Abstract
An operon containing the genes purD and purE and part of the purK gene was cloned from the facultative anaerobic gram-positive bacterium Lactococcus lactis by complementation of the purD mutation in Escherichia coli SØ609. The genes encode enzymes in the de novo pathway of purine nucleotides. The expression of the genes was regulated approximately 35-fold at the transcription level by the availability of purines in the growth medium. Deletion analysis of the nucleotide region upstream of purD indicated that a region of 145 bp is enough to give regulated expression of the reporter lacLM genes, which encode β-galactosidase. Deletion of a region 79 bp upstream of the transcription start point reduced the promoter activity 33-fold when incubated in a purine-free medium and to values below the detection limit when incubated in a purine-containing medium. No secondary transcription start points were mapped in or close to this region, indicating that a putative activator site and not a promoter was deleted or partly destroyed.
Lactic acid bacteria are used in the production of fermented foods. The facultative anaerobe bacterium Lactococcus lactis is the best studied of lactic acid bacteria but is also increasingly being used as a model organism representing gram-positive anaerobes. L. lactis is primarily used for the production of various cheeses and other fermented milk products, such as buttermilk. These fermentation processes are very complex and not easily controlled, and it is therefore of interest to modify L. lactis genetically in a controlled manner to improve and control lactate formation, bacteriophage resistance, flavor formation, etc. To obtain such modifications we want to increase our knowledge about regulated expression of genes in L. lactis. Several reports describe various regulated expressions triggered by external stimuli like change in pH (11, 27, 31), temperature (1, 6), osmotic pressure (14), Cl− ion concentration (32), and sugar composition of the medium (25, 37).
In a previous study we showed that a purine-requiring mutant of L. lactis cannot grow in milk, whereas the wild-type strain can (5). Apparently there are not sufficient purine compounds in milk to support growth, and L. lactis therefore depends on its own de novo synthesis of purine nucleotides. The de novo synthesis of purine nucleotides requires in general 10 enzymatic steps leading to IMP, which functions as a precursor for both AMP and GMP nucleotides. The purine nucleotides can also be formed by salvage reactions from purine nucleosides and bases (39). Whereas the de novo pathway seems conserved among various organisms, the salvage pathways can vary more between organisms (23). The organization of genes involved in purine metabolism and the regulation of the expression of these genes have been primarily described for Escherichia coli and Bacillus subtilis (7, 30, 33, 36, 38, 39). Very little is presently known about purine metabolism in L. lactis. However, as mentioned above, purine-requiring mutants have been described previously as has the gene hpt, which encodes hypoxanthine guanine phosphoribosyltransferase, involved in the salvaging of purine bases (5, 21).
This work describes the isolation and characterization of a putative operon from L. lactis consisting of three genes, purD, purE, and purK, encoding enzymes in the de novo pathway of purine nucleotides. Also, we show that the expression of the operon is regulated, and it is suggested that the regulation is mediated at the transcription level by an activator.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Table 1 shows the bacterial strains and plasmids used in this work. L. lactis was grown routinely at 30°C in M17 (35), the purine-free defined DN medium (5), or SA medium (12). E. coli was grown in Luria-Bertani medium or the phosphate-buffered AB salt medium (3) as previously described (21). When necessary, erythromycin was added to a final concentration of 1 mg/liter for L. lactis or 300 mg/liter for E. coli and ampicillin was added at 50 mg/liter for E. coli. As purine sources the following were added at the indicated final concentrations: adenine (15 mg/liter) hypoxanthine (15 mg/liter), and guanosine (30 mg/liter).
TABLE 1.
