Hybridization of in vitro RNA transcripts from retrieved 16S rDNA sequences with matching sequence-specific probes at increasing levels of formamide.
RESULTS
Total assemblage.
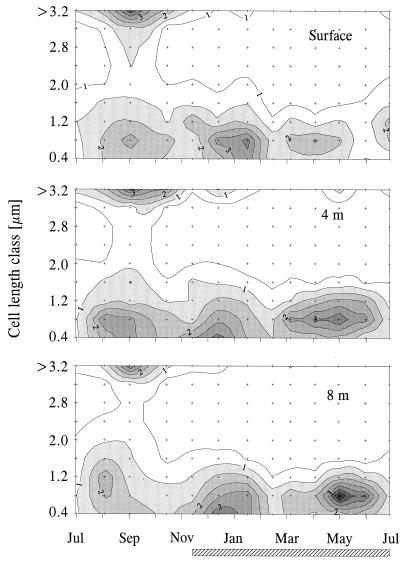
Cell densities determined by DAPI staining ranged from 0.11 × 106 to 0.55 × 106 cells ml−1 (Table 3). Two peaks of total numbers in early autumn and midwinter were separated by distinct minima in July, October, and February. Filamentous cells were present in the plankton throughout the season at densities of 0.076 × 104 to 1.33 × 104/ml−1, reaching maximal abundances in early autumn and continuously decreasing in number during the period of ice coverage. In early winter, filaments were more frequent in the lower water layers. Three clearly separated maxima of nonfilamentous bacterial biomass were found in late summer, midwinter, and spring (Fig. 1). Between July and January, a considerable amount (>35% in September and October) of biomass was present in cells >3.2 μm in length, and during August and September, the biomass was distributed more evenly over bacterial size classes. Between March and June, biomass maxima were formed exclusively by small bacteria (0.4 to 1.2 μm) and were more pronounced in the deeper water layers.
TABLE 3.
Total picoplankton and filament abundances in Gossenköllesee and detection by probe EUB338
| Date | Total cell concn (105 ml−1)
|
% detected by EUB338a
|
Filament concn (103 ml−1)
|
% detected by EUB338a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Surface | 4 m | 8 m | Surface | 4 m | 8 m | Surface | 4 m | 8 m | Surface | 4 m | 8 m | |
| 4 July 1996 | 1.90 | 2.07 | 1.95 | 77 | 82 | 78 | 6.69 | 6.33 | 4.66 | 83 | 93 | 89 |
| 5 August 1996 | 3.29 | 3.93 | 3.28 | 58 | 45 | 63 | 6.77 | 5.09 | 2.16 | 85 | 88 | 92 |
| 3 September 1996 | 4.52 | 4.96 | 3.36 | 63 | 57 | 55 | 13.35 | 11.78 | 11.00 | 72 | 87 | 80 |
| 15 October 1996 | 2.91 | 2.86 | 2.31 | 53 | 52 | 43 | 8.29 | 9.22 | 6.22 | 72 | 69 | 68 |
| 13 November 1996 | 2.78 | 4.28 | 3.67 | 61 | 59 | 58 | 7.70 | 8.55 | 5.76 | 73 | 70 | 74 |
| 13 December 1996 | 3.78 | 5.54 | 5.12 | 50 | 40 | 36 | 5.92 | 9.18 | 3.62 | 86 | 79 | 82 |
| 15 January 1997 | 4.94 | 3.24 | 4.03 | 43 | 44 | 26 | 5.38 | 8.21 | 4.28 | 89 | 89 | 76 |
| 13 February 1997 | 2.05 | 2.78 | 2.44 | 42 | 44 | 44 | 2.79 | 4.88 | 1.40 | 87 | 88 | 89 |
| 5 March 1997 | 2.70 | 3.45 | 3.04 | 50 | 53 | 59 | 2.35 | 3.26 | 2.49 | 89 | 84 | 92 |
| 2 April 1997 | 2.51 | 3.49 | 2.59 | 48 | 44 | 43 | 1.51 | 2.27 | 1.09 | 98 | 96 | 89 |
| 29 April 1997 | 2.30 | 3.82 | 3.59 | 61 | 49 | 65 | 2.60 | 2.00 | 1.27 | 95 | 99 | 98 |
| 29 May 1997 | 1.13 | 2.36 | 3.14 | 64 | 65 | 64 | 1.69 | 1.13 | 0.76 | 95 | 98 | 100 |
| 25 June 1997 | 1.85 | 2.15 | 1.93 | 77 | 70 | 59 | 0.79 | 1.90 | 0.86 | 92 | 97 | 95 |
a Percentage of cells stained by DAPI.
