Abstract
Growth of Thermus thermophilus HB27 was inhibited by a proline analog, 3,4-dehydroproline (DHP). This result suggested that the γ-glutamyl kinase (the product of the proB gene) was inhibited by feedback inhibition in T. thermophilus. DHP-resistant mutants were reported previously for Escherichia coli (A. M. Dandekar and S. L. Uratsu, J. Bacteriol. 170:5943–5945, 1988) and Serratia marcescens (K. Omori, S. Suzuki, Y. Imai, and S. Komatsubara, J. Gen. Microbiol. 138:693–699, 1992), and their mutated sites in the proB gene were identified. Comparison of the amino acid sequence of T. thermophilus γ-glutamyl kinase with those of E. coli and S. marcescens mutants revealed that the DHP resistance mutations occurred in the amino acids conserved among the three organisms. For eliminating the feedback inhibition, we first constructed a DHP-resistant mutant, TH401, by site-directed mutagenesis at the proB gene as reported for the proline-producing mutant of S. marcescens. The mutant, TH401, excreted about 1 mg of l-proline per liter at 70°C after 12 h of incubation. It was also suggested that T. thermophilus had a proline degradation and transport pathway since it was able to grow in minimal medium containing l-proline as sole nitrogen source. In order to disrupt the proline degradation or transport genes, TH401 was mutated by UV irradiation. Seven mutants unable to utilize l-proline for their growth were isolated. One of the mutants, TH4017, excreted about 2 mg of l-proline per liter in minimal medium at 70°C after 12 h of incubation.
Thermus thermophilus, a gram-negative aerobic eubacterium, is one of the most widely studied species of extremely thermophilic microorganisms. We have been working on the molecular genetics and molecular reproduction of T. thermophilus HB27. We have already cloned and sequenced three proline biosynthetic genes, proB, proA, and proC, and reported that the proB and proA genes exist in tandem (7, 9).
We have also constructed physical maps of the HB27 chromosome and of a large plasmid, pTT27, and determined the locations of all proline biosynthetic genes on the chromosomal DNA (20, 21). We have already succeeded in overproducing carotenoids in T. thermophilus HB27 (6), but at present there is no report about extracellular production of amino acids in extreme thermophiles. We have elucidated the consensus sequences for strong promoters of T. thermophilus (11) and developed a thermostable antibiotic resistance gene (12). It is also easy to disrupt or mutate genes on chromosomal DNA in T. thermophilus HB27 (8). Among the extreme thermophiles, a host-vector system has been established only in T. thermophilus. Generally, the reaction rate of thermostable enzymes which are produced from T. thermophilus is higher than those of enzymes from mesophiles. In a fermentation process such as amino acid production, T. thermophilus may contribute to the improvement of amino acid productivity since fermentation at a high temperature eliminates the problems of contamination and cooling procedures. So, we decided to attempt excretion of proline at a high temperature with T. thermophilus mutants.
l-Proline is synthesized from glutamate by the sequential reaction of γ-glutamyl kinase, γ-glutamyl phosphate reductase, and pyrroline-5-carboxylate reductase in bacteria (1). Genes proB and proA, which encode γ-glutamyl kinase and γ-glutamyl phosphate reductase, respectively, were found to comprise an operon in T. thermophilus (9), Escherichia coli (5), and Serratia marcescens (13). In E. coli and S. marcescens, γ-glutamyl kinase is subject to feedback control by l-proline (3, 13), but γ-glutamyl phosphate reductase and pyrroline-5-carboxylate reductase are not inhibited by proline (3, 15). Meanwhile, E. coli and S. marcescens rapidly degrade proline by proline dehydrogenase (proline oxidase), encoded by the putA gene (3, 14, 22). So far, it has been reported that E. coli mutants resistant to proline analogs, dl-3,4-dehydroproline and l-azetidine-2-carboxylic acid, excreted l-proline into the medium. But the amount of l-proline excreted was too small for practical use because of the existence of the proline degradation pathway (2). For S. marcescens, Sugiura et al. (18, 19) have constructed a proline-overproducing strain, SP126, as a double mutant resistant to 3,4-dehydroproline and thiazolidine-4-carboxylate and derived from a proline dehydrogenase-deficient mutant (18). Strain SP126 produced about 20 mg of l-proline per ml in the fermentation medium (18).
