Abstract
Endozoicomonas are often predominant bacteria and prominently important in coral health. Their role in dimethylsulfoniopropionate (DMSP) degradation has been a subject of discussion for over a decade. A previous study found that Endozoicomonas degraded DMSP through the dddD pathway. This process releases dimethyl sulfide, which is vital for corals coping with thermal stress. However, little is known about the related gene regulation and metabolic abilities of DMSP metabolism in Endozoicomonadaceae. In this study, we isolated a novel Endozoicomonas DMSP degrader and observed a distinct DMSP metabolic trend in two phylogenetically close dddD-harboring Endozoicomonas species, confirmed genetically by comparative transcriptomic profiling and visualization of the change of DMSP stable isotopes in bacterial cells using nanoscale secondary ion spectrometry. Furthermore, we found that DMSP cleavage enzymes are ubiquitous in coral Endozoicomonas with a preference for having DddD lyase. We speculate that harboring DMSP degrading genes enables Endozoicomonas to successfully colonize various coral species across the globe.
Coral Endozoicomonas are widespread DMSP degraders monopolized by the DddD enzyme and display various degradation mechanisms.
INTRODUCTION
Bacteria from the genus Endozoicomonas (Gammaproteobacteria, Oceanospirillales, and Endozoicomonadaceae) are predominantly and commonly found in a wide array of animal hosts living in coral reefs (1–6) and are considered important for the health of their hosts, especially corals (7–9). Putative metabolic interactions between Endozoicomonas and coral hosts have been proposed based on genomic information (2, 10–12). For example, genes involved in amino acid metabolism (2, 11, 13) and the enrichment of type III and type IV secretion system effectors (11, 12, 14) suggest that the bacteria might play a role in supplying amino acids to the coral holobiont (15) and use effectors to communicate with other associated organisms, especially their coral hosts. In addition, some eukaryotic-like proteins (ELPs) are found in Endozoicomonas (11, 14), suggesting a coevolution with their eukaryotic hosts.
In addition to these genomic factors suggesting symbiosis, evidence from more than a decade ago indicates that Endozoicomonas could be a putative dimethylsulfoniopropionate (DMSP) degrader (16, 17). Only recently has this function been experimentally confirmed in three Endozoicomonas acroporae strains with the discovery of a complete dddD gene–based operon, combined with biochemical analyses (11). DMSP is a ubiquitous sulfur-containing compound in marine environments that, once broken down, produces dimethyl sulfide (DMS). DMS is the precursor of sulfur-containing aerosol particles which indirectly affect Earth’s climate and link the marine and terrestrial sulfur cycles by participating in cloud formation (18). Beyond this crucial role in the geochemical cycle, DMSP also serves multiple other functions. In marine algae, DMSP functions as an osmolyte (19), cryoprotectant (20), scavenger of reactive oxygen species (21), and antioxidant (22). The increase in DMSP concentration in stressed coral (23, 24) makes DMSP a stress indicator (25) and is associated with its function as an antioxidant (26).
Bacteria are key players in DMSP degradation, which involves two metabolic pathways: demethylation and cleavage. The demethylation pathway, involving the enzymes DmdA, DmdB, DmdC, and DmdD, is widely distributed in marine bacteria and allows the cells to use both sulfur and carbon derived from DMSP. In contrast, the cleavage pathway, in which bacteria retain carbon but release sulfur into the ambient environment, generates DMS. There are eight DMSP lysis enzymes that can be grouped into three categories based on the byproducts they produce [see detailed review in (27) for more information about bacterial DMSP catabolism]. The largest category of DMSP lysis enzymes, which generate cytotoxic acrylate (28), contains six DMSP lyases, including five from the Cupin superfamily (DddL, DddQ, DddW, DddK, and DddY) and one (DddP) from the M24 metallopeptidase family. The second category, which produces acryloyl–coenzyme A (CoA), consists of a novel adenosine 5′-triphosphate (ATP)–dependent lyase (DddX) from the acyl-CoA synthetase superfamily (29). The third category, which mediates the additional acyl coenzyme (CoA-transferase) before the lyase activity and generates 3-hydroxypropionate (30, 31), contains one lyase (DddD) from the type III acyl-CoA transferase family and is called the CoA-DMSP lysis pathway. The DMSP catabolic operon, facilitated by the dddD gene, was first identified in Marinomonas MWYL (31) and consists of five genes, including detS, adh, mms, gcv, and dddD. First, DMSP is imported through detS, which belongs to the betaine-carnitine-choline transporter family. It is followed by the enzymes adh and mms, belonging to families of Fe-containing alcohol dehydrogenases and methylmalonatesemialdehyde dehydrogenases, respectively. DMS will not be cleaved without its transcriptional regulator, gcv, which belongs to the LysR family.
