Abstract
A trehalose synthase (TSase) that catalyzes the synthesis of trehalose from d-glucose and α-d-glucose 1-phosphate (α-d-glucose 1-P) was detected in a basidiomycete, Grifola frondosa. TSase was purified 106-fold to homogeneity with 36% recovery by ammonium sulfate precipitation and several steps of column chromatography. The native enzyme appears to be a dimer since it has apparent molecular masses of 120 kDa, as determined by gel filtration column chromatography, and 60 kDa, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Although TSase catalyzed the phosphorolysis of trehalose to d-glucose and α-d-glucose 1-P, in addition to the synthesis of trehalose from the two substrates, the TSase equilibrium strongly favors trehalose synthesis. The optimum temperatures for phosphorolysis and synthesis of trehalose were 32.5 to 35°C and 35 to 37.5°C, respectively. The optimum pHs for these reactions were 6.5 and 6.5 to 6.8, respectively. The substrate specificity of TSase was very strict: among eight disaccharides examined, only trehalose was phosphorolyzed, and only α-d-glucose 1-P served as a donor substrate with d-glucose as the acceptor in trehalose synthesis. Two efficient enzymatic systems for the synthesis of trehalose from sucrose were identified. In system I, the α-d-glucose 1-P liberated by 1.05 U of sucrose phosphorylase was linked with d-glucose by 1.05 U of TSase, generating trehalose at the initial synthesis rate of 18 mmol/h in a final yield of 90 mol% under optimum conditions (300 mM each sucrose and glucose, 20 mM inorganic phosphate, 37.5°C, and pH 6.5). In system II, we added 1.05 U of glucose isomerase and 20 mM MgSO4 to the reaction mixture of system I to convert fructose, a by-product of the sucrose phosphorylase reaction, into glucose. This system generated trehalose at the synthesis rate of 4.5 mmol/h in the same final yield.
Trehalose (1-α-d-glucopyranosyl-α-d-glucopyranoside) is a nonreducing disaccharide with an α,α-1,1 glycosidic linkage and is widely distributed in plants, insects, fungi, yeast, and bacteria (7). Due to the absence of reducing ends in trehalose, it is highly resistant to heat, pH, and Maillard’s reaction (24). In trehalose-producing organisms, this compound may serve as an energy reserve, a buffer against stresses such as desiccation and freezing, and a protein stabilizer (5, 7, 26, 31, 32). If trehalose can be produced economically, then it has potential commercial applications as a sweetener, a food stabilizer, and an additive in cosmetics and pharmaceuticals (6, 25). Recently, trehalose production through fermentation of yeast (17) and Corynebacterium (30), enzymatic processes from starch (18, 34) and maltose (19, 22, 23, 33), and extraction from transformed plants (10) has been reported.
Our approach to trehalose production is to use an enzymatic process to produce trehalose from sucrose, one of the least expensive sugars. Since sucrose is efficiently converted to α-d-glucose 1-phosphate (α-d-glucose 1-P) and fructose by sucrose phosphorylase (SPase), we screened microorganisms for an enzyme that converts α-d-glucose 1-P to trehalose on the assumption that the combination of the putative trehalose synthase (TSase) and SPase would convert sucrose into trehalose. Although similar enzyme activities have been reported in the basidiomycete Flammulina velutipes (11) and in the yeast Pichia fermentans (27), these enzymes have not been well characterized.
Our objectives were (i) to screen microorganisms, primarily fungi, for TSase activity; (ii) to purify and characterize the TSase; (iii) to identify the enzymatic process by which trehalose is produced from sucrose; and (iv) to identify an enzymatic process for production of trehalose from sucrose in which the fructose component is also converted to trehalose.
MATERIALS AND METHODS
Materials.
α-d-Glucose 1,6-bisphosphate (glucose 1,6-P), phosphoglucomutase (PGM; from rabbit muscle) (EC 5.4.2.2), and glucose-6-phosphate 1-dehydrogenase (G6PDH; from yeast) (EC 1.1.1.49) were purchased from Boehringer Mannheim Co., Ltd. (Tokyo, Japan). SPase (from Leuconostoc mesenteroides) (EC 2.4.1.7) was purchased from Kikkoman Co. Ltd. (Tokyo, Japan). Glucose isomerase (GIase; from Streptomyces phaeochromogenes) (EC 5.3.1.18) was purified from a commercially available immobilized enzyme solution (Nagase Biochemical, Osaka, Japan) by DEAE column chromatography and gel filtration chromatography on a high-performance liquid chromatograph to remove glucose 1-P-degrading activity and glucose oxidase-like activity. β-NADP+, trehalose, and other sugars were obtained from Wako Pure Chemicals (Tokyo, Japan).
Microorganisms and culture conditions.
Microorganisms were obtained mainly from the Institute of Fermentation, Osaka, Japan (IFO), and the Institute of Applied Microbiology. Some of the microorganisms tested were obtained from a culture collection in the Nishiki Research Laboratory. Basidiomycetes were grown at 26°C for 7 to 10 days in GYM medium (pH 5.5), which contained the following, in grams per liter: yeast extract (Difco, Detroit, Mich.), 7.5; malt extract (Difco), 2; KH2PO4, 5; MgSO4 · 7H2O, 0.5; and glucose or trehalose, 40. Other fungi were grown at 28°C for 3 to 5 days in GYMB medium (pH 6.2), which contained the following, in grams per liter: yeast extract, 6; malt extract, 6; Bacto Peptone (Difco), 10; glucose, 10; and trehalose, 20. Yeasts were grown in YPT medium (pH 6.0), which contained the following, in grams per liter: trehalose, 20; yeast extract, 10; and Bacto Peptone, 20.