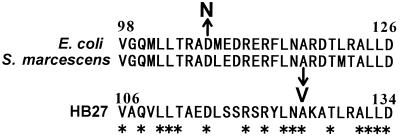
In T. thermophilus, the control system in proline biosynthesis has not been elucidated. However, we thought that the feedback control of proline biosynthesis in T. thermophilus should be similar to that of E. coli and S. marcescens, since the amino acid sequences of proline biosynthetic enzymes in T. thermophilus show a high similarity to sequences of those of E. coli and S. marcescens (7, 9). E. coli and S. marcescens mutants resistant to 3,4-dehydroproline have already been determined to be proB mutants (4, 14). The comparison of the amino acid sequences of γ-glutamyl kinases in E. coli, S. marcescens, and T. thermophilus showed that these mutations occurred in the positions conserved among the three microorganisms (Fig. 1). We thought that it was possible to construct a 3,4-dehydroproline-resistant mutant of T. thermophilus by introducing the same mutations into the proB gene found in the mutants of E. coli and S. marcescens. We determined the strategy for construction of a proline-producing strain of T. thermophilus by following two steps: first, construction of a 3,4-dehydroproline-resistant mutant by introduction of mutations into the proB gene, and second, isolation of a mutant which cannot utilize proline for its growth by mutagenizing the dehydroproline-resistant mutant.
FIG. 1.
Comparison of the amino acid sequences of γ-glutamyl kinases in E. coli, S. marcescens, and T. thermophilus. The amino acid substitutions found in E. coli (4) and S. marcescens (14) are shown by arrows. Asterisks show the amino acid residues conserved in the three microorganisms.
MATERIALS AND METHODS
Chemicals.
Restriction endonucleases and DNA modification enzymes were purchased from Toyobo (Tokyo, Japan) or Takara Shuzo (Kyoto, Japan). l-Proline and dl-3,4-dehydroproline were purchased from Sigma Chemical Co. (St. Louis, Mo.). All the other reagents used were of the purest grade available.
Bacterial strains and growth conditions.
T. thermophilus HB27 (17) and its proline auxotrophic mutant, TH104 (7), were used. TM medium (10) was used for routine cultivation of T. thermophilus. Minimal medium (MM) (10) was also used. When necessary, 3,4-dehydroproline and l-proline were added to MM at the concentration of 1 mM. MM-proline plates which contained 10 mM l-proline instead of (NH4)2SO4 were used to isolate proline-producing mutants. The growth curve of T. thermophilus was measured as follows. T. thermophilus HB27 was grown in 10 ml of TM medium at 70°C for 16 h. A 0.1-ml aliquot of the culture was inoculated into 10 ml of a fresh medium. The growth was monitored by measuring absorbance at 580 nm. E. coli JM109 (23) was also used in the cloning experiments.
Preparation of the fragment containing the mutated proB genes.
Four primers, PROBDNF (5′-GTCCTCCTCACCGCCGAGAACCTC-3′), PROBDNR (5′-GAGGTTCTCGGCGGTGAGGAGGAC-3′), PROBAVF (5′-AGGAGCCGCTACCTGAACGTCAAG-3′), and PROBAVR (5′-CTTGACGTTCAGGTAGCGGCTCCT) (Fig. 2), were prepared to introduce the mutations into the proB gene. Two primers, PROB_F (5′-GGGAATTCCCGAGGCCATGCCGGAGGC-3′) and PROB_R (5′-GAAGCTTTCATGCCTCCTCCTTCAAGGC-3′) (Fig. 2), were prepared to amplify the fragment containing the entire mutated proB gene. The primers contained restriction endonuclease sites for EcoRI (italics) and HindIII (underlined).