Among the eight lyase enzymes, DddD exhibits a greater abundance than DddP in the coastal seawater of Sanriku, Japan (32). This suggests that dddD might be a primary DMS producer via DMSP cleavage in the coastal seawater, especially when lysed by a member of the Oceanospirillales order (33). However, until now, most of the studies related to DMSP have focused on the pelagic SAR11 clades of Alphaproteobacteria and phytoplankton-related Rhodobacterales, which are the main degraders of DMSP via demethylation in the ocean (33). Little is known about the DMSP-degrading bacteria in other DMSP-producing hotspots, such as coral reefs. Being one of the predominant coral-associated bacteria from the order Oceanospirillales, E. acroporae harbors one copy of the functional DMSP lyase DddD (11), suggesting that Endozoicomonas might play a role in DMSP degradation of coral holobionts. However, there is not enough evidence to represent the cleavage ability, features, and gene regulation of DMSP metabolism in Endozoicomonas.
In this study, we successfully isolated a novel dddD-harboring DMSP degrader, Candidatus Endozoicomonas ruthgatesiae strain 8E (hereafter “8E”). The phylogenetic characteristics of DddD proteins were compared in dddD-harboring Endozoicomonas. Through direct gas chromatography–mass spectrometry (GC-MS) measurement of DMS, we observed a distinct metabolic profile within two Endozoicomonas species and visually confirmed this for 8E on a single-cell level using NanoSIMS. Gene expression results indicated that 8E uses DMSP as a carbon source, as evidenced by the up-regulation of main energy and carbohydrate-related genes, whereas no similar up-regulation was observed in E. acroporae. On the basis of these results, we propose that the distinct gene regulation and metabolic profiles observed in the utilization of DMSP by the two bacterial species reflect different functional roles, including differences in adaptation and evolution within DMSP metabolism. Our findings illuminate the mysterious role of Endozoicomonas in DMSP metabolism. Furthermore, on the basis of the remarkable DMSP cleavage capacity in 8E and the metabolic differences between two Endozoicomonas species, we propose that the contribution to and functionality of Endozoicomonas in the coral sulfur cycle are more important and diverse than previously thought.
RESULTS
Phenotypic and taxonomic traits of a novel coral-associated bacterium Candidatus Endozoicomonas ruthgatesiae 8E
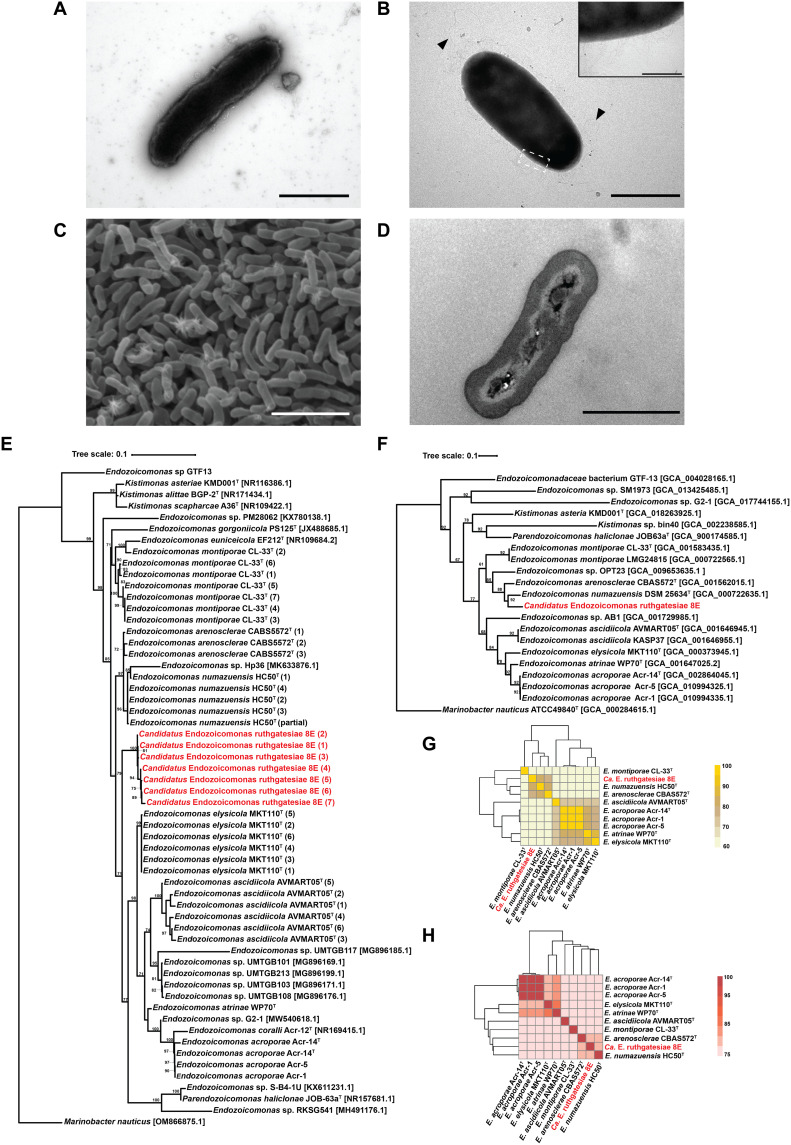
8E was isolated from the tissue-mucus mixture of Acropora sp. coral from Kenting, Taiwan. Bacteria are rod-shaped (Fig. 1A), but their size and morphology change on different media (Fig. 1, A to C). Peritrichous flagella-like and fimbria-like structures were only observed in minimal medium cultures with 0.1 mM DMSP (Fig. 1B) but not in modified marine broth version 4 (mmbv4) medium. Otherwise, a few small granule-like structures in the middle of the cells were observed (Fig. 1D. A similar structure has been found in another Endozoicomonas species (see the Supplementary Materials and table S1 for detailed morphological and biochemical features) (13).