FIG. 1.
Seasonal dynamics of the biomass size distribution of nonfilamentous pelagic procaryotes in Gossenköllesee at different depths. Values on contour lines indicate micrograms of bacterial dry mass per liter. The hatched bar outlines the period of ice cover.
FISH with domain- and group-specific probes.
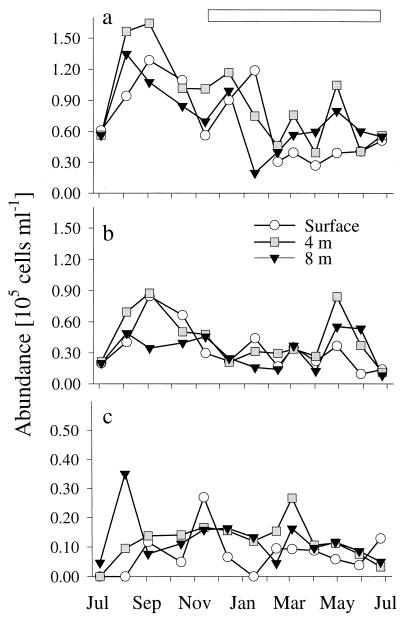
Between 26 and 82% of all DAPI-stained objects (mean, 55%) could be visualized with bacterial probe EUB338, and between 68 and 100% of filamentous cells (mean, 87%) were detected by EUB338 (Table 3). Detection rates were lowest at a depth of 8 m (mean, 53%) between December and February for all cells and from October to December for filamentous forms. Archaea represented from less than 1% to a maximum of 5% of all DAPI-stained cells (mean, 1.4%) and reached their highest densities in September and shortly after ice cover formation in late November (Fig. 2). During September and October, there was a distinct maximum of the filamentous fraction of archaea throughout the water column. On an annual average, beta-proteobacteria formed 24% (range, 5 to 41%) of all DAPI-stained cells and 45% (range, 20 to 89%) of the cells detected by the bacterial probe. At all three depths, the beta-proteobacteria reached their maximal abundances in late summer (August to September) (Fig. 3a). Another maximum during midwinter (December and January) was followed by a sharp decline in numbers. The alpha-proteobacteria showed a similar peak in early autumn in the upper water layers (Fig. 3b). A second peak was observed in May at a depth of 4 m, succeeding an annual maximum of chlorophyll a which occurred at the same depth during early spring (Table 1). Alpha-proteobacteria accounted for 4 to 23% of all DAPI-stained cells (mean, 11%) and, on average, 21% of the EUB338 counts (range, 7 to 45%). The ratio of alpha- to beta-proteobacteria ranged from 0.15 in midwinter and early summer to 0.90 during April and May (annual mean, 0.50), with distinct maxima in autumn (September to November) and spring (March to May) at the surface and at a depth of 4 m. Gamma-proteobacteria always accounted for less than 2% of the total cell density (data not shown). Bacteria of the Cytophaga-Flavobacterium cluster never exceeded 4 × 104 ml or−1 10% of all DAPI-stained cells (mean, 3.5%) (Fig. 3c). Threadlike morphotypes formed up to 80% of all of the bacteria hybridizing with CF319a in the surface layer during October but were virtually undetectable during early summer (June and July; annual mean, 20%) (Fig. 4). Members of this phylogenetic cluster were a dominant fraction of all filamentous bacteria for most of the year (mean, 53%), except for two distinct minima in summer and winter, respectively.
FIG. 3.
Seasonal fluctuations in the abundance of beta (a) and alpha (b) proteobacteria and (c) bacteria in the Cytophaga-Flavobacterium cluster in Gossenköllesee. Note the different scale in panel c. The horizontal bar indicates the period of ice cover.
Populations monitored with clone-specific probes.