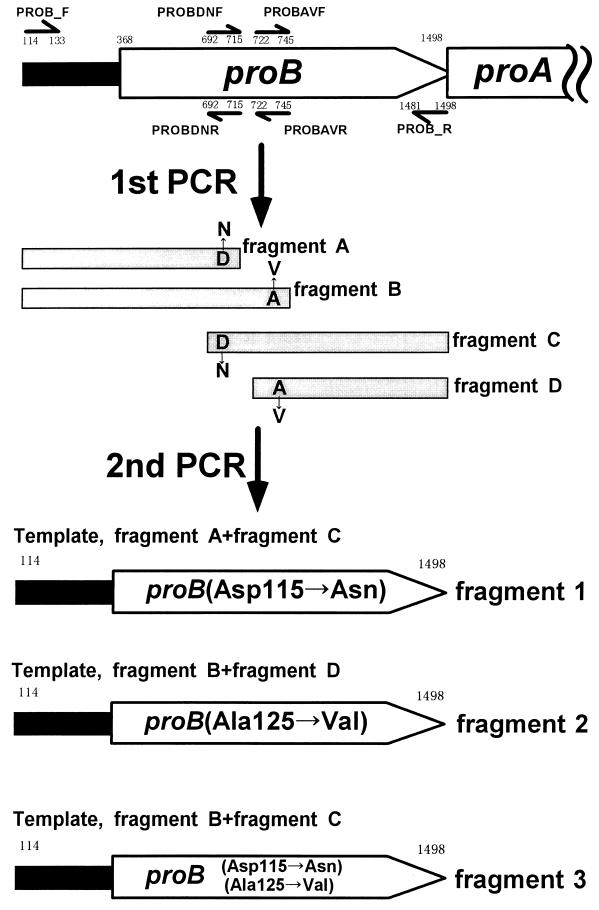
FIG. 2.
Outline of the preparation of DNA fragments which contain three types of mutated proB genes. The detailed explanations are given in Materials and Methods. The numbers shown below or above each primer correspond to the nucleotide numbers of the proBA region.
In the first PCR, the four DNA fragments which would be the DNA template for construction of the mutated proB gene were prepared (Fig. 2). For the preparation of fragments A, B, C, and D, primers PROB_F and PROBDNR, PROB_F and PROBAVR, PROB_R and PROBDNF, and PROB_R and PROBAVF were used, respectively. The PCR mixture contained PCR buffer with 2 mM MgCl2 (Takara Shuzo), 0.2 mM deoxynucleoside triphosphates, 0.1 μM (each) primer, 0.1 μg of template (pUC-pro3,5+ [9]), and 2.5 U of rTaq DNA polymerase (Takara Shuzo). A DNA thermal cycler (Perkin-Elmer Cetus) was used with the following conditions: melting temperature, 96°C (30 s); annealing temperature, 62°C (1 s); and polymerization at 72°C (150 s). Twenty-five cycles were run with a subsequent polymerization period of 10 min at 72°C. The amplified fragments A (610 bp), B (637 bp), C (814 bp), and D (787 bp) were recovered from 1.0% agarose gels and used for the second PCR.
In the second PCR, three types of DNA fragments containing the mutated proB genes were prepared. For the amino acid change of Asp-115 to Asn in the proB gene, fragments A and C were used for the DNA template (Fig. 2, fragment 1). For the amino acid change of Ala-125 to Val (Fig. 2, fragment 2), fragments B and D were used. For the two amino acid changes of Asp-115 and Ala-125 to Asn and Val, respectively, fragments B and C were used (Fig. 2, fragment 3). The second PCR mixture contained PCR buffer with 2 mM MgCl2 (Takara Shuzo), 0.2 mM deoxynucleoside triphosphates, 0.1 μM (each) primer PROB_F and PROB_R, 10 ng of each amplified fragment, and 2.5 U of rTaq DNA polymerase (Takara Shuzo). PCR was performed under the following conditions: 96°C for 30 s, gradual decrease of the temperature to 62°C for 45 s, and immediate increase of the temperature from 62 to 72°C, and maintenance for 150 s. Twenty-five cycles were run with a subsequent polymerization period of 10 min at 72°C. The three types of amplified DNA fragments were recovered from 1.0% agarose gels and filled and phosphorylated with T4 DNA polymerase and T4 polynucleotide kinase. Each fragment was ligated to SmaI-digested and dephosphorylated pUC19, and the ligation mixtures were used for E. coli JM109 transformation. Three types of plasmids which contained D115N, V125A, and D115N and V125A replacements were prepared, and their nucleotide sequences were determined.