The G. frondosa strain that we studied has been deposited with the National Institute of Bioscience and Human Technology, Agency of Industrial Science and Technology, Tsukuba, Japan, under accession no. FERM BP-35.
Enzyme assay.
The mycelia of fungi and basidiomycetes were collected, disrupted with an HG-30 homogenizer (Hitachi Co., Ltd., Tokyo, Japan) and centrifuged to give a supernatant for the trehalose phosphorolysis assay. For the trehalose synthesis assay, the crude sample was applied to a 20-ml DEAE-Toyopearl 650C column (Tosoh Co. Ltd., Tokyo, Japan) equilibrated with 20 mM potassium phosphate buffer (pH 7.0) and proteins were eluted with a linear gradient of 20 to 500 mM KCl in a total volume of 200 ml. Fractions showing trehalose phosphorolysis activity were collected and concentrated with a hollow fiber ultrafiltration apparatus (Minimodule nM-3; Asahi Kasei Co. Ltd., Tokyo, Japan) to give a concentrated crude enzyme sample. For preparation of crude enzyme samples from yeasts, a similar procedure was used except that cells were disrupted with a high-speed shaking-disrupting apparatus (Michael Co. Ltd.).
TSase was routinely measured as trehalose phosphorolysis activity (trehalose + inorganic phosphate [Pi]→d-glucose + α-d-glucose 1-P). The amount of the α-d-glucose 1-P produced was determined by coupling the TSase activity with PGM (α-d-glucose 1-P→α-d-glucose 6-P) and G6PDH (α-d-glucose 6-P + NADP+→6-phospho-d-gluconolactone + NADPH + H+). NADPH generation was measured by the change in absorbance at 340 nm and was assumed to be proportional to TSase activity. The reaction mixture contained 200 mM trehalose, 40 mM potassium phosphate buffer (pH 7.0), 10 mM glutathione, 16 μM EDTA, 1 mM NADP+, 1.3 mM MgCl2 · 6H2O, 67 μM glucose 1,6-P, 1.55 U of PGM per ml, 1.75 U of G6PDH per ml, and an enzyme sample, in a total volume of 2 ml. During incubation at 30°C, the change in absorbance at 340 nm was monitored with an automated spectrophotometer (Shimadzu model UV-2200). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of α-glucose 1-P per min, and specific activity was expressed as the number of units of enzyme per milligram of protein. Protein concentrations were measured with a protein assay kit (Bio-Rad Laboratories, Tokyo, Japan) using bovine serum albumin as the standard.
TSase was also assayed as trehalose synthesis from glucose and α-glucose 1-P. The reaction mixture contained 100 mM α-glucose 1-P, 100 mM α-d-glucose 1-P, 100 mM HEPES buffer (pH 7.0), and an enzyme sample, in a total volume of 250 μl. Reaction mixtures were incubated at 35°C for 3 h and stopped by boiling for 5 min. The trehalose produced was measured by high-performance liquid chromatography (HPLC) on a Shimadzu LC-10A system equipped with a YMC-pack polyamine II column (4.6 by 250 mm) and a refraction index detector (Shimadzu RID-6A). The solvent system was 70% acetonitrile–30% H2O with a flow rate of 1.0 ml/min.
Gel electrophoresis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (13) was done with the Pharmacia Phast system. After electrophoresis, proteins were stained with the Phast gel protein silver stain kit. For measurement of the molecular mass of TSase, the following standard proteins were used; phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase B (30 kDa), soybean trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa).
Purification of TSase from G. frondosa.
TSase was monitored by assaying trehalose phosphorolysis activity. All the operations were done in a cold room (4°C) unless otherwise specified.
(i) Step 1. Preparation of cell extract.
Fruiting bodies of G. frondosa (wet weight, 611 g) were suspended in 1.2 liters of cold 20 mM potassium phosphate buffer (pH 7.0) containing 20% (vol/vol) glycerol, 1 mM EDTA, and 1 mM dithiothreitol and disrupted with a Waring blender. Cell debris was removed by centrifugation at 12,000 × g for 20 min, and the supernatant was used as the cell extract.
(ii) Step 2. Ammonium sulfate fractionation.
Solid ammonium sulfate was added to the cell extract to give a concentration of 40% (wt/vol), and the mixture was left standing overnight. The next morning, the mixture was centrifuged to remove insoluble material, yielding 1,560 ml of the enzyme solution.
(iii) Step 3. Butyl-Toyopearl column chromatography.
We applied the enzyme solution to a butyl-Toyopearl 650C column (2.4 by 24 cm; Tosoh) equilibrated with 20 mM potassium phosphate buffer (pH 7.0) containing 40% ammonium sulfate. The adsorbed enzyme was eluted with a linear gradient of 40 to 0% ammonium sulfate in the same buffer in a total volume of 800 ml. The enzyme activity was eluted at approximately 10% ammonium sulfate. The active fractions (250 ml) were pooled and dialyzed against two changes of 5 liters of 20 mM potassium phosphate buffer (pH 7.0).
(iv) Step 4. SuperQ-Toyopearl column chromatography.
The dialyzed solution was applied to a SuperQ-Toyopearl 650M column (2.5 by 17 cm; Tosoh) equilibrated with 20 mM potassium phosphate buffer (pH 7.0). The column was washed and proteins were eluted with a linear gradient of 0 to 0.2 M KCl in the same buffer in a total volume of 600 ml. Active fractions (160 ml) that were eluted at approximately 0.1 M KCl were combined and dialyzed against 5 liters of 20 mM potassium phosphate buffer (pH 6.0). The pH was adjusted for the next column chromatography step.
(v) Step 5. AF-Blue-Toyopearl column chromatography.
