Abstract
Cyclic tetrapeptides exhibit high cellular permeability and a wide range of biological properties and thus have gained great interest in the field of medicinal chemistry. We synthesized highly strained 12-membered head to tail cyclic peptides with varying reactive amino acids, without oligomerization using the exclusively intramolecular CyClick chemistry. This occurs by a two-step process involving the low-energy formation of a 15 atom-containing cyclic imine, followed by a chemoselective ring contraction of the peptide backbone generating a highly strained 12 atom-containing cyclic tetrapeptide. This reaction exhibited high substrate scope and generated head to tail cyclic tetrapeptides with varying amino acids at the N-terminus, showing chemoselectivity without the need for side group protection.
Keywords: Tetrapeptides, Macrocyclic peptides, CyClick chemistry, 4-imidazolidinone ring
1. Introduction
A wide range of biological activities, high protease stability, and ability to selectively inhibit protein-protein interactions (PPIs) make cyclic peptides highly attractive in the field of medicinal chemistry [1-6]. Despite these significant advantages, there are severe challenges particularly associated with the synthesis of small head-to-tail cyclic peptides. Tetrapeptides, which contain 12 atoms in their backbone, are often generated in poor yields [7-12]. This is due to the high ring strain associated with additional transannular interactions between substituents of cyclic head-to-tail-tetrapeptides. Current methods for chemical synthesis of head-to-tail cyclic tetrapeptides in solution are limited due to the formation of dimers or oligomers [9,10] and the requirement of multiple turn inducers [11] that reduces overall yields and restricts chemical diversity of cyclic peptides.
To avoid the formation of oligomers, cyclization reactions are carried out at very low concentrations (0.001–0.005 M) [10-12] leading to significantly slow reaction rates (~3 days) [11,12]. Therefore, the synthesis of cyclic tetrapeptides comprising any of the regular l-amino acids is still considered highly challenging.
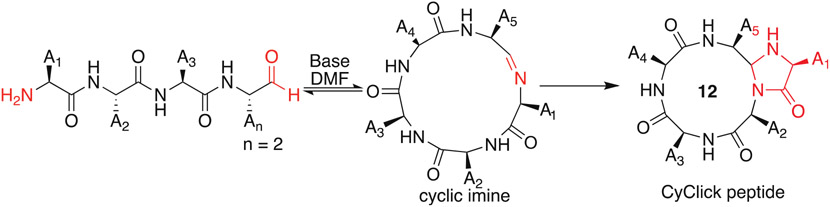
Herein, we describe the synthesis of head-to-tail cyclic tetrapeptides through an exclusively intramolecular CyClick strategy (Fig. 1) that involves formation of an imine between a C-terminal aldehyde and the N-terminus of the linear pentapeptide followed by ring-closure with the amide backbone chain to form a lactam bridge and a 4-imidazolidinone moiety at the site of cyclization [13-15]. We propose that the ring-contraction of cyclic imines of pentapeptides by the CyClick approach is responsible for decreasing the activation energy for the synthesis of cyclic tetrapeptides. Since the CyClick reaction is exclusively intramolecular, we hypothesized that cyclization can be carried out at high concentrations (25 mM) without the formation of any linear dimers or oligomers while concomitantly reducing the overall rate of the reaction.
Fig. 1.
Scheme for the synthesis of cyclic tetrapeptides by CyClick chemistry.
