Abstract
Members of the 4-oxalocrotonate tautomerase (4-OT) subgroup in the tautomerase superfamily (TSF) are constructed from a single β unit and form homo- or heterohexamers, whereas those of the other four subgroups are composed of two consecutively joined β-α-β units and form trimers. A subset of sequences, double the length of the short 4-OTs, is found in the 4-OT subgroup. These “fused” 4-OTs form a separate subgroup that connects to the short 4-OTs in a sequence similarity network (SSN). The fused gene can be a template for the other four subgroups, resulting in diversification of activity. Analysis of the SSN shows that multiple nodes in the fused 4-OTs connect to five linker nodes, which in turn, connect to the short 4-OTs. Some fused 4-OTs are symmetric trimers and others are asymmetric trimers. The origin of this asymmetry was investigated by subjecting the sequences in three linker nodes and a closely associated fourth node to kinetic, mutagenic, and structural analysis. The results show that each sequence corresponds to the α- or β-subunit of a heterohexamer that functions as a 4-OT. Mutagenesis indicates that the key residues in both are αPro-1 and βArg-11, like that of a typical 4-OT. Crystallographic analysis shows that both heterohexamers are asymmetric, where one heterodimer is flipped 180° relative to the other two heterodimers. Fusion of two subunits (α and β) of one asymmetric heterohexamer generates an asymmetric trimer with 4-OT activity. Hence, asymmetry can be introduced at the heterohexamer level and then retained in the fused trimers.
Graphical Abstract
INTRODUCTION
The 4-oxalocrotonate tautomerase (4-OT) subgroup is the largest of the five subgroups in the tautomerase superfamily (TSF).1 TSF members (>11,000 members) are distinctive because they share a simple β-α-β building block and frequently have a catalytic amino-terminal proline.2,3 Most TSF members are constructed from a single β-α-β unit (58–84 amino acids) or two consecutively joined β-α-β units (110–150 amino acids).4 Oligomers with both arrangements are found in the 4-OT subgroup, whereas only oligomers made up of two consecutively fused β-α-β units are found in the other four subgroups.1 Accordingly, there are homo- and heterohexamers (consisting of short subunits) as well as symmetric and asymmetric trimers (consisting of long subunits) in the 4-OT subgroup, but only symmetric trimers have been found, thus far, in the other four subgroups.1,5,6 Three representative oligomers in the TSF are shown in Figure 1.
Figure 1.
Three representative oligomers in the TSF. (A) Representative symmetric trimer (PDB code 5UNQ).5,6 (B) Representative asymmetric trimer (PDB code 6BLM).5,6 (C) The canonical 4OT, a homohexamer (PDB code 1OTF).1 The catalytic proline residue is shown with a space-filling sphere.
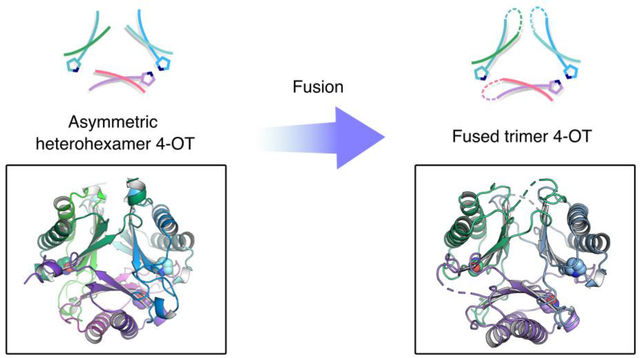
The predominance of this structural theme has long suggested that a gene fusion event took place early in the evolutionary history of the TSF leading to the diversification of function that is seen today. Analysis of the sequence similarity network (SSN) uncovered a potential clue with the discovery of the “fused 4-OTs” in the 4-OT subgroup, which had previously been thought to consist of only short 4-OTs.1,5,6 The fused 4-OTs, double the length of the short 4-OTs, still connect to the short 4-OTs in the SSN. Since the fused 4-OTs share features (i.e., the trimeric arrangement) with possible progenitors for the other four subgroups, they were explored in detail.
Crystallographic characterization along with bioinformatics analysis identified two clusters with distinct oligomeric arrangements. Both clusters consisted of trimers, but in one, the trimers were symmetric and in the other, the trimers were asymmetric.5,6 In the asymmetric trimer, one of the subunits is flipped 180° relative to the other two subunits. As a result, there are three unique interfaces composed of different residues. The two structures provide two different scaffolds that might provide two different possibilities for diversification.
In a Level 2 network (where the sequences are grouped together at higher stringency with an increased minimum edge alignment score)1,7 for the 4-OT subgroup, multiple nodes in the fused 4-OTs (red nodes) connect to three linker nodes (magenta and cyan nodes,) which, in turn, connect to the short 4-OTs (blue nodes) (Figure 2). Along with a closely associated fourth node (the α-subunit in the magenta node, which connects to the α-subunit in the cyan node), the sequences make two putative heterohexamers. Both heterohexamers are found in closely related hosts, Herbaspirillum sp. CF444 and Herbaspirillum sp. YR522. The genes for the α- and β-subunits are found adjacent to each other in the genome. The α-subunits begin with a proline and the β-subunits begin with a serine.
Figure 2.
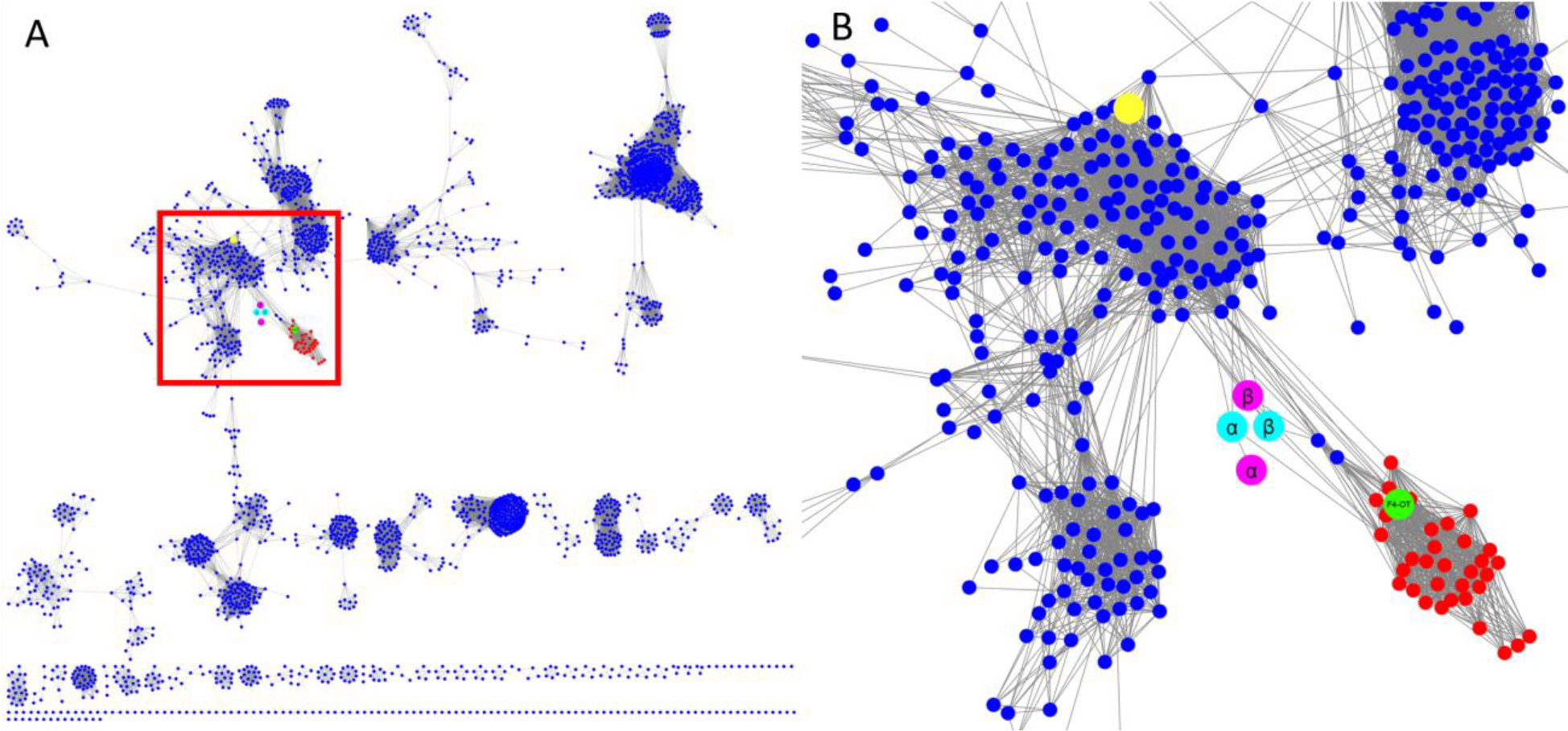
The full level 2 network (90% representative network) for the (A) 4-OT subgroup and (B) blow-up of the region with the nodes of interest. (A) The level 2 network for the 4-OT subgroup where the region of interest is shown in the red box. (B) Detailed view of the nodes of interest. The nodes containing the short 4-OTs are shown in blue and those containing fused 4-OTs are shown in red. The node shown in green contains a fused 4-OT from Burkholderia lata (ATCC 17760, UniProt accession: Q392K7), which is an asymmetric trimer.5,6 The two putative heterohexamer 4-OTs are shown in cyan and magenta, where the subunits for the YR and CF heterohexamers are shown respectively in cyan and magenta. Each subunit has one node. The large yellow node at the top contains the 62-amino acid sequence for the canonical 4-OT.1
The α- and β-subunits in the four representative nodes that connect the short 4-OTs and the fused 4-OT were cloned, co-expressed, and the resulting proteins were subjected to sequence, kinetic, mutagenic, and crystallographic analysis. Both proteins are heterohexamers that function as 4-OTs.1 Sequence and mutagenic analysis showed that the αPro1 and βArg11 are key residues in both heterohexamers. Crystallographic analysis showed that both heterohexamers are asymmetric. Fusion of the α- and β-subunits of the YR heterohexamer by a short linker generated a fused asymmetric trimer. This observation suggests that asymmetry might have been introduced in the heterohexamer before fusion and that the fused protein retained this property.