Strain and plasmid list
| Strain or plasmid | Genotype and/or relevant feature(s) | Source or reference |
|---|---|---|
| L. lactis | ||
| CHCC285 | Wild type | 21 |
| MG1363 | Plasmid free | 9 |
| MG1614 | MG1363 Rifr Strr | 9 |
| E. coli | ||
| DH5α | Φ80 lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | 10 |
| LN3 | SØ609/λZAPII-purDEK | This study |
| SØ609 | ara thi rpsL Δ(pro-gpt-lac) hpt deoD purD | 13 |
| Plasmids | ||
| pBluescript KS(+/−) | Cloning vector Apr | Stratagene |
| pIC19H | Cloning vector Apr | ATCC 37408b |
| pAK80 | Eryr; promoterless lacLM | 11 |
| pLN48 | 7.2-kb KpnI fragment from LN3 inserted into pBluescript KS(−) | This study |
| pLN51 | 3.2-kb KpnI-SpeI fragment from pLN48 inserted into pBluescript KS(+) | This study |
| pLN67 | 0.9-kb EcoRI fragment from pLN51 inserted into pBluescript KS(+) | This study |
| pLN71 | 0.9-kb HindIII fragment from pLN67 (purD′) fused to lacLM in pAK80 | This study |
| pLN72 | 0.9-kb HindIII fragment from pLN67 (orf1′) fused to lacLM in pAK80 | This study |
| p10a | Exonuclease III deletion derivative of pLN71 | This study |
| pLN82 | 0.9-kb EcoRI fragment from pLN67 inserted in pIC19H | This study |
| pLN84 to pLN90 | Exonuclease III deletion derivatives of pLN82 | This study |
| pLN93 to pLN99 | HindIII-BamHI fragments from pLN84-90 fused to lacLM in pAK80 | This study |
p19, p22, p27, and p30 are also exonuclease III deletion derivatives of pLN71 (this study).
American Type Culture Collection, Rockville, Md.
DNA isolation, manipulations, and sequencing.
Plasmid isolation from L. lactis was done as described previously (24). For E. coli the Qiagen (Hilden, Germany) plasmid purification kit was used. Plasmid transformation of L. lactis and E. coli was performed as described previously (21). Isolation of bacteriophage DNA was done as previously described (34). Procedures for the use of restriction endonucleases, T4 DNA ligase, and calf intestine alkaline phosphatase were performed as recommended by the suppliers (Boehringer, Mannheim, Germany; Stratagene, La Jolla, Calif., and Promega, Madison, Wis.). The nucleotide sequence of both strands of DNA was determined with the Sequenase 2.0 sequencing kit (United States Biochemicals) together with the universal primers (Stratagene) or customized primers (Pharmacia). All nucleotide sequence data were processed, and the deduced amino acid sequences were compared by using the Genetics Computer Group software package (version 8.1) (4) and the SwissProt Protein Database (release 35.0).
Cloning of purDEK.
The bacteriophage vector λZAPII cI857(Ts) (Stratagene), containing partial Sau3A-digested chromosomal DNA from L. lactis CHCC285, was provided by Egon Bech Hansen, Chr. Hansen A/S, Hørsholm, Denmark, as in vitro-packaged phage particles (21). The E. coli strain SØ609 purD (Table 1) was infected with phage particles containing the L. lactis DNA and plated on minimal medium agar plates containing glucose, Casamino Acids, and thiamine at 30°C. Two lysogens that grew without purines added (Pur+) were isolated. From both these strains, phage lysates that could transduce SØ609 to Pur+ at a high frequency were produced. The bacteriophage strain λLN3 was isolated from a single plaque of the transducing lysate. All bacteriophage handling was done as described previously (34). DNA from λLN3 was isolated; digested with KpnI, giving fragments containing the insert in the λLN3-derived plasmid pBluescript SK(−); ligated; and transformed into SØ609. Ampicillin-resistant colonies that grew on purine-free agar plates were selected. These colonies were shown to have obtained the plasmid pLN48 [pBluescript SK(−) with a 3.2-kb insert] originating from λLN3. The insert was subsequently cloned as a KpnI-SpeI fragment into the KpnI-SpeI sites of pBluescript KS(+). The resulting plasmid, pLN51, could transform SØ609 to Pur+.
Construction of plasmids containing various parts of the promoter region of purDEK fused to lacLM.