2. Results and discussion
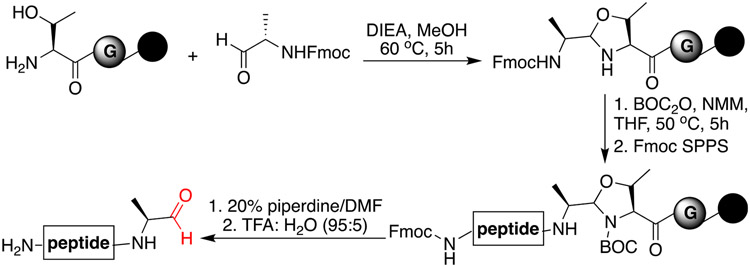
Our first goal was to optimize the cyclization reaction with a peptide aldehyde using different bases. We synthesized C-terminal peptide aldehydes by the reaction scheme in Fig. 2 involving coupling of Fmoc-Ala-CHO on free threonine on solid support, followed by successive residue attachment via Fmoc-SPPS (Fig. 2, Figure S1, Supplementary data) [13-16]. The cleavage of a peptide from solid support using trifluoroacetic acid generated an aldehyde at the C-terminus. Our previous study showed very high cyclization efficiency (ca. 80–99%) with medium-sized head-to-side-chain macrocycles (20 atoms ring size) with varying amino acids at the N-terminus except with bulky residues such as Val (~37%). Therefore in this study, we proceeded with a peptide aldehyde VVGPFEY 1a containing Val at the N-terminus for the head-to-side chain cyclization to evaluate the role of different bases in the efficiency of cyclization (Fig. 3, Figure S2, Supplementary data).
Fig. 2.
Synthetic scheme for the synthesis of C-terminal peptide aldehyde.
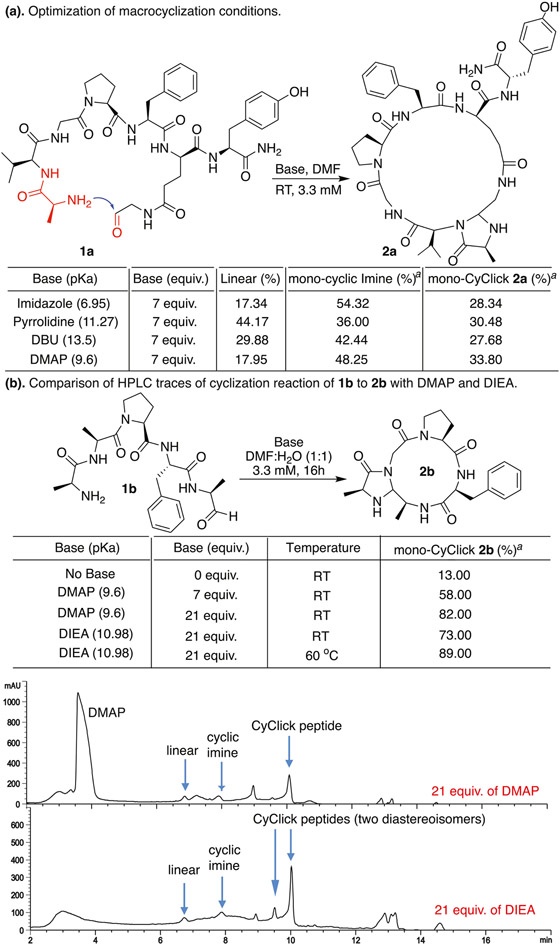
Fig. 3.
(a) Optimization of the cyclization by varying bases. aConversion (%) is determined by sodium cyanoborohydride reduction followed by HPLC. Reaction conditions: 3.3 mM of peptide 1a in DMF was treated with different bases and reaction was left stirring at room temperature for 16 h. (b) Comparison of the HPLC traces of the cyclization reaction with 21 equiv. of DMAP and DIEA. The use of volatile DIEA simplifies analysis of reaction mixtures.
We used bases with different pKa values [17,18] (DMAP(4-Dimethylaminopyridine), pKa ~9.6), (imidazole, pKa ~6.95), (pyrrolidine, pKa ~11.27) and (DBU (1,8-Diazabicyclo[5.4.0]undec-7-ene), pKa ~13.5) but did not observe any significant change in the amount of cyclization product (28–34%, Fig. 3a, Figure S2, Supplementary data). None of the bases led to the formation of any dimers or oligomers with a hexapeptide aldehyde VVGPFEY 1a, thus further confirming the exclusive intramolecular nature of CyClick Chemistry.