The dialyzed solution was applied to an Affinity (AF)-Blue-Toyopearl 650ML column (1.0 by 17 cm; Tosoh) equilibrated with 20 mM potassium phosphate buffer (pH 6.0). After the column had been washed, proteins were eluted with a linear gradient of 0 to 2 M KCl in the same buffer in a total volume of 400 ml. TSase activity was eluted at approximately 0.8 M KCl. The active fractions (220 ml) were combined and dialyzed against 5 liters of 20 mM potassium phosphate buffer (pH 6.0).
(vi) Step 6. AF-Red-Toyopearl column chromatography.
An AF-Red-Toyopearl 650ML column (1.0 by 17 cm; Tosoh) was equilibrated with 20 mM potassium phosphate buffer (pH 7.0). TSase was eluted with a linear gradient (0 to 2 M) of KCl in the same buffer in a total volume of 400 ml. TSase activity (210 ml) that was eluted at 0.8 M KCl was pooled and dialyzed against 5 liters of 20 mM potassium phosphate buffer (pH 7.0).
(vii) Step 7. Rechromatography on a SuperQ-Toyopearl column.
TSase was further purified by rechromatography on a SuperQ-Toyopearl 650M column (0.7 by 4.5 cm). The buffer was the same as that used in step 4. TSase was eluted with a linear gradient of 0 to 100 mM KCl in a total volume of 400 ml. The active fractions (170 ml) that were eluted at approximately 30 mM KCl were combined and dialyzed against the same buffer. In step 4, the enzyme was eluted at 100 mM KCl in a steeper linear gradient, which was probably higher than the true concentration.
(viii) Step 8. TSK gel G3000SW column chromatography.
The dialyzed solution was applied to a TSK gel G3000SW column (0.75 by 30 cm) equilibrated with 20 mM potassium phosphate buffer (pH 7.0) in an HPLC system. Proteins were eluted with the same buffer at a constant flow rate of 0.5 ml/min. TSase was detected at 16 min, giving a single sharp peak.
Characterization of TPase.
For determination of optimal pH, pH stability, heat stability, and substrate specificity of TSase, the above-described reaction and assay conditions for trehalose phosphorolysis and synthesis were used, except where otherwise specified. The following buffer systems were used: acetate-Na buffer (pH 4.0–5.5), morpholineethanesulfonic acid (MES) (pH 5.5 to 7.0), HEPES (pH 7.0 to 8.0), Tris-HCl (pH 7.5 to 9.0), and glycine-NaOH (pH 8.5 to 12.0).
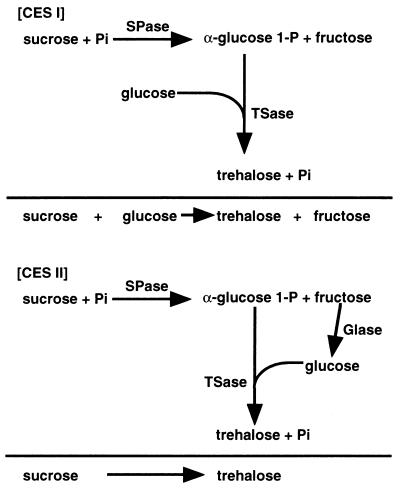
CES I.
SPase liberates α-d-glucose 1-P and fructose from sucrose. In coupled enzyme system I (CES I), the α-d-glucose 1-P liberated from sucrose by the action of SPase was linked with glucose to produce trehalose by the trehalose synthesis activity of TSase. The standard reaction mixture contained 300 mM sucrose, 300 mM glucose, 10 mM Pi, 1.05 U of SPase per ml, and 1.05 U of TSase per ml in 100 mM MES buffer (pH 6.5). For maximal production of trehalose, the mixture was incubated at various temperatures and pHs.
CES II.
Coupled enzyme system II (CES II) was designed to convert the fructose liberated from sucrose by SPase in CES I to glucose by the action of GIase, which was then linked with α-d-glucose 1-P to yield trehalose. The reaction mixture for CES II consisted of 300 mM sucrose, 20 mM Pi, 20 mM MgSO4, and 1.05 U of TSase, SPase, and GIase each per ml in 100 mM HEPES (pH 7.0). The amounts of trehalose, sucrose, glucose, and fructose were determined by HPLC, and the amount of α-d-glucose 1-P was determined by the enzyme assay described above.
RESULTS
Screening of microorganisms.
We collected ascomycetes, basidiomycetes, zygomycetes, and imperfect (mitosporic) fungi from various culture collections to determine if they produced TSase. Among 132 strains tested, most strains had at least some trehalose phosphorolysis activity. We then examined trehalose phosphorolysis-positive strains to determine if they also could synthesize trehalose from α-d-glucose 1-P and d-glucose (Table 1). Among these strains, G. frondosa CM 236 had the highest activity, although the phosphorolysis activity of this strain was not very high. This strain had the same activity in both glucose- and trehalose-containing media, suggesting that TSase was produced constitutively.
TABLE 1.
