Abstract
Ataxia-telangiectasia mutated (ATM) plays a central role in the cellular response to DNA damage and ATM alterations are common in several tumor types including bladder cancer. However, the specific impact of ATM alterations on therapy response in bladder cancer is uncertain. Here, we combine preclinical modeling and clinical analyses to comprehensively define the impact of ATM alterations on bladder cancer. We show that ATM loss is sufficient to increase sensitivity to DNA-damaging agents including cisplatin and radiation. Furthermore, ATM loss drives sensitivity to DNA repair–targeted agents including poly(ADP-ribose) polymerase (PARP) and Ataxia telangiectasia and Rad3 related (ATR) inhibitors. ATM loss alters the immune microenvironment and improves anti-PD1 response in preclinical bladder models but is not associated with improved anti-PD1/PD-L1 response in clinical cohorts. Last, we show that ATM expression by immunohistochemistry is strongly correlated with response to chemoradiotherapy. Together, these data define a potential role for ATM as a predictive biomarker in bladder cancer.
ATM loss drives sensitivity to DNA damaging and DNA repair targeted agents but has mixed impact on anti-PD1 sensitivity.
INTRODUCTION
The ataxia-telangiectasia mutated (ATM) gene encodes a serine/threonine kinase that plays a critical role in maintaining genomic integrity (1). Following DNA damage, the ATM protein is activated and is responsible for coordinating DNA repair with cell cycle arrest and other cellular activities in a cell- and context-dependent manner (2). Reflecting its central role in the DNA damage response (DDR), biallelic germline mutations in ATM cause ataxia telangiectasia (AT), a rare inherited syndrome characterized by sensitivity to DNA-damaging agents, increased cancer predisposition, as well as immune and neurologic defects (3).
ATM is recurrently lost or mutated in multiple tumor types including bladder cancer (4). Tumor ATM loss can result from an inherited germline ATM mutation accompanied by loss-of-heterozygosity (LOH) of the wild-type (WT) allele, via acquired (somatic) loss or mutation, or by nongenetic mechanisms (5). Given its critical role in the DDR, there is substantial interest in targeting ATM deficiency in cancer. In bladder cancer, several studies have identified an association between alterations in DNA repair genes and improved response to cisplatin-based chemotherapy (6–9). However, studies have used different criteria to define DDR gene alterations, and the specific contribution of ATM versus other DDR genes in driving this association is unclear.
In addition to driving sensitivity to DNA-damaging agents such as cisplatin, there is also notable interest in targeting tumor ATM deficiency using DNA repair–directed agents (10). Poly(ADP-ribose) polymerase (PARP) inhibitors preferentially target tumors with homologous recombination (HR) deficiency and are U.S. Food and Drug Administration–approved in several clinical contexts. However, the association among ATM loss, HR function, and PARP inhibitor sensitivity is not well defined in bladder cancer or other tumor types and may be context-dependent (11). Several completed and ongoing PARP inhibitor clinical trials in advanced bladder cancer have enrolled patients with tumor ATM alterations (12–15); however, the trial designs vary markedly and the total number of ATM-altered cases is low. Therefore, it is currently uncertain whether an association exists between ATM mutational status and PARP inhibitor response in bladder cancer.
DNA damage and the ensuing cellular response can lead to changes in the immune microenvironment and therefore can affect tumor sensitivity to anti-PD1/PD-L1 agents (16). Several studies have linked ATM loss or inhibition with immune activation via both tumor cell–intrinsic and –extrinsic mechanisms (17–19). In bladder cancer, DDR gene alterations have been associated with improved anti-PD1/PD-L1 response (20), but the specific contribution of ATM alterations to tumor immune contexture and anti-PD1/PD-L1 response is unclear.
Here, we combine clinical and functional analyses to define the effect of ATM deficiency on cellular properties and therapeutic sensitivities in bladder cancer. We find that ATM loss is sufficient to confer increased sensitivity to clinically relevant DNA-damaging agents and DNA repair–targeted agents including PARP and ATR inhibitors. We observe changes in immune contexture including an increase in CD4+ T cells and improved anti-PD1 response in an ATM-deficient syngeneic preclinical model, but we did not identify statistically significant differences in anti-PD1/PD-L1 response in ATM-mutant versus nonmutant clinical cohorts. Last, we performed an ATM immunohistochemistry (IHC) assay on several cohorts of clinical bladder tumors and found that in a chemoradiotherapy-treated cohort, ATM IHC is more strongly correlated with response than ATM mutational status. Together, these data demonstrate that ATM deficiency is sufficient to confer a DNA repair–deficient phenotype and increase sensitivity to DNA-damaging and DNA repair–targeted agents in bladder cancer and suggest that ATM IHC is a more reliable biomarker of functional ATM loss than ATM mutations identified by sequencing. These data have important implications for multiple ongoing clinical trials that are investigating the association between alterations in DDR genes and clinical response to chemotherapy, chemoradiotherapy, and PARP inhibitors.
RESULTS
Predicted deleterious ATM alterations are present in a subset of bladder tumors
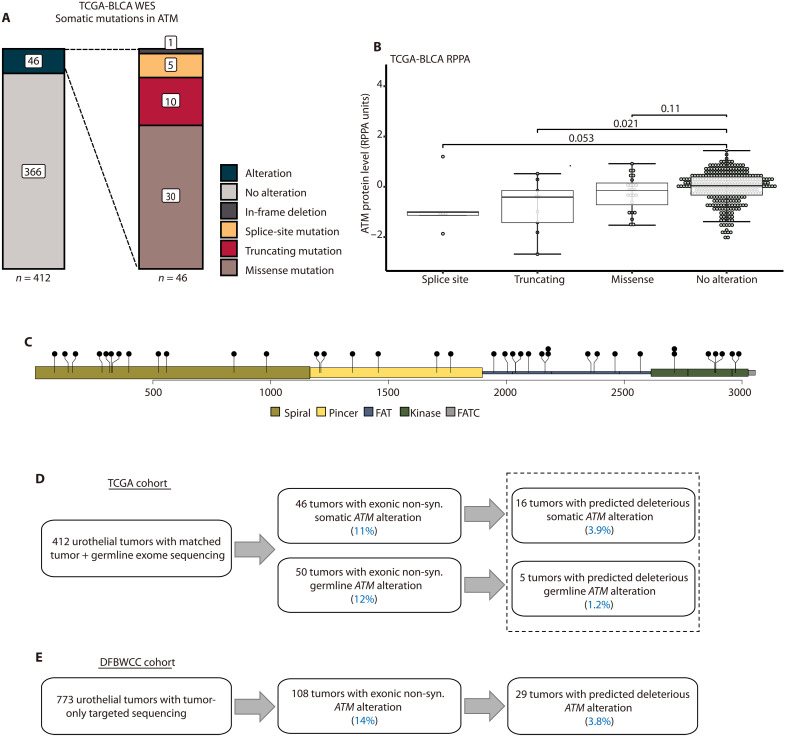
We first characterized the frequency of different classes of ATM alterations in bladder tumors. The Cancer Genome Atlas (TCGA) bladder cancer (BLCA) study comprehensively profiled 412 muscle-invasive bladder cancers (MIBCs) (4). Among tumors with available whole-exome sequencing (WES) data, 46 (11%) had at least one reported somatic ATM alteration including in-frame (n = 1), missense (n = 30), truncating (nonsense/frameshift, n = 10), or splice site mutation (n = 5; Fig. 1A). The presence of a truncating or splice site mutation was associated with lower ATM protein levels compared to tumors without an ATM mutation (P = 0.021 and P = 0.053, respectively), whereas the presence of a missense mutation was not (P = 0.11; Fig. 1B).
Fig. 1. ATM alterations in bladder cancer.
(A) Frequency and classes of somatic ATM alterations in the TCGA-BLCA WES cohort. (B) Tumors with ATM splice sites or truncating alterations had lower protein levels (as quantified by RPPA) in the TCGA-BLCA cohort (Wilcoxon rank sum test). (C) Distribution of all observed non-synonymous somatic missense mutations in TCGA-BLCA cohort. FAT, FRAP-ATM-TRRAP domain; FATC, FRAP-ATM-TRRAP C-terminal domain. (D) ATM alterations in TCGA-BLCA WES cohort. (E) ATM alterations in the DFBWCC urothelial tumor cohort. RPPA, reverse phase protein array; DFBWCC, Dana-Farber/Brigham and Women’s Cancer Center.
The most common class of alteration in the BLCA cohort was missense mutation. However, the observed missense mutations were distributed across the ATM gene rather than clustered in specific locations (Fig. 1C). Similarly, in the TCGA pan-cancer cohort (n = 10,967 tumors), nearly 500 distinct ATM missense mutations are observed (fig. S1A), but only seven amino acid positions have been identified as missense mutational hotspots (fig. S1B) (21). Given the uncertainty in predicting the functional impact of most ATM missense mutations, we restricted our subsequent analyses to cases with missense mutations or other alterations that met stringent filtering criteria and were predicted to be functionally deleterious (see Materials and Methods). This approach identified 16 TCGA cases (3.9%) with a predicted deleterious ATM alteration (Fig. 1D). We used similar methods to analyze available germline data for the TCGA cohort and identified an additional five cases with a predicted deleterious germline ATM alteration (three truncating and two missense mutations; Fig. 1D and fig. S2). Therefore, the overall frequency of predicted deleterious ATM alterations in the TCGA cohort was 21 of 412 (5.1%; Fig. 1D). Approximately half (10 of 21; 48%) of the tumors with a predicted deleterious ATM alteration had accompanying ATM LOH. There were no differences in age, sex, or stage at diagnosis between patients with a predicted deleterious ATM alteration versus other patients in the TCGA cohort (fig. S3). Patients with a predicted deleterious ATM alteration had significantly improved overall survival (OS) and progression-free survival (PFS) compared to patients with WT ATM tumors or tumors with ATM variants of uncertain significance (VUS; fig. S4). Last, we investigated whether alterations in other significantly mutated genes in TCGA cohort co-occurred or were mutually exclusive with ATM alterations. Using a similar filtering strategy to identify predicted deleterious alterations, the only gene significantly co-mutated with ATM after correcting for multiple hypothesis testing was KDM6A [P = 0.042, Fisher’s exact test, Benjamini-Hochberg (BH) correction]. There were no genes with mutations that were mutually exclusive with ATM alterations. Unlike reports from some other tumor types, ATM and TP53 alterations were not mutually exclusive in this cohort (P = 0.21, Fisher’s exact test).
We next analyzed targeted tumor DNA sequencing data from a cohort of 773 patients with urothelial cancer from the Dana-Farber/Brigham and Women’s Cancer Center (DFBWCC; table S1) (22). Applying similar filtering methods as were used for the TCGA cohort, we identified 29 cases (3.8%) with a predicted deleterious ATM alteration (Fig. 1E and fig. S5). There were no genes that were significantly co-altered with ATM in the DFBWCC cohort. ATM and TP53 alterations were mutually exclusive (P = 0.008, Fisher’s exact test), but this difference was not significant after multiple-hypothesis correction (P = 0.24, BH correction). Together, these analyses indicate that when stringent filtering is applied, the frequency of predicted deleterious ATM alterations is approximately 5% in bladder cancer.