Construction of a 3,4-dehydroproline-resistant mutant.
T. thermophilus HB27 was transformed with 3 μg each of three types of plasmid (containing fragment 1, 2, or 3 [Fig. 2]) as described previously (10). The transformed cells were washed with 0.85% NaCl (saline) and resuspended in saline. The diluted suspensions were spread on MM plates containing dl-3,4-dehydroproline at the concentration of 1 mM. When the plasmid containing fragment 2 which included the amino acid change in the proB gene of Ala-125 to Val was used for the transformation, many 3,4-dehydroproline-resistant mutants were obtained. Eight transformants among them were randomly selected, and total DNAs were prepared by the method of Saito and Miura (16). Total DNA from the eight transformants was digested with SphI, and DNA fragments including the proBA genes of 2.8 kb in size were recovered by agarose gel electrophoresis. Each DNA fragment was ligated with SphI-digested and dephosphorylated pUC19 and introduced into E. coli JM109. The clones containing the entire mutant proB genes were screened by colony hybridization using the inserted fragment of pUC-pro3,5+ as a probe. The entire proB genes of eight transformants were sequenced and checked for the amino acid change of Ala-125 to Val. The eight transformants showed the same growth curves in MM containing 1 mM 3,4-dehydroproline. We named one of the eight transformants TH401 and used it for further experiments.
Construction of a proline-producing strain.
Strain TH401 was grown in TM medium for 3 h at 70°C. The culture was diluted 1/10 with saline and irradiated for 3 min under a 15-W UV lamp. Five milliliters of the culture was added to 5 ml of fresh TM medium and incubated at 70°C for 60 min in the dark. This culture was diluted and spread on TM plates. A total of 8,217 colonies on TM plates were replica plated onto MM and MM-proline plates. Seven strains which were able to grow on MM plates but not on MM-proline plates were isolated. We named them TH4011, TH4012, TH4013, TH4014, TH4015, TH4016 and TH4017.
Bioassay of proline production.
An overnight TM culture of each strain was diluted 1/10 with saline (0.85% NaCl), and 5 μl was spotted onto the MM plates onto which TH104 had been spread. The plates were incubated at 70°C for 2.5 days. After incubation, the growth of TH104 around the spots was checked.
Analysis of amino acid production.
Each strain was incubated in 10 ml of MM at 70°C for 12 h. All cells were removed by centrifugation (18,000 × g, 10 min), and the amino acids in the culture supernatant were analyzed by ion-exchange chromatography with a Hitachi L-8500 amino acid analysis system (Hitachi, Tokyo, Japan).
Nucleotide sequence accession number.
The nucleotide sequence of the proBA genes in T. thermophilus is in the EMBL, GenBank, and DDBJ databases under accession no. D29973.
RESULTS AND DISCUSSION
Analysis of the regulation system of proline biosynthesis in T. thermophilus.
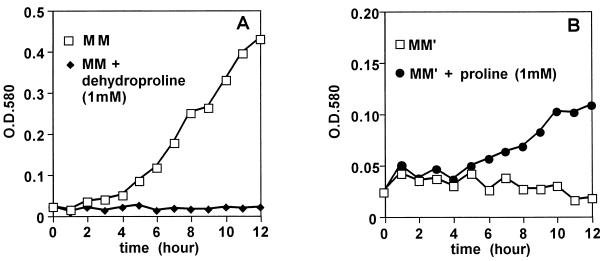
The growth of T. thermophilus in 3,4-dehydroproline-containing MM was first assayed. As shown in Fig. 3A, the growth of T. thermophilus was clearly inhibited by dehydroproline at the concentration of 1 mM. This result indicated that proline biosynthesis was controlled by feedback inhibition and that γ-glutamyl kinase was inhibited by proline in T. thermophilus.