Monitoring the formation of cyclic imine by the reaction between the N-terminus and C-terminal aldehyde is problematic using LCMS because the masses of the monocyclic imine and 4-imidazolidinone monoCyClick product are identical (Figure S2, Supplementary data). Therefore, we distinguished the formation of CyClick cyclization product cyc-4-Imz-V(VGPFEY) 2a with 4-imidazolidinone moiety at the site of cyclization as compared to the cyclic imine by adding sodium cyanoborohydride in the reaction for 16 h followed by analysis using LCMS. No change in the mass of CyClick peptide cyc-4-Imz-V(VGPFEY) 2a was observed after incubation with sodium cyanoborohydride as compared to the to the cyclic imine which underwent reduction to generate reduced cyclic imine-product 2a′ that showed an increase in mass by +2. We analyzed all the reactions by HPLC and MS (Figure S2, Supplementary data).
From base studies, we discovered that DMAP (7 equiv.) at room temperature gave the highest conversion to monocyclic 4-imidazolidinone product cyc-4-Imz-V(VGPFEY) 2a (33.8%) and we moved to cyclizing head-to-tail linear peptide AGPFA 1b under optimized conditions. We also carried out the head-to-tail cyclization in the absence of base but a very small amount of the cyclic product was observed thus confirming the importance of base in this reaction (13%, Fig. 3b, Figure S3, Supplementary data). We observed (58%) conversion to head-to-tail monocyclic 4-imidazolidinone product cyc-4-Imz-A(GPFA) 2b under the reaction conditions (Figure S3, Supplementary data). To further increase the cyclization efficiency, we increased the amount of DMAP to 21 equiv. and observed a significant increase in the conversion to head-to-tail monocyclic product cyc-4-Imz-A(GPFA) 2b (82%). The use of excess DMAP raises an issue in the HPLC data analysis. The large DMAP peak in the HPLC chromatogram can overshadow peptide peaks, making purification and analysis challenging (Fig. 3b). To avoid this problem, we switched to a volatile base, diisopropylethylamine (DIEA, pKa ~10.98), for the cyclization so that it can be removed before the analysis of a reaction mixture by HPLC (Fig. 3b, Figure S3, Supplementary data). The cyclization reaction of AGPFA 1b with DIEA (21 equiv.) at 60 °C proceeded smoothly and generated monocyclic product cyc-4-Imz-A(GPFA) 2b (89%) without the formation of dimers, and DIEA was removed by rotary evaporator, resulting in a clean chromatogram for the reaction mixture (Fig. 3b). Based on the high cyclization efficiency and facile HPLC analysis, we chose to use DIEA, 60 °C for the synthesis of head-to-tail cyclic tetrapeptides. We confirmed the structure of monocyclic tetrapeptide cyc-4-Imz-A(GPFA) 2b by NMR spectroscopy after synthesizing it on a large scale (81% conversion) from linear pentapeptide aldehyde AGPFA 1b (Figure S4, Supplementary data). Next, we conducted variable temperature (VT) NMR studies on 2b to determine intramolecular hydrogen bonding in the cyclic product. Based on chemical shift temperature coefficients (ΔδNH/ΔT > −4.6 ppb/K) [19], both Ala and Gly amide NHs of 2b are involved in intramolecular hydrogen bonds (Figure S5, Supplementary data).