ATM loss sensitizes to DNA-damaging agents
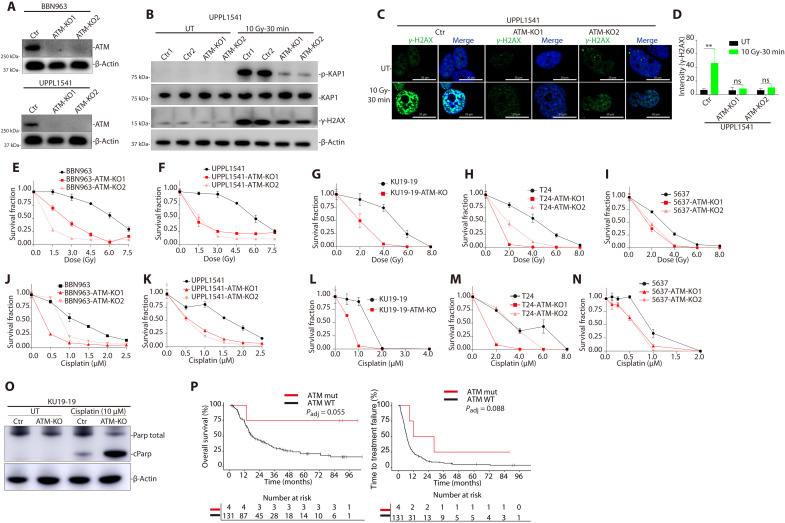
To further investigate the impact of ATM loss on bladder tumor properties, we used CRISPR-Cas9 techniques to delete the ATM gene in a panel of human and mouse bladder cancer cell lines (Fig. 2A and fig. S6A). We first tested the impact of ATM loss on DNA damage signaling. Kruppel-associated box (KRAB)–associated protein 1 [(KAP1) encoded by the TRIM28 gene] is a primary target of ATM phosphorylation following DNA damage, and we found that radiation-induced KAP1 phosphorylation was significantly reduced in ATM-deleted bladder cancer cells (Fig. 2B and fig. S6B). Similarly, radiation-induced phospho-H2AX (γ-H2AX) foci were significantly lower in ATM-deleted cells compared to control cells (Fig. 2, B to D, and fig. S6, C to E). Together, these results demonstrate that ATM loss abrogates normal ATM-mediated DNA damage signaling in bladder cancer cells.
Fig. 2. ATM deficiency results in altered DNA damage signaling and increased sensitivity to DNA-damaging agents in bladder cancer preclinical models.
(A) Immunoblot demonstrating loss of ATM protein in ATM-deleted murine bladder cancer cell lines BBN963 and UPPL1541 (see also fig. S6). (B) Immunoblot demonstrating a decrease in radiation-induced KAP1 and γ-H2AX phosphorylation in ATM-deleted compared to WT ATM bladder cancer cells. (C) Immunofluorescence images demonstrating a decrease in radiation-induced γ-H2AX foci formation in ATM-deleted compared to WT ATM cells. (D) Quantification of γ-H2AX foci. There is a significant increase in γ-H2AX foci following radiation in WT ATM cells but not in ATM-deleted cells (**P < 0.01, Student’s t test). (E to I) Cell survival curves for WT ATM and ATM-deleted human and mouse bladder cancer cell lines following ionizing radiation. There was a significant increase in radiation sensitivity in ATM-deleted cells compared to WT ATM lines across all models (see also fig. S7). (J to N) Cell survival curves for WT ATM and ATM-deleted human and mouse bladder cancer cell lines following cisplatin treatment. There was a significant increase in cisplatin sensitivity in ATM-deleted cells compared to WT ATM lines across all models. (O) Cisplatin treatment resulted in higher levels of cleaved PARP in ATM-deleted compared to WT ATM bladder cancer cells, consistent with increased cisplatin-induced apoptosis. (P) Time to treatment failure (TTF; left) and OS (right) of patients with ATM-mutant and nonmutant DFBWCC metastatic urothelial cancer treated with cisplatin-based chemotherapy. All cell line data are plotted as the means ± SD (n = 5) unless otherwise specified. Ctr, control; KO, knockout; UT, untreated; Gy, gray; ns, not significant.
We next tested the impact of ATM loss on sensitivity to DNA-damaging agents commonly used to treat bladder cancer. We observed a significant increase in radiation sensitivity across human and mouse bladder cancer cell lines in vitro and in vivo (Fig. 2, E to I, and fig. S7, A to D). Similarly, cisplatin sensitivity was also significantly higher in all ATM-deleted bladder cancer cell lines compared to WT ATM controls (Fig. 2, J to N, and fig. S7, E to H). Cisplatin treatment resulted in a larger increase in cleaved PARP levels in the ATM-deleted cells compared to WT ATM cells, consistent with an increase in cisplatin-induced apoptosis (Fig. 2O) (23). In a DFBWCC cohort of 131 patients with metastatic urothelial cancer treated with cisplatin-based chemotherapy, there was a trend toward longer time to treatment failure and improved OS in patients with an ATM alteration that met our stringent filtering criteria (P = 0.088 and P = 0.055, respectively; Fig. 2P); however, the total number of ATM-altered cases was small (n = 4). Together, these data demonstrate that ATM deficiency is sufficient to drive increased sensitivity to DNA-damaging agents across multiple bladder cancer preclinical models.
ATM loss sensitizes to DNA repair–directed agents
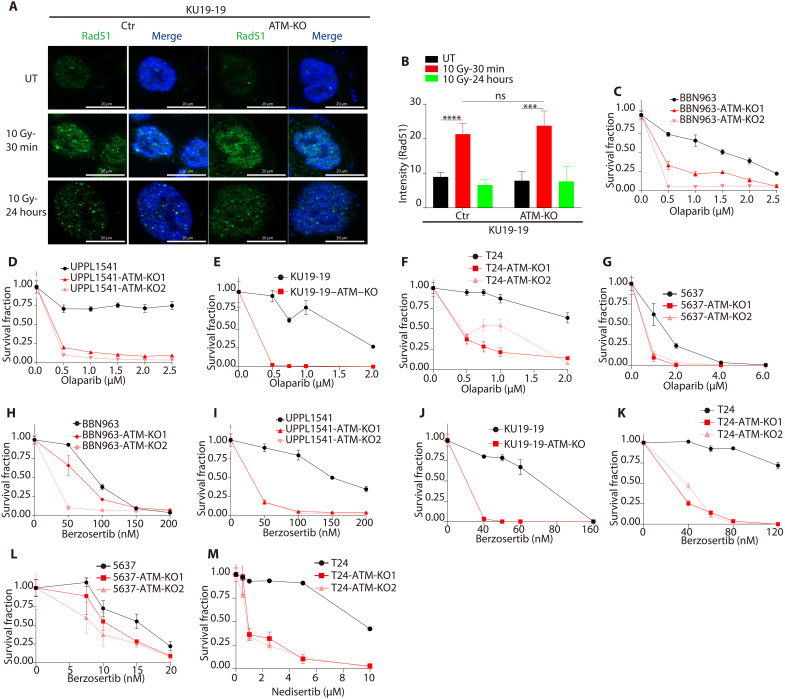
We next tested the impact of ATM loss on sensitivity to several DNA repair–targeted agents. PARP inhibitors are approved in several HR-deficient tumor settings, and we first wished to determine the impact of ATM loss on HR function in bladder cancer cells. We previously found that ATM loss does not affect HR function in the DR-GFP reporter cell line (24), and here, we observed that ATM loss did not inhibit radiation-induced Rad51 foci formation (Fig. 3, A and B, and fig. S8A). However, despite the lack of an HR defect noted in either the DR-GFP or Rad51 foci formation assays, we observed a significant increase in PARP inhibitor sensitivity in all ATM-deleted cell lines compared to WT ATM parental lines (Fig. 3, C to G, and fig. S8, B to D). These results suggest that ATM loss sensitizes to PARP inhibition in bladder cancer, but that the mechanism of sensitization is distinct from the mechanism conferred by loss of canonical HR genes such as BRCA1/2.
Fig. 3. ATM loss drives sensitivity to inhibitors of PARP, ATR, and DNA-PK in bladder cancer preclinical models.
(A) Immunofluorescence images showing radiation-induced Rad51 foci formation in ATM-deleted and WT ATM KU19-19 bladder cancer cells. (B) Quantification of radiation-induced Rad51 foci shows no difference in ATM-deleted compared to WT ATM KU19-19 cells. Data are displayed as means ± SD, n = 5, ***P < 0.001 and ****P < 0.0001. (C to G) Cell survival curves for WT ATM and ATM-deleted human and mouse bladder cancer cell lines following olaparib treatment. There was a significant increase in olaparib sensitivity in ATM-deleted cells compared to WT ATM lines across all models (see also fig. S8). (H to L) Cell survival curves for WT ATM and ATM-deleted human and mouse bladder cancer cell lines following berzosertib treatment. There was a significant increase in berzosertib sensitivity in ATM-deleted cells compared to WT ATM lines across nearly all models. (M) Cell survival curves for WT ATM and ATM-deleted bladder cancer cells following nedisertib treatment.
ATM and ATR kinases have partially redundant functions in cellular DDR signaling, and there is substantial interest in ATR inhibition as a synthetic lethal strategy to target ATM-deficient tumors (5). We tested the differential sensitivity of ATM-deleted versus WT ATM bladder cancer cell lines to the ATR inhibitor berzosertib. In nearly all ATM-deleted cell lines tested, there was a significant increase in ATR inhibitor sensitivity relative to WT ATM cells (Fig. 3, H to L, and fig. S8, E to H). Last, we tested the sensitivity of ATM-deleted and WT ATM cells to an inhibitor of DNA protein kinase (DNA-PK). DNA-PK plays a critical role in end joining repair of double-strand breaks, and ATM loss sensitizes to DNA-PK inhibition in some contexts (25, 26). We observed a marked increase in sensitivity to the DNA-PK inhibitor nedisertib in ATM-deleted compared to WT ATM bladder cancer cells (Fig. 3M and fig. S8I). Together, these data suggest that ATM deletion is sufficient to increase sensitivity to multiple DNA repair–directed agents across bladder cancer preclinical models.
ATM loss affects immune contexture and augments anti-PD1 response in preclinical BC models but is not associated with anti-PD1/PD-L1 response in BC clinical cohorts
We next sought to investigate the relationship among ATM mutational status, immune contexture, and immune checkpoint inhibitor (ICI) response in bladder cancer. A previous study identified an association between DDR gene alterations and improved response to anti-PD1/PD-L1 therapy in patients with advanced bladder cancer (20), but the specific impact of ATM deficiency on immune properties in bladder cancer is unknown.