FIG. 3.
Growth curves of T. thermophilus HB27. HB27 was grown in MM with or without 3,4-dehydroproline (A) and in (NH4)2SO4-minus MM (MM′) with or without l-proline (B). O.D.580, optical density at 580 nm.
Analysis of the existence of a proline degradation system in T. thermophilus.
The proline degradation system in T. thermophilus was examined since it disturbs overproduction of proline. In E. coli, two proline degradative genes, the putA and putP genes, have been reported previously. The putA gene encodes proline dehydrogenase, which catalyzes the conversion of proline to pyrroline-5-carboxylate. The putP gene encodes proline permease, which is necessary for uptake of extracellular proline into the cell. If T. thermophilus has PutA and PutP activity, it is able to grow in MM which contains proline as a sole nitrogen source. We checked whether T. thermophilus was able to utilize proline for its growth. As shown in Fig. 3B, T. thermophilus was able to grow in MM containing proline as sole nitrogen source. This result indicated that T. thermophilus had a proline degradation system.
Site-directed mutagenesis of the chromosomal proB gene in T. thermophilus.
The mutation sites of the proline-overproducing mutants have been determined previously in E. coli and S. marcescens (3, 4, 14). These proline-overproducing mutants showed 3,4-dehydroproline resistance, and their γ-glutamyl kinases were not inhibited by proline. We compared the amino acid sequences of γ-glutamyl kinases among T. thermophilus and two proline-overproducing mutants. As shown in Fig. 1, the mutations occurred at the sites which were conserved in the three γ-glutamyl kinases.
Three kinds of plasmids including the mutated proB gene were prepared by recombinant PCR (Fig. 2). Fragment 1 had the amino acid substitution of Asn for Asp115 in the proB gene, fragment 2 had the amino acid substitution of Val for Ala125, and fragment 3 had both amino acid substitutions (Fig. 2). We transformed T. thermophilus HB27 by using these plasmids. Many dehydroproline-resistant transformants were obtained when the plasmid including fragment 2 was used for the donor DNA whereas no colony was obtained when the plasmid including fragment 1 or fragment 3 was used. No dehydroproline-resistant colonies appeared on the MM containing dehydroproline when no DNA was added. This result showed that only the A125V substitution led to dehydroproline resistance in HB27 and that the D115N substitution might impair the heat stability of γ-glutamyl kinase of T. thermophilus. The eight transformants were randomly selected, and their growth was assayed in MM containing 3,4-dehydroproline or proline as sole nitrogen source. Every mutant showed the same growth curve in MM containing 3,4-dehydroproline or proline as sole nitrogen source. From this result, we confirmed that the eight mutants had the same mutations only in the proB gene. One of the eight 3,4-dehydroproline-resistant mutants was named TH401. The growth of TH401 was not inhibited in MM containing 1 mM 3,4-dehydroproline. The growth curve of TH401 in MM containing 1 mM 3,4-dehydroproline was the same as that of wild type in MM (data not shown). This fact suggested that TH401 was not subjected to feedback inhibition in proline biosynthesis.
Construction of a proline-producing mutant.
Strain TH401 was further mutated by UV irradiation. Seven strains, TH4011, TH4012, TH4013, TH4014, TH4015, TH4016, and TH4017, which were unable to grow on MM-proline plates containing proline as a sole nitrogen source were isolated. The growth curves of these seven mutants in MM were the same as those of TH401 and HB27 wild type. The proline production levels of seven mutants and TH401 were tested by bioassay (see Materials and Methods). The growth of the proline auxotrophic mutant was observed only around the spot of TH4017. This result showed that TH4017 excreted proline into the medium. The detailed amino acid production levels of T. thermophilus HB27, TH401, and TH4017 were measured. The amino acid production was measured twice, and a little difference was observed in two experiments. As shown in Table 1, TH4017 produced about 2 mg of proline per liter at 70°C after 12 h of incubation in MM but the wild type did not. TH401 also produced about 1 mg of proline per liter. The proline production levels of other mutants (TH4011, TH4012, TH4013, TH4014, TH4015, and TH4016) were the same as that of TH401 (data not shown). TH4017 showed the highest production of proline among the mutants. This result indicated that TH4017 was the first T. thermophilus strain excreting proline into the medium.