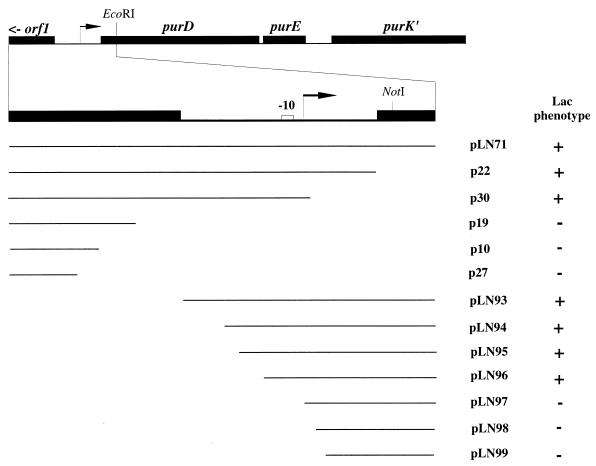
An EcoRI fragment containing 846 bp of purD and upstream region including the promoter from the plasmid pLN51 was cloned into the EcoRI site of pBluescript KS(+), giving pLN67. The 846-bp fragment, including a part of the polylinker region, was removed from pLN67 by digestion with HindIII. The HindIII fragment was ligated to HindIII-digested promoter probe vector pAK80 containing the promoterless lacLM genes from Leuconostoc mesenteroides, which encode β-galactosidase (11). The resulting plasmid, pLN71, is shown in Fig. 1. To map the location of the promoter, a series of nested deletions of the 846-bp fragment was made with exonuclease III, which was included in the Erase-a-Base kit (Amersham). The plasmid pLN71 was digested with BamHI. Following protection of the BamHI overhangs from digestion with exonuclease III by using α-thio-deoxynucleoside triphosphates and Klenow enzyme, the plasmid was further digested with NotI, which cleaves within purD (Fig. 1). The plasmid was then digested with exonuclease III from the NotI site at 21°C for various times ranging from 1 to 10 min. All other steps recommended for the Erase-A-Base kit were performed as suggested by the supplier (Amersham). After ligation of the various deleted variants of the plasmid pLN71, the plasmids were transformed into L. lactis MG1363 and plated on DN medium agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) for screening of β-galactosidase activity. Only a few plasmids were selected for further characterization (Fig. 1).
FIG. 1.
Mapping the purD promoter by exonuclease III deletions. The structure of the purDEK region from L. lactis is shown on the top. Filled boxes correspond to the structural genes, and the open box corresponds to a putative −10 region. The localizations of DNA fragments present in the promoter probe plasmid pAK80 are shown below the physical map and are labelled by the plasmid designation. The Lac-positive (+) or -negative (−) phenotype (defined by the presence or absence, respectively, of blue color development on DN medium agar plates containing X-Gal) was analyzed in MG1363 transformants of the respective plasmid.
Another series of nested deletions of the 846-bp fragment was constructed from the opposite end. The EcoRI fragment of pLN67 was cloned into the EcoRI site of pIC19H in both orientations, giving pLN81 and pLN82 (Table 1). The plasmid pLN82 was cut with SacI (3′ overhang) and ClaI (5′ overhang) and digested unidirectionally with exonuclease III from the ClaI site, at 30°C for 1 to 4 min. Following S1 nuclease digestion and ligation as recommended for the kit (Erase-A-Base; Amersham), the plasmid derivatives were transformed into E. coli DH5α selecting for ampicillin resistance. Plasmids were purified, and inserts were screened for size by double digestion with HindIII and BamHI. Plasmids with inserts of 500 to 240 bp were further analyzed (pLN84 to pLN90). The actual deletion points in plasmids pLN84 to pLN90 were determined by nucleotide sequencing using the reverse primer (Stratagene). The inserts of pLN84 to pLN90 were cut out by digestion with HindIII and BamHI and ligated into HindIII- and BamHI-digested pAK80. Upon electroporation of the ligation mixes into MG1614 and selection for erythromycin resistance, plasmids were purified. The inserts in plasmids pLN93 through pLN99 (Fig. 1) correspond, one to one, to the inserts in plasmids pLN84 through pLN90.
Determination of β-galactosidase activity.