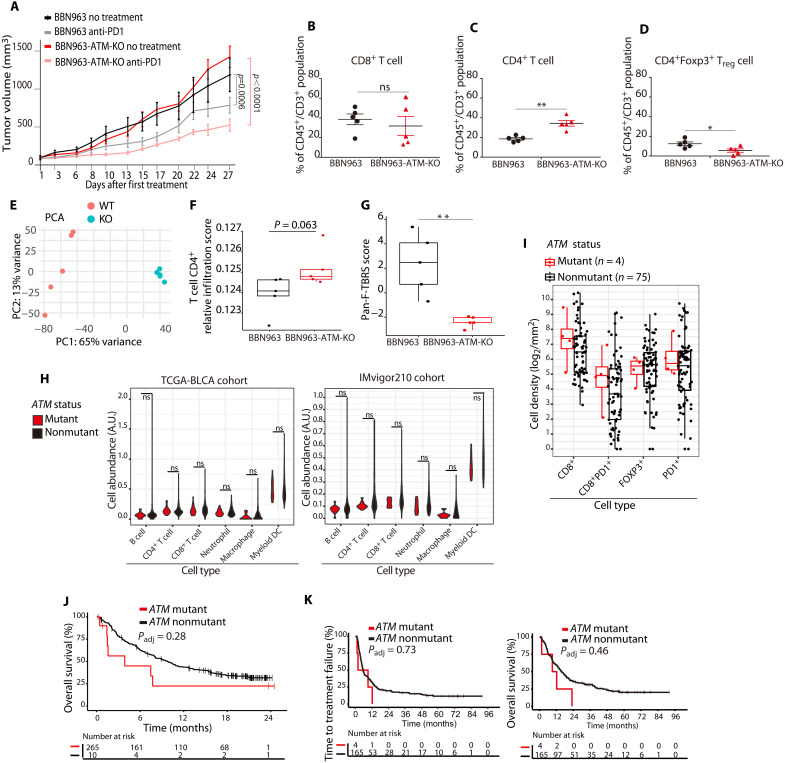
To directly test the impact of ATM loss on immune contexture and anti-PD1 response, we leveraged the isogenic BBN963 and BBN963-ATM-KO mouse bladder cancer models. We established a cohort of mice bearing BBN963 or BBN963-ATM-KO tumors and randomized to treatment with an anti-PD1 antibody or vehicle. Mice were treated with anti-PD1 antibody (10 mg/kg) or vehicle delivered intraperitoneally (ip) twice weekly for 4 weeks. BBN963 and BBN963-ATM-KO tumors displayed similar growth kinetics in vehicle-treated mice. Although anti-PD1 treatment led to significant growth delay in both models, the growth delay was more pronounced in BBN963-ATM-KO tumors (Fig. 4A and fig. S9, A and B).
Fig. 4. ATM loss is sufficient to drive immune changes and improve anti-PD1 response in preclinical bladder models but ATM mutations are not associated with immune properties or anti-PD1 response in clinical bladder cohorts.
(A) Growth curves for ATM-deleted and WT ATM tumor xenografts. The ATM-deleted tumors had a larger response to anti-PD1 treatment than WT ATM xenografts (linear regression model). (B) Flow cytometry showed no differences in the number of CD8+ T cells in ATM-deleted versus WT ATM tumors. Data are plotted as the means ± SD, n = 5 tumors. (C) Flow cytometry showed a significantly higher number of CD4+ T cells in ATM-deleted compared to WT ATM tumors, **P < 0.01. (D) Flow cytometry showed significantly fewer FOXP3+ CD4+ T cells in ATM-deleted compared to WT ATM tumors, *P < 0.05. (E) Principal components analysis (PCA) of RNA sequencing (RNA-seq) counts from ATM-deleted and WT ATM tumors. (F) RNA-seq–based immune cell fraction estimation using TIMER revealed a trend toward increased CD4+ T cells in ATM-deleted compared to WT ATM tumors (Student’s t test; BH correction). (G) A gene expression signature of transforming growth factor–β signaling in fibroblasts was higher in WT ATM tumors than in ATM-deleted tumors. (H) RNA-seq analysis of immune cell subsets in ATM-mutant versus ATM nonmutant tumors from the TCGA (left) and IMvigor210 (right) bladder cancer cohorts. There were no significant differences in any immune cell subsets (Wilcoxon rank sum test). (I) Quantification of multiplexed immunofluorescence data from institutional bladder cancer cases showed no difference in CD8, PD1, or FOXP3 staining between ATM-mutant and nonmutant bladder tumors. (J) OS of patients with ATM-mutant and nonmutant bladder tumors in the Imvigor210 cohort (Kaplan-Meier method). (K) TTF (left) and OS (right) for ATM-mutant and ATM nonmutant institutional bladder cancer cases following anti-PD1/PD-L1 treatment (Kaplan-Meier method).
To further dissect the impact of ATM loss on immune properties, we established a second cohort of BBN963 and BBN963-ATM-KO tumor–bearing mice. When tumors reached 500 mm3 in size, mice were euthanized and tumors were harvested with half of the tumor snap-frozen for RNA sequencing (RNA-seq) and half processed into single cells for flow cytometry (fig. S9C). Flow cytometric analysis showed no difference in the number of CD8+ T cells in BBN963 versus BBN963-ATM-KO tumors (Fig. 4B); however, there were significantly more CD4+ T cells in the BBN963-ATM-KO tumors compared to BBN tumors (Fig. 4C). The increase in CD4+ T cells in the BBN963-ATM-KO tumors was not driven by an increase in immunosuppressive CD4+/FOXP3+ cells, which were significantly lower in BBN963-ATM-KO tumors than in BBN963 tumors (Fig. 4D).
RNA-seq analysis revealed that BBN963 and BBN963-ATM-KO tumors were characterized by distinct gene expression profiles (Fig. 4E; table S2). Using TIMER to infer immune cell subsets, we observed no differences in CD8+ cells but a trend toward increased CD4+ cells in the BBN963-ATM-KO tumors compared to the BBN963 tumors (Fig. 4F and fig. S9E). In addition, BBN963-ATM-KO samples showed a trend toward increased T cell receptor (TCR) diversity, which has been associated with improved anti-PD1/PD-L1 responses in bladder cancer (27), as well as increased immune score and higher expression of granzyme B (GZMB) and other genes that have been associated with improved anti-PD1/PD-L1 response (fig. S9, F to Q). Last, a signature of transforming growth factor–β signaling in fibroblasts that has been associated with poor anti–PD-L1 response in bladder tumors was higher in the BBN963 tumors than in the BBN963-ATM-KO tumors (Fig. 4G) (28). Together, these data suggest that ATM loss promotes CD4+ cytotoxic T cell infiltration and increases sensitivity to anti-PD1 therapy in an immune-competent bladder cancer preclinical model.
We next compared the immune properties of ATM-mutant versus nonmutant bladder tumors in several clinical cohorts. Using available RNA-seq data to infer cellular phenotypes, we did not observe a difference in immune cell populations between ATM-mutant and nonmutant tumors in the TCGA bladder cohort or in patients enrolled on IMVigor210 (29), a randomized phase 2 clinical trial of the anti–PD-L1 agent atezolizumab in advanced bladder cancer (Fig. 4H). Similarly, using a multiplexed immunofluorescence (mIF) assay to assess immune markers in bladder tumors from patients treated at DFBWCC (30), we did not observe differences in CD8, FOXP3, PD1, or PD-L1 staining between ATM-mutant versus nonmutant tumors (Fig. 4I). There was also no difference in OS in ATM-mutant versus nonmutant patients who received anti-PD1/PD-L1 therapy in the IMVigor210 cohort (Fig. 4J) or time to treatment failure or OS in a DFBWCC cohort of 169 patients with bladder cancer treated with anti-PD1/PD-L1 therapy (Fig. 4K). Together, these analyses suggest that the presence of a predicted deleterious ATM alteration is not significantly associated with immune properties or anti-PD1/PD-L1 response in clinical bladder cancer cohorts.
ATM immunohistochemistry correlates with ATM mutation status and chemoradiotherapy response in MIBC
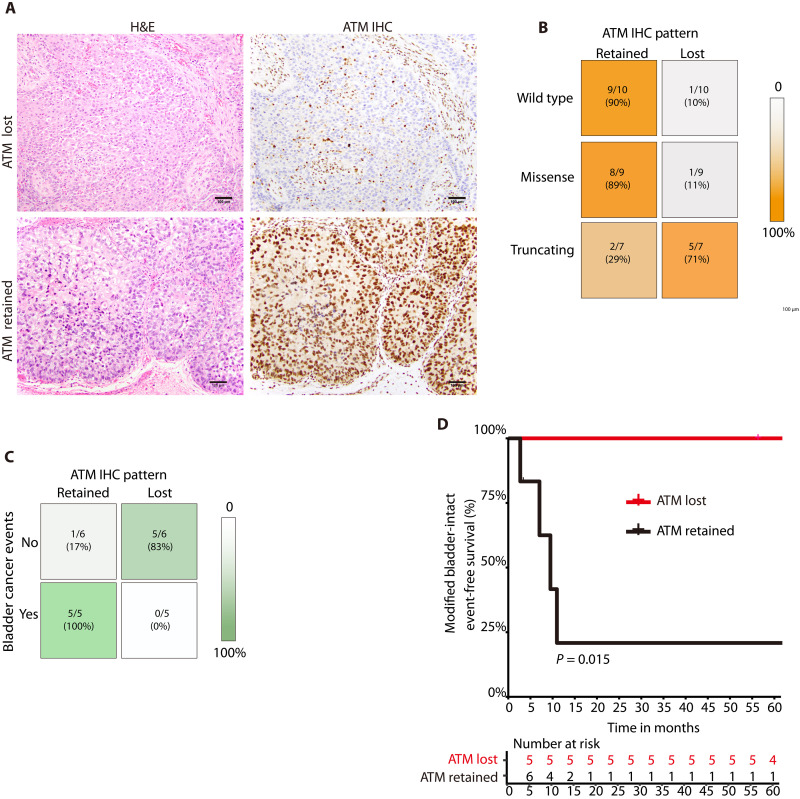
Given the challenge of predicting the functional impact of clinically observed ATM alterations, several recent efforts in other tumor types have focused on developing an ATM IHC assay to identify tumors with loss of ATM protein (31–33). IHC is an attractive method because it has the potential to identify functional ATM deficiency conferred by a wide variety of genetic or epigenetic alterations, and ATM IHC has been performed in several PARP and ATR inhibitor trials (11, 34, 35). However, ATM IHC has not been investigated specifically in bladder cancer. We identified bladder cancer cases from DFBWCC with available targeted DNA sequencing and selected a cohort that included tumors with WT ATM as well as tumors with either ATM missense or truncating (nonsense/frameshift) mutations (table S3). We then performed ATM IHC using a commercially available monoclonal antibody (see Materials and Methods). All ATM IHC cases were reviewed independently by two pathologists blinded to ATM mutational status and each case was assigned by consensus as having ATM expression either retained or lost (Fig. 5A). There was a significant correlation between ATM mutational status and ATM IHC expression (P = 0.008, two-sided chi-square test; Fig. 5B). Most tumors with WT or missense mutations in ATM had retained ATM expression, whereas most tumors with an ATM truncating alteration had loss of ATM expression.
Fig. 5. Correlations among ATM mutational status, ATM IHC expression, and chemoradiotherapy response.