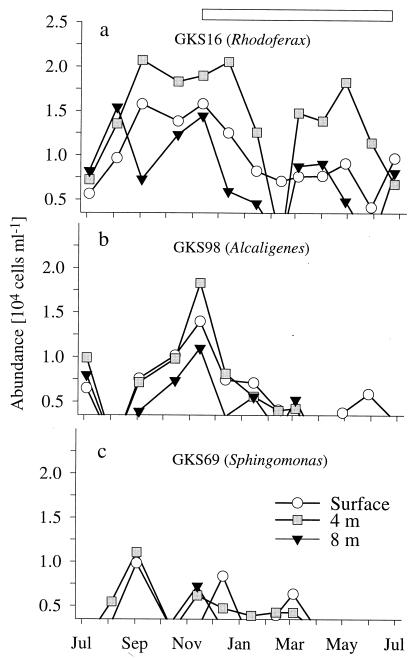
Each pair of probes was found to define identical populations, i.e., two probes targeted at different positions of one rRNA sequence (Table 2) detected bacterial cells that could not be distinguished in terms of their numbers in parallel samples and their cell morphologies. As the pairs of probes appeared to be equivalent in terms of detected population sizes, evaluation of the seasonal samples was performed by using the probe that gave the brighter hybridization signal (probes GKS16-442, GKS69-218, and GKS98-1459). The hybridization of RNA transcripts from the 16S rDNAs with these probes at various concentrations of formamide resulted in typical sigmoidal melting curves (Fig. 5). Probes targeted to three of the four clones detected bacterial populations that at least seasonally exceeded 1% of the DAPI-stained cells (Fig. 6a to c). The sequence of the fourth clone, GKS59, affiliated with alpha-proteobacteria, was associated with a filamentous morphotype (cell length, 11 to 20 μm) that occurred sporadically throughout the year at low densities (data not shown). The bacteria hybridizing with probes GKS16-61 and GKS16-442 were rod-shaped cells (lengths, 1.5 to 3.5 μm) and short filaments (8 to 13 μm). The phylotype GKS16 is affiliated with beta-proteobacteria, and Rhodoferax fermentans is its closest known relative with a 16S rRNA similarity of 93.9%. These populations were most abundant between September and January, and the populations were seasonally more stable at the surface than in the lower water layers (Fig. 6a). The maximal relative densities were 6% of DAPI-stained cells. At 4 m, we observed a biphasic annual population development: a distinct spring bloom between March and May was set apart from the previous autumn to early winter maximum by a decline to a level below the detection limit during February. A similar drop was also observed at a depth of 8 m. On both dates in July, when the lake mixes, population densities were very similar at all three depths.
FIG. 6.
Population dynamics of cells hybridizing with probes targeted at beta-proteobacteria (GKS16 [a] and GKS98 [b]) and alpha-proteobacteria (GKS69 [c]) The genera with the highest 16S rRNA sequence similarity with the respective clones are in parentheses. The horizontal bar indicates the period of ice cover.
The second beta-proteobacterial population hybridizing with GKS98-826 and GKS98-1459 consisted of rods with cell lengths of 2 to 4.5 μm. They showed a clearly parallel seasonal development at all depths (Fig. 6b). After a decline below the detection densities in August, the populations reached maximal abundances during the period of ice layer formation (4% of DAPI-stained objects) and then declined below the limit of detection during the early spring. On a seasonal average, cells hybridizing with probes for clones GKS16 and GKS98 constituted about one-quarter of the beta-proteobacteria, but this fraction fluctuated between >50% in the surface layer during the formation of the winter ice cover and less than 10% in the lower water layers during February.
Bacteria targeted by probe GKS69-218 or GKS69-1451 were brightly fluorescent rods with cell lengths of 2.8 to 9.5 μm. The phylotype GKS69 clusters with the genus Sphingomonas within the alpha-proteobacteria. The respective population appeared sporadically at densities of 5,000 to 10,000 cells ml−1 between August and March, mainly in the upper water layers (Fig. 6c).
DISCUSSION
Seasonal community changes.