EXPERIMENTAL PROCEDURES
Materials.
Chemicals, biochemicals, buffers, and solvents were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO), Fisher Scientific Inc. (Pittsburgh, PA), Fluka Chemical Corp. (Milwaukee, WI), or EM Science (Cincinnati, OH). The syntheses of 2-hydroxymuconate (2-HM)8, 5-(methyl)-2-hydroxymuconate (5-Me-2-HM)9, 5-(carboxymethyl)-2-hydroxymuconate (CHM)10, and 2-hydroxy-2,4-pentadienoate (HPD)11 are reported in the indicated references. Phenylenolpyruvate (PP) was purchased from Fluka Chemical Corp. (Milwaukee, WI). The Phenyl Sepharose 6 Fast Flow and DEAE-Sepharose resins, and the prepacked PD-10 Sephadex G-25 columns were obtained from GE Healthcare (Piscataway, NJ). The Sephadex G-75 resin was obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO). The Econo-Column chromatography columns were obtained from Bio-Rad Laboratories, Inc. (Hercules, CA). The Amicon stirred cell concentrators, the ultrafiltration membranes (10,000 Da, MWCO), and the Amicon Ultra (3 kDa MWCO) centrifugal filter units were purchased from EMD Millipore Inc. (Billerica, MA). Enzymes and reagents used for molecular biology procedures were obtained from New England Biolabs, Inc. (Ipswich, MA). The QIAquick Gel Extraction Kit was obtained from Qiagen (Venlo, NL).
Bacterial Strains, Plasmids, and Growth Conditions.
Escherichia coli strain BL21(DE3) was obtained from Agilent Technologies (Santa Clara, CA). The genes encoding the α and β subunits of the heterohexamer 4-OT from Herbaspirillum sp. YR522 (UniProt accession J2VT77 and J3HY51, respectively) were codon-optimized for expression in E. coli, synthesized, and cloned into the expression vector pJ411 by ATUM (Newark, CA). In one plasmid, the two subunits are under the control of separate promoters and express separately creating a heterohexamer designated “heterohexamer-YR522–4OT” (hhYR-4OT). In a separate plasmid, the stop codon of the α subunit and the start codon of the β subunit are removed, the two genes are fused using a short linker, and the two subunits are expressed as a single gene product designated “fused-YR522–4OT” (fYR-4OT). The linker used in fYR-4OT (SAADGAPPSL) mimics that of the fused 4-OT from Burkholderia lata (UniProt accession Q392K7)5. The genes encoding the α and β subunits of a putative heterohexamer 4-OT from Herbaspirillum sp. CF444 (UniProt accession J3DHL6 and J2M343, respectively) were synthesized in the same fashion and the gene products are designated “heterohexamer-CF444–4OT” (hhCF-4OT) and “fused-CF444–4OT” (fCF-4OT). Plasmids were isolated from cell cultures using the Sigma GenElute Plasmid Miniprep Kit. Cells for cloning and overexpression were grown on Lysogeny broth (LB) agar plates or in LB medium supplemented with kanamycin (Kn) (30 μg/mL).
General Methods.
Techniques for restriction enzyme digestion, ligation, transformation, and other standard molecular biology manipulations were based on methods described previously.12 Oligonucleotide primers were synthesized by Sigma-Aldrich. DNA sequencing was performed at the DNA core facility of the Institute for Cellular and Molecular Biology (ICMB) at the University of Texas. Electrospray ionization mass spectrometry (ESI-MS) and LC-MS/MS analysis were carried out respectively on an Orbitrap Fusion 1 or an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific, San Jose, CA) in the Proteomics Facility in the ICMB. Steady-state kinetic assays were performed on an Agilent 8453 diode-array spectrophotometer at 22 °C. Nonlinear regression data analysis was performed using the program Grafit (Erithacus Software Ltd., Staines, U.K.). Protein concentrations were determined by the Bradford method.13 SDS-PAGE was carried out on denaturing gels containing 12% polyacrylamide.14
Construction of the 4OT Variants.
Four hhYR-4OT variants (αP1A, αR39A, βR11A and βR62A) and five hhCF-4OT variants (αP1A, αL11Y, αR39A, βR11A and βR62A) were constructed using a whole-plasmid PCR technique.15 Wild-type hhYR-4OT or wild-type hhCF-4OT in the pJ411 vector served as the template along with the primer pairs listed in Table S1. The PCR mixture (200 μL) contained the wild-type plasmid (50 ng), forward and reverse primers (0.5 μM each), 10 × ThermoPol buffer (20 μL), dNTPs (0.2 mM each), and Vent DNA polymerase (4 units). The cycling parameters were 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s, a variable annealing temperature for 20 s, and 72 °C for 4.5 min, with a final elongation step of 72 °C for 5 min. Following the PCR, the methylated parent plasmid was removed by the addition of DpnI restriction enzyme (1 μL of a 20 unit/μL solution) and incubation at 37 °C for 30 min. An aliquot of the DpnI-digested product was transformed into E. coli BL21(DE3) cells via the heat shock method.12 Transformants were selected at 37 °C on LB/Kn plates overnight. Plasmid DNA was isolated from a single colony and the incorporation of the mutation was confirmed using DNA sequencing.
The βR39A hhYR-4OT variant was constructed using overlap extension PCR16 with the wild-type hhYR-4OT gene in the pJ411 vector serving as the template. The variant primers (listed in Table S1) were used in combination with the universal T7 promoter and terminator primers. PCR concentrations and the thermal cycling parameters were the same as those described above. The PCR product and an aliquot of the pJ411 vector containing the wild-type hhYR-4OT gene were digested using the XbaI and XhoI restriction enzymes. The digested products were electrophoresed on an agarose gel and the desired bands were excised and purified using a QIAquick Gel Extraction Kit. The purified digested products were ligated using T4 DNA ligase at room temperature overnight. An aliquot of the ligated DNA was transformed into E. coli BL21(DE3) cells via the heat shock method.12 Transformants were selected at 37 °C on LB/Kn plates overnight. Plasmid DNA was isolated from a single colony and the incorporation of the mutation was confirmed using DNA sequencing.
Expression and Purification of Wild-Type, Fused, and 4-OT Variant.
A single colony of E. coli BL21(DE3) containing the desired gene was used to inoculate LB/Kn medium (~20 mL). The cells were shaken overnight at 37 °C and an aliquot was used to inoculate LB/Kn medium (4 × 400 mL, 1.6 L total) so that the initial OD600 reading was ~0.05. Cells were grown at 37 °C with shaking until the OD600 reached ~0.6 (~2 h). The temperature was lowered to 15 °C and protein expression was induced by the addition of isopropyl β-D-thiogalactoside (IPTG) (final concentration of 0.2 mM). After growing overnight (~16 h), the cells were collected by centrifugation (15 min at 11000g) at 4 °C, suspended in ~40 mL of 20 mM Na2HPO4 buffer (pH 7) containing 15 mM NaCl (buffer A). The cells were sonicated and centrifuged as described previously.4,6 The clarified supernatant was applied to a DEAE Sepharose column (~7 mL bed volume), equilibrated in buffer A. The column was washed with buffer A (20 mL), and retained proteins were subsequently eluted with a linear gradient (100 mL total, 50 mL buffer A to 50 mL buffer A made 500 mM NaCl). Fractions (~1.4 mL) were collected and analyzed by SDS-PAGE. The fractions containing the desired protein were pooled, placed on ice, and stirred while solid (NH4)2SO4 was added to make a final concentration of 2.5 M. After stirring for 30 min, the sample was centrifuged (15 min at 18000g), and the pellet was discarded. The supernatant was loaded onto a Phenyl-Sepharose column (~7 mL bed volume) equilibrated with buffer A made 3M (NH4)2SO4. The column was washed with buffer A made 3M (NH4)2SO4 (20 mL) followed by a linear gradient [100 mL total, 50 mL of buffer A made 3M (NH4)2SO4 to 50 mL of buffer A]. Fractions (~1.4 mL) were collected and analyzed by SDS-PAGE. The fractions containing the desired protein were pooled and concentrated (~0.3 mL) using an Amicon stirred cell concentrator equipped with a 3 kDa membrane followed by an Amicon Ultra (3 kDa MWCO) centrifugal filter unit. The concentrated sample was loaded onto a Sephadex G-75 size-exclusion column (30 mL bed volume) and proteins were eluted with buffer A using gravity flow at 22 °C. Fractions (~0.4 mL) were collected and analyzed using SDS-PAGE. The fractions containing near-homogeneous target protein were pooled and stored at −80 °C. A sample of the purified protein was prepared as described elsewhere and analyzed by electrospray ionization mass spectrometry (ESI-MS).17,18
Steady-State Kinetics of the Heterohexamer- and Fused 4-OTs.