The plasmid-containing strains were grown in 30 ml of DN medium supplemented with 1% glucose, erythromycin, guanosine, adenine, and hypoxanthine without shaking (purine excess). To obtain conditions of partial purine starvation, the bacterial culture was grown in the presence of excess purines as described above. At an optical density at 600 nm (OD600) of 0.3 to 0.4, determined with a Spectronic Genesys 5 spectrophotometer, 1.5 ml of bacterial culture was centrifuged and washed twice in DN medium and resuspended in 1.5 ml of fresh medium without purines. Growth was continued for 1.5 h to an OD600 of ∼1. A volume of 1 ml of bacterial culture was withdrawn at an OD600 of ∼0.3 for the purine excess conditions and at an OD600 of ∼1 for the purine-free conditions. Immediate addition of chloramphenicol to a concentration of 100 μg/ml stopped protein synthesis. The samples were put on ice until the β-galactosidase activity was determined and normalized to OD600. The activity is given in Miller units (20).
RNA extraction.
Cultures of CHCC285 were grown in 50 ml of SA medium containing 1% glucose with slow magnetic stirring. For purine excess conditions, the medium was supplemented with guanosine, adenine, and hypoxanthine. At an OD600 of ∼0.4, 5 ml of culture was mixed with 5 ml of an EPS solution (60% ethanol, 2% phenol, and 0.9% NaCl) prechilled at −30°C. After centrifugation at 5,000 × g for 5 min at 4°C, the pellets were washed in EPS solution mixed 1:1 with 0.9% NaCl. The pellet was frozen at −80°C and lyophilized in a vacuum centrifuge without heating. The lyophilized pellet was ground with acid-washed glass beads (100-μm diameter; Sigma) on the tip of a melted Pasteur pipette. After dissolving the pellets in a buffer containing 10 mM sodium acetate and 300 mM sucrose at pH 4.8 (0°C), the solution was added to a solution of sodium dodecyl sulfate in 10 mM sodium acetate (pH 4.8), to a final sodium dodecyl sulfate concentration of 1%. The solution was equilibrated at 65°C and extracted three times with phenol-acetate, pH 4.8, at 65°C. The RNA was precipitated at −18°C after addition of sodium acetate to 300 mM and ethanol at 70%, centrifuged, dried, and redissolved in 25 μl of water.
Primer extension analysis.
Approximately 5 pmol of oligonucleotide (5′-CCAAACGCTTTTTATATACACAGAC-3′) was radioactively labelled at the 5′ end with 10 pmol of [32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase. The reaction was performed in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol, 0.1 mM spermidine, 0.1 mM EDTA, and 10 U of T4 polynucleotide kinase in a total volume of 50 μl for 20 min at 37°C. The reaction was stopped by the addition of EDTA to 25 mM and heating at 70°C for 10 min. After mixing 10 μl of labelled primer with 10 μg of total RNA in a total volume of 20 μl, annealing was performed in the presence of 100 mM KCl, by heating to 90°C followed by a slow cooling to room temperature over approximately 30 min. Elongation was performed in 30 μl at 41°C for 30 min, with a SuperScript II reverse transcriptase (GibcoBRL) and the buffer conditions recommended by the manufacturer. The elongation products were precipitated with 3 volumes of ethanol, dried, and resuspended in 8 μl of the formamide loading buffer supplied with the ThermoSequenase DNA sequencing kit (Amersham). The sizes of the elongation products were determined by separation on a polyacrylamide DNA sequencing gel with a DNA sequencing ladder as a size marker followed by exposure to X-ray film.
Nucleotide sequence accession number.
The nucleotide sequence of the L. lactis purDEK operon, shown in Fig. 2, is available from the EMBL nucleotide sequence database under accession no. AJ000883.
FIG. 2.
Nucleotide sequence of the purDEK region from L. lactis CHCC285. The nucleotide sequence of the 3,573-bp Sau3AI fragment present in pLN51. The translated amino acid sequences of four open reading frames are shown below the nucleotide sequence and are labelled by the gene designation. Putative ribosomal binding sites (SD) are shown in boldface type. The transcription start point is underlined and marked +1, and the presence of a putative −10 region is likewise indicated by underlining. The locations of various endpoints from exonuclease deletions are indicated by underlining of the last nucleotide present in the deletion plasmids and an arrow above the nucleotide sequence pointing toward the other end of the DNA fragment in the particular plasmid (indicated beside the arrow).
RESULTS
Cloning of L. lactis purD gene.