Bacterial in situ identification and the determination of total biomasses appear to describe supplementary features of microbial communities. Either analysis established that the microbial assemblage of Gossenköllesee continuously transformed, even during the months when the lake was sealed by the winter ice cover. The icebreak period between June and July appeared to be a major promoter of community transition (c.f. reference 26). This is easily conceivable by considering, e.g., the allochthonous input from the winter ice cover (35a), the subsequent thermal mixing of the water body (Table 1) and the increased exposition to sunlight. Recently, Sommaruga and coworkers demonstrated that UV A and B radiation readily penetrates the water column of Gossenköllesee and that this is sufficient to directly and indirectly affect bacterial 3[H]thymidine and 14[C]leucine incorporation (40) and to influence the bacterivory of a heterotrophic nanoflagellate (39). It was not until September that alpha- and beta-proteobacteria (Fig. 3), filamentous bacteria (Table 2) and archaea (Fig. 2), and cells hybridizing with clone-specific probes (Fig. 6) formed population maxima at the lake surface. During the autumn thermal mixing and the period of ice cover formation, the total microbial biomass declined in all layers (Fig. 1), yet at the same time, small cells of the archaeoplankton (Fig. 2) and bacteria hybridizing with GKS98-1459 (Fig. 6b) produced distinct annual maxima. Interestingly, Murray et al. (25) reported strong seasonality of archaeal rRNA concentrations in Antarctic coastal waters, with the highest values occurring during the austral winter. This corresponds to our observation that pelagic archaea were abundant in the lake’s plankton only during autumn and hardly present thereafter.
Successions beneath the winter cover, when the water temperatures ranged between 0 and 4.2°C (Table 1), were characterized not only by two clearly separated blooms of microbial biomass in the smaller cell size classes during midwinter and spring (Fig. 1) but also by distinct changes in community composition (Fig. 2, 3, and 6a and b). Between January and March, rates of detection with the bacterial probe dropped (Table 3), the numbers of beta-proteobacteria and filamentous members of the Cytophaga-Flavobacterium cluster declined significantly (Fig. 3a and 4), and bacteria related to clone GKS16 dropped below the limit of detection in all but the surface water layers (Fig. 6a). Simultaneously, the biomass of very small bacteria increased (Fig. 1), concomitant with the winter maxima of chlorophyll a concentrations (Table 1). The presence of an active microbial community beneath the winter ice cover was also indicated by the gradual decline of oxygen saturation in the lowest water layers (Table 1 in reference 44). The pelagic bacteria in Gossenköllesee formed a second biomass maximum in late spring in the lower water layers (Fig. 1), in parallel with a density increase of both the alpha- and beta-proteobacteria (Fig. 3a and b) and of the population hybridizing with GKS16-442 (Fig. 6a). This development coincided with the decline of the phytoplankton bloom at a depth of 4 m (Table 1) but also with the onset of the snowmelt in the catchment area (44), as reflected by decreased lake water alkalinity at 0 m (Table 1). This water influx might be the cause for the reduced bacterial biomass at the lake surface prior to the ice break (Fig. 1).
Bacteria of the Cytophaga-Flavobacterium cluster bloomed in deeper water layers in summer, at the surface during ice formation, and at the depth with the highest chlorophyll a concentrations in early spring (Table 1). In summary, this hints at phylogenetic diversity within this group masked by the generality of the probe. This conclusion is also supported by the contrasting dynamics of the filamentous and nonfilamentous morphotypes (Fig. 3c and 4). A fraction of these threadlike bacteria might indeed genotypically resemble small cells with a high potential for morphological plasticity, as has been shown, e.g., for marine psychrophiles (23) and a Comamonas acidovorans strain isolated from a lake (18). Since a prominent part of the planktonic filaments in Gossenköllesee may reach cell lengths of >50 μm (31), they form a considerable fraction of the total microbial biomass. During extended periods, most of the threadlike bacteria were affiliated with the Cytophaga-Flavobacterium cluster (Fig. 4), so that this phylogenetic group at least seasonally constituted the largest pool of bacterial biomass in the system.
Studying microbial population dynamics by FISH.
It has been shown that releasing allochthonous microbes into the plankton may result in their rapid disappearance (see, e.g., reference 7), but the fate and dynamics of autochthonous bacterioplankton populations is much less well understood. Gordon and Giovannoni (17) monitored the seasonal density variation of a bacterial rDNA sequence in the Atlantic, yet they did not attempt to estimate absolute cell abundances. Recently, Pinhassi et al. (28) presented a protocol to determine the in situ densities of individual bacterial species which, however, requires prior cultivation of the target organisms. Our approach differs from these in that it analyzes single bacterial cells rather than nucleic acid extracts. Thus, we could take into account the microscopically obvious difference between filamentous and nonfilamentous bacteria, which is certainly of ecological relevance, e.g., considering growth rates (18) or biomass contribution to the community. Recently, FISH has been successfully applied in Antarctic coastal water (25). A similar but indirectly also cultivation-dependent strategy used to monitor bacterial populations in a lake based on fluorescently labeled monoclonal antibodies has been described by Faude and Höfle (10). The two populations investigated in that study together formed less than 0.1% of the total bacterial community, which demonstrates the difficulty of specifically isolating those microbes from the plankton that form larger populations in situ.