The activities of hhYR-4OT, fYR-4OT, hhCF-4OT, and fCF-4OT were examined with 2-hydroxymuconate (2-HM), 5-(methyl)-2-hydroxymuconate (5-Me-2-HM), phenylenolpyruvate (PP), 2-hydroxy-2,4-pentadienoate (HPD), and 5-(carboxymethyl)-2-hydroxymuconate (CHM) by assays described elsewhere.19,20 The kinetic parameters were measured for the substrates processed in 20 mM Na2HPO4 buffer (pH 7) containing 15 mM NaCl (buffer A) as follows. Stock solutions of 2-HM (60 mM), 5-Me-2-HM (30mM), PP (140 mM), HPD (130 mM), and CHM (20 mM) were made in ethanol. The enol–keto tautomerization of 2-HM was monitored by following the increase at 236 nm (ε = 9775 M−1 cm−1) using substrate concentrations ranging from 5–440 μM with an enzyme concentration of 50 nM for hhYR-4OT, 40 nM for fYR-4OT, 22 nM for hhCF-4OT, and 7 nM for fCF-4OT. The enol–keto tautomerization of 5-Me-2-HM was monitored by following the increase at 236 nm (ε = 18500 M−1cm−1) using substrate concentrations ranging from 2–280 μM with an enzyme concentration of 50 nM for hhYR-4OT, 40 nM for fYR-4OT, 130 nM for hhCF-4OT, and 50 nM for fCF-4OT. The enol–keto tautomerization of PP was monitored by following the decrease at 284 nm (ε = 11500 M−1cm−1) using substrate concentrations ranging from 4–380 μM with an enzyme concentration of 50 nM for hhYR-4OT, 90 nM for fYR-4OT, 440 nM for hhCF-4OT, and 50 nM fCF-4OT. The enol–keto tautomerization of HPD was monitored by following the decrease at 267 nm (ε = 12700 M−1cm−1) using substrate concentrations ranging from 10–380 μM with an enzyme concentration of 140 nM for hhYR-4OT, 90 nM for fYR-4OT, 440 nM for hhCF-4OT, and 480 nM fCF-4OT. The enol–keto tautomerization of CHM was monitored by following the increase at 236 nm (ε = 7070 M−1cm−1) using a substrate concentration ranging from 18–340 μM with an enzyme concentration of 1.6 mM for fCF-4OT. Initial rates were determined, plotted against the substrate concentrations, and fit to the Michaelis–Menten equation using Grafit. The estimated errors for kcat and Km are included in the Tables. The estimated errors for kcat/Km are estimated to range from 10–20%.
Steady-State Kinetics of the 4-OT Variants.
The activities of the 4-OT variants were examined using 2-HM. The kinetic parameters were measured as described above for wild-type. The enol–keto tautomerization of 2-HM (18 mM stock) was monitored by following the increase at 236 nm (ε = 9775 M−1cm−1) using substrate concentrations ranging from 7–30 μM. The hhYR-4OT variant concentrations were 4 μM for αP1A, 0.2 μM for αR39A, 4 μM for βR11A, 0.3 μM for βR39A, and 12 μM for βR62A. The hhCF-4OT variant concentrations were 1 μM for αP1A, 0.05 μM for αL11Y, 1 μM for αR39A, 8 μM for βR11A, 0.1 μM for βR39A, and 0.7 μM for βR62A. Initial rates were determined, plotted against the substrate concentration, and fit to the Michaelis–Menten equation using Grafit. The estimated errors for kcat and Km are included in the Tables. The estimated errors for kcat/Km are estimated to range from 10–20%. For some variants, the concentration of enzyme nears the concentration of substrate and complicates the analysis. We chose to take these parameters at face value (in other words, assume that steady-state kinetics is operative) and not pursue a more complex analysis.
Crystallization.
Initial crystallization conditions for hhYR-, hhCF-, and fYR-4OT were identified using sparse-matrix screening using a Phoenix crystallization robotic system (Art Robbins Instruments). The identified hits for crystallization were optimized systematically using the sitting drop vapor diffusion technique. The hhYR-4OT was crystallized in 54–64% 2-methyl-2,4-pentanediol (MPD), 0.1M sodium cacodylate pH 6.8, and 5% polyethylene glycol (PEG) 8000. The hhCF-4-OT was crystallized in 15–25% PEG 3350, 0.1M HEPES buffer (pH 7.0–8.0) and 0.2M magnesium formate. fYR-4OT was crystallized in 1.5–2M (NH4)2SO4, 0.1M HEPES buffer (pH 7.2–8.0), and 2% PEG 400. In all crystallization setups, protein solution (~9–12 mg/mL) was mixed with an equal volume of the reservoir solution and equilibrated against 500 μL of the reservoir at room temperature. All crystals were cryoprotected by soaking in mother liquor supplemented with 30% glycerol, followed by flash-freezing in liquid nitrogen. Although fCF-4OT was pure, crystallization efforts did not result in high-quality crystals.
Data collection, Processing, Structure Determination and Refinement.
X-ray diffraction data for the hhYR-, hhCF-, and fYR-4OT were collected at Advanced Photon Source (APS) beamline 23-ID-D (Argonne National Laboratories). The data sets were indexed, integrated, and scaled using HKL-2001.21 The structures were determined by molecular replacement (MR) using Phase-MR2 from the PHENIX Suite of program.22 One protomer of the symmetric heterohexamer 4-OT (PDB: 3MB2)19 was used as a search model for the initial phases for hhYR- and hhCF-4OT structure factors. Using high-resolution electron density maps, chain identity was assigned as the α- and β-subunits of hhYR- and hhCF-4OT based on the differences in sequences between α- and β-subunits. One protomer of the asymmetric fused 4-OT (PDB: 6BLM)5,6 crystal structure was used as a search model for the initial phases for the fYR-4OT structure. Structure refinement was performed using Phenix Refine23 along with iterative model building in COOT24. TLS parameters were included in the refinement of all structures. The fYR-4OT has considerable twinning as detected by the PHENIX software, Xtriage25. Initial rounds of refinement were performed using Phenix with manual adjustments with COOT and a final round of refinement using REFMAC5 as part of the CCP4 package26. Hence, fYR-4OT was refined using the twin operators l, h, k and k, l, h against intensity amplitudes. The final structures were evaluated after refinement using Molprobity7.27 The refinement statistics for the structures are summarized in Table 1. All figures were prepared with PyMol (The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC).28
Table 1:
Crystallographic Data Collection and Refinement Statistics
| YR | FYR | CF | |
|---|---|---|---|
|
| |||
| Data collection | |||
| Space group | C121 | P1 | C121 |
| Cell dimensions | |||
| a, b, c (Å) | 110.19, 61.03, 92.14 | 49.09, 49.41, 49.32 | 112.751, 87.480, 86.698 |
| α, β, γ (°) | 90.00, 123.82, 90.00 | 120.37, 102.16, 102.11 | 90.00, 125.12, 90.00 |
| Resolution (Å) | 50.00–2.05 (2.09–2.05)* | 50.00–2.30 (2.34–2.30) | 50.00– 2.70 (2.75 – 2.70) |
| Rsym/ Rpim | 0.075(0.508)/0.045(0.314) | 0.040(0.274)/0.037(0.256) | 0.069(0.426)/0.042(0.122) |
| CC ½ ϒ | 0.997 (0.791) | 0.993(0.886) | 0.994 (0.793) |
| I / σ | 23.2 (2.0) | 19.5(2.2) | 6.7 (1.6) |
| Completeness (%) | 99.59 (97.99) | 96.0 (96.2) | 95.0(80.7) |
| Redundancy | 3.6 (3.4) | 2.0 (2.0) | 3.4 (2.4) |
| Wilson B-factor (Å2) | 36.9 | 60.7 | 37.9 |
| Refinement | |||
| Resolution (Å) | 26.29–2.04 (2.11–2.04) | 46.42–2.30 (2.35 – 2.29) | 43.74– 2.70 (2.78 – 2.70) |
| No. reflections | 32444 (3076) | 18084 (1828) | 18289 (1352) |
| Rwork | 0.1923 (0.2550) | 0.2693 (0.4170) | 0.2126 (0.2667) |
| Rfree± | 0.2302 (0.3165) | 0.3031 (0.4420) | 0.2689 (0.3558) |
| No. atoms | 3156 | 2552 | 5643 |
| Protein | 2984 | 2520 | 5579 |
| Water | 172 | 32 | 64 |
| B-factors (Å2) | |||
| Protein | 46.6 | 68.8 | 41.3 |
| Water | 47.3 | 60.1 | 36.9 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.007 |
| Bond angles (°) | 0.99 | 1.57 | 1.08 |
| Ramachandran plot | |||
| Favored | 98.72% | 91.81% | 98.33% |
| Allowed | 1.28% | 8.19% | 1.53% |
| Outliers | 0.00% | 0.00% | 0.14% Δ |
| Molprobity score | 1.47α / 97th percentileβ | 3.11 / 18th percentile | 1.74 / 99th percentile |
Values for the corresponding parameters in the outermost shell in parenthesis.
CC1/2 is the Pearson correlation coefficient for a random half of the data, the two numbers represent the lowest and highest resolution shell, respectively.
Rfree is the Rwork calculated for about 10% of the reflections randomly selected and omitted from refinement.
The Molprobity score is calculated by combining the Clashscore with rotamer and Ramachandran percentage and scaled on the basis of X-ray resolution.
The 100th percentile is considered the best and the 0th percentile as the worst among structures of similar resolution.
There is one Ramachandran outliers in hhCF, which corresponds to Lys-29 in chain F. It has a disordered side chain due to flexibility.
RESULTS AND DISCUSSION
Discovery of hhYR-4OT and hhCF-4OT.
The hhYR-4OT and hhCF-4OT were discovered during a SSN analysis of the 4-OT subgroup of the TSF.1 Due to the diversity of the 4-OT subgroup, this analysis shows several Level 2 subgroups.1,5 The Level 2 subgroup containing canonical 4-OT divides into many clusters. The hhYR-4OT and hhCF-4OT representative nodes are connected by multiple linker-edges between the clusters containing the “fused 4-OT” and the “4-OT” representative nodes (Figure 2).