TABLE 1.
Amino acids production levels of HB27, TH401, and TH4017
| Amino acid | Production level (mg/liter) for strain:
|
||
|---|---|---|---|
| HB27 | TH401 | TH4017 | |
| Proline | 1.01 | 1.99 | |
| Aspartate | 0.59 | 1.56 | |
| Threonine | 1.86 | 3.44 | |
| Serine | 0.28 | 0.73 | |
| Glutamate | 0.43 | 1.35 | |
| Glycine | 0.36 | 0.47 | |
| Alanine | 0.11 | 6.38 | 4.81 |
| Cystine | |||
| Valine | 1.75 | 3.86 | |
| Methionine | 0.28 | 0.63 | |
| Isoleucine | 0.23 | 0.44 | |
| Leucine | 0.55 | 1.19 | |
| Tyrosine | 0.52 | 1.33 | |
| Phenylalanine | 0.40 | 0.62 | |
| Lysine | 0.13 | 0.02 | |
| Histidine | |||
| Arginine | 0.17 | ||
| Tryptophan | |||
| Methionine sulfoxide | |||
| Cysteic acid | |||
| Total | 0.41 | 14.7 | 22.4 |
As shown in Table 1, TH4017 excreted various amino acids into the medium. The amounts of some of them were larger than that of proline. Since the production of various amino acids was also observed for TH401, it is highly likely that this fact is due to the mutation in the proB gene. In E. coli, proline biosynthesis is linked to arginine biosynthesis through the interconversion of pyrroline-5-carboxylate to ornithine (1). No other linkages of proline biosynthesis have been reported. TH401 and TH4017 did not excrete arginine into the medium, but they produced relatively larger amounts of alanine, threonine, and valine. It will be interesting if proline biosynthesis in T. thermophilus has some relationship to these amino acid biosynthetic pathways.
Until now, the level of l-proline produced from TH4017 has been very low. We thought this was mainly due to the low expression of the proline biosynthetic genes and the remaining activity of the l-proline degradation system in T. thermophilus. We believed that TH4011, TH4012, TH4013, TH4014, TH4015, TH4016, and TH4017 were defective for proline uptake but not for proline dehydrogenase because all mutants could not grow in MM containing 10 mM proline as sole nitrogen source but could grow in MM containing 100 mM proline. As already reported, there were no strong promoter sequences upstream of the proBA and proC genes in T. thermophilus (7, 9). Increasing l-proline production by replacing the proline biosynthetic promoters should be effective. We also plan to attempt to increase l-proline production by disrupting the putA gene.
ACKNOWLEDGMENT
We thank Hiroshi Matsui (Ajinomoto Co., Inc., Kawasaki, Japan) for analysis of the amino acid production levels of various mutants of T. thermophilus.