(A) Representative photomicrographs of urothelial carcinoma and corresponding ATM IHC demonstrating loss of ATM expression (top row) or retained expression (bottom row). In tumors with ATM loss, non-neoplastic cells (e.g. endothelial cells and fibroblasts) retain ATM expression and serve as an internal positive control. (B) Correlation between ATM mutational status and ATM IHC pattern in a cohort of bladder tumors from the DFBWCC. Most tumors with WT ATM or an ATM missense mutation have retained ATM staining by IHC, whereas most tumors with an ATM nonsense or frameshift mutation have ATM loss by IHC. (C) Correlation between ATM IHC pattern and bladder cancer events in the MGH cohort. (D) Modified bladder-intact event-free survival (mBI-EFS) in MGH cases with ATM protein expression retained versus lost (Kaplan-Meier method). There was significantly longer mBI-EFS in patients with loss of ATM expression. MGH, Massachusetts General Hospital.
We next wished to assess whether ATM IHC patterns correlated with clinical outcomes. Trimodality therapy (TMT) is a curative treatment approach for MIBC that involves transurethral tumor resection followed by concurrent chemoradiotherapy (36, 37). Recently, Kamran et al. (38) performed WES of MIBCs from patients treated with TMT at the Massachusetts General Hospital (MGH) and correlated genomic features with clinical outcomes. Thirteen of 76 (17%) tumors harbored at least one ATM alteration and four (5.2%) had an ATM alteration that was predicted to be deleterious using our stringent filtering criteria (table S4). Tumor tissue was available for 11 of the 13 ATM-mutant cases and ATM IHC was performed as described above. Six of 11 ATM-mutant tumors had retained ATM expression by IHC, while five had ATM protein loss. Among the four tumors with predicted deleterious ATM alterations, two had retained ATM expression and two had loss of expression. Among the 11 MGH cases with ATM IHC, 5 patients had a bladder cancer event during follow-up (defined as an incomplete response to induction TMT, an invasive bladder or metastatic recurrence, or cystectomy for recurrent disease), whereas 6 patients did not have a bladder cancer event. There was a significant association between ATM expression and the likelihood of a bladder cancer event (P = 0.015, Fisher’s exact test; Fig. 5C), whereas there was no association between any ATM alteration or a predicted deleterious ATM alteration and a bladder cancer event (table S4). Modified bladder-intact event-free survival (mBI-EFS), a composite endpoint defined as lack of invasive bladder recurrence, pelvic recurrence, metastatic recurrence, cystectomy, or death from bladder cancer, was significantly longer in the subset of TMT patients with loss of ATM expression than in patients with retained ATM expression (P = 0.015; Fig. 5D). Together, these data suggest that loss of ATM expression by IHC is associated with improved clinical outcomes in patients with MIBC treated with TMT and that ATM IHC is a stronger correlate of clinical outcomes than ATM mutational status.
DISCUSSION
ATM plays a critical role in the DDR in normal cells, but ATM is recurrently lost or mutated in a subset of bladder and other tumor types (5). The specific contribution of ATM loss to tumorigenesis and tumor evolution across cellular contexts is poorly understood; however, functional ATM deficiency might increase DNA damage tolerance, thereby avoiding damage-induced cell death while also promoting protumorigenic changes in the tumor microenvironment (39, 40).
A variety of ATM alterations are present in bladder tumors, and the observed mutational frequency in a cohort can vary markedly based on the criteria used to nominate alterations. Approximately 12% of cases in the bladder cancer TCGA cohort have at least one ATM alteration, including missense, nonsense, or splice site mutations as well as in-frame or frameshift insertions/deletions (indels). While splice site, nonsense, or frameshift mutations are likely to result in the loss of a functional ATM protein, many of the missense mutations—particularly those outside the kinase domain—may not affect ATM function. To maximize the likelihood of identifying only those alterations most likely to affect protein function, we used stringent filtering criteria and found that only 5% of tumors in the TCGA cohort and 4% of tumors in a separate institutional bladder cancer cohort harbored an ATM alteration that was likely to result in loss of ATM function. Clinical studies, including several completed and ongoing clinical trials, have used different criteria to define ATM alterations, and it is likely that these differences may contribute to heterogeneity in findings across studies. We suggest using stringent filtering criteria to maximize the potential to nominate only those ATM alterations most likely to lead to functional deficiency.
As an alternative approach to inferring ATM functional status in bladder tumors, we also applied an ATM IHC assay to directly assess ATM protein expression. There was a correlation between the mutation class identified by tumor sequencing and the IHC pattern: Most tumors with WT ATM or an ATM missense mutation had retained ATM staining, whereas most tumors with truncating ATM alterations had loss of ATM staining. However, there were clear exceptions to this association, as some tumors with WT ATM or an ATM missense mutation had a loss of ATM protein expression by IHC, suggesting that alternative mechanisms such as epigenetic silencing may result in loss of ATM protein in a subset of bladder tumors. Analysis of larger cohorts with WT and mutant ATM cases will be required to further define this relationship.
The ability to reliably identify ATM alterations resulting in functional ATM deficiency is of notable clinical interest because ATM deficiency may be therapeutically targetable. Given ATM’s central role in the DDR, ATM-deficient tumors may have increased sensitivity to DNA-damaging agents including cisplatin and radiation. We find that ATM loss is sufficient to alter DNA damage signaling and drive increased cisplatin and radiation sensitivity in multiple preclinical human and mouse bladder cancer models, and sensitivity has also been observed in other non-bladder cancer preclinical models (24, 41, 42). However, clinical sensitivity to DNA-damaging agents has been more difficult to demonstrate, likely reflecting the heterogeneity of clinical cohorts and treatment regimens as well as differences in ATM mutation–calling approaches as discussed above. Nonetheless, we identified a trend toward improved outcomes in ATM-mutant cases in a cohort of patients with platinum-treated advanced bladder cancer. In addition, a recent study of patients with advanced cancer receiving palliative radiation did demonstrate increased local control of irradiated sites in patients with ATM-mutant tumors (43), thus providing important clinical evidence that ATM alterations may be associated with increased radiation sensitivity.
TMT is a curative treatment approach for MIBC and is supported by long-term safety and efficacy data (36, 37, 44). Cisplatin is one of the preferred concurrent chemotherapy agents delivered with radiation as part of TMT, and our preclinical cisplatin and radiation sensitivity data presented here support ATM loss as a predictor of increased sensitivity to cisplatin-based chemoradiotherapy regimens. In addition, in a clinical cohort of TMT-treated patients, functional ATM deficiency as identified by loss of ATM protein expression by IHC was more strongly associated with TMT outcomes than ATM mutational status alone. This is the first demonstration of an association between ATM functional status and TMT outcomes, and future studies are needed to validate this finding in larger cohorts as well as further explore the relationship among ATM mutational status, ATM IHC pattern, and clinical outcomes in patients with MIBC treated with TMT or other approaches.
An alternative bladder-sparing treatment option for MIBC is currently being investigated in several ongoing phase 2 clinical trials (45). MIBC patients with tumor DDR gene alterations who experience a complete clinical response (cCR) to cisplatin-based neoadjuvant chemotherapy forego standard-of-care radical cystectomy and instead undergo close surveillance. Interim analyses of two of these trials have been reported and did not find an association between ATM alterations and increased likelihood of cCR (46, 47). Potential explanations for the lack of signal include differences in methods used to nominate ATM alterations, the limited follow-up to date, and the small number of ATM-mutant cases. Additional follow-up and secondary analyses, perhaps including ATM IHC, may be useful in further investigating the association between ATM status and outcomes in these MIBC clinical trials.
PARP inhibitors are approved for use in several clinical settings and are under active investigation in bladder cancer. We observed a marked increase in PARP inhibitor sensitivity in multiple ATM-deleted preclinical bladder cancer models. However, this increase in PARP inhibitor sensitivity was not accompanied by features frequently associated with HR deficiency such as loss of radiation-induced Rad51 foci, suggesting that PARP inhibitor sensitivity conferred by ATM loss is mediated via different mechanism(s) than sensitivity conferred by loss of canonical HR genes such as BRCA1/2. Preclinical studies have identified toxic levels of nonhomologous end joining (NHEJ) as one potential mechanism of PARP inhibitor sensitivity in ATM-deficient cells (48), but the role of toxic NHEJ or other mechanisms in clinical cohorts remains to be determined. Two recent clinical trials [BAYOU (14) and ATLANTIS (15)] identified the activity of PARP inhibitors in subsets of bladder cancer patients with presumed HR deficiency based on alterations in one or more of a panel of putative HR genes. ATM was included in the HR gene list in both trials, but the number of ATM-altered cases was very low, making it difficult to meaningfully assess the relationship between ATM status and clinical PARP inhibitor sensitivity. Additional data from ongoing and planned bladder cancer PARP inhibitor trials are needed.
ATR and DNA-PK are related DDR kinases that also play key roles in DNA damage signaling. Multiple ATR and DNA-PK inhibitors are in various phases of clinical development in biomarker-selected and unselected populations. A randomized phase 2 clinical trial in advanced bladder cancer did not show benefit to the addition of the ATR inhibitor berzosertib to gemcitabine and cisplatin in a biomarker unselected population (49). Tumors with ATM deficiency may be more dependent on ATR signaling for survival, and ATM loss is one of the biomarkers under investigation in ATR clinical trials (10). We observed a marked increase in sensitivity to both ATR and DNA-PK inhibition in ATM-deficient models, consistent with increased dependence on these related DDR kinases for survival. Focusing clinical investigations on patients with tumor ATM loss or other biomarkers of ATR/DNA-PK inhibitor sensitivity may maximize the opportunity to observe the activity of these agents in bladder cancer.
ICIs have transformed the treatment landscape of advanced bladder cancer, but optimal predictive biomarkers of ICI sensitivity in bladder cancer have not been defined. Tumor mismatch repair deficiency (MMRd)/microsatellite instability (MSI) is associated with increased ICI response rates and pembrolizumab (anti-PD1) is approved for use in MMRd/MSI tumors across histologic subtypes. Alterations in DNA repair genes beyond the MMR pathway have also been associated with ICI response, although the data are less robust. In one study, advanced bladder cancer patients with a mutation in at least one gene across a panel of DDR genes had longer PFS following ICI treatment than patients lacking a DDR gene mutation (20). Although ATM was included in the DDR gene list, only four patients had a predicted deleterious ATM alteration. Our ATM-deleted syngeneic model was modestly more sensitive to anti-PD1 therapy than its WT ATM counterpart and had higher levels of infiltrating cytotoxic CD4+ T cells, which have been associated with immune-mediated cytotoxicity and ICI response in bladder cancer (50). However, despite these differences in the preclinical models, we did not observe differences in immune contexture or clinical outcomes by ATM status in several cohorts of patients with bladder cancer treated with ICIs. A recent study using a network-based approach found that ATM alterations were associated with improved ICI response and outcomes only among tumors with high TMB (51), which may explain the lack of association observed in the non-TMB stratified cohorts analyzed in this study. Therefore, although ATM loss is sufficient to drive changes in immune contexture and improve ICI response in isogenic preclinical systems, a predicted deleterious ATM alteration does not appear to be sufficient to significantly increase ICI sensitivity in clinical settings, likely due to the increased tumor and treatment heterogeneity that exists in clinical cohorts compared to preclinical systems.