A plasmid, pLN51, containing L. lactis chromosomal DNA was isolated as described in Materials and Methods by the ability to complement the purD mutation in E. coli SØ609. The nucleotide sequence of the insert of pLN51 is shown in Fig. 2. Four open reading frames can be found and are designated orf1′ (nucleotide 364 to <1 [Fig. 2]), purD (nucleotide 720 to 1958 [Fig. 2]), purE (nucleotide 1991 to 2476 [Fig. 2]), and purK′ (nucleotide 2527 to >3573 [Fig. 2]). Each of the reading frames is preceded by a good putative ribosome binding site (Fig. 2) except orf1′ (18). Further downstream in the putative orf1′ is located a GTG codon (nucleotide 250 [Fig. 2]) preceded by a better putative ribosome binding site (nucleotide 260 to 257 [Fig. 2]). Comparisons of the deduced amino acid sequences with amino acid sequences from the SwissProt Protein Database revealed high identity for the last three reading frames with the PurD, PurE, and PurK amino acid sequences from B. subtilis, E. coli, and S. cerevisiae (Table 2). A high degree of identity (30%) was found between the deduced amino acid sequence of the partially cloned orfA′ and the amino acid sequence of the TetR protein from the E. coli transposon Tn10. However, this identity is not obvious if the start codon is located further downstream (nucleotide 250 [Fig. 2]).
TABLE 2.
Comparison of homologous Pur proteins
| Origin of Pur homolog | % Identity to L. lactis proteina
|
||
|---|---|---|---|
| PurD | PurE | PurKb | |
| B. subtilis | 54 | 61 | 45 |
| E. coli | 49 | 59 | 30 |
| S. cerevisiae | 39 | 52 | 34 |
Percent identical amino acid residues after alignment of the deduced amino acid sequence of the indicated L. lactis Pur proteins with homologous sequences. Accession numbers for B. subtilis, E. coli, and S. cerevisiae sequences, respectively, are as follows: for PurD, P12039, P15640, and P07244; for PurE, P12044, P09028, and P21264; and for PurK, P12045, P09029, and P21264.
The amino acid sequence deduced from L. lactis purK′ is not complete.
Regulation of purDEK expression.
The plasmid pAK80 contains a promoterless copy of the lacLM genes of Leuconostoc mesenteroides, which encode a β-galactosidase enzyme that is active in L. lactis (11). An 846-bp fragment (nucleotide 1 to 846 [Fig. 2]) containing the upstream region of purD was cloned in both orientations into plasmid pAK80 (Table 1) in front of the lacLM genes. The resulting plasmids pLN71 (purD-lacLM) (Fig. 1; Table 1), pLN72 (orf1-lacLM) (Table 1), and pAK80 were used for transformation of L. lactis MG1363. The strains MG1363/pLN71 and MG1363/pLN72 both gave blue colonies on X-Gal-containing agar plates, whereas MG1363/pAK80 gave white colonies. This suggested that there is promoter activity in each direction of the 846-bp fragment. The strains MG1363/pLN71, MG1363/pLN72, and MG1363/pAK80 were grown exponentially in the presence of adenine, hypoxanthine, and guanosine in the defined DN medium. After a shift to purine-free medium, β-galactosidase activity was measured (Table 3). The results showed that the expression of the β-galactosidase directed from the purDEK promoter in pLN71 was regulated approximately 35-fold depending on the available purines in the medium.
TABLE 3.
β-Galactosidase production from fusion plasmids
| Plasmid | β-Galactosidase activity (Miller units) from plasmid with:
|
|||
|---|---|---|---|---|
| Fusion pointa
|
Addition to mediumb
|
|||
| Upstream | Downstream | No Pur | Pur | |
| pAK80 | <1 | <1 | ||
| pLN71 | 1 | 846 | 390 | 13 |
| pLN72 | 846 | 1 | 22 | 11 |
| pLN95 | 450 | 846 | 270 | 6 |
| pLN96 | 497 | 846 | 8 | <1 |
| pLN97 | 588 | 846 | <1 | <1 |
| p30 | 1 | 594 | 530 | 13 |
Fusion points correspond to the nucleotide numbers shown in Fig. 2.
The basal growth medium was DN medium containing erythromycin (No Pur) or further supplemented with guanosine, adenine, and hypoxanthine (Pur).