Fungi that showed trehalose phosphorylating activity
| Fungusa | Sp act (U)b |
|---|---|
| Ascomycota | |
| Byssochlamys nivea IFO 30569 | 0.26 |
| Chaetomium thermophilum IFO 30072 | 0.016 |
| Chaetomium thermophilum var. coprophilum IFO 9679 | 0.013 |
| Glomerella cingulata IFO 7478 | 0.018 |
| Rosellinia necatrix IFO 5954 | 0.10 |
| Thermoascus aurantiacus IFO 31693 | 0.015 |
| Thielavia arenaria IFO 31061 | 0.026 |
| Thielavia terrestris IFO 9732 | 0.019 |
| Basidiomycota | |
| Agaricus bisporus IFO 30774 | 0.13 |
| Coriolus consors ATCC 20565 | 0.22 |
| Grifola frondosa CM 236 | 0.02 |
| Lenzites betulina CM 207 | 0.35 |
| Lyophyllum ulmarium IFO 9637 | 0.21 |
| Pholiota adiposa IFO 9779 | 0.0017 |
| Pleurotus cornucopiae IFO 30528 | 0.02 |
| Pleurotus ostreatus CM 566 | 0.01 |
| Rhodophyllus clypeatus CM 809 | 0.024 |
| Schizophyllum commune CM 556 | 0.091 |
| Sporidiobolus johnsonii IFO 6903 | 0.09 |
| Tyromyces palustris IFO 30339 | 0.03 |
| Zygomycota | |
| Rhizomucor miehei IFO 9740 | 0.30 |
| Rhizomucor pusillus IFO 4579 | 0.043 |
| Rhizopus azygosporus IFO 31989 | 0.021 |
| Rhizopus chinensis IFO 4768 | 0.025 |
| Rhizopus chinensis var. liquefaciens IFO 4737 | 0.015 |
| Rhizopus microsporus IFO 31988 | 0.029 |
| Mitosporic fungi | |
| Acremonium alabamense IFO 32241 | 0.014 |
| Cercospora beticola IFO 7398 | 0.010 |
| Humicola grisea var. thermoidea IFO 9854 | 0.045 |
| Myceliophthora thermophila IFO 31843 | 0.001 |
| Sterigmatomyces halophilus IFO 1488 | 0.02 |
a Fungi are classified in accordance with Ainsworth & Bisby’s Dictionary of the Fungi (9).
b Trehalose phosphorolysis activity expressed in the presence of trehalose.
Purification of TSase from G. frondosa.
We purified TSase 106-fold with 36% recovery (Table 2). The purified TSase gave a single protein band with an apparent molecular mass of 60 kDa on SDS-polyacrylamide gel electrophoresis (Fig. 1). The specific activity of the purified enzyme was 4.2 U.
TABLE 2.
Purification of trehalose synthase from G. frondosa
| Step | Protein (mg) | Total activity (U) | Sp act (U) | Yield (%) | Purifi-cation (fold) |
|---|---|---|---|---|---|
| Crude extract | 8,100 | 320 | 0.04 | 100 | 1 |
| (NH4)2SO4 (0–40%) | 6,400 | 270 | 0.04 | 85 | 1 |
| Butyl-Toyopearl | 1,100 | 200 | 0.19 | 63 | 5 |
| SuperQ-Toyopearl | 190 | 200 | 1.0 | 61 | 26 |
| AF-Blue-Toyopearl | 68 | 170 | 2.5 | 53 | 62 |
| AF-Red-Toyopearl | 49 | 160 | 3.2 | 49 | 80 |
| SuperQ-Toyopearl | 31 | 140 | 4.4 | 43 | 111 |
| HPLC | 27 | 110 | 4.2 | 36 | 106 |
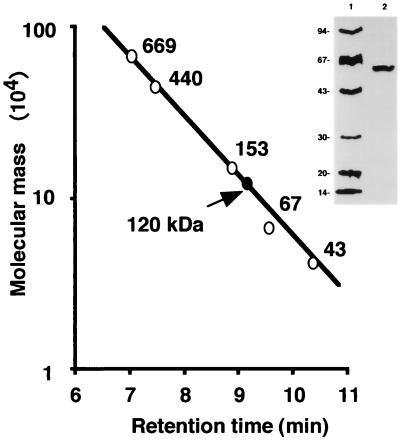
FIG. 1.
SDS-polyacrylamide gel electrophoretic analysis of TSase from G. frondosa and molecular mass determination of native TSase by gel filtration. The final TSase sample (lane 2) was electrophoresed together with the standard proteins (lane 1). The molecular mass of the native TSase was calibrated by TSK gel G3000SW gel filtration in an HPLC system. The reference proteins were thyroglobulin (669 kDa), ferritin (440 kDa), glucose oxidase (153 kDa), albumin (67 kDa), and ovalbumin (43 kDa). TSase eluted at a position corresponding to 120 kDa.
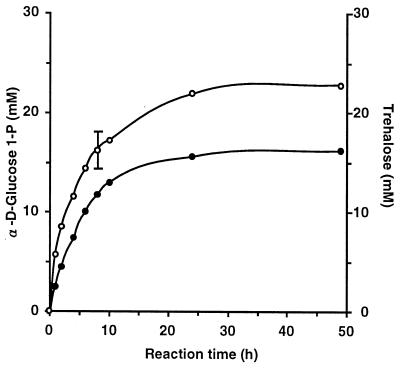
To determine if the purified TSase could be used for enzymatic production of trehalose, we monitored trehalose synthesis and phosphorylation. The reactions were carried out under almost optimum conditions (see below). After 48 h of incubation at 30°C and pH 7.0, the reaction mixture containing 0.2 U of TSase per ml and a 0.1 M concentration each of trehalose and Pi produced 23 mM α-d-glucose 1-P (23% conversion; equilibrium constant, i.e., [α-d-glucose 1-P][glucose]/[trehalose][Pi], 0.085) (Fig. 2). When a reaction mixture containing 0.2 U of TSase per ml and a 25 mM concentration each of α-d-glucose 1-P and glucose was incubated at 35°C and pH 7.0 for 48 h, 16 mM trehalose (65% conversion; equilibrium constant, i.e., [trehalose][Pi]/[α-d-glucose 1-P][glucose], 3.5) was produced (Fig. 2). We concluded that the equilibrium of TSase under these reaction conditions was toward the synthesis of trehalose.
FIG. 2.