Purification of hhYR-4OT, hhCF-4OT, fYR-4OT and fCF-4OT.
The hhYR-4OT and hhCF-4OT were overexpressed in E. coli BL21 (DE3) and purified to near homogeneity. Purified hhYR-4OT and hhCF-4OT were analyzed using ESI-MS and gave two major peaks and two minor peaks each. For hhYR-4OT, the two major peaks correspond to the expected masses of the α- and β-subunits (6740 and 7838 Da, respectively), with the initiating methionine removed (Table S2). The two minor peaks correspond to the masses of the α- and β-subunits (6871 and 7969 Da, respectively), with the initiating methionine retained. For hhCF-4OT, the two major peaks correspond to the expected masses of the α- and β-subunits (7160 and 7497 Da, respectively), with the initiating methionine removed. The two minor peaks correspond to the masses of the α- and β-subunits (7291 and 7628 Da, respectively), with the initiating methionine retained.
For both hhYR-4OT and hhCF-4OT, the α-subunit has proline in the N-terminal position, whereas the β-subunit has serine in the N-terminal position. As noted elsewhere, when proteins are expressed in E. coli, the removal of the initiating methionine correlates with the identity of the second amino acid.29 For Pro and Ser the initiating Met is likely removed (but only enzyme with an unblocked Pro1 functions in the 4-OT reaction1). The mass spectral analysis shows that the ~41% of hhYR-4OT begins with proline and ~68% of hhCF-4OT begins with proline (Table S3). Compared with other TSF members, this is low as usually almost 100% of a protein species shows a free and unblocked proline.30 An explanation for this observation is not apparent. However, the kinetic parameters (next section) were corrected using the percent of active enzyme (with the unblocked prolyl nitrogen).
The genes encoding the α- and β-subunits for hhYR- or hhCF-4OT were fused together by removing the stop codon of the α-subunit and the start codon of the β-subunit and inserting a short amino acid linker (SAADGAPPSL) that mimics that of fused 4-OT (from Burkholderia lata)5 between the two genes. The resulting fused proteins were overexpressed in E. coli BL21 (DE3) and purified to near homogeneity. Purified fYR-4OT and fCF-4OT were analyzed using ESI-MS and fYR-4OT gave a major and minor peak, but fCF-4OT only showed one peak. For fYR-4OT, the major species has a mass of 15,426 Da and the minor species has a mass of 15558 Da (Table S2). For fCF-4OT, the only major species has a mass of 15,515 Da. The major peaks correspond to the expected masses with the initiating methionine removed and the minor peak correspond to the expected mass with the initiating methionine retained. The mass spectral analysis shows that the ~59% of fYR-4OT begins with proline and ~100% of hhCF-4OT begins with proline (Table S3). The kinetic parameters (next section) were corrected using the percent of active enzyme (with the unblocked prolyl nitrogen).
Kinetic Characterization of hhYR-4OT, hhCF-4OT, fYR-4OT and fCF-4OT.
The activities of heterohexamers (hhYR-4OT and hhCF-4OT) and fused 4-OTs (fYR-4OT and fCF-4OT) were examined using 2-HM, 5-Me-2-HM, PP, HPD, and CHM (reactions shown in Scheme 1).
Scheme 1.
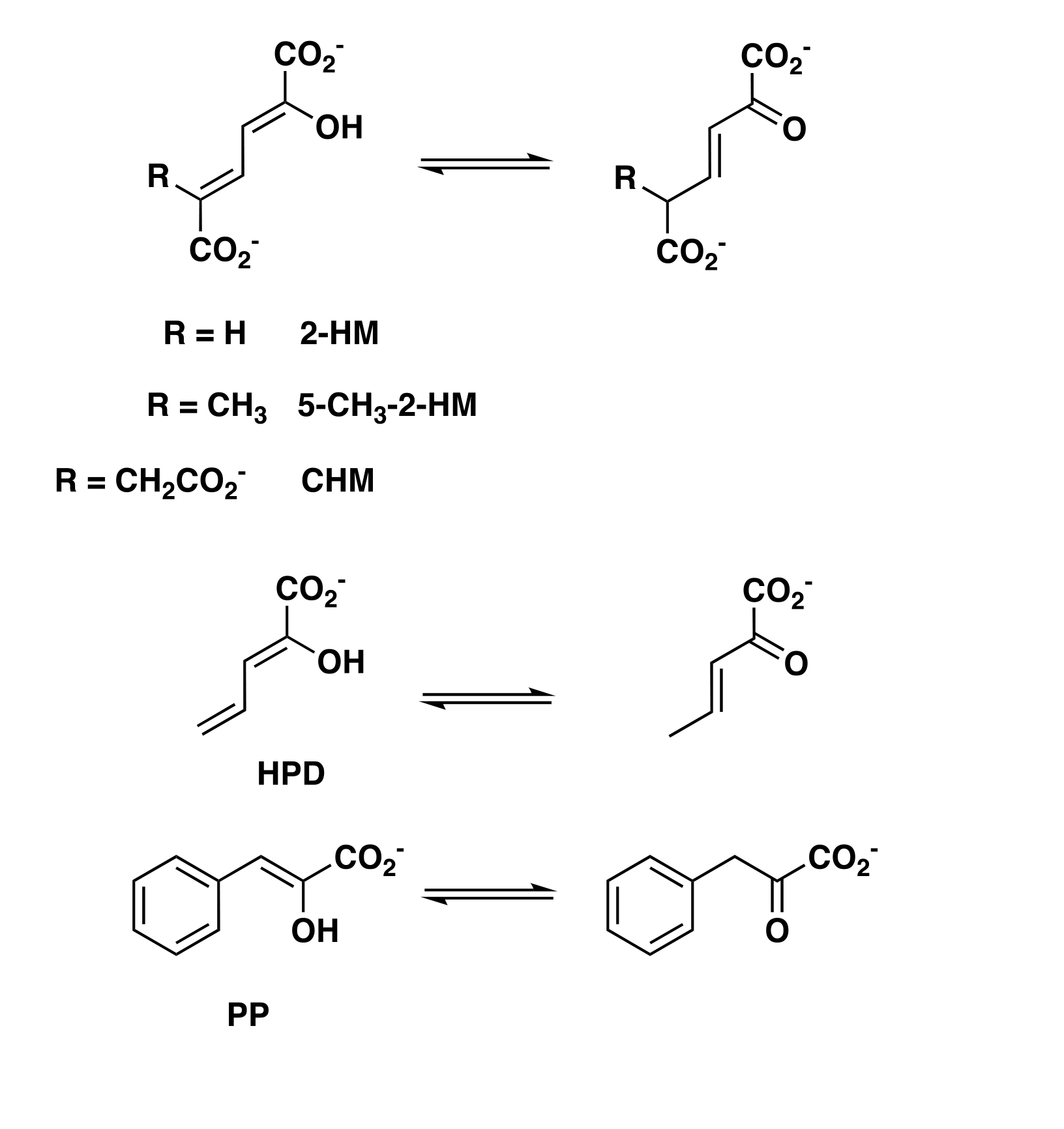
Enzyme-catalyzed Tautomerization Reactions
All process 2-HM, 5-Me-2-HM, PP, and HPD, but only fCF-4OT processes CHM. The kinetic parameters for hhYR-4OT, fYR-4OT, hhCF-4OT, and fCF-4OT are collected in Tables 2 and 3.
Table 2.
Steady-state kinetic parameters for hhYR-4OT and fYR-4OT.
| Substrate | Enzyme | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
|
| ||||
| 2-HM | hhYR-4OT | 30 ± 1 | 170 ± 10 | 1.8 × 105 |
| fYR-4OT | 80 ± 5 | 260 ± 30 | 3.1 × 105 | |
|
| ||||
| 5-Me-2-HM | hhYR-4OT | 2.0 ± 0.1 | 110 ± 10 | 1.8 × 104 |
| fYR-4OT | 4.6 ± 0.2 | 60 ± 10 | 7.7 × 104 | |
|
| ||||
| PP | hhYR-4OT | 80 ± 15 | 450 ± 100 | 1.8 × 105 |
| fYR-4OT | 60 ± 10 | 220 ± 40 | 2.7 × 105 | |
|
| ||||
| HPD | hhYR-4OT | 20 ± 5 | 510 ± 150 | 3.9 × 104 |
| fYR-4OT | 30 ± 3 | 300 ± 50 | 1.0 × 105 | |
Table 3.
Steady-state kinetic parameters for hhCF-4OT and fCF-4OT.
| Substrate | Enzyme | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
|
| ||||
| 2-HM | hhCF-4OT | 30 ± 1 | 80 ± 7 | 3.7 × 105 |
| fCF-4OT | 180 ± 10 | 290 ± 25 | 6.2 × 105 | |
|
| ||||
| 5-Me-2-HM | hhCF-4OT | 6 ± 1 | 100 ± 20 | 6.0 × 104 |
| fCF-4OT | 11 ± 0.5 | 110 ± 10 | 1.0 × 105 | |
|
| ||||
| PP | hhCF-4OT | 60 ± 10 | 390 ± 65 | 1.5 × 105 |
| fCF-4OT | 140 ± 20 | 300 ± 50 | 4.7 × 105 | |
|
| ||||
| HPD | hhCF-4OT | 30 ± 3 | 620 ± 90 | 4.8 × 104 |
| fCF-4OT | 40 ± 5 | 330 ± 50 | 1.2 × 105 | |
|
| ||||
| CHM | - | - | - | - |
| fCF-4OT | 0.03 ± 0.004 | 250 ± 70 | 1.2 × 102 | |
The kcat/Km values (104-105) for hhYR-4OT indicates that the enzyme shows a preference for 2-HM and PP followed by HPD and then 5-Me-2-HM. The hhYR-4OT has a 10-fold lower kcat/Km with 5-Me-2-HM than it does with 2-HM and PP, mostly due to a low kcat value with this substrate. The hhCF-4OT also shows a preference for 2-HM and PP (~105), followed by 5-Me-2-HM and HPD. The kcat/Km value for hhCF-4OT with HPD is 8-fold lower than that of 2-HM, mostly due to a nearly 8-fold increase in Km.