Mapping of the purD promoter by deletion analysis and primer extension.
To locate the promoter with the purine-regulated activity on the 846-bp fragment in pLN71, a series of nested deletions of the insert were constructed as described in Materials and Methods. In Fig. 1 and Table 3 are shown the results of the analysis. The smallest plasmid with an upstream deletion that still has full regulation and activity is pLN95, whereas the smallest plasmid with a downstream deletion that has full regulation and activity is p30. This result indicates that only the region from nucleotide 450 to 594 (Fig. 2) is necessary for the regulation and activity and therefore contains the operator site and promoter. Deletion from nucleotide 450 to 497 (pLN95 and pLN96) (Table 3; Fig. 2) severely reduced expression, which indicates that either a binding site for an activator or the promoter is located within or near this region.
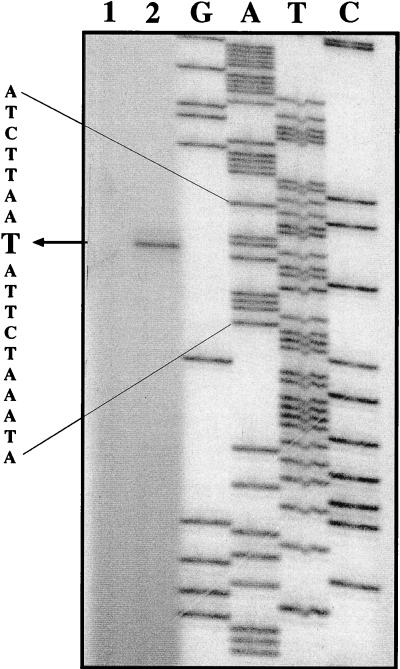
In order to map the transcription start point, we performed primer extension analysis on RNA extracted from the wild-type strain CHCC285 grown in the presence and the absence of purines (Fig. 3). A single band appears in the primer extension, at a position corresponding to a start point at the adenine nucleotide at position 575 (Fig. 2). This transcription start point is located far from the region between nucleotide 450 and 497, which is necessary for high expression (Table 3). However, the band shows up under conditions where the cells had been grown in the absence of purines (Fig. 3, lane 2), whereas in the presence of purines no band could be seen (Fig. 3, lane 1). These results are in accordance with the regulation of the specific β-galactosidase activities of MG1363/pLN71 under the same growth conditions (Table 3). Upstream of the mapped transcription start point (nucleotide 575 [Fig. 2]) a putative −10 region (TAAGAT, nucleotide 563-568 [Fig. 2]), −35 region (CTTTTTC, nucleotide 539-545 [Fig. 2]), and −44 region (TGTT, nucleotide 531-534 [Fig. 2]) are located. None of these regions are highly conserved in comparison with the consensus of some strong promoters (−44, AGTT; −35, CTTGACA; −15, TG; −10, TATAAT) found in L. lactis (22). Other sequences found between nucleotide 450 and the mapped transcription start point at nucleotide 575 show more resemblance to the consensus −35 and −10 regions; however, none of these are located near the transcription start point.
FIG. 3.
Primer extension analysis of purC and purD transcription start points. Primer extension experiments were performed with 10 μg of RNA extracted from CHCC285 with primer MKP57 (5′-CCAAACGCTTTTTATATACACAGA-3′). RNA was extracted from cells growing exponentially in SA medium (lane 2) or in the same medium supplemented by purines (lane 1). Lanes G, A, T, and C contain sequencing reaction mixtures using the same primer as in the primer extension experiments and PCR-generated template DNA. The nucleotide sequence of the region around the transcription start point is shown, with the starting nucleotide in boldface type. The picture was scanned at 300 dots per in. by using a Scan Jet 4c/T device (Hewlett-Packard) and the DeskScan II version 2.3 software. The TIF file was imported into Top Draw (version 3.1) for the addition of text.
DISCUSSION
The purDEK operon has been cloned from the L. lactis subsp. lactis strain CHCC285, and its nucleotide sequence has been determined. Three reading frames showed extensive similarity to the purD, purE, and purK reading frames from other organisms. Unfortunately we did not obtain the last part of the purK gene, so we do not know whether purK is the last gene in the operon. A terminator structure is located after the purK gene in the L. lactis subsp. cremoris strain MG1363 (15), so we expect that purK might be the last gene in the purDEK operon in CHCC285.