Probes designed for single retrieved 16S rRNA sequences of uncultivated microorganisms cannot be easily tested for specificity. We used pairs of probes targeted to distant positions of the 16S rRNA and compared abundances and morphologies of hybridized cells from parallel samples in order to minimize the risk of detecting multiple populations. In addition, melting curve equivalents were determined by hybridizing the sequence-specific probes against RNA transcripts from the retrieved 16S rDNAs (Fig. 5), which have been shown to resemble the melting behavior of native rRNA (29). On these grounds, it was possible to set hybridization conditions that ensured the specificity of the probes for the respective rRNA targets. Yet, for definitive confirmation of the identity of the targeted 16S rDNA (and not just the target region), it might be necessary to, e.g., flow sort the respective microbes and then amplify and sequence their 16S rDNAs (45).
Knowing that some bacterial species might have practically identical 16S rDNA sequences (12), can one actually study the population dynamics of a single bacterial species by in situ hybridization? Considering macroecological principles of competitive exclusion, it is probably unlikely to find closely related microorganisms co-occurring in a nutrient-poor system dominated by strong physical and chemical fluctuations. One must, however, be aware of the physiological variability that may be found within different DNA-DNA homology groups of one single species (35). We thus caution against premature conclusions about the ecology of microorganisms based exclusively on their in situ occurrence if not backed up by taxonomic and ecophysiological studies on isolates.
However, the knowledge of in situ distributions of microbial populations at our level of analysis provides a basis for such research, as it allows us to set the pelagic bacteria into the traditional framework of plankton ecology. By applying FISH for single 16S rDNA sequences retrieved from the site, we could readily demonstrate that heterotrophic bacterial species (phylotypes) show distinct vertical abundance patterns (Fig. 6a) and that their relative importance in the community may be limited to particular seasons (Fig. 6b). We envisage the microbial assemblage in the lake to be of a “seed bank” nature in that numerous species might be permanently present in the system at low abundances but form larger populations only during periods of optimal competitiveness. However, the causes of the population dynamics observed remain unknown. We suggest utilization of the diversity information available in the increasing number of planktonic bacterial 16S rDNA clone libraries (e.g., 6, 15, 19) to study the in situ abundances of individual microorganisms at different sites and during different seasons. Such investigations might eventually lead to a better understanding of the function of individual bacterial populations in aquatic environments and of the forces governing their abundances, biomasses, and cell morphologies.
ACKNOWLEDGMENTS
We thank Ramon Rosselló-Mora for fruitful discussions; Hansjörg Thies, Birgit Sattler, Anton Wille, and Ruben Sommaruga for providing background data; Birgit Rattunde for skillful laboratory assistance; and Kerstin Sahm, Lutgarde Raskin, and Daniel Oerther for their suggestions on in vitro transcription.
This study was supported by the Austrian Science Foundation (Proj. P11685-MOB), the European Community (MOLAR, ENV4-CT95-0007), the Deutsche Forschungsgemeinschaft (Am 73/2-4), and the Max Planck Society.