In summary, our preclinical and clinical analyses inform the link between ATM biology and clinical translation in bladder cancer. First, the mutational landscape of ATM in bladder cancer is complex, but only alterations that result in loss of function are likely to confer an ATM-deficient phenotype; therefore, the criteria used to categorize ATM alterations in clinical cohorts can substantially affect the apparent association between ATM alterations and relevant clinical phenotypes. Second, complete loss of ATM protein in well-defined isogenic bladder cancer preclinical models is sufficient to induce sensitivity to a variety of clinically relevant drug classes including DNA-damaging agents, DNA repair–targeted agents, and anti-PD1 agents. However, clinical tumors are far more heterogeneous, and therefore, the association between ATM loss of function and clinical sensitivity to these agents remains to be determined. The best opportunity to identify an association between ATM loss and therapeutic effect will require stringent criteria to identify true loss-of-function ATM alterations in a clinical cohort treated with one or more agents that directly target ATM deficiency, such as cisplatin-based chemoradiotherapy.
MATERIALS AND METHODS
Experimental design
The objective of this study was to combine genomic, clinical, and preclinical (in vitro and in vivo) approaches to define the biological impact and clinical relevance of ATM alterations in bladder cancer.
TCGA cohort analyses
TCGA-BLCA WES tumor and matched normal BAM files were downloaded from the GDC data portal (https://portal.gdc.cancer.gov/; release number 12.0). Germline variants were called via GATK (version 3.8) HaplotypeCaller using the GRCh38 reference genome. Somatic variant calls generated by the MuTect2 pipeline were downloaded from the GDC data portal (https://portal.gdc.cancer.gov/; release number 12.0) in VCF formatted files. Germline and somatic variants that did not pass the default filters of MuTect2 were removed to filter out likely false positive calls. Germline and somatic mutations were annotated by ANNOVAR (52), and the pathogenicity of the variants was assessed by InterVar (version 2.2.2) (53). InterVar classifies variants into five categories: “Benign,” “Likely Benign,” “Uncertain Significance,” “Likely Pathogenic,” and “Pathogenic.” Germline and somatic mutations were classified as pathogenic if InterVar labeled them as “Likely Pathogenic” or “Pathogenic” or ClinVar (54) evidence suggested that they are likely pathogenic or pathogenic. Germline and somatic mutations were classified as variants of uncertain significance if both InterVar and ClinVar annotated the mutations as “Uncertain Significance.”
Reverse phase protein array data were downloaded with the TCGAbiolinks R package. The available samples (n = 343) were divided into four groups according to their somatic ATM alteration status and class (splice site n = 5, truncating n = 10, missense n = 24, no somatic alteration n = 304). The Wilcoxon test was used to compare the protein levels of ATM in each ATM-altered group to the group without somatic mutations in ATM. No correction was performed for multiple testing.
Mutual exclusivity and co-occurrence between somatic pathogenic gene mutations were tested using Fisher’s exact test. Pairwise testing was performed between ATM and each gene that was listed among the significantly mutated genes in the TCGA-BLCA cohort (4). The BH procedure was used to correct for multiple testing.
The Kaplan-Meier method was used to estimate the survival curves after dividing the patients into two groups: patients with pathogenic ATM mutations (ATM MUT) and patients with WT ATM or ATM variants of uncertain significance (ATM VUS). The log-rank test was used to compare the survival curves. Cox’s proportional hazards model was used to calculate the hazard ratios and to adjust for covariates such as age at diagnosis, sex, and race.
TIMER2.0 was used to estimate immune cell abundance in the TCGA-BLCA cohort based on expression levels of predetermined marker genes for each of the following immune cell phenotypes: B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and myeloid dendritic cells (55). Wilcoxon rank sum test was used to compare ATM-mutant and WT ATM samples and generate two-sided P values.
IMvigor210 cohort analyses
IMvigor210 was a single-arm phase 2 study investigating atezolizumab in patients with metastatic urothelial cancer (NCT02108652 and NCT02951767) (28). Patients had tumor tissue harvested within 2 years before study entry which was used for correlative studies including RNA-seq (for immune cell infiltration assessment) and targeted tumor DNA sequencing with the FoundationOne panel (Foundation Medicine, Cambridge, MA). A total of 275 patients had both RNA-seq and DNA sequencing data available and were used for the correlative analyses. ATM mutational status was determined from FoundationOne data using the same approach as was applied to tumor DNA from the TCGA cohort described above. For survival analysis, the Kaplan-Meier method was used and Cox-proportional hazards were estimated as above. Covariates included in the analysis were immune phenotype and tumor mutational burden (non-synonymous mutations per megabase). TIMER2.0 was used as above to estimate immune cell abundance. The Wilcoxon rank sum test was used to compare ATM-mutant and WT samples and generate two-sided P values.
Institutional datasets and analyses
The DFBWCC cohort consisted of 773 patients diagnosed with urothelial cancer with available targeted tumor DNA sequencing performed using the OncoPanel assay (22, 56). Tumor specimens and clinicopathologic information were collected from patients who consented to Institutional Review Board (IRB)–approved protocol nos. 11-104 or 17-000. Samples were filtered to include bladder urothelial carcinomas and upper tract urothelial carcinomas, and primary biopsy sites were used when multiple sequenced samples were available from a single patient. ATM mutational status was determined from OncoPanel data using the same approach as was applied to tumor DNA from the TCGA cohort described above. The Kaplan-Meier method and Cox-proportional hazards model were used as above. Covariates included for the ICI cohort were the number of prior lines of systemic therapy, tumor mutational burden, and therapy (single agent ICI versus combination). Covariates for the platinum cohort were a number of lines of prior systemic therapy and the type of platinum agent (cisplatin versus carboplatin).
The MGH cohort consisted of 76 patients with MIBC treated with curative-intent TMT with available WES and clinical data (38). IHC was performed on all ATM-mutant cases with available formalin-fixed, paraffin-embedded (FFPE) tumor tissue. This study was approved by the Mass General Brigham IRB (protocol no. 2008P001128) and conducted in accordance with recognized ethical guidelines. mBI-EFS is defined as muscle-invasive recurrence, locoregional progression, distant metastasis, radical cystectomy for either cancer recurrence or toxicity, or death from bladder cancer.
Cell culture and ATM KO cell lines
The BBN963 and UPPL1541 murine bladder cancer cell lines were gifts from the William Kim laboratory (57). BBN963 and UPPL1541 were cultured and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. Human bladder cancer cell lines T24 and 5637 were purchased from American Type Culture Collection and KU19-19 was purchased from DSMZ. The cell lines were cultured in RPMI 1640 media supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin. All cell lines were grown at 37°C in a 5% CO2 incubator.
The Alt-R CRISPR-Cas9 System (IDT Technologies) was used to delete the ATM gene in mouse and human bladder cancer cell lines. CRISPR RNA (crRNA) oligonucleotides targeting ATM are listed in table S5. Cas9 nuclease was purchased from Horizon Discovery. The crRNA was annealed with ATTO 550 tracrRNA, and ribonucleoparticles were then assembled by adding Cas9. RNPs were delivered into cells using electroporation-based nucleofection (Lonza system). Flow cytometry was used to sort ATTO 550 positive single cells 24 hours following nucleofection. Single cells were expanded and clonal populations were screened by immunoblot to identify clones with complete loss of expression of the ATM protein.
Reagents
Chemical, single guide RNA, and antibody details are provided in table S5.
Immunoblotting
Cells were trypsinized with Tryp-LE Express (Gibco), washed with phosphate-buffered saline (PBS), and lysed with radioimmunoprecipitation assay buffer (Pierce) supplemented with protease inhibitors cocktail (Roche), phosphatase inhibitors (phosSTOP, Roche), and phenylmethylsulfonyl fluoride (Cell Signaling Technology). Cells were then sonicated and centrifuged. The supernatant was isolated and protein concentration was quantified using the Bio-Rad Protein Assay. The protein lysate was then denatured in Laemmli buffer (Bio-Rad) supplemented with reducing agent NuPAGE (Invitrogen) and heated at 95°C for 10 min. Samples were loaded into 3 to 8% gradient gels and run for 1 to 3 hours at 90 to 110 V on ice. Gels were then transferred to 0.22-μm polyvinylidene fluoride membrane (Millipore) for 2 to 3 hours at 90 V on ice. Membranes were blocked with 5% bovine serum albumin (BSA)/tris-buffered saline with Tween 20 and incubated with primary antibody, rinsed in PBS, and exposed to secondary antibody (see table S5 for antibody details). Membranes were developed using enhanced chemiluminescent substrate (PerkinElmer) and imaged using an Amersham Imager 600.
Rad51 foci formation
Glass coverslips were placed into wells of a 12-well plate and cells were plated in the wells. The following day, cells were treated with 10 grays (Gy) using a cell irradiator (Rad Source Technologies). Thirty minutes and 24 hours after irradiation, the glass slides were transferred to a new plate and fixed in 4% paraformaldehyde for 20 min, and then permeabilized for 5 min on ice using 0.2% Triton X-100 in 1% BSA/PBS. Cells were then blocked in 1% BSA/PBS for 1 hour at room temperature and incubated with an anti-Rad51 antibody (Cell Signaling Technology, clone d4b10, catalog no. 8875) overnight at 4°C. A fluorescence-conjugated anti-Rabbit secondary antibody was added and slides were incubated in the dark at 4°C for 1 hour. Last, the glass coverslips were mounted on slides with ProLong Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Fluorescent images were obtained using a Zeiss microscope and were quantified using ImageJ software.
In vitro drug and radiation sensitivity assays
Clonogenic survival assays were used to evaluate drug and radiation sensitivity. Briefly, 3000 to 5000 cells were plated per well into six-well plates, and on the following day, cells were treated with drugs or irradiated. The cells were then allowed to grow for 7 to 10 days and then fixed with 10% formalin for 30 min and stained with a 1% crystal violet (CV) solution for 30 min. Plates were imaged using an Amersham Imager 600 instrument. One milliliter of 1% SDS solution was then added to each well to solubilize the CV, and the solutions were quantified at 570 nm on a microplate reader (BioTek). Survival was calculated as the absorbance at a given dose or concentration relative to the absorbance of untreated samples. Data were plotted and analyzed using GraphPad Prism.
Xenograft experiments
All experiments were performed in accordance with the Dana-Farber Cancer Institute (DFCI) IACUC guidelines at the DFCI Longwood Center Animal Resource Facility under an approved protocol. Eight mice were randomized into each of four groups: (i) BBN963 untreated, (ii) BBN963-ATM-KO untreated, (iii) BBN963 irradiated, and (iv) BBN963-ATM-KO irradiated. Five million BBN963 or BBN963-ATM-KO cells in 75 μl of PBS were mixed with 75 μl of Matrigel (Corning), and then injected into the flanks of 6-week-old C57BL/6 mice (Jackson Laboratory) subcutaneously. Radiation treatments were performed using a small animal radiation research platform (Xstrahl Inc). Mice were anesthetized with isoflurane and imaged by computed tomography. Image-guided radiation therapy was performed with a field size designed to completely encompass the xenografted tumor while minimizing exposure to adjacent normal structures. Each mouse was treated with three fractions of 4 Gy over 5 to 6 days. Tumor volume was measured every 2 or 3 days. InVivoPlus anti-mouse PD1 (CD279) (Bio-X-cell, clone J43, #BP0033-2) was delivered via intraperitoneal injection at a dose of 10 mg/kg twice per week for 4 weeks. Mice began treatment when the average tumor volume reached 150 mm3. Tumor volumes were measured twice per week using a digital caliper and growth curves were generated using GraphPad Prism 7. A linear regression model was used to compare the anti-PD1 treatment effect in ATM-KO and WT ATM groups.