The expression of the purDEK operon was regulated at the transcriptional level, giving approximately 35-fold higher expression in the absence of purines in the growth medium. This regulation of expression was shown by fusing the promoter to a reporter gene on a plasmid and would perhaps have been more pronounced if single copy expression were measured. However, it was possible to detect a transcript from the wild-type strain only when the strain was grown in the absence of purines and not in their presence, also showing the regulated expression in single copy.
Using two divergent sets of nested deletions we were able to localize the purD promoter and regulatory region between nucleotides 450 and 593. Deletions in this region (nucleotides 450 to 497) severely lowered the expression (Table 3). This indicated that a part of a promoter or a binding site for an activator is located within or near nucleotide 450 to 497 (Fig. 2). However, our primer extension analysis mapped the transcription start point to nucleotide 575 (Fig. 3), which makes it unlikely that the region contains the promoter. Thus, the region likely contains a binding site for an activator (26). A transcription activator was also found to be involved in the regulation of the expression of the L. lactis purC gene (17).
If the purDEK promoter is controlled by a transcription activator as it seems, it is a unique feature compared to other examined bacteria. In E. coli the regulation of purine de novo genes is by a repressor, PurR (16, 28, 29), which in the presence of the corepressors guanine or hypoxanthine (2, 19) binds to Pur box sequences in the promoter regions. The B. subtilis pur operon is regulated by two independent mechanisms, an attenuation mechanism utilizing an RNA binding protein (7) and repression of transcription initiation by a repressor (8, 36). The effector molecule for the attenuation control appears to be ATP (7), while 5-phosphoritosyl-α-1-pyrophosphate is an inducer of the PurR repressor (36). L. lactis has apparently evolved yet another mechanism for regulating its purine de novo genes in response to the purine availability.
ACKNOWLEDGMENTS
We acknowledge the Lundbeck Foundation for financial support.
We thank Per Nygaard for E. coli SØ609, Egon Bech Hansen for the bacteriophage library of genomic DNA from L. lactis, and Jan Martinussen for the help with the primer extension analysis and sharing unpublished results. We are also grateful for the technical assistance from Anette Ager Lauridsen and Kristina Brandborg Jensen.
REFERENCES
- 1.Arnau J, Sørensen K I. The isolation of novel heat shock genes in Lactococcus lactis using RNA subtractive hybridization. Gene. 1997;188:229–234. doi: 10.1016/s0378-1119(96)00812-8. [DOI] [PubMed] [Google Scholar]
- 2.Choi K Y, Zalkin H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J Bacteriol. 1992;174:6207–6214. doi: 10.1128/jb.174.19.6207-6214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 4.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickely F, Nilsson D, Hansen E B, Johansen E. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol Microbiol. 1995;15:839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x. [DOI] [PubMed] [Google Scholar]
- 6.Duwat P, Ehrlich S D, Gruss A. The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol Microbiol. 1995;17:1121–1131. doi: 10.1111/j.1365-2958.1995.mmi_17061121.x. [DOI] [PubMed] [Google Scholar]
- 7.Ebbole D J, Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987;262:8274–8287. [PubMed] [Google Scholar]
- 8.Ebbole D J, Zalkin H. Interaction of a putative repressor protein with an extended control region of the Bacillus subtilis pur operon. J Biol Chem. 1989;264:3553–3561. [PubMed] [Google Scholar]
- 9.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jochimsen B, Nygaard P, Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Mol Gen Genet. 1975;143:85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- 14.Kilstrup M, Jacobsen S, Hammer K, Vogensen F K. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilstrup, M., and J. Martinussen. Unpublished data.