The fYR-4OT and fCF-4OT have higher kcat and kcat/Km values than those for hhYR-4OT and hhCF-4OT for every substrate assayed with one exception. The kcat value for the fYR-4OT with PP is 25% lower than its heterohexamer counterpart. The fYR-4OT and fCF-4OT have Km values equal to or lower than those for hhYR-4OT and hhCF-4OT for every substrate assayed except for 2-HM. The fCF-4OT is the only enzyme that processes CHM. In all cases, the fused is more efficient than the corresponding heterohexamer 4-OTs (as assessed by the kcat/Km values).
Purification and Kinetic Characterization of hhYR-4OT Variants.
A host of studies identified Pro-1, Arg11, Arg39, and Phe50 (from different protomers) as critical residues in the canonical 4-OT-catalyzed reaction.1,3 Pro1 has a pKa of ~6.4 (determined by 15N NMR titration)31 and transfers a proton from the 2-hydroxy group of 2-HM to C-5 of the product. The interaction between Arg11 and the C-6 carboxylate group of 2-HM binds substrate and draws electron density to C-5 to facilitate protonation at C-5.32 Arg39 interacts with the 2-hydroxy group and a carboxylate oxygen at C-1. The role of Arg39 is more catalytic, where the positively charged guanidinium moiety stabilizes the developing carbanion (after deprotonation of the 2-OH group).32 The proximity of Phe50 to the prolyl nitrogen is partially responsible for the lowered pKa value.33 (The role of Phe50 was not examined in the hhYR- and hhCF-4OTs.)
To determine the importance of conserved residues implicated in activity or binding in these 4-OTs, the αP1A, αR39A, βR11A, βR39A and βR62A variants of hhYR-4OT and hhCF-4OT were constructed. Additionally, the αL11Y variant for hhCF-4OT was constructed. Tyr is found in place of a Leu in the α-subunit of the hhYR-4-OT. The mutation had no effect. The variants were overexpressed in E. coli BL21(DE3) and purified to near homogeneity. Purified variants were analyzed using ESI-MS giving two major peaks and two minor peaks each. The two major peaks correspond to the expected masses of the α- and β- subunits with the initiating methionine removed. The two minor peaks correspond to the mass of the α- and β- subunits with the initiating methionine retained. In all cases, the CF constructs have a higher percent of enzyme with an unblocked amino-terminal proline (Table S3). The kinetic parameters were corrected for the percentage of active enzyme where the initiating methionine was removed.
The activities of the variants were examined using 2-HM. All of the variants processed 2-HM to varying degrees and the kinetic parameters for the hhYR-4OT and hhCF-4OT variants are given in Tables 4 and 5, respectively. The fused constructs (fYR-4OT and fCF-4OT) are included for comparative purposes, but no variants were constructed.
Table 4.
Steady-state kinetic parameters for hhYR-4OT, fYR-4OT and hhYR-4OT variants using 2-HM.
| Enzyme | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
|
| |||
| hhYR-4OT | 30 ± 10 | 170 ± 10 | 1.8 × 105 |
| fYR-4OT | 80 ± 5 | 260 ± 30 | 3.1 × 105 |
| αP1A | 0.4 ± 0.03 | 250 ± 30 | 1.6 × 103 |
| αR39A | 2.2 ± 0.1 | 300 ± 30 | 7.3 × 103 |
| βR11A | 0.030 ± 0.003 | 140 ± 20 | 2.1 × 102 |
| βR39A | 20 ± 1 | 260 ± 20 | 7.7 × 104 |
| βR62A | 1.3 ± 0.1 | 770 ± 100 | 1.7 × 103 |
Table 5.
Steady-state kinetic parameters for hhCF-4OT, fCF-4OT, and hhCF-4OT variants using 2-HM.
| Enzyme | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
|
| |||
| hhCF-4OT | 30 ± 1 | 80 ± 10 | 3.8 × 105 |
| fCF-4OT | 180 ± 10 | 290 ± 25 | 6.2 × 105 |
| CFαP1A | 0.40 ± 0.04 | 120 ± 20 | 3.3 × 103 |
| CFαL11Y | 80 ± 4 | 190 ± 20 | 4.2 × 105 |
| CFαR39A | 15 ± 5 | 750 ± 210 | 2.0 × 104 |
| CFβR11A | 0.07 ± 0.004 | 140 ± 20 | 5.0 × 102 |
| CFβR39A | 40 ± 3 | 230 ± 25 | 1.7 × 105 |
| CFβR62A | 9 ± 1 | 400 ± 60 | 2.3 × 104 |
For the hhYR-4OT variants, the most drastic reductions in the values of kcat/Km are seen for the βR11A- (down 860-fold), αP1A- (down 112-fold), and the βR62A-variants (down 105-fold). For these same variants, the kcat values are down 1000-fold, 75-fold, and 23-fold, respectively. The Km values are mostly comparable except that for the βR62A variant increased 4.5-fold. The only other significant decrease in kcat/Km is seen for the αR39A variant (down 24-fold). The results for the αP1A-, βR11A- and αR39A-variants are consistent with previous results (with the canonical 4OT). The decrease in activity for βR62A variant is unusual: the same variant for the canonical 4OT (covalently modified by an irreversible inhibitor) showed no effect.32 Examination of the hhYR-4OT crystal structure shows that Arg62 is part of a disordered loop with no obvious interactions.
For the hhCF-4OT variants, the most drastic reductions in the values of kcat/Km are seen for the βR11A- (down 760-fold) and the αP1A-variants (down 115-fold). For these same variants, the kcat values are down 430-fold and 75-fold, respectively. The Km values are mostly comparable except that for the αR39A variant increased 9-fold. This same variant showed a 19-fold decrease in kcat/Km. The results for the αP1A-, βR11A- and αR39A-variants are generally consistent with previous results (with the canonical 4OT).32 There is a 16-fold decrease in kcat/Km for the βR62A variant, but it is not as drastic as that observed for the hhYR-4OT variant.
Determination of the Structures of the hhYR- and hhCF-4OT and the fYR-4OT.
To understand the origins of the asymmetry in the fused 4-OTs, crystallographic analysis was employed to determine the configuration of two heterohexamers, YR- and CF-4OT from Herbaspirillum sp. One protomer of a symmetric heterohexamer 4-OT (PDB: 3MB2)19, where each residue was converted to alanine, was used as a model for molecular replacement to assign the oligomerization state without bias. The structure for hhYR-4OT (2.0 Å resolution) contains continuous electron density for all residues from Pro1 to Gln64 for the α-subunit and from Ser1 to Gly72 for the β-subunit in the heterohexamer. The structure for hhCF (2.7 Å resolution) shows continuous electron density from Pro1 to Val57 for the α -subunit with the final six residues (IAAGQE) disordered and from Ser1 to Arg63 for the β-subunit, with only the C-terminal Thr64 missing. Both structures crystallized in space group C121 with one molecule in the asymmetric unit. Crystal parameters, data collection statistics, and final refinement statistics are in Table 1.
As anticipated, the hhYR- and CF-4OT have the signature TSF fold where each protomer (in the heterohexamers) shows the β-α-β fold (Figures 3A,B). The α-subunit of each heterohexamer contains the conserved N-terminal proline moiety that accounts for the activity of 4-OT subgroup members.1 The β-subunits contain an N-terminal serine residue that has not been previously implicated in a known activity. Both heterohexamers have an asymmetric arrangement similar to the fused 4OT trimer where the ββ interface lacks a Pro1 and the αα interface accounts for this loss with two N-terminal prolines (Figures 3C,D).5,6 How the individual interfaces contribute to the overall activity is not known, but the enzyme catalyzes the same reactions as the symmetric ones with nearly comparable catalytic efficiencies.5,6
Figure 3.