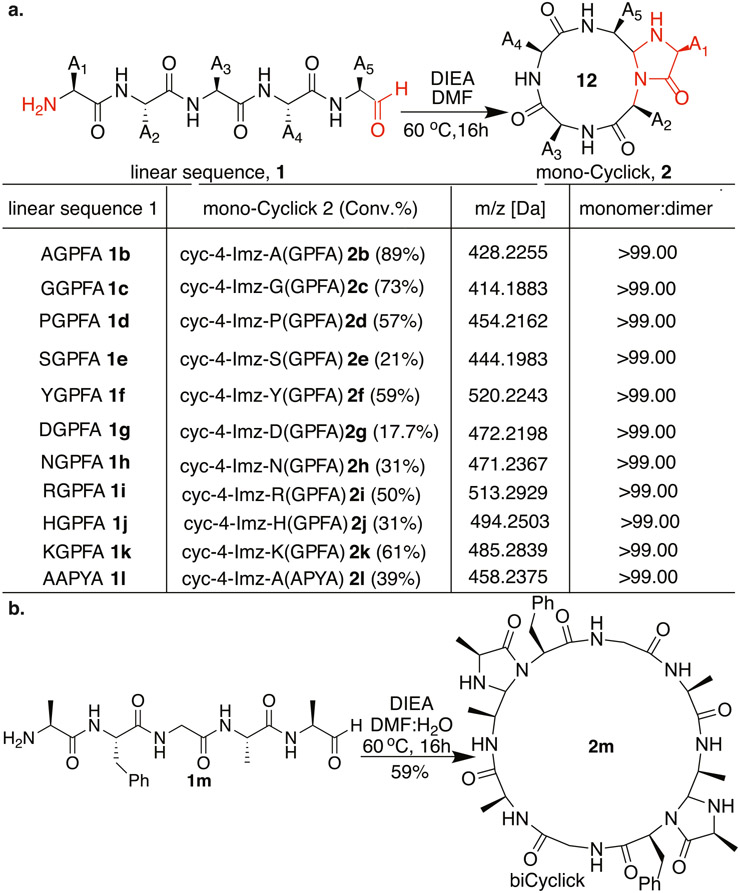
We further proceeded with cyclization of random linear pentapeptide aldehydes XGPFA 1b-1k with Ala at the C-terminus and varying amino acid residues at the N-terminus (X = A, G, P, S, Y, D, N, R, H, K) (Fig. 4a, Figure S6 and Table S1, Supplementary data). The peptide aldehydes XGPFA 1b-1k were dissolved in a DMF:water mixture (molar ratio 1:1) at the concentration of 3.3 mM along with DIEA under optimized conditions. Gratifyingly, all the peptide aldehydes XGPFA 1b-1k cyclized smoothly and generated monocyclic tetrapeptides cyc-4-Imz-X(GPFA) (2b-2k) within 16 h (Fig. 4a, Figure S6 and Table S1, Supplementary data). Based on previous cyclization methods for making cyclic tetrapeptides, we recognized that there could be a possibility of the formation of cyclic dimers due to the high strain associated with small ring sizes. We carefully analyzed all the reactions by LC–MS and the desired monocyclic tetrapeptide products were exclusively observed which is in contrast to the other cyclic tetrapeptide formation strategies leading to the formation of cyclodimers [7-9]. The unprotected amino acid residues, including tyrosine, serine, asparagine, and aspartic acid, did not cause any apparent side reactions during the cyclization, confirming the chemoselective nature of the CyClick Chemistry. Notably, we did not observe any side product with a peptide KGPFA 1k containing lysine at the N-terminus. In comparison, the conventional lactamization conditions (HATU, DIEA, DMF; DEPBT, DIEA, DMF; PyBOP, DIEA, DMF) have been reported to form linear dimers [20] along with modifications to the reactive side chains (Asp, Ser, Lys) of the amino acids [20]. Next, we changed amino acid residues in the middle of the chain and cyclized peptide AAPYA 11 with 39% conversion to the monocyclic product 21 without the formation of any linear dimers (Fig. 4a, Figure S6 and Table S1, Supplementary data). We also attempted to synthesize a cyclic tetrapeptide from linear pentapeptide aldehyde AFGAA 1m without a turn inducer. Mono-CyClick product was not detected, but formation of biCyClick dimers (59%) was confirmed by the reduction with sodium cyanoborohydride (Fig. 4b, Figure S7, Table S1, Supplementary data).
Fig. 4.
(a) Scope of peptide cyclization for the synthesis of strained head-to-tail cyclic tetrapeptides. Reaction conditions: 3.3 mM of peptides 1b-1l in H2O:DMF were treated with DIEA (21 equiv.) and reaction mixtures were left for stirring at 60 °C for 16 h and analysis by HPLC to determine the (%) conversion of cyclic tetrapeptides 2b-2l. (b) Novel biCyClick peptide structure 2m from linear pentapeptide 1m without the turn inducer.
Since the CyClick method works in an exclusively intramolecular fashion, we attempted head-to-tail cyclization of linear peptide AGPFA 1b at high concentrations (25 mM, 25 times of other methods reported in literature) [10] under the optimized conditions. The reaction resulted in the formation of monocyclic tetrapeptide cyc-4-Imz-A(GPFA) 2b with high conversion (89.5%) in 16 h with minor monoCyClick to cyclodimer ratio of >17:1. These results indicate that the CyClick strategy with a two-step process involving the formation of the cyclic imine with a pentapeptide (15 atoms, low energy barrier) followed by contraction of the ring by the amide backbone to generate highly strained monocyclic tetrapeptides (12 atoms) can compensate for the high energy barrier in synthesizing head-to-tail cyclic tetrapeptides by conventional approaches.