- 16.Kilstrup M, Meng L M, Neuhard J, Nygaard P. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J Bacteriol. 1989;171:2124–2127. doi: 10.1128/jb.171.4.2124-2127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilstrup M, Jessing S G, Wichmand-Jørgensen S B, Madsen M, Nilsson D. Activation control of pur gene expression in Lactococcus lactis: proposal of a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions. J Bacteriol. 1998;180:3900–3906. doi: 10.1128/jb.180.15.3900-3906.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig W, Seewaldt E, Klipper-Balz R, Schleifer K H, Magrum L, Woese C R, Fox G E, Stackebrandt E. The phylogenetic position of Streptococci and Enterococcus. J Gen Microbiol. 1985;131:543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 19.Meng L M, Nygaard P. Identification of hypoxanthine and guanine as the corepressors for the purine regulon genes of Escherichia coli. Mol Microbiol. 1990;487:2187–2191. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller J M. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Nilsson D, Lauridsen A A. Isolation of purine auxotrophic mutants of Lactococcus lactis and characterisation of the gene hpt encoding hypoxanthine guanine phosphoribosyltransferase. Mol Gen Genet. 1992;235:359–364. doi: 10.1007/BF00279381. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson D, Johansen E. A conserved sequence in tRNA and rRNA promoters of Lactococcus lactis. Biochim Biophys Acta. 1994;1219:141–144. doi: 10.1016/0167-4781(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 23.Nygaard P. Utilization of preformed purine bases and nucleosides. In: Munch-Petersen A, editor. Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. New York, N.Y: Academic Press; 1983. pp. 27–93. [Google Scholar]
- 24.Pedersen M L, Arnved K R, Johansen E. Genetic analysis of the minimal replicon of the Lactococcus lactis subsp. lactis biovar. diacetylactis citrate plasmid. Mol Gen Genet. 1994;244:374–382. doi: 10.1007/BF00286689. [DOI] [PubMed] [Google Scholar]
- 25.Qian N, Stanley G A, Bunte A, Rådström P. Product formation and phosphoglucomutase activities in Lactococcus lactis: cloning and characterisation of a novel phosphoglucomutase gene. Microbiology. 1997;143:855–865. doi: 10.1099/00221287-143-3-855. [DOI] [PubMed] [Google Scholar]
- 26.Raibaut O, Schwartz M. Positive regulation of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 27.Rallu F, Gruss A, Maguin E. Lactococcus lactis and stress. Antonie Leeuwenhoek. 1996;70:243–251. doi: 10.1007/BF00395935. [DOI] [PubMed] [Google Scholar]
- 28.Rolfes R, Zalkin H. Escherichia coli gene purR encoding a repressor protein for purine nucleotide synthesis. Cloning, nucleotide sequence and interaction with the purF operator. J Biol Chem. 1988;263:19653–19661. [PubMed] [Google Scholar]
- 29.Rolfes R, Zalkin H. Regulation of Escherichia coli purF. Mutations that define the promoter, operator, and purine repressor gene. J Biol Chem. 1988;263:19649–19652. [PubMed] [Google Scholar]
- 30.Rolfes R, Zalkin H. Purification of the Escherichia coli purine regulon repressor and identification of corepressors. J Bacteriol. 1990;172:5637–5642. doi: 10.1128/jb.172.10.5637-5642.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders J W, Leenhouts K, Haandrikman A, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders J W. Environmental stress response in Lactococcus lactis: identification of genes and use of expression signals. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 33.Saxild H H, Nygaard P. Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools. J Gen Microbiol. 1991;137:2387–2394. doi: 10.1099/00221287-137-10-2387. [DOI] [PubMed] [Google Scholar]
- 34.Silhavy T J. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 35.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng M, Nagy P L, Zalkin H. Identification of the Bacillus subtilis pur operon repressor. Proc Natl Acad Sci USA. 1995;92:7455–7459. doi: 10.1073/pnas.92.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J J, Reizer J, Milton H S., Jr Regulation of 2-deoxyglucose phosphate accumulation in Lactococcus lactis vesicles by metabolite-activated, ATP-dependent phosphorylation of serine-46 in HPr of the phosphotransferase system. Microbiology. 1994;140:3421–3429. doi: 10.1099/13500872-140-12-3421. [DOI] [PubMed] [Google Scholar]
- 38.Zalkin H. Organization and regulation of genes for de novo purine nucleotide synthesis in Bacillus subtilis. Res Microbiol. 1991;142:765–769. doi: 10.1016/0923-2508(91)90053-d. [DOI] [PubMed] [Google Scholar]
- 39.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 561–579. [Google Scholar]