Fig. 1. Morphological and taxonomic characteristics of the genus Endozoicomonas.
(A) Transmission electron microscopy (TEM) image. Bacteria were incubated in mmbv4 medium. Negative stained by 2% phosphotungstic acid. Scale bar, 1 μm. (B) TEM image. Bacteria were incubated in minimal medium with 0.1 mM DMSP. Scale bar, 1 μm. Triangles indicate the peritrichous flagella-like structures, and the insert is the magnification (×10) of fimbriae-like structure. Insert bar = 0.1 μm. Negative stained by 2% uranyl acetate. (C) Bacterial colony surface observed by SEM. Scale bar, 10 μm. (D) Bacterial TEM thin section. Scale bar, 1 μm. (E) The maximum likelihood tree was calculated with MEGA11.09 and tested by bootstrap method with 1000 replications using the Tamura 3-parameter model. The bootstrap values are shown at the corners of branching points. Bacteria used for the analysis in this study are noted with GenBank accession numbers, and the asterisk “*” indicates that the 16S ribosomal RNA (rRNA) sequence was annotated by Prokka 1.14.6. For each strain, only one copy of 16S rRNA underwent analysis. (F) Phylogenetic tree built by up-to-date bacterial core gene sets (concatenated alignment of 92 core genes). The phylogenetic tree is based on whole genome sequences, and RAxML (Randomized Axelerated Maximum Likelihood) was used for phylogeny reconstruction. The length of concatenated alignments was 84741, the GenBank accession numbers are shown in parentheses, and gene support indices are given at the corner of branching points. (G) Average nucleotide identity (ANI). (H) Average amino acid identity (AAI).
The phylogenetic analysis based on the 16S ribosomal RNA (rRNA) gene sequences (Fig. 1E) and the 92 bacterial core gene sets (Fig. 1F) of Endozoicomonadaceae both indicated that the closest species to 8E was Endozoicomonas numazuensis HC50T (97.40% identity). An average nucleotide identity (ANI) of 80.56% (Fig. 1G) and an average amino acid identity (AAI) of 81.35% (Fig. 1H) also supported the conclusion that E. numazuensis HC50T is phylogenetically close to 8E.
The objective threshold for defining a species requires a 16S rRNA similarity of 97% (34), an ANI greater than 95%, and an AAI greater than 62% (35, 36). 8E shared more than 80% of AAI with E. numazuensis HC50T and Endozoicomonas arenosclerae CABS5572T, but only around 62% of its AAI with other available Endozoicomonas genomes. This wide range of 62 to 100% suggests interspecies variation and puts 8E at the edge of a new genus classification (35, 36). These results, as well as the phenotypic characteristics, 16S rRNA gene phylogenetic tree, and ANI calculation, lead us to propose that 8E is a novel species, Candidatus Endozoicomonas ruthgatesiae, in the genus Endozoicomonas.