Time course of phosphorolysis of trehalose into α-d-glucose 1-P and d-glucose and synthesis of trehalose from the two substrates. For trehalose phosphorolysis, the reaction mixture, which contained 0.1 M trehalose, 0.1 M Pi, and 0.2 U of TSase per ml in 100 mM potassium phosphate buffer (pH 7.0), was incubated at 30°C and the amounts of the α-d-glucose 1-P (○) produced were measured by the enzymatic method. For trehalose synthesis, the reaction mixture, which contained 25 mM α-d-glucose 1-P, 25 mM d-glucose, and 0.2 U of TSase per ml in 100 mM HEPES buffer (pH 7.0), was incubated at 35°C for various periods and the amount of trehalose (•) was measured. An error bar is included for the point for which the standard deviation was greater than 10% of the point’s value.
Characterization of TSase.
The following enzymatic properties of TSase were determined with the purified enzyme: (i) molecular mass; (ii) optimum temperature and heat stability; (iii) optimum pH and pH stability; (iv) effects of cations on TSase activity; and (v) substrate specificity.
(i) Molecular mass.
The relative molecular mass of TSase was 60 kDa by SDS-polyacrylamide gel electrophoresis and that of the native enzyme was 120 kDa by HPLC gel filtration (Fig. 1), suggesting that TSase is composed of two identical 60-kDa subunits. We did not study dilution dissociation of TSase any further.
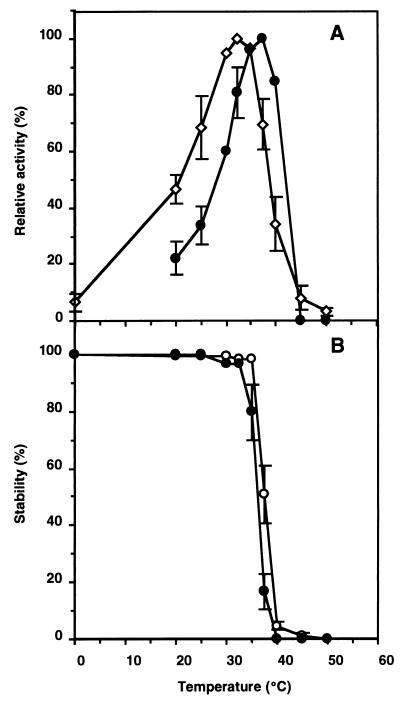
(ii) Optimum temperature and heat stability.
Trehalose phosphorylation and synthesis activities measured as a function of temperature from 0 to 50°C showed that the former and latter activities were highest at 32.5 and 37.5°C, respectively (Fig. 3A). When these reaction mixtures were preincubated for 30 min at various temperatures, the trehalose phosphorolysis activity was stable up to 35°C but decreased rapidly at temperatures above that (Fig. 3B). The trehalose synthesis activity was stable up to 32.5°C (Fig. 3B).
FIG. 3.
Effects of temperature on trehalose phosphorolysis and synthesis of TSase. Error bars are included for the points for which the standard deviation was greater than 10% of the point’s value. (A) For determination of the optimum temperatures, the trehalose phosphorolysis (○) and synthesis (•) activities at the indicated temperatures were determined. The TSase activity for phosphorolysis at the optimal temperature was 0.011 U and TSase synthesized trehalose at 5.03 μmol/h at the optimal temperature. (B) For determination of heat resistance, the remaining phosphorolysis (○) and synthesis (•) activities were measured after the enzyme had been placed at the indicated temperatures for 30 min. The remaining synthesis activity was also measured at 35°C under the standard reaction conditions after the same treatment. A value of 0.01 U is equivalent to 100% activity for phosphorolysis, and a value of 2.99 μmol of trehalose per h is equivalent to 100% activity for synthesis.
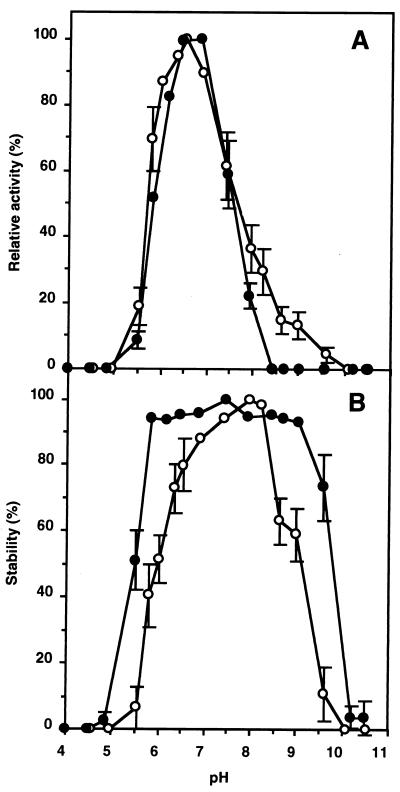
(iii) Optimum pH and pH stability.
The trehalose phosphorylation and synthesis activities of TSase were measured at various pHs in buffers that had the same ionic concentrations. The phosphorolysis and synthesis activities were the highest at pH 6.5 and pH 6.5 to 6.8, respectively (Fig. 4A). After a 24-h incubation of the enzyme solution in buffer, the phosphorylation activity was highest at pH 8.3 (Fig. 4B), and the synthesis activity was highest between pH 5.5 and 9.0 (Fig. 4B).
FIG. 4.