3. Conclusions
In conclusion, we demonstrated the applicability of exclusively intramolecular CyClick strategy in cyclizing small peptides using a volatile base for easy analysis by HPLC. We first synthesized linear pentapeptides containing C-terminal aldehydes with varying reactive amino acids at the N-terminus. We showed the formation of the desired monocyclic tetrapeptide products with all the peptides without the formation of any byproducts due to the reaction at the reactive side chains and any linear dimers or oligomers. We confirmed the formation of stable 4-imidazolidinone cyclic tetrapeptide by NMR analysis and carried out VT-NMR studies to determine the intramolecular H-bonding pattern in cyclic tetrapeptides. We cyclized linear peptides at high concentrations and generated desired cyclic tetrapeptides in high conversion, which is in contrast to the traditional strategies. The results from these studies concluded that the high activation energy barrier associated with the head-to-tail cyclization of small tetrapeptides can be overcome by cyclic imine capture (15 atoms, less activation barrier) followed by chemoselective ring contraction due to intramolecular amide backbone addition to generate strained head-to-tail cyclic tetrapeptide (12 atoms). Such chemoselective ring contraction is not possible with linear imine due to the high-energy barrier thus our CyClick method provides a novel approach for generating small cyclic tetrapeptides without the formation of any dimers or oligomers. In our laboratory, we are exploring these small cyclic tetrapeptides as inhibitors for targeting protein-protein interactions responsible for diseases such as MDM2-P53 and nonstructural proteins (nsps) responsible for SARS-Cov2 infections.
4. Experimental
The materials, compound characterizations, and additional data are described in detail in Appendix A: supporting data. All environmental and safety precautions were followed according to the material safety data sheets of the chemicals utilized in this work.
4.1. General
All commercial materials (Sigma-Aldrich, Fluka, and Nova-biochem) were used without further purification. All solvents were reagent or HPLC (Fisher) grade. All reactions were performed under air in glass vials. Conversions refer to chromatographically pure compounds; % conversions were obtained by comparison of HPLC peak areas of products and starting materials. HPLC was used to monitor reaction progress.
4.2. Analytical methods
HPLC:
Peptide compositions were evaluated by high performance liquid chromatography (HPLC) on an Agilent 1100/1200 series HPLC equipped with a 5.0 mm C-18 reversed-phase column. All separations used mobile phases of 0.1% formic acid (v/v) in water (solvent A) and 0.1% formic acid (v/v) in acetonitrile (solvent B). A linear gradient of 0–80% solvent B in 30 min at room temperature with a flow rate of 1.0 mL min−1 was used. The eluent was monitored by UV absorbance at 220 nm unless otherwise noted.
LC-MS:
Mass spectrometry to check reaction mixtures was performed using an Agilent 1100 Series HPLC with MSD VL mass spectrometer using positive polarity electrospray ionization (+ESI).
HRMS:
High resolution MS data were acquired on Thermo Exactive Plus using a heated electrospray source. The solution was infused at a rate of 10–25 μL/min/Electrospray using 3.3 KV. The typical settings were Capillary temp 320 °C. S-lens RF level was between 30 and 80 with an AGC setting of 1 E−6. The maximum injection time was set to 50 ms. Spectra were taken at 140,000 resolution at m/z 200 using Tune software and analyze with Thermo's Freestyle software.