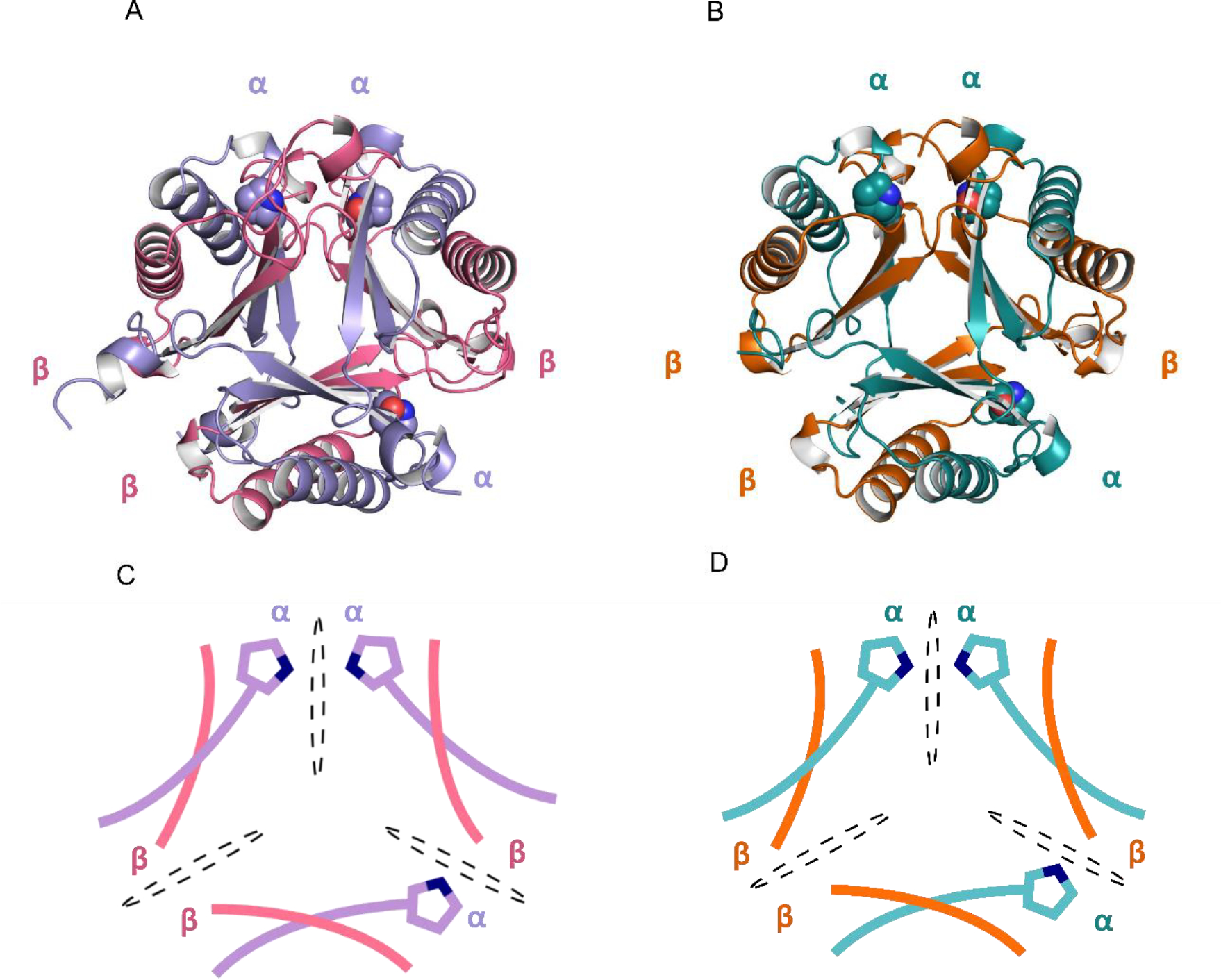
Asymmetric heterohexamers YR- and CF-4OT (A) Ribbon diagram of Herbaspirillum sp. hhYR-4OT. The α- and β-domains are labeled and colored in purple and pink. (B) Ribbon diagram of Herbaspirillum sp. hhCF-4OT. The α-domain is colored in teal and the β-domain is colored in orange. (C-D) Schematic representation of the asymmetric arrangement of the two heterohexamers. Asymmetry is defined by three unique interfaces and are labeled as αα, αβ, and ββ. Each interface is divided by a dotted black oval. Pro1 is shown as a space-filling sphere in the cyan and gray at each interface. The color scheme is identical to that in A and B.
To understand the asymmetric oligomerization state of the two heterohexamers, an in-depth analysis of the electrostatic interactions between the three unique interfaces of each heterohexamer was conducted (Figure 4A and Table S6). In our previous crystallographic analysis of fused 4OT6, we found that salt bridge interactions dominate the subunit interface interactions. Both hhYR- and hhCF-4OT share ~50% sequence identity to fused 4OT from Burkholderia lata and most of the residues that form salt bridges are conserved (Figure 4A)5. The salt bridge networks are highly similar between hhYR- and hhCF-4OT (Figure 4A). At the αα interface in hhYR-4OT, for example, there are electrostatic interactions between Arg58/Asp13 (Glu13 in hhCF-4OT), Lys16/Asp49 (Glu49 in hhCF-4OT), and Asp35/Arg38 for a total of six salt bridge interactions (Figure 4B). At the ββ interface, there are two salt bridges between reciprocal Arg39/Glu37 residues from two β-subunit chains (Figure 4B). Within the central core, a network of salt bridges form between three Lys43 (Arg43 in hhCF-4OT) residues of adjacent β-subunits and three Glu4 residues within the corresponding α-chains (Figures 4B,C). Lastly, at the αβ interface, Arg39 of the α-subunit and Glu37(Asp37 in hhCF-4OT) of the β-subunit form a salt bridge pair near the surface. This salt bridge is not conserved in fused 4-OT6 and is unique to the two heterohexamers as the charged residues (Glu37/Asp37) are replaced by alanine in fused 4OT. The numerous salt bridge interactions between the α- and β- subunits of hhYR- and hhCF-4OT underscore the importance of these contacts in favoring the asymmetric configurations of these two 4-OT subgroup members.5,6
Figure 4.
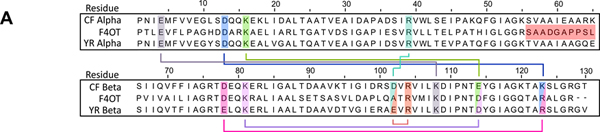
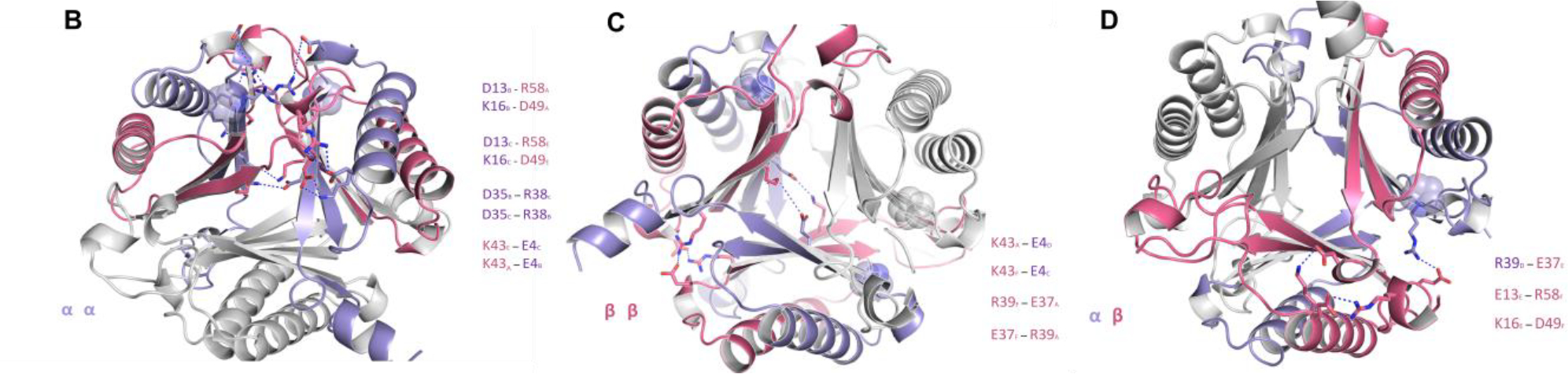
Conservation between hhYR/hhCF and fused 4OT, a representative asymmetric 4-OT trimer.5 (A) Multiple sequence alignment between the α- and β-chains of hhYR/hhCF and one protomer of the representative fused 4OT. Conserved salt bridge partners are highlighted and outlined between the α and β subunits. Highlighted in red is the linker region in fused 4OT Alignment was conducted with Clustal Omega.34 (B) Ribbon diagram of the salt bridge interactions in the ββ interface. Participating subunits are colored in purple (α-subunit) and pink (β-subunit) while chains that are not within the interface are in gray. Salt bridge interactions are shown with dotted blue lines and residues in stick form. Pro1 is shown as a sphere. (C) Salt bridge interactions within the αα interface. (D) Salt bridge interactions within the αβ interface.