Effects of pH on trehalose phosphorolysis and synthesis of TSase. Error bars are included for the points for which the standard deviation was greater than 10% of the point’s value. (A) For determination of optimum pHs, the trehalose phosphorolysis (○) and synthesis (•) activities at the indicated pHs were measured. The TSase activity at the optimal pH for phosphorolysis was 0.011 U, and TSase synthesized trehalose at 5.39 μmol/h at the optimal pH. (B) For determination of pH resistance, the remaining phosphorolysis (○) and synthesis (•) activities were measured at pH 6.0 after the enzyme had been placed at the indicated pHs at 4°C for 24 h. The remaining synthesis activity was measured at pH 7.0 after the same treatment. A value of 0.011 U is equivalent to 100% activity for phosphorolysis, and a value of 5.39 μmol of trehalose per h is equivalent to 100% activity for synthesis.
(iv) Effects of cations on TSase activity.
The presence of EDTA did not affect trehalose phosphorylation or synthesis activities, which showed that TSase required no cations for the activity. Cu2+ at 1 mM completely abolished both phosphorylation and synthesis activities. At 0.1 mM Cu2+, phosphorylation and synthesis activities were reduced to 76 and 48%, respectively. Zn2+ at 1 mM also reduced the phosphorolysis and synthesis activities to 40 and 0%, respectively. Other cations, Mn2+, Fe3+, Co2+, Ca2+, Mg2+, Ba2+, K+, Na+, Al3+, Rb+, and Cs+, showed no significant effect on TSase.
(v) Substrate specificity.
TSase phosphorylation was specific for trehalose, and no activity was detected with other disaccharides, such as neotrehalose, palatinose, cellobiose, lactose, sucrose, maltose, or isomaltose. TSase synthesis activity was also very specific. When α-d-glucose 1-P was used as a substrate donor, only d-glucose served as the acceptor. l-Glucose, d-galactose, d-mannose, d-xylose, d-fructose, d-sorbitol, d-mannitol, or d-fucose did not serve as the acceptor. When β-glucose 1-P, α-d-galactose 1-P, α-d-mannose 1-P, or α-d-xylose 1-P was used as a substrate donor and d-glucose was used as the acceptor, no disaccharide synthesis was detected.
Production of trehalose from sucrose by CES I.
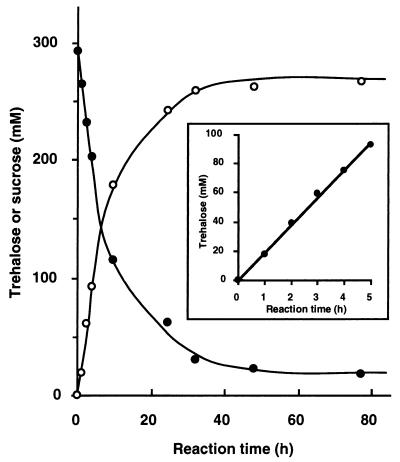
α-d-Glucose 1-P is liberated from sucrose by SPase (Fig. 5). The SPase equilibrium favors phosphorolysis and yields α-d-glucose 1-P (12, 29). TSase catalyzes the condensation of α-d-glucose 1-P and d-glucose to trehalose. When the concentrations of the substrates, sucrose and glucose, were 300 mM each, the optimum temperature was 37.5°C, the optimum pH was 6.5, and the optimum concentration of Pi was 20 mM. Under these conditions, trehalose was generated from sucrose at a 90 mol% yield (Fig. 6). The rate of trehalose synthesis, as determined by the conversion rates in the initial reaction periods (0 to 4 h), was 18 mmol/h under these conditions (Fig. 6).
FIG. 5.
Schematic representation of coupled enzyme systems, CES I and CES II, for the synthesis of trehalose from sucrose.
FIG. 6.
Conversion of sucrose into trehalose by CES I. The reaction conditions were as follows: 300 mM each sucrose and glucose, 20 mM Pi, and 1.05 U of SPase and TSase each per ml in 100 mM MES buffer (pH 6.5); temperature, 37.5°C. The standard deviation for all the points was within 10% of the point’s value.
Production of trehalose from sucrose by CES II.
In the CES I system, fructose is generated as a by-product. We expected the fructose to be converted to d-glucose by the action of GIase, which in turn might be incorporated into trehalose by the action of TSase (Fig. 5). We added 20 mM MgSO4 because the GIase used requires Mg2+ and conducted the reaction in an Ar gas atmosphere since weak glucose oxidase-like activity was contained in the GIase. The optimum conditions were 30 to 32.5°C and pH 6.5 to 7.1 (data not shown). The concentration of Pi was not tested extensively, but 20 mM Pi gave an acceptable yield (see below).
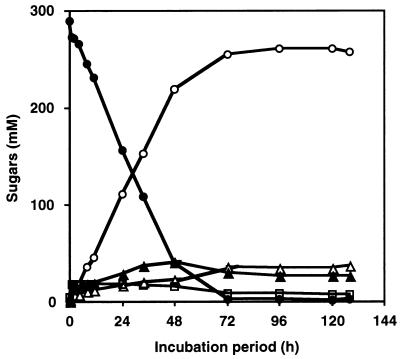
The course of trehalose production by CES II under the optimum conditions is shown in Fig. 7. The fructose liberated from the sucrose was rapidly incorporated into trehalose without significant accumulation. Sucrose was converted into trehalose at a 90 mol% yield, the same conversion efficiency as that for CES I. The conversion rate was 4.5 mmol/h (data not shown), which was lower than that in CES I. This slower reaction is probably due to the difference in the concentration of glucose as the substrate in the reaction mixtures; the concentration of glucose in CES I is much higher than that in CES II. In CES I, all of the glucose is immediately available, while the glucose in CES II is gradually supplied by the action of GIase.
FIG. 7.
Conversion of sucrose into trehalose by CES II. The reaction mixture, which contained 300 mM sucrose, 20 mM MgSO4, 1.05 U of SPase, TSase, and GIase each per ml, and 20 mM Pi in 100 mM HEPES buffer (pH 7.0), was incubated at 30°C for the indicated periods, and the amounts of trehalose (○), sucrose (•), glucose (▵), and fructose (▴) were measured by HPLC. α-d-Glucose 1-P (□) was measured by the enzymatic method described in Materials and Methods. The standard deviation for all the points was within 10% of the point’s value.