4.3. Fmoc solid-phase peptide synthesis (Fmoc-SPPS)
Peptides were synthesized manually on a 0.25 mM scale using Rink amide resin. Resin was swollen with DCM for 1 h at room temperature. Fmoc was deprotected using 20% piperidine–DMF for 5 min to obtain a deprotected peptide-resin. The first Fmoc-protected amino acid (1.25 mmol/5 equiv.) was coupled using HOAt (1.25 mm/5 equiv.) and DIC (1.25 mmol/5 equiv.) in DMF (final conc. 0.05 M) for 15 min at room temperature. Fmoc-protected amino acids (0.75 mmol/3 equiv.) were sequentially coupled on the resin using HBTU (0.75 mmol/3 equiv.) and DIEA (1.5 mm/6 equiv.) in DMF (final conc. 0.05 M) for 5 min at room temperature. Peptides were synthesized using standard protocols. Any Fmoc-protected amino acid added after Fmoc-proline was subjected to the conditions of the first amino acid coupling. Peptides were cleaved from the resin using a cocktail of 95:5, trifluoroacetic acid: water for 2 h. The resin was removed by filtration and the resulting solution was concentrated. The residue was diluted with ACN/water mixture. The resulting solution was purified by semi-preparative chromatography.
4.4. Procedure for synthesis of C-terminal peptide aldehydes
Fmoc-Gly-OH and Fmoc-Thr-OH were coupled with the general peptide synthesis procedure to swollen Rink resin (0.5 mmol/g). Following Fmoc-deprotection, the resin (500 mg) was added to a solution of Fmoc-AA-CHO (alanine aldehyde) (1 mmol/4 equiv.) in 1% DIEA v/v in MeOH (2.5 mL/final conc. 0.1 M) and rocked for 5 h at 60 °C. The resin was washed with MeOH (5 × 3 mL), DMF (5 × 3 mL), DCM (5 × 3 mL), and THF (5 × 3 mL). The resin was rocked for 5 h at 50 °C in a solution of Boc anhydride BOC2O (1.25 mmol/5 equiv.) and NMM (1.25 mmol/5 equiv.) in THF (2.5 mL/final conc. 0.1 M). The resin was washed with THF (5 x 3 mL), DCM (5 × 3 mL), and DMF (5 x 3 mL) followed by coupling of the rest of Fmoc-amino acid residues using normal Fmoc SPPS. After Fmoc-SPPS, the resulting resin was washed three times with DMF, DCM and MeOH. This was followed by the cleavage of the C-terminal peptide aldehyde from solid support using cleavage cocktail TFA:H2O (95:5) for 2 h at room temperature. The peptide aldehyde was analyzed by LCMS and purified by HPLC followed by lypholization to obtain pure white solid peptide aldehyde.
4.5. Procedure for macrocyclization in solution
Lyophilized peptide aldehyde (2 mg) was mixed with 21 equiv. DIEA in a 1:1 DMF/H2O solution (final conc. 3 mM). The reaction was shaken at room temperature for 16 h. The product was analyzed with HPLC, NMR, and MS. The presence of [M+2H]2+ of a cyclic dimer will give the same nominal mass as the [M+H]+ for the monocyclic compound; however, the doubly-charged dimer will have a different isotopic pattern with 0.5 Da separation compared to the monocyclic compound, making the two compounds discernible.
4.6. Procedure for base studies
To a lyophilized peptide aldehyde VVGPFEY (2 mg) 1a in 1:1 DMF/H2O solution (final conc. 3 mM), different bases such as DMAP, imidazole, pyrrolidine, or DBU were added (7 equiv.) and the reactions were shaken at room temperature for 16 h. To differentiate the mono-cyclic imine from mono-CyClick product, sodium cyanoborohydride (10 equiv.) was added to the reaction mixtures and reactions were shaken at room temperature for another 16 h. The reactions were analyzed with HPLC and MS.
4.7. Procedure for base optimization
To a lyophilized peptide aldehyde 1b (1 mg) in 1:1 DMF/H2O solution (final conc. 3 mM), DIEA or DMAP at different equivalents (7–21 equiv.) were added and the reactions were shaken at different temperatures (room temperature or 60 °C) for 16 h. The reactions were analyzed with HPLC and MS.
4.8. Procedure for cyclization at high concentration
Lyophilized peptide aldehyde AGPFA 1b (4 mg) was solubilized in a 1:1 DMF/H2O (362 μL) solution to make a concentration of 25 mM, DIEA (21 equiv.) was added, and the reaction was shaken at 60 °C for 16 h. The product was analyzed with HPLC and MS. The reaction proceeded smoothly with conversion of 89.5% to mono-CyClick product with cyclodimer ratio of >17:1.