REFERENCES
- 1.Acinas S G, Rodríguez-Valera F, Pedrós-Alió C. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rDNA. FEMS Microbiol Ecol. 1997;24:27–40. [Google Scholar]
- 2.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake using in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R, Glöckner F O, Neef A. Modern methods in subsurface microbiology: in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol Rev. 1997;20:191–200. [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahr M, Hobbie J E, Sognin M L. Bacterial diversity in an antarctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 7.Boyle M, Ahl T, Molin S. Application of a strain-specific rRNA oligonucleotide probe targeting Pseudomonas fluorescens Ag1 in a mesocosm study of bacterial release into the environment. Appl Environ Microbiol. 1995;61:1384–1390. doi: 10.1128/aem.61.4.1384-1390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organisation and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 9.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 10.Faude U, Höfle M G. Development and application of monoclonal antibodies for in situ detection of indigenous bacterial strains in aquatic ecosystems. Appl Environ Microbiol. 1997;63:4534–4542. doi: 10.1128/aem.63.11.4534-4542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felip M, Sattler B, Psenner R, Catalan J. Highly active microbial communities in the ice and snow cover of a high mountain lake. Appl Environ Microbiol. 1995;61:2394–2401. doi: 10.1128/aem.61.6.2394-2401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrman J A, Sleeter T D, Carlson C A, Proctor L M. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar Ecol Prog Ser. 1989;57:207–217. [Google Scholar]
- 14.Fuhrman J A, Comeau D E, Hagström A, Chan A M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni S J, Mullins T D, Field K G. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes. NATO ASI (Adv Sci Inst) Ser Ser G Ecol Sci. 1995;38:217–248. [Google Scholar]
- 16.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K H, Schleifer K H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 17.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn M W, Höfle M G. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl Environ Microbiol. 1998;64:1910–1918. doi: 10.1128/aem.64.5.1910-1918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiorns W D, Methé B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jürgens K, Arndt H, Rothhaupt K O. Zooplankton-mediated changes of bacterial community structure. Microb Ecol. 1994;27:27–42. doi: 10.1007/BF00170112. [DOI] [PubMed] [Google Scholar]
- 21.Loferer-Krößbacher M, Klima J, Psenner R. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl Environ Microbiol. 1998;64:688–694. doi: 10.1128/aem.64.2.688-694.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 23.McCallum K, Inniss W E. Thermotolerance, cell filamentation, and induced protein synthesis in psychrophilic and psychrotrophic bacteria. Arch Microbiol. 1990;153:585–590. [Google Scholar]
- 24.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 25.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernthaler J, Alfreider A, Posch T, Andreatta S, Psenner R. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllesee, Austria) Appl Environ Microbiol. 1997;63:4778–4783. doi: 10.1128/aem.63.12.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernthaler J, Sattler B, S̆imek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 28.Pinhassi J, Zweifel U L, Hagström A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polz M F, Cavanaugh C M. A simple method for quantification of uncultured microorganisms in the environment based on in vitro transcription of 16S rRNA. Appl Environ Microbiol. 1997;63:1028–1033. doi: 10.1128/aem.63.3.1028-1033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 31.Posch T, Pernthaler J, Alfreider A, Psenner R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, Cyto-Clear slides, and image analysis. Appl Environ Microbiol. 1997;63:867–873. doi: 10.1128/aem.63.3.867-873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Psenner R. Determination of size and morphology of aquatic bacteria by automated image analysis. In: Kemp P, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publications; 1993. pp. 339–345. [Google Scholar]
- 34.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Marianger Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosselló-Mora R A, Lalucat J, Garciá-Valdés E. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl Environ Microbiol. 1994;60:966–972. doi: 10.1128/aem.60.3.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Sattler, B., and A. Wille. Personal communication.
- 36.Shut F, de Vries E J, Gotschal J C, Robertsen B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S̆imek K, Vrba J, Pernthaler J, Posch T, Hartmann P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sommaruga R, Oberleitner A, Psenner R. Effect of UV radiation on the bacterivory of a heterotrophic nanoflagellate. Appl Environ Microbiol. 1996;62:4395–4400. doi: 10.1128/aem.62.12.4395-4400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smmaruga R, Obernosterer I, Herndl G J, Psenner R. Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Appl Environ Microbiol. 1997;63:4178–4184. doi: 10.1128/aem.63.11.4178-4184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommaruga R, Psenner R. Permanent presence of grazing-resistant bacteria in a hypertrophic lake. Appl Environ Microbiol. 1995;61:3457–3459. doi: 10.1128/aem.61.9.3457-3459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckmann N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. 1998. http://www.mikro.biologie.tu-muenchen.de/pub/ARB http://www.mikro.biologie.tu-muenchen.de/pub/ARB. Department of Microbiology, Technische Universität München, Munich, Germany. . Department of Microbiology, Technische Universitt Mnchen, Munich, Germany. [Google Scholar]
- 44.Thies, H., U. Nickus, C. Arnold, R. Schnegg, A. Wille, and R. Psenner. Biogeochemistry of a high mountain lake in the Austrian Alps. Verh. Int. Verein Limnol., in press.
- 45.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]