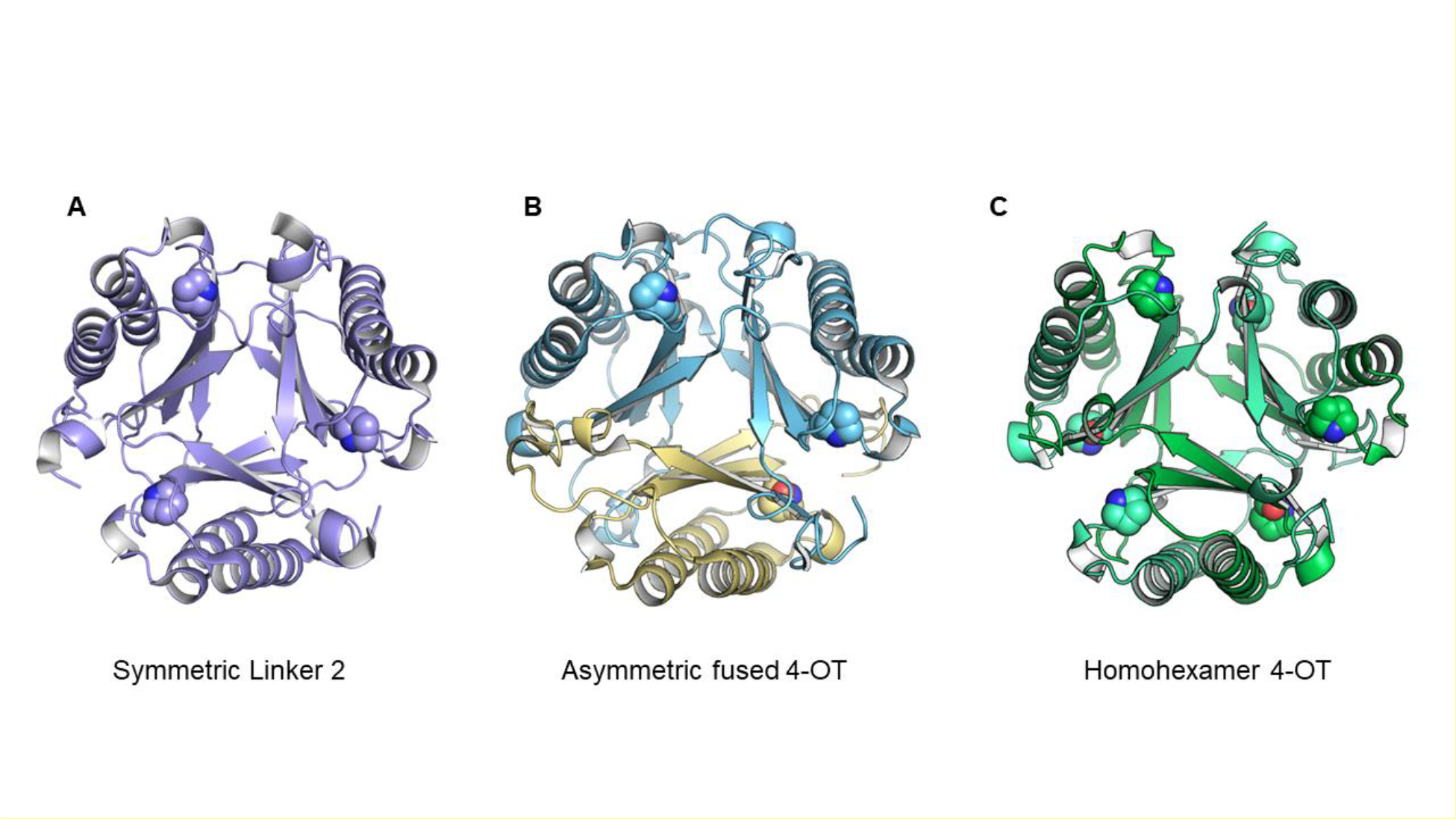
If the unique, asymmetric oligomerization state of hhYR and hhCF is in part due to favorable hydrophilic interactions across the three distinctive interfaces, we then asked whether these interactions would still be maintained in a symmetric state.5,6 To test this, an artificial symmetric model was constructed by flipping two subunit chains in hhYR and hhCF. We generated the model by superimposing each subunit to a representative symmetric fused 4OT, that of Linker 25, which shares ~38% similarity with hhYR and 37% with hhCF (Figure S1A). The resulting artificial model has no significant steric clashes. However, only one unique salt bridge pair is conserved at each of the αβ interfaces between Glu37and Arg39 (Figure S1B,C). In the natural asymmetric state, hhYR and hhCF both have 15 salt bridges. Therefore, when nature “flipped” a pair of α/β subunits to generate an asymmetric heterohexamer, many favorable electrostatic interactions were gained.
To evaluate the evolutionary origins of the asymmetric 4-OT trimers, the hhYR and hhCF enzymes were “fused” through the addition of the linker loop region found in that of fused 4OT (highlighted in Figure 4A) to mimic the fused 4OT found in the superfamily (Figure 5).
Figure 5.
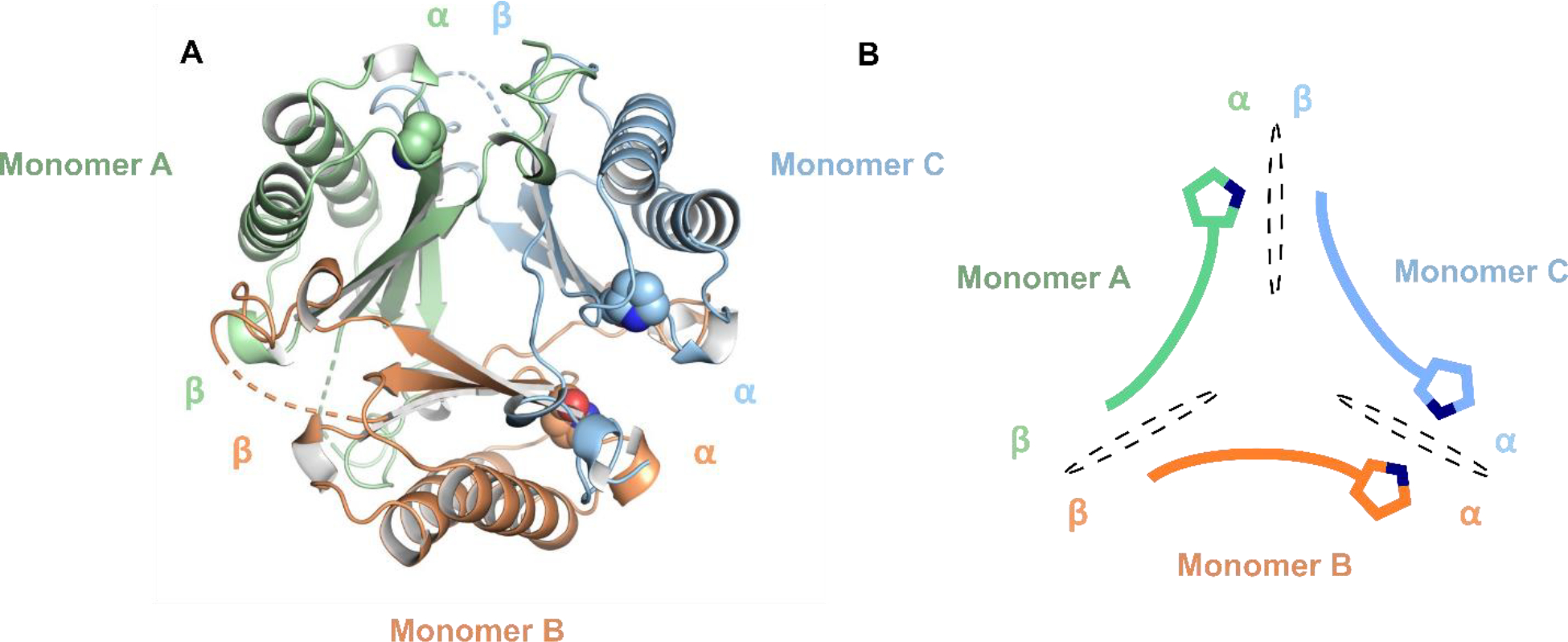
X-ray crystal structure of the asymmetric fYR trimer. (A) Ribbon diagram of the fused trimer determined at 2.3 Å resolution where each protomer is denoted by distinct colors. Pro1 is shown as space-filling spheres for each protomer. (B) Schematic representation of each protomer within the asymmetric fused trimer. Each interface is shown a black dotted oval.
The structure of the fused YR trimer (fYR) was determined by X-ray crystallography to a resolution of 2.3 Å. One protomer of the asymmetric fused 4-OT (PDB: 6BLM)5,6 was used as a search model to identify the solution for molecular replacement. There is electron density from Pro1 to Tyr125 for each protomer in the fused trimer. As the loop region (residues Ala60 to Gln64) is highly flexible, the electron density is not visible here. The overall architecture of fYR reveals the same asymmetric conformation as the hhYR counterpart where one pair of α/β subunits is flipped 180° relative to the other two protomers. The addition of the linker loop region does not alter the asymmetric arrangement. Likely, asymmetry was first seen in the heterohexamer and later preserved in the fused 4-OTs.
Upon the discovery of the asymmetric fused 4-OT from B. lata, an extensive investigation of the factors responsible for the asymmetry was undertaken.5,6 The fused 4-OT is in a cluster of asymmetric 4-OTs (86 total) where a multiple sequence alignment (MSA) shows that they are characterized by two “4-OT like” sequences that are joined in the middle by a short linker (SAADGAPPSL).5 The N-terminal section has the conserved (and catalytically active) Pro1, positively charged residues at positions 11 (His) and 39 (Arg), and a nonpolar residue at position 50 (generally Phe), followed by the GxGG-motif (important for oligomerization in 4-OT hexamers).3 The C-terminal section is comparable where there are two arginines (positions 82 and 110), a nonpolar residue at position 121, and a conserved Pro66 (10 positions before the first conserved arginine). This would be the exact position of an N-terminal (and catalytically active) Pro had this C-terminal section not been fused to the N-terminal section.
Linker 2, a symmetric fused 4-OT found in a cluster of 47 sequences, has most of same conserved features (Pro1, His11, Arg39, a hydrophobic residue at position 50, i.e., Ile50, Arg71, Arg99, and a hydrophobic residue at position 110, i.e., Met110.)5 In both “halves” of linker 2, the GxGG motif is not as pronounced. Moreover, there is no gap in the MSA and no obvious proline that might represent the start of a 4-OT sequence.
On the basis of these two observations, (Pro66, in the asymmetric sequences, aligns with Pro1 in the canonical 4-OT and the sequences for the symmetric fused 4OTs did not show an obvious proline that might represent the start of a 4-OT sequence), an “unfused” version of the fused 4OT was constructed.5 The resulting heterohexamer, where each protomer now has an N-terminal residue was examined by X-ray crystallography and found to be an asymmetric heterohexamer. Hence, the linker is not responsible for the asymmetry.5 A subsequent study showed that the number of salt bridges governed whether the trimer was asymmetric or symmetric.6 For the most part, members of the fused 4-OT cluster have more salt bridges if an asymmetric trimer forms and members of the Linker 2 cluster have more salt bridges if a symmetric trimer forms.6
Here, the converse experiment was carried out: the α- and β-protomers of two asymmetric heterohexamers were fused to produce the corresponding asymmetric trimers. Hence, asymmetry is introduced at the heterohexamer level and then propagated into the trimers when fused. Once again, subsequent crystallographic analysis of both asymmetric heterohexamers and one asymmetric trimer showed that the number of salt bridges is the determining factor in trimer arrangement. These results reinforce the current hypothesis for the evolution of the TSF members where a gene duplication event took place early in the evolutionary history of the TSF leading to the diversification of activity seen today.1 One scenario involves the duplication of a 4-OT-like gene followed by fusion of the two genes to generate a fused 4-OT. The symmetric and asymmetric fused 4-OTs could further diverge to generate the new activities in the other four subgroups.