DISCUSSION
We have described a novel TSase from a basidiomycete, G. frondosa, that uses α-d-glucose 1-P as one of the substrates. The results of screening suggested that basidiomycetes had relatively higher trehalose phosphorolysis and synthesis activities than ascomycetes, zygomycetes, and mitosporic fungi did. An enzyme activity similar to TSase detected in F. velutipes (11) and P. fermentans (27) was very unstable, and the enzyme was not purified. Several trehalose phosphorylases reported to date decompose trehalose to β-glucose 1-P and glucose (4, 15, 17, 34). The TSase in this study shows no activity toward β-glucose 1-P.
TSase from G. frondosa shows differences in optimum temperature (Fig. 3) and pH (Fig. 4) for the forward and reverse reactions. A similar difference in optimum temperature of the forward and reverse reactions was observed for the trehalose phosphorylase from Euglena (15) and the SPase from Leuconostoc (12), the reason for which is unclear. The great difference in the equilibrium constants for the reactions of the two directions (Fig. 2) may result from this unusual characteristic. TSase from G. frondosa also shows differences in temperature and pH stability for the two directions. It is possible that the activity in one direction can be damaged in some unknown way more than the activity in the other direction.
We found that TSase activity was widespread in the fungi we examined. Trehalose is believed to be synthesized mainly from trehalose 6-phosphate by the dephosphorylating activity of trehalose 6-P phosphatase (16). Trehalose 6-P is synthesized from UDP-glucose and glucose 6-P by the action of trehalose 6-P synthase (14). Trehalose is also synthesized from β-d-glucose 1-P and glucose by the reversible reaction of trehalose phosphorylases (1, 4, 15, 19, 33). In addition, trehalose is synthesized from maltose by the intramolecular transglucosylating activity of the TSase in Thermus aquaticus (22) and from malto-oligosaccharide by the combination of malto-oligosyl trehalose synthase and malto-oligosyl trehalose trehalohydrolase in an Arthrobacter sp. (20, 21). It is thus apparent that trehalose is synthesized by several pathways in a wide variety of microorganisms. Although the physiological role of this TSase in G. frondosa is unclear, it is quite possible that the trehalose synthesized from α-d-glucose 1-P and glucose by the TSase in G. frondosa in the present study has some physiological role in this strain, which is at present unclear.
For the enzymatic synthesis of trehalose, the TSase from G. frondosa can be combined with any enzyme that generates α-d-glucose 1-P. These enzymes include starch phosphorylase (28), β-1,3-glucan phosphorylase (2), cellobiose phosphorylase (3), and laminaribiose phosphorylase (8). Enzymatic processes such as these often are commercially useful because they are simple, and the mixture of sugars after the enzyme reaction can often be used directly without further purification. From the commercial point of view, starch is the most promising substrate. We have conducted preliminary experiments in which TSase and potato starch phosphorylase are combined in a starch-containing reaction mixture. The yield of trehalose was significant but lower than that in the CES I and CES II systems (data not shown). Trehalose produced by such a process could be used in various applications. For example, trehalose is preferred over sucrose and sorbitol as a food additive for moisturizing deep-frozen fish, meats, ham, sausage, and noodles. Trehalose improves the elasticity of these foods and helps prevent discoloration caused by Maillard’s reaction. Greater availability of trehalose at a reduced price would further increase its use in food processing and preservation.
REFERENCES
- 1.Aisaka K, Masuda T. Production of trehalose phosphorylase by Catellatospora ferruginea. FEMS Microbiol Lett. 1995;131:47–51. doi: 10.1016/0378-1097(95)00233-u. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht G J, Kauss H. Purification, crystallization and properties of a β-(1→3)-glucan phosphorylase from Ochromonas malhamensis. Phytochemistry. 1971;10:1293–1298. [Google Scholar]
- 3.Alexander J K. Cellobiose phosphorylase from Clostridium thermocellum. Methods Enzymol. 1972;28:944–948. [Google Scholar]
- 4.Belocopitow E, Maréchal L R. Trehalose phosphorylase from Euglena gracilis. Biochim Biophys Acta. 1970;198:151–154. doi: 10.1016/0005-2744(70)90045-8. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter J F, Crowe J H. Modes of stabilization of a protein by organic solutes during desiccation. Cryobiology. 1988;25:459–470. [Google Scholar]
- 6.Colaco C, Sen S, Thangavelu M, Pinder S, Roser B. Extraordinary stability of enzymes dried in trehalose: simplified molecular biology. Bio/Technology. 1992;10:1007–1011. doi: 10.1038/nbt0992-1007. [DOI] [PubMed] [Google Scholar]
- 7.Elbein A D. The metabolism of α,α-trehalose. Adv Carbohydr Chem Biochem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- 8.Goldemberg S H, Maréchal L R. Laminaribiose phosphorylase and β-1,3 oligoglucan phosphorylase from Euglena gracilis. Methods Enzymol. 1972;28:953–960. [Google Scholar]
- 9.Hawksworth D L, Kirk P M, Sutton B C, Pegler D N. Ainsworth & Bisby’s dictionary of the fungi. 8th ed. Wallingford, United Kingdom: CAB International; 1995. [Google Scholar]
- 10.Hoekema, A., J. Pen, M. P. Does, and P. J. M. van den Elzen. January 1995. Production of trehalose in plants. World Intellectual Property Organization patent 9501446.