REFERENCES
- 1.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 2.Baich A, Pierson D J. Proline-producing strain in Escherichia coli. Biochim Biophys Acta. 1965;104:397–402. doi: 10.1016/0304-4165(65)90345-4. [DOI] [PubMed] [Google Scholar]
- 3.Csonka L N, Gelvin S B, Goodner B W, Orser C S, Siemieniak D, Slightom J L. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene. 1988;64:199–205. doi: 10.1016/0378-1119(88)90335-6. [DOI] [PubMed] [Google Scholar]
- 4.Dandekar A M, Uratsu S L. A single base pair change in proline biosynthesis genes causes osmotic stress tolerance. J Bacteriol. 1988;170:5943–5945. doi: 10.1128/jb.170.12.5943-5945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutch A H, Rushlow K E, Smith C J. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984;12:6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshino T, Fujii R, Nakahara T. Overproduction of carotenoids in Thermus thermophilus. J Ferment Bioeng. 1994;77:423–424. [Google Scholar]
- 7.Hoshino T, Kosuge T, Hidaka Y, Tabata K, Nakahara T. Molecular cloning and sequence analysis of the proC gene encoding Δ1-pyrroline-5-carboxylate reductase from an extremely thermophilic eubacterium, Thermus thermophilus. Biochem Biophys Res Commun. 1994;199:410–417. doi: 10.1006/bbrc.1994.1244. [DOI] [PubMed] [Google Scholar]
- 8.Kosuge T, Hoshino T. Molecular cloning and sequencing analysis of the lysR gene from the extremely thermophilic eubacterium, Thermus thermophilus HB27. FEMS Microbiol Lett. 1997;157:73–79. doi: 10.1111/j.1574-6968.1997.tb12755.x. [DOI] [PubMed] [Google Scholar]
- 9.Kosuge T, Tabata K, Hoshino T. Molecular cloning and sequence analysis of the proBA operon from an extremely thermophilic eubacterium, Thermus thermophilus. FEMS Microbiol Lett. 1994;123:55–62. doi: 10.1111/j.1574-6968.1994.tb07201.x. [DOI] [PubMed] [Google Scholar]
- 10.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maseda H, Hoshino T. Screening and analysis of DNA fragments that show promoter activities in Thermus thermophilus. FEMS Microbiol Lett. 1995;128:127–134. doi: 10.1111/j.1574-6968.1995.tb07511.x. [DOI] [PubMed] [Google Scholar]
- 12.Maseda H, Hoshino T. Fusion with ribosomal protein L32 increased the in vivo thermostability of kanamycin nucleotidyltransferase in Thermus thermophilus. J Ferment Bioeng. 1996;82:525–530. [Google Scholar]
- 13.Omori K, Suzuki S, Imai Y, Komatsubara S. Analysis of the Serratia marcescens proBA operon and feedback control of proline biosynthesis. J Gen Microbiol. 1991;137:509–517. doi: 10.1099/00221287-137-3-509. [DOI] [PubMed] [Google Scholar]
- 14.Omori K, Suzuki S, Imai Y, Komatsubara S. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J Gen Microbiol. 1992;138:693–699. doi: 10.1099/00221287-138-4-693. [DOI] [PubMed] [Google Scholar]
- 15.Rossi J J, Vender J, Berg C M, Coleman W H. Partial purification and some properties of Δ1-pyrroline-5-carboxylate reductase from Escherichia coli. J Bacteriol. 1977;129:108–114. doi: 10.1128/jb.129.1.108-114.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 17.Sakaki Y, Oshima T. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J Virol. 1975;15:1449–1453. doi: 10.1128/jvi.15.6.1449-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiura M, Kisumi M. Proline-hyperproducing strains of Serratia marcescens: enhancement of proline analog-mediated growth inhibition by increasing osmotic stress. Appl Environ Microbiol. 1985;49:782–786. doi: 10.1128/aem.49.4.782-786.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiura M, Takagi T, Kisumi M. Proline production by regulatory mutants of Serratia marcescens. Appl Microbiol Biotechnol. 1985;21:213–219. [Google Scholar]
- 20.Tabata K, Hoshino T. Mapping of 61 genes on the refined physical map of the chromosome of Thermus thermophilus HB27 and comparison of genome organization with that of T. thermophilus HB8. Microbiology. 1996;142:401–410. doi: 10.1099/13500872-142-2-401. [DOI] [PubMed] [Google Scholar]
- 21.Tabata K, Kosuge T, Nakahara T, Hoshino T. Physical map of the extremely thermophilic bacterium Thermus thermophilus HB27 chromosome. FEBS Lett. 1993;331:81–85. doi: 10.1016/0014-5793(93)80301-a. [DOI] [PubMed] [Google Scholar]
- 22.Wood J M. Genetics of l-proline utilization in Escherichia coli. J Bacteriol. 1981;146:895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]