Flow cytometry and immunophenotyping
Tumors were harvested, minced, and then digested with enzyme cocktail [DMEM medium with collagenase/hyaluronidase (1 mg/ml; STEMCELL), 10 mM Hepes, 2% FBS, and deoxyribonuclease (0.1 mg/ml; STEMCELL)] for 45 min at 37°C. Digested samples were filtered through a 70-μm pore cell strainer (Fisherbrand). Single-cell suspensions were centrifuged at 350g for 5 min and washed in pre-cooled flow buffer (PBS with 2% FBS). Samples were incubated with RBC Lysis Buffer (BioLegend) at room temperature for 5 min, washed with flow buffer, and centrifuged. Cell pellets were resuspended in 1% BSA/PBS and 1.5 million cells were aliquoted to tubes for staining. Single-cell suspensions were incubated with Fc block (1:500; anti-CD16/CD32 antibody; BioLegend) for 10 min at 4°C and samples were stained with fixable viability dye eFluor780 (Invitrogen) before staining with markers. UltraComp eBeads Compensation Beads (Invitrogen) were stained with one-tenth concentration of antibody as tumor cells and used for compensation. Tumor cells stained with a single fluorescence-conjugated antibody were used for gating and confirming compensation. See table S5 for antibody details. Surface antibodies were incubated with samples on ice for 30 min and antibodies for intracellular markers were incubated with samples for 40 min after fixing and permeabilization (eBioscience). Samples were washed and suspended with a flow buffer, and flow cytometry data were obtained using a BD LSRFortessa analyzer (BD Biosciences) with data analysis using FlowJo 10 software.
RNA sequencing
Snap-frozen tumors were sent to Novogene (Durham, NC) for RNA extraction and sequencing (20 million reads per sample). The Nextflow RNA-seq pipeline (58) (nf-core/rnaseq; version 3.9) was used to perform quality control, and STAR aligner and Salmon (59) were used for quantification. Counts were normalized using the median of ratios method and differential expression analysis was performed with DESeq2 (60). A general assessment of full profiles was performed using principal components analysis. Immune cell fractions as well as immune and stroma scores were derived using TIMER (61) and ESTIMATE (62) algorithms. TCR receptor diversity was calculated on the basis of RNA-seq results using MiXCR (version 4.1.2; MiLaboratories Inc) (63). Calculated scores and fractions were compared between groups using Student’s t tests. P values were corrected using the BH method. Raw and processed sequencing files will be deposited in GEO.
Multiplex immunofluorescence
ImmunoProfile is a mIF assay performed at the Center for Immuno-Oncology at DFCI. A 5-μm-thick section from the FFPE tumor was mounted on a glass slide and stained for PD-L1, PD1, FOXP3, CD8, cytokeratin (AE1/AE3), and DAPI (nuclear counterstain). Slides were then scanned at 20× using a Vectra Polaris imaging platform and regions of interest (ROIs) were defined for each image. Within each ROI, quantitative image analysis was performed using InForm Image Analysis software (PerkinElmer/Akoya) to phenotype and score cells based on a signal from each marker. Cell count was calculated per ROI and averaged (unweighted) across ROIs, reported as count per millimeter squared ± SE. The statistical significance of differential cell-type enrichment between groups was estimated with the Wilcox rank sum test.
ATM immunohistochemistry
Bladder cancer cases with banked FFPE tumor tissue were reviewed by a pathologist to identify the block with the highest tumor density. IHC was performed on 5-μm-thick FFPE tissue sections from bladder cancer cases using an anti-ATM antibody (dilution: 1:100; Abcam, Cambridge, UK, clone Y170, catalog no. ab32420) following EDTA antigen retrieval. Staining was performed using the Leica BOND-III automated staining platform and the Leica Biosystems BOND Polymer Refine Detection kit. Positive control slides were stained in parallel. ATM IHC slides were reviewed independently by two pathologists blinded to the clinical and genomic data. A consensus interpretation of ATM expression as retained or lost was assigned for each case. Loss of ATM expression was defined as a complete lack of staining for ATM in the tumor cell nuclei. Retained ATM expression was defined as positive staining for ATM (moderate to strong intensity). Adjacent nonneoplastic endothelial cells and fibroblasts were used as an internal positive control.
Statistical analyses
Genomic, clinical, and functional data were analyzed as detailed above.
Acknowledgments
We thank the William Kim laboratory (University of North Carolina) for the BBN963 and UPPL1541 cell lines. We thank Dana-Farber/Harvard Cancer Center for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry service. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516.
Funding: This work was supported by NCI (R01CA272657 to K.W.M., R01CA259007 to D.T.M., C06CA059267 to J.A.E., and U01CA260369 to P.A.), the Research and Technology Innovation Fund (KTIA_NAP_13-2014-0021 and 2017-1.2.1-NKP-2017-00002 to Z.Sza.), the Breast Cancer Research Foundation (BCRF-21-159 to Z.S.), Kræftens Bekæmpelse (R281-A16566 to Z.S.), the Department of Defense through the Prostate Cancer Research Program (W81XWH-18-2-0056 to Z.S.), Det Frie Forskningsråd Sundhed og Sygdom (7016-00345B to Z.S.), NIH (P01CA228696-01A1 to Z.S.), and the Radiation Oncology Institute (to D.T.M.)
Author contributions: Conceptualization: Y.Z., J.B., Z.S., and K.W.M. Methodology: Y.Z., J.B., E.A., A.J.N., K.S., D.R.S., A.N., R.T.B., W.A., Z.S., and K.W.M. Investigation: Y.Z., J.B., E.A., S.C.K., A.J.N., K.S., D.F., D.R.S., Z.S., A.S., A.N., R.T.B., T.H., I.E., B.S., K.F., and C.-L.W. Visualization: Y.Z., J.B., E.A., S.C.K., W.A., and K.W.M. Supervision: J.A.E., D.T.M., W.A., Z.S., and K.W.M. Writing—original draft: Y.Z., J.B., and K.W.M. Writing—review and editing: All authors.
Competing interests: Judit Börcsök, Zsofia Sztupinszki, Zoltan Szallasi, and Kent W Mouw are listed as co-inventors on a pending patent for a mutational signature based method to identify nucleotide excision repair deficiency. D.R.S. has independent research support from Pfizer and Merck and speaking fees from MSD. K.W.M. declares advisory/consulting fees from EMD Serono, Pfizer, UroGen, and Riva Therapeutics; declares research support from Pfizer and Novo Ventures; is an equity stockholder in Riva Therapeutics; and declares writing/speaking fees from UpToDate and OncLive. J.A.E. declares advisory/consulting fees from Blue Earth Diagnostics, Boston Scientific, AstraZeneca, Merck, EMD Serono, Roivant Pharma, Myovant Sciences, Janssen, Bayer Healthcare, Progenics Pharmaceuticals, Pfizer, Astellas, Genentech, Gilead, IBA, Elekta, Lantheus, and Angiodynamics; and declares writing/speaking fees from UpToDate. P.A. declares advisory/consulting fees from Merck, Natera, Protara, Imvax, and Abyost Pharmaceuticals. The other authors declare that they have no competing interests.
Data and materials availability: RNA sequencing data have been uploaded to the GEO database (www.ncbi.nlm.nih.gov/geo/) at accession number GSE232850. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for tables S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Y. Shiloh, Y. Ziv, The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013). [PubMed] [Google Scholar]
- 2.A. Ciccia, S. J. Elledge, The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. Rothblum-Oviatt, J. Wright, M. A. Lefton-Greif, S. A. McGrath-Morrow, T. O. Crawford, H. M. Lederman, Ataxia telangiectasia: A review. Orphanet J. Rare Dis. 11, 159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. G. Robertson, J. Kim, H. Al-Ahmadie, J. Bellmunt, G. Guo, A. D. Cherniack, T. Hinoue, P. W. Laird, K. A. Hoadley, R. Akbani, M. A. A. Castro, E. A. Gibb, R. S. Kanchi, D. A. Gordenin, S. A. Shukla, F. Sanchez-Vega, D. E. Hansel, B. A. Czerniak, V. E. Reuter, X. Su, B. de Sa Carvalho, V. S. Chagas, K. L. Mungall, S. Sadeghi, C. S. Pedamallu, Y. Lu, L. J. Klimczak, J. Zhang, C. Choo, A. I. Ojesina, S. Bullman, K. M. Leraas, T. M. Lichtenberg, C. J. Wu, N. Schultz, G. Getz, M. Meyerson, G. B. Mills, D. J. M. Conkey; TCGA Research Network, J. N. Weinstein, D. J. Kwiatkowski, S. P. Lerner, Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556.e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Choi, T. Kipps, R. Kurzrock, ATM mutations in cancer: Therapeutic implications. Mol. Cancer Ther. 15, 1781–1791 (2016). [DOI] [PubMed] [Google Scholar]