Supplementary Material
Acknowledgements
This work was supported by grants (Grant No. CHE-1752654 and CHE-2108774) from National Science Foundation (NSF). Also, this work was supported by a grant (1R35GM133719-01) from the National Institute of Health (NIH) and Winship Pilot Grant by Emory.
Footnotes
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Monika Raj reports was provided by Emory University. Monika Raj reports a relationship with Emory University that includes:. Monika Raj has patent #17/502,506 issued to Monika Raj. N/A
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tet.2022.133071.
Data availability
Data will be made available on request.
References
- [1].Chiba H, Agematu H, Dobashi K, Yoshioka T, J. Antibiot 52 (1999) 700–709. [DOI] [PubMed] [Google Scholar]
- [2].Aldrich JA, Senadheera SN, Ross NC, Ganno ML, Eans SO, McLaughlin JP, J. Nat. Prod 76 (2013) 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ong YS, Gao L, Kalesh KA, Yu Z, Wang J, Liu C, Li Y, Sun H, Lee SS, Curr. Top. Med. Chem 17 (2017) 2302–2318. [DOI] [PubMed] [Google Scholar]
- [4].Zhang H, Chen S, RSC Chem. Biol 3 (2022) 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jing X, Jin K, Med. Res. Rev 40 (2020) 753–810. [DOI] [PubMed] [Google Scholar]
- [6].Buckton LK, Rahimi MN, McAlpine SR, Chem. Eur J 27 (2020) 1487–1513. [DOI] [PubMed] [Google Scholar]
- [7].Sarojini V, Cameron AJ, Varnava KG, Denny WA, Sanjayan G, Chem. Rev 119 (2019) 10318–10359. [DOI] [PubMed] [Google Scholar]
- [8].Meutermans WDF, Bourne GT, Golding SW, Horton DA, Campitelli MR, Craik D, Scanlon M, Smythe ML, Org. Lett 5 (2003) 2711–2714. [DOI] [PubMed] [Google Scholar]
- [9].El Haddadi M, Cavelier F, Vives E, Azmani A, Verducci J, Martinez J, J. Pept. Sci 6 (2000) 560–570. [DOI] [PubMed] [Google Scholar]
- [10].Wong CTT, Lam HY, Song T, Chen G, Li X, Angew. Chem. Int. Ed 52 (2013) 10212–10215. [DOI] [PubMed] [Google Scholar]
- [11].Fairweather KA, Sayyadi N, Luck IJ, Clegg JK, Jolliffe KA, Org. Lett 12 (2010) 3136–3139. [DOI] [PubMed] [Google Scholar]
- [12].Skropeta D, Jolliffe KA, Turner P, J. Org. Chem 69 (2004) 8804–8809. [DOI] [PubMed] [Google Scholar]
- [13].Adebomi V, Cohen RD, Wills R, Chavers HAH, Martin GE, Raj M, Angew. Chem. Int. Ed 58 (2019) 19073–19080. [DOI] [PubMed] [Google Scholar]
- [14].Wills R, Adebomi V, Raj M, Chembiochem 22 (2021) 52–62. [DOI] [PubMed] [Google Scholar]
- [15].Wills R, Adebomi V, Raj M, Synlett 31 (2020) 1537–1542. [Google Scholar]
- [16].Moulin A, Martinez J, Fehrentz J-A, J. Pept. Sci 13 (2007) 1–15. [DOI] [PubMed] [Google Scholar]
- [17].Williams R, pKa Data Compiled by R. Williams, 2011. http://www.chem.wisc.edu/areas/organic/index-chem.htm. [Google Scholar]
- [18].Kaupmees K, Trummel A, Leito I, Croat. Chem. Acta 87 (2014) 385–395. [Google Scholar]
- [19].Cierpicki T, Otlewski J, Biomol J. NMR 21 (2001) 249–261. [DOI] [PubMed] [Google Scholar]
- [20].Bechtler C, Lamers C, RSC Med. Chem 12 (2021) 1325–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.