The process of gene duplication and fusion is how new proteins (with new activities) are generated from simpler ancestral proteins. Poelarends and co-workers replicated this process in the laboratory to optimize the biocatalytic potential of 4-OT.35 In the experiment, two 4-OT protomers (in the canonical 4-OT)1 were fused by a short linker (GGGAG) that connected the C-terminus of one protomer to the N-terminus of a second protomer. After 11 rounds of directed evolution, crystallographic analysis showed that the resulting construct was an asymmetric trimer. Hence, asymmetry can be introduced in laboratory evolution or natural evolution.
Supplementary Material
ACKNOWLEDGEMENTS
The protein mass spectrometry analysis was conducted in the Institute for Cellular and Molecular Biology Protein and Metabolite Analysis Facility at the University of Texas at Austin. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Funding
This research was supported by the National Institutes of Health Grants GM-129331 to C.P.W. and GM-104896 to Y.J.Z. and the Robert A. Welch Foundation Grants F-1334 and 2125 to C.P.W.
ABBREVIATIONS
- APS
Advanced Photon Source
- CHM
5-(carboxymethyl)-2-hydroxymuconate
- DEAE
diethylaminoethyl
- ESI-MS
electrospray ionization mass spectrometry
- fCF-4OT
fused-CF444–4-oxalocrotonate tautomerase
- hhCF-4OT
heterohexamer CF444–4-oxalocrotonate tautomerase
- fYR-4OT
fusedYR522–4-oxalocrotonate tautomerase
- hhYR-4OT
heterohexamerYR522–4-oxalocrotonate tautomerase
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid
- 2-HM
2-hydroxymuconate
- HPD
2-hydroxy-2,4-pentadienoate
- ICMB
Institute for Cellular and Molecular Biology
- IPTG
isopropyl α-D-thiogalactoside
- Kn
kanamycin
- LB
Lysogeny broth
- 5-Me-2-HM
5-(methyl)-2-hydroxymuconate
- MPD
2-methyl-2,4-pentanediol
- MR
molecular replacement
- MWCO
molecular weight cutoff
- MME
monomethyl ether
- MSA
multiple sequence alignment
- 4-OT
4-oxalocrotonate tautomerase
- 2-OP
2-oxo-3-pentynoate
- PDB
protein data bank
- PP
phenylenolpyruvate
- PMSF
phenylmethylsulfonyl fluoride
- PEG
polyethylene glycol
- SSN
sequence similarity network
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TSF
tautomerase superfamily
Footnotes
The authors declare no competing financial interest.
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank: PDB entry 8T9P for the hhYR-4OT, 8T9O for hhCF-4OT, and 8T9Q for fCF-4OT.
UniProt accession ID
The α and β subunits of a heterohexamer 4-OT from Herbaspirillum sp. YR522 (UniProt accession J2VT77 and J3HY51, respectively)
The α and β subunits of a heterohexamer 4-OT from Herbaspirillum sp. CF444 (UniProt accession J3DHL6 and J2M343, respectively)
REFERENCES
- (1).Davidson R, Baas B-J, Akiva E, Holliday G, Polacco BJ, LeVieux JA, Pullara CR, Zhang YJ, Whitman CP, and Babbitt PC (2018) A global view of structure-function relationships in the tautomerase superfamily. J. Biol. Chem. 293, 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Murzin AG (1996) Structural classification of proteins: new superfamilies. Curr. Opin. Struct. Biol. 6, 386–394. [DOI] [PubMed] [Google Scholar]
- (3).Poelarends GJ, Veetil VP, and Whitman CP (2008) The chemical versatility of the β-α-β fold: Catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cell. Mol. Life Sci. 65, 3606–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).LeVieux JA, Baas B-J, Kaoud TS, Davidson R, Babbitt PC, Zhang YJ, Whitman CP (2017) Kinetic and structural characterization of a cis-3-chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily. Arch Biochem Biophys, 636, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Baas B-J, Medellin BP, LeVieux JA de Ruijter M, Zhang YJ, Brown SD, Akiva E, Babbitt PC, and Whitman CP (2019) Structural, kinetic, and mechanistic analysis of an asymmetric 4-oxalocrotonate tautomerase trimer. Biochemistry 58, 2617–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Medellin BP, Lancaster EB, Brown SD, Rakhade S, Babbitt PC, Whitman CP, and Zhang YJ (2020) Structural basis for the asymmetry of a 4-oxalocrotonate tautomerase trimer. Biochemistry 59, 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zallot R, Oberg N, and Gerlt JA (2019) The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes, and metabolic pathways. Biochemistry 58, 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Whitman CP, Aird BA, Gillespie WR, and Stolowich NJ (1991) Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J. Am. Chem. Soc. 113, 3154–3162. [Google Scholar]
- (9).Lian H, Czerwinski RM, Stanley TM, Johnson WH Jr., Watson RJ, and Whitman CP (1998) The contribution of the substrate’s carboxylate group to the mechanism of 4-oxalocrotonate tautomerase. Bioorg. Chem. 26, 141–156. [Google Scholar]
- (10).Hajipour G, Johnson WH Jr., Dauben PD, Stolowich NJ, and Whitman CP (1993) Chemical and enzymatic ketonization of 5-(carboxymethyl)-2-hydroxymuconate. J. Am. Chem. Soc. 115, 3533–3542. [Google Scholar]
- (11).Stanley TM, Johnson WH Jr., Burks EA, Whitman CP, Hwang C-C, and Cook PF (2000) Expression and stereochemical and isotope effect studies of active 4-oxalocrotonate decarboxylase. Biochemistry 39, 718–726. [DOI] [PubMed] [Google Scholar]
- (12).Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd ed, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- (13).Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- (14).Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- (15).Laible M, and Boonrod K (2009) Homemade site directed mutagenesis of whole plasmids. J Vis Exp. 27, e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ho SN, Hunt HD, Horton RM, Pullen JK, and Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- (17).Wang SC, Person MD, Johnson WH Jr., and Whitman CP (2003) Reactions of trans-3-chloroacrylic acid dehalogenase with acetylene substrates: consequences of and evidence for a hydration reaction. Biochemistry 42, 8762–8773. [DOI] [PubMed] [Google Scholar]
- (18).Lancaster EB, Johnson WH Jr., LeVieux JA, Hardtke HA, Zhang YJ, and Whitman CP (2023) A mutagenic analysis of NahE, a hydratase-aldolase in the naphthalene degradative pathway. Arch. Biochem. Biophys. 733, 109471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Burks EA, Fleming CD, Mesecar AD, Whitman CP, and Pegan SD (2010) Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily. Biochemistry 49, 5016–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Burks EA, Yan W, Johnson WH Jr., Li W, Schroeder GK, Min C, Gerratana B, Zhang Y, and Whitman CP (2011) Kinetic, crystallographic, and mechanistic characterization of TomN: elucidation of a function for a 4-oxalocrotonate tautomerase homologue in the tomaymycin biosynthetic pathway. Biochemistry 35, 7600–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution - - from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866. [DOI] [PubMed] [Google Scholar]
- (22).Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ,Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macro-molecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- (25).Murshudov GN, Vagin AA, and Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- (26).Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P [Google Scholar]
- Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).De Lano WL (2002) The PyMol molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- (29).Hirel P-H, Schmitter J-M, Dessen P, Fayat G, and Blanquet S (1989) Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Czerwinski RM, Johnson WH Jr., Whitman CP, Harris TK, Abeygunawardana C, and Mildvan AS (1997) Kinetic and structural effects of mutations of the catalytic amino-terminal proline in 4-oxalocrotonate tautomerase. Biochemistry 36, 14551–14560. [DOI] [PubMed] [Google Scholar]
- (31).Stivers JT, Abeygunawardana C, Mildvan AS, Hajipour G, and Whitman CP (1996) 4-Oxalocrotonate tautomerase: pH dependence of catalysis and pKa values of active site residues. Biochemistry 35, 814–823. [DOI] [PubMed] [Google Scholar]
- (32).Harris TK, Czerwinski RM, Johnson WH Jr., Legler PM, Abeygunawardana C, Massiah MA, Stivers JT, Whitman CP, and Mildvan AS (1999) Kinetic, stereochemical, and structural effects of mutations of the active site arginine residues in 4-oxalocrotonate tautomerase. Biochemistry 38, 12343–12357. [DOI] [PubMed] [Google Scholar]
- (33).Czerwinski RM, Harris TK, Massiah MA, Mildvan AS, and Whitman CP (2001) The structural basis for the perturbed pKa of the catalytic base in 4-oxalocrotonate tautomerase: kinetic and structural effects of mutations of Phe-50. Biochemistry 40, 1984–1995. [DOI] [PubMed] [Google Scholar]
- (34).Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, and Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Xu G, Kunzendorf A, Crotti M, Rozeboom HJ, Thunnissen A-MWH, and Poelarends GJ (2022) Gene fusion and directed evolution to break structural symmetry and boost catalysis by an oligomeric C-C bond-forming enzyme. Angew. Chem. Int. Ed. 61, e202113970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