- 11.Kitamoto Y, Akashi H, Tanaka H, Mori N. α-Glucose-1-phosphate formation by a novel phosphorylase from Flammulina velutipes. FEMS Microbiol Lett. 1988;55:147–150. [Google Scholar]
- 12.Kitaoka K, Takahashi H, Kara K, Hashimoto H, Sasaki T, Taniguchi H. Purification and characterization of sucrose phosphorylase from Leuconostoc mesenteroides ATCC 12291 cells, and disaccharide synthesis by the enzyme. Oyo Toshitsu Kagaku. 1994;41:165–172. . (In Japanese.) [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lapp D F, Patterson B W, Elbein A D. Trehalose phosphate synthetase from Mycobacterium smegmatis. Methods Enzymol. 1972;28:515–519. [Google Scholar]
- 15.Maréchal L R, Belocopitow E. Metabolism of trehalose in Euglena gracilis. I. Partial purification and some properties of trehalose phosphorylase. J Biol Chem. 1972;247:3223–3228. [PubMed] [Google Scholar]
- 16.Matula M, Mitchell M, Elbein A D. Partial purification and properties of a highly specific trehalose phosphate phosphatase from Mycobacterium smegmatis. J Bacteriol. 1971;107:217–222. doi: 10.1128/jb.107.1.217-222.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki, J., K. Miyagawa, and Y. Sugiyama. November 1993. Process for production of trehalose. Japan Kokai Tokkyo Koho (Japan patent) JP05292986.
- 18.Mukai K, Tabuchi A, Nakada T, Shibuya T, Chaen H, Fukuda S, Kurimoto M, Tsujisaka Y. Production of trehalose from starch by thermostable enzymes from Sulfolobus acidocaldarius. Starch. 1997;49:26–30. [Google Scholar]
- 19.Murao S, Nagano H, Ogura S, Nishino T. Enzymatic synthesis of trehalose from maltose. Agric Biol Chem. 1985;49:2113–2118. [Google Scholar]
- 20.Nakada T, Maruta K, Mitsuzumi H, Kubota M, Chaen H, Sugimoto T, Kurimoto M, Tsujisaka Y. Purification and characterization of a novel enzyme, maltooligosyl trehalose trehalohydrolase, from Arthrobacter sp. Q36. Biosci Biotechnol Biochem. 1995;59:2215–2218. doi: 10.1271/bbb.59.2215. [DOI] [PubMed] [Google Scholar]
- 21.Nakada T, Maruta K, Tsusaki K, Kubota M, Chaen H, Sugimoto T, Kurimoto M, Tsujisaka Y. Purification and properties of a novel enzyme, maltooligosyl trehalose synthase, from Arthrobacter sp. Q36. Biosci Biotechnol Biochem. 1995;59:2210–2214. doi: 10.1271/bbb.59.2210. [DOI] [PubMed] [Google Scholar]
- 22.Nishimoto T, Nakada T, Chaen H, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y. Purification and characterization of a thermostable trehalose synthase from Thermus aquaticus. Biosci Biotechnol Biochem. 1996;60:835–839. doi: 10.1271/bbb.60.835. [DOI] [PubMed] [Google Scholar]
- 23.Nishimoto T, Nakano M, Nakada T, Chaen H, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y. Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp. R48. Biosci Biotechnol Biochem. 1996;60:640–644. doi: 10.1271/bbb.60.640. [DOI] [PubMed] [Google Scholar]
- 24.Ohtani M, Usui N. Production of trehalose by fermentation and its application. Shokuhin Kagaku (Food Chemicals) 1994;1994:91–95. . (In Japanese.) [Google Scholar]
- 25.Roser B. Trehalose, a new approach to premium dried foods. Trends Food Sci Technol. 1991;2:166–169. [Google Scholar]
- 26.Rudolph A S, Crowe J H. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology. 1985;22:367–377. doi: 10.1016/0011-2240(85)90184-1. [DOI] [PubMed] [Google Scholar]
- 27.Schick I, Haltrich D, Kulbe K D. Trehalose phosphorylase from Pichia fermentans and its role in the metabolism of trehalose. Appl Microbiol Biotechnol. 1995;43:1088–1095. [Google Scholar]
- 28.Shimomura S, Fukui T. A comparative study on α-glucan phosphorylases from plant and animal interrelationship between the polysaccharide and pyridoxal phosphate binding sites by affinity electrophoresis. Biochemistry. 1980;19:2287–2294. doi: 10.1021/bi00552a001. [DOI] [PubMed] [Google Scholar]
- 29.Silverstein R, Voet J, Reed D, Abeles R H. Purification and mechanism of action of sucrose phosphorylase. J Biol Chem. 1967;242:1338–1346. [PubMed] [Google Scholar]
- 30.Tsuchida, T., Y. Murakami, and Y. Nishimoto. August 1993. Process for production of trehalose by Corynebacterium, Brevibacterium, Microbacterium, and Arthrobacterium. Japan Kokai Tokkyo Koho (Japan patent) JP05211882.
- 31.van Laere A. Trehalose, reserve and/or stress metabolite? FEMS Microbiol Rev. 1989;63:201–210. [Google Scholar]
- 32.Wiemken A. Trehalose in yeast stress protectant rather than reserve carbohydrate. Antonie Leeuwenhoek. 1990;58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida M, Nakamura N, Horikoshi K. Production and application of maltose phosphorylase and trehalose phosphorylase by a strain of Plesiomonas. Oyo Toshitsu Kagaku. 1995;42:19–25. . (In Japanese.) [Google Scholar]
- 34.Yoshida M, Nakamura N, Horikoshi K. Production of trehalose from starch by maltose phosphorylase and trehalose phosphorylase from a strain of Plesiomonas. Starch. 1997;49:21–26. [Google Scholar]