- 6.G. Iyer, A. V. Balar, M. I. Milowsky, B. H. Bochner, G. Dalbagni, S. M. Donat, H. W. Herr, W. C. Huang, S. S. Taneja, M. Woods, I. Ostrovnaya, H. al-Ahmadie, M. E. Arcila, J. C. Riches, A. Meier, C. Bourque, M. Shady, H. Won, T. L. Rose, W. Y. Kim, B. E. Kania, M. E. Boyd, C. K. Cipolla, A. M. Regazzi, D. Delbeau, A. S. McCoy, H. A. Vargas, M. F. Berger, D. B. Solit, J. E. Rosenberg, D. F. Bajorin, Multicenter prospective phase II trial of neoadjuvant dose-dense gemcitabine plus cisplatin in patients with muscle-invasive bladder cancer. J. Clin. Oncol. 36, 1949–1956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.E. R. Plimack, R. L. Dunbrack, T. A. Brennan, M. D. Andrake, Y. Zhou, I. G. Serebriiskii, M. Slifker, K. Alpaugh, E. Dulaimi, N. Palma, J. Hoffman-Censits, M. Bilusic, Y. N. Wong, A. Kutikov, R. Viterbo, R. E. Greenberg, D. Y. T. Chen, C. D. Lallas, E. J. Trabulsi, R. Yelensky, D. J. McConkey, V. A. Miller, E. A. Golemis, E. A. Ross, Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur. Urol. 68, 959–967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Y. Teo, R. M. Bambury, E. C. Zabor, E. Jordan, H. Al-Ahmadie, M. E. Boyd, N. Bouvier, S. A. Mullane, E. K. Cha, N. Roper, I. Ostrovnaya, D. M. Hyman, B. H. Bochner, M. E. Arcila, D. B. Solit, M. F. Berger, D. F. Bajorin, J. Bellmunt, G. Iyer, J. E. Rosenberg, DNA damage response and repair gene alterations are associated with improved survival in patients with platinum-treated advanced urothelial carcinoma. Clin. Cancer Res. 23, 3610–3618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E. M. Van Allen, K. W. Mouw, P. Kim, G. Iyer, N. Wagle, H. Al-Ahmadie, C. Zhu, I. Ostrovnaya, G. V. Kryukov, K. W. O’Connor, J. Sfakianos, I. Garcia-Grossman, J. Kim, E. A. Guancial, R. Bambury, S. Bahl, N. Gupta, D. Farlow, A. Qu, S. Signoretti, J. A. Barletta, V. Reuter, J. Boehm, M. Lawrence, G. Getz, P. Kantoff, B. H. Bochner, T. K. Choueiri, D. F. Bajorin, D. B. Solit, S. Gabriel, A. D’Andrea, L. A. Garraway, J. E. Rosenberg, Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 4, 1140–1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. M. Cleary, A. J. Aguirre, G. I. Shapiro, A. D. D'Andrea, Biomarker-guided development of DNA repair inhibitors. Mol. Cell 78, 1070–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Carreira, N. Porta, S. Arce-Gallego, G. Seed, A. Llop-Guevara, D. Bianchini, P. Rescigno, A. Paschalis, C. Bertan, C. Baker, J. Goodall, S. Miranda, R. Riisnaes, I. Figueiredo, A. Ferreira, R. Pereira, M. Crespo, B. Gurel, D. Nava Rodrigues, S. J. Pettitt, W. Yuan, V. Serra, J. Rekowski, C. J. Lord, E. Hall, J. Mateo, J. S. de Bono, Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B Trial. Cancer Discov. 11, 2812–2827 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. Grivas, Y. Loriot, R. Morales-Barrera, M. Y. Teo, Y. Zakharia, S. Feyerabend, N. J. Vogelzang, E. Grande, N. Adra, A. Alva, A. Necchi, A. Rodriguez-Vida, S. Gupta, D. H. Josephs, S. Srinivas, K. Wride, D. Thomas, A. Simmons, A. Loehr, R. L. Dusek, D. Nepert, S. Chowdhury, Efficacy and safety of rucaparib in previously treated, locally advanced or metastatic urothelial carcinoma from a phase 2, open-label trial (ATLAS). BMC Cancer 21, 593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.T. Powles, D. Carroll, S. Chowdhury, G. Gravis, F. Joly, J. Carles, A. Fléchon, P. Maroto, D. Petrylak, F. Rolland, N. Cook, A. V. Balar, S. S. Sridhar, M. D. Galsky, P. Grivas, A. Ravaud, R. Jones, J. Cosaert, D. Hodgson, I. Kozarewa, R. Mather, R. McEwen, F. Mercier, D. Landers, An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat. Med. 27, 793–801 (2021). [DOI] [PubMed] [Google Scholar]
- 14.J. E. Rosenberg, S. H. Park, T. V. Dao, D. E. Castellano, J. R. Li, S. Mukherjee, K. Howells, H. Dry, M. C. Lanasa, R. Stewart, D. F. Bajorin, BAYOU: A phase II, randomized, multicenter, double-blind, study of durvalumab (D) in combination with olaparib (O) for the first-line treatment of platinum-ineligible patients with unresectable, stage IV urothelial carcinoma (UC). J. Clin. Oncol. 40, 437–437 (2022). [Google Scholar]
- 15.S. J. Crabb, S. A. Hussain, E. Soulis, S. Hinsley, L. Dempsey, A. Trevethan, Y. P. Song, J. Barber, J. A. Frew, J. Gale, G. Faust, S. J. Brock, U. B. McGovern, O. Parikh, D. Enting, S. Sundar, G. Ratnayake, K. Lees, T. Powles, R. J. Jones, A randomized, double blind, biomarker selected, phase II clinical trial of maintenance PARP inhibition following chemotherapy for metastatic urothelial carcinoma (mUC): Final analysis of the ATLANTIS rucaparib arm. J. Clin. Oncol. 40, 436–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. W. Mouw, M. S. Goldberg, P. A. Konstantinopoulos, A. D. D'Andrea, DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 7, 675–693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R. Yi, A. Lin, M. Cao, A. Xu, P. Luo, J. Zhang, ATM Mutations benefit bladder cancer patients treated with immune checkpoint inhibitors by acting on the tumor immune microenvironment. Front. Genet. 11, 933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Hu, M. Zhou, X. Bao, D. Pan, M. Jiao, X. Liu, F. Li, C. Y. Li, ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. J. Clin. Invest. 131, e139333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Q. Zhang, M. D. Green, X. Lang, J. Lazarus, J. D. Parsels, S. Wei, L. A. Parsels, J. Shi, N. Ramnath, D. R. Wahl, M. Pasca di Magliano, T. L. Frankel, I. Kryczek, Y. L. Lei, T. S. Lawrence, W. Zou, M. A. Morgan, Inhibition of ATM increases interferon signaling and sensitizes pancreatic cancer to immune checkpoint blockade therapy. Cancer Res. 79, 3940–3951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Y. Teo, K. Seier, I. Ostrovnaya, A. M. Regazzi, B. E. Kania, M. M. Moran, C. K. Cipolla, M. J. Bluth, J. Chaim, H. al-Ahmadie, A. Snyder, M. I. Carlo, D. B. Solit, M. F. Berger, S. Funt, J. D. Wolchok, G. Iyer, D. F. Bajorin, M. K. Callahan, J. E. Rosenberg, Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J. Clin. Oncol. 36, 1685–1694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. T. Chang, T. S. Bhattarai, A. M. Schram, C. M. Bielski, M. T. A. Donoghue, P. Jonsson, D. Chakravarty, S. Phillips, C. Kandoth, A. Penson, A. Gorelick, T. Shamu, S. Patel, C. Harris, J. J. Gao, S. O. Sumer, R. Kundra, P. Razavi, B. T. Li, D. N. Reales, N. D. Socci, G. Jayakumaran, A. Zehir, R. Benayed, M. E. Arcila, S. Chandarlapaty, M. Ladanyi, N. Schultz, J. Baselga, M. F. Berger, N. Rosen, D. B. Solit, D. M. Hyman, B. S. Taylor, Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. H. Nassar, R. Umeton, J. Kim, K. Lundgren, L. Harshman, E. M. van Allen, M. Preston, F. Dong, J. Bellmunt, K. W. Mouw, T. K. Choueiri, G. Sonpavde, D. J. Kwiatkowski, Mutational analysis of 472 urothelial carcinoma across grades and anatomic sites. Clin. Cancer Res. 25, 2458–2470 (2019). [DOI] [PubMed] [Google Scholar]
- 23.P. Mullen, PARP cleavage as a means of assessing apoptosis. Methods Mol. Med. 88, 171–181 (2004). [DOI] [PubMed] [Google Scholar]
- 24.S. Rafiei, K. Fitzpatrick, D. Liu, M. Y. Cai, H. A. Elmarakeby, J. Park, C. Ricker, B. S. Kochupurakkal, A. D. Choudhury, W. C. Hahn, S. P. Balk, J. H. Hwang, E. M. van Allen, K. W. Mouw, ATM loss confers greater sensitivity to atr inhibition than PARP inhibition in prostate cancer. Cancer Res. 80, 2094–2100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. H. L. Fok, A. Ramos-Montoya, M. Vazquez-Chantada, P. W. G. Wijnhoven, V. Follia, N. James, P. M. Farrington, A. Karmokar, S. E. Willis, J. Cairns, J. Nikkilä, D. Beattie, G. M. Lamont, M. R. V. Finlay, J. Wilson, A. Smith, L. O. O’Connor, S. Ling, S. E. Fawell, M. J. O’Connor, S. J. Hollingsworth, E. Dean, F. W. Goldberg, B. R. Davies, E. B. Cadogan, AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 10, 5065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. J. Davis, B. P. Chen, D. J. Chen, DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair 17, 21–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. Snyder, T. Nathanson, S. A. Funt, A. Ahuja, J. Buros Novik, M. D. Hellmann, E. Chang, B. A. Aksoy, H. al-Ahmadie, E. Yusko, M. Vignali, S. Benzeno, M. Boyd, M. Moran, G. Iyer, H. S. Robins, E. R. Mardis, T. Merghoub, J. Hammerbacher, J. E. Rosenberg, D. F. Bajorin, Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 14, e1002309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Mariathasan, S. J. Turley, D. Nickles, A. Castiglioni, K. Yuen, Y. Wang, E. E. Kadel III, H. Koeppen, J. L. Astarita, R. Cubas, S. Jhunjhunwala, R. Banchereau, Y. Yang, Y. Guan, C. Chalouni, J. Ziai, Y. Şenbabaoğlu, S. Santoro, D. Sheinson, J. Hung, J. M. Giltnane, A. A. Pierce, K. Mesh, S. Lianoglou, J. Riegler, R. A. D. Carano, P. Eriksson, M. Höglund, L. Somarriba, D. L. Halligan, M. S. van der Heijden, Y. Loriot, J. E. Rosenberg, L. Fong, I. Mellman, D. S. Chen, M. Green, C. Derleth, G. D. Fine, P. S. Hegde, R. Bourgon, T. Powles, TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. V. Balar, M. D. Galsky, J. E. Rosenberg, T. Powles, D. P. Petrylak, J. Bellmunt, Y. Loriot, A. Necchi, J. Hoffman-Censits, J. L. Perez-Gracia, N. A. Dawson, M. van der Heijden, R. Dreicer, S. Srinivas, M. M. Retz, R. W. Joseph, A. Drakaki, U. N. Vaishampayan, S. S. Sridhar, D. I. Quinn, I. Durán, D. R. Shaffer, B. J. Eigl, P. D. Grivas, E. Y. Yu, S. Li, E. E. Kadel3rd, Z. Boyd, R. Bourgon, P. S. Hegde, S. Mariathasan, A. Thåström, O. O. Abidoye, G. D. Fine, D. F. Bajorin; IMvigor210 Study Group , Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 389, 67–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. D. Carey, D. Gusenleitner, M. Lipschitz, M. G. M. Roemer, E. C. Stack, E. Gjini, X. Hu, R. Redd, G. J. Freeman, D. Neuberg, F. S. Hodi, X. S. Liu, M. A. Shipp, S. J. Rodig, Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 130, 2420–2430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Neeb, N. Herranz, S. Arce-Gallego, S. Miranda, L. Buroni, W. Yuan, A. Athie, T. Casals, J. Carmichael, D. N. Rodrigues, B. Gurel, P. Rescigno, J. Rekowski, J. Welti, R. Riisnaes, V. Gil, J. Ning, V. Wagner, I. Casanova-Salas, S. Cordoba, N. Castro, M. D. Fenor de la Maza, G. Seed, K. Chandran, A. Ferreira, I. Figueiredo, C. Bertan, D. Bianchini, C. Aversa, A. Paschalis, M. Gonzalez, R. Morales-Barrera, C. Suarez, J. Carles, A. Swain, A. Sharp, J. Gil, V. Serra, C. Lord, S. Carreira, J. Mateo, J. S. de Bono, Advanced prostate cancer with ATM loss: PARP and ATR inhibitors. Eur. Urol. 79, 200–211 (2021). [DOI] [PubMed] [Google Scholar]
- 32.H. Kaur, D. C. Salles, S. Murali, J. L. Hicks, M. Nguyen, C. C. Pritchard, A. M. de Marzo, J. S. Lanchbury, B. J. Trock, W. B. Isaacs, K. M. Timms, E. S. Antonarakis, T. L. Lotan, Genomic and clinicopathologic characterization of atm-deficient prostate cancer. Clin. Cancer Res. 26, 4869–4881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. M. Miller, C. Nworu, L. McKee, D. Balcerzak, L. Pham, J. Pugh, Y. Z. Liu, H. Gustafson, E. Marwah, T. Lamb, J. Clements, Development of an immunohistochemical assay to detect the ataxia-telangiectasia mutated (ATM) protein in gastric carcinoma. Appl. Immunohistochem. Mol. Morphol. 28, 303–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.T. A. Yap, D. S. P. Tan, A. Terbuch, R. Caldwell, C. Guo, B. C. Goh, V. Heong, N. R. M. Haris, S. Bashir, Y. Drew, D. S. Hong, F. Meric-Bernstam, G. Wilkinson, J. Hreiki, A. M. Wengner, F. Bladt, A. Schlicker, M. Ludwig, Y. Zhou, L. Liu, S. Bordia, R. Plummer, E. Lagkadinou, J. S. de Bono, First-in-human trial of the oral ataxia telangiectasia and RAD3-related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov. 11, 80–91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.P. A. Konstantinopoulos, A. A. B. A. da Costa, D. Gulhan, E. K. Lee, S.-C. Cheng, A. E. Wahner Hendrickson, B. Kochupurakkal, D. L. Kolin, E. C. Kohn, J. F. Liu, E. H. Stover, J. Curtis, N. Tayob, M. Polak, D. Chowdhury, U. A. Matulonis, A. Färkkilä, A. D. D’Andrea, G. I. Shapiro, A Replication stress biomarker is associated with response to gemcitabine versus combined gemcitabine and ATR inhibitor therapy in ovarian cancer. Nat. Commun. 12, 5574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.N. J. Giacalone, W. U. Shipley, R. H. Clayman, A. Niemierko, M. Drumm, N. M. Heney, M. D. Michaelson, R. J. Lee, P. J. Saylor, M. F. Wszolek, A. S. Feldman, D. M. Dahl, A. L. Zietman, J. A. Efstathiou, Long-term outcomes after bladder-preserving tri-modality therapy for patients with muscle-invasive bladder cancer: An updated analysis of the Massachusetts General Hospital experience. Eur. Urol. 71, 952–960 (2017). [DOI] [PubMed] [Google Scholar]
- 37.D. J. Konieczkowski, J. A. Efstathiou, K. W. Mouw, Contemporary and emerging approaches to bladder-preserving trimodality therapy for muscle-invasive bladder cancer. Hematol. Oncol. Clin. North Am. 35, 567–584 (2021). [DOI] [PubMed] [Google Scholar]
- 38.S. C. Kamran, Y. Zhou, K. Otani, M. Drumm, Y. Otani, S. Wu, C.-L. Wu, A. S. Feldman, M. Wszolek, R. J. Lee, P. J. Saylor, J. Lennerz, E. Van Allen, H. Willers, T. S. Hong, Y. Liu, E. Davicioni, E. A. Gibb, W. U. Shipley, K. W. Mouw, J. A. Efstathiou, D. T. Miyamoto, Genomic tumor correlates of clinical outcomes following organ-sparing chemoradiation therapy for bladder cancer. Clin. Cancer Res. 10.1158/1078-0432.CCR-23-0792 (2023). [DOI] [PMC free article] [PubMed]
- 39.R. Russell, L. Perkhofer, S. Liebau, Q. Lin, A. Lechel, F. M. Feld, E. Hessmann, J. Gaedcke, M. Güthle, M. Zenke, D. Hartmann, G. von Figura, S. E. Weissinger, K. L. Rudolph, P. Möller, J. K. Lennerz, T. Seufferlein, M. Wagner, A. Kleger, Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat. Commun. 6, 7677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Squatrito, C. W. Brennan, K. Helmy, J. T. Huse, J. H. Petrini, E. C. Holland, Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell 18, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Perkhofer, A. Schmitt, M. C. Romero Carrasco, M. Ihle, S. Hampp, D. A. Ruess, E. Hessmann, R. Russell, A. Lechel, N. Azoitei, Q. Lin, S. Liebau, M. Hohwieler, H. Bohnenberger, M. Lesina, H. Algül, L. Gieldon, E. Schröck, J. Gaedcke, M. Wagner, L. Wiesmüller, B. Sipos, T. Seufferlein, H. C. Reinhardt, P. O. Frappart, A. Kleger, ATM deficiency generating genomic instability sensitizes pancreatic ductal adenocarcinoma cells to therapy-induced dna damage. Cancer Res. 77, 5576–5590 (2017). [DOI] [PubMed] [Google Scholar]
- 42.C. Wang, N. Jette, D. Moussienko, D. G. Bebb, S. P. Lees-Miller, ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Transl. Oncol. 10, 190–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. L. Pitter, D. L. Casey, Y. C. Lu, M. Hannum, Z. Zhang, X. Song, I. Pecorari, B. M. Millan, J. Ma, R. M. Samstein, I. X. Pei, A. J. Khan, L. Z. Braunstein, L. G. T. Morris, C. A. Barker, A. Rimner, K. M. Alektiar, P. B. Romesser, C. H. Crane, J. Yahalom, M. J. Zelefsky, H. I. Scher, J. L. Bernstein, D. L. Mandelker, B. Weigelt, J. S. Reis-Filho, N. Y. Lee, S. N. Powell, T. A. Chan, N. Riaz, J. Setton, Pathogenic ATM mutations in cancer and a genetic basis for radiotherapeutic efficacy. J. Natl. Cancer Inst. 13, 266–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.J. A. Efstathiou, K. Bae, W. U. Shipley, D. S. Kaufman, M. P. Hagan, N. M. Heney, H. M. Sandler, Late pelvic toxicity after bladder-sparing therapy in patients with invasive bladder cancer: RTOG 89-03, 95-06, 97-06, 99-06. J. Clin. Oncol. 27, 4055–4061 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.P. Grivas, DNA damage response gene alterations in urothelial cancer: Ready for practice? Clin. Cancer Res. 25, 907–909 (2019). [DOI] [PubMed] [Google Scholar]
- 46.M. D. Galsky, S. Daneshmand, K. G. Chan, T. B. Dorff, J. P. Cetnar, B. O Neil, A. D'souza, R. Mamtani, C. Kyriakopoulos, P. Garcia, S. Izadmehr, M. Yu, Q. Zhao, R. Mehrazin, S. C. Lewis, J. Sfakianos, S. K. Pal, Phase 2 trial of gemcitabine, cisplatin, plus nivolumab with selective bladder sparing in patients with muscle- invasive bladder cancer (MIBC): HCRN GU 16-257. J. Clin. Oncol. 39, 4503–4503 (2021). [Google Scholar]
- 47.D. M. Geynisman, P. Abbosh, E. A. Ross, M. R. Zibelman, P. Ghatalia, F. Anari, K. Ansel, J. R. Mark, L. Stamatakis, J. H. Hoffman-Censits, R. Viterbo, E. M. Horwitz, M. A. Hallman, R. K. Alpaugh, R. E. Greenberg, M. C. Smaldone, R. G. Uzzo, D. Chen, A. Kutikov, E. R. Plimack, A phase II trial of risk enabled therapy after initiating neoadjuvant chemotherapy for bladder cancer (RETAIN BLADDER): Interim analysis. J. Clin. Oncol. 39, 397–397 (2021).33434060 [Google Scholar]
- 48.G. Balmus, D. Pilger, J. Coates, M. Demir, M. Sczaniecka-Clift, A. C. Barros, M. Woods, B. Fu, F. Yang, E. Chen, M. Ostermaier, T. Stankovic, H. Ponstingl, M. Herzog, K. Yusa, F. M. Martinez, S. T. Durant, Y. Galanty, P. Beli, D. J. Adams, A. Bradley, E. Metzakopian, J. V. Forment, S. P. Jackson, ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 10, 87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. K. Pal, P. H. Frankel, A. Mortazavi, M. Milowsky, U. Vaishampayan, M. Parikh, Y. Lyou, P. Weng, R. Parikh, B. Teply, R. Dreicer, H. Emamekhoo, D. Michaelson, C. Hoimes, T. Zhang, S. Srinivas, W. Y. Kim, Y. Cui, E. Newman, P. N. Lara Jr., Effect of cisplatin and gemcitabine with or without berzosertib in patients with advanced urothelial carcinoma: A phase 2 randomized clinical trial. JAMA Oncol. 7, 1536–1543 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D. Y. Oh, S. S. Kwek, S. S. Raju, T. Li, E. McCarthy, E. Chow, D. Aran, A. Ilano, C. C. S. Pai, C. Rancan, K. Allaire, A. Burra, Y. Sun, M. H. Spitzer, S. Mangul, S. Porten, M. V. Meng, T. W. Friedlander, C. J. Ye, L. Fong, Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 181, 1612.e13–1625.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.W. H. Weir, P. J. Mucha, W. Y. Kim, A bipartite graph-based expected networks approach identifies DDR genes not associated with TMB yet predictive of immune checkpoint blockade response. Cell Rep Med 3, 100602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.K. Wang, M. Li, H. Hakonarson, ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Q. Li, K. Wang, InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100, 267–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. J. Landrum, J. M. Lee, M. Benson, G. R. Brown, C. Chao, S. Chitipiralla, B. Gu, J. Hart, D. Hoffman, W. Jang, K. Karapetyan, K. Katz, C. Liu, Z. Maddipatla, A. Malheiro, K. McDaniel, M. Ovetsky, G. Riley, G. Zhou, J. B. Holmes, B. L. Kattman, D. R. Maglott, ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.T. Li, J. Fu, Z. Zeng, D. Cohen, J. Li, Q. Chen, B. Li, X. S. Liu, TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.L. M. Sholl, K. Do, P. Shivdasani, E. Ceramiet al. , Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1, e87062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. Saito, C. C. Smith, T. Utsumi, L. M. Bixby, J. Kardos, S. E. Wobker, K. G. Stewart, S. Chai, U. Manocha, K. M. Byrd, J. S. Damrauer, S. E. Williams, B. G. Vincent, W. Y. Kim, Molecular subtype-specific immunocompetent models of high-grade urothelial carcinoma reveal differential neoantigen expression and response to immunotherapy. Cancer Res. 78, 3954–3968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.P. A. Ewels, A. Peltzer, S. Fillinger, H. Patel, J. Alneberg, A. Wilm, M. U. Garcia, P. di Tommaso, S. Nahnsen, The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020). [DOI] [PubMed] [Google Scholar]
- 59.R. Patro, G. Duggal, M. I. Love, R. A. Irizarry, C. Kingsford, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.T. Li, J. Fan, B. Wang, N. Traugh, Q. Chen, J. S. Liu, B. Li, X. S. Liu, TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.K. Yoshihara, M. Shahmoradgoli, E. Martinez, R. Vegesna, Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. A. Bolotin, S. Poslavsky, A. N. Davydov, F. E. Frenkel, L. Fanchi, O. I. Zolotareva, S. Hemmers, E. V. Putintseva, A. S. Obraztsova, M. Shugay, R. I. Ataullakhanov, A. Y. Rudensky, T. N. Schumacher, D. M. Chudakov, Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 35, 908–911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for tables S1 to S5
References
Tables S1 to S5