Genome assembly and features of 8E
We recovered a single contig with a length of 7,377,917 bp using a hybrid sequencing strategy of Oxford Nanopore Technology (ONT) and Illumina sequencing platforms. This contig was larger than most of the other genomes detected in the genus Endozoicomonas (table S2). On the basis of the analysis by CheckM, only 2 of 507 markers [c_Gammaproteobacteria (UID4444)] were missing, 502 genes were detected once, and 3 genes appeared twice, which resulted in 99.14% genome completeness, 0.54% contamination, and 0.00% strain heterogeneity. Overall, the quality of the 8E genome is “finished” according to the guidelines established by the Genomic Standards Consortium (37).
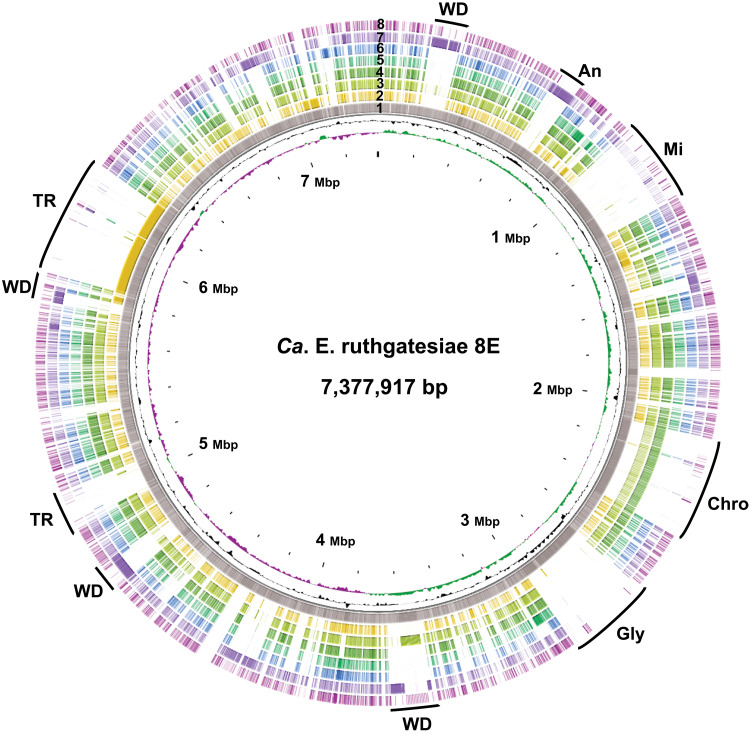
To clarify genome characteristics, comparative genomic analyses were conducted to detect unique features of 8E. Seven Endozoicomonas type strain genomes were included in the genomic comparison. Ten regions in the genome of 8E shared low similarity with other Endozoicomonas species (Fig. 2). In these 10 regions, we identified different clusters of genes which included WD40 domain protein (WD40 superfamily), ankyrin-related protein, midasin AAA adenosine triphosphatase (ATPase), chromosome segregation protein/ATPase, glycotransferase family, and tandem repeat (TR). Notably, these copies of genes usually gather adjacently in 8E, especially the ELPs that we define as proteins found in the bacterial genome that may have originated in eukaryotes. We detected 160 WD40 domain proteins in the 8E genome (table S2). Most of these were disseminated into four regions (Fig. 2) located at 0.197 to 0.331 Mbps (47 copies), 3.49 to 3.64 Mbps (34 copies), 4.62 to 4.78 Mbps (38 copies), and 5.50 to 5.79 Mbps (23 copies) (Fig. 2 and fig. S1A). A total of 66 gene copies of ankyrin-related proteins were found and annotated as ankyrin repeat (3 copies) or ankyrin-like protein (63 copies). Unlike the WD40 proteins, most of the ankyrin-related proteins were clustered in a single region around 0.436 to 0.744 Mbps (Fig. 2 and fig. S1B). Both types of aforementioned proteins shared similar arrangements with E. acroporae Acr-14T (ring 7), which had 23 WD40 proteins and 97 ankyrin repeat proteins (table S2). Midasin AAA ATPase is a member of the MDN1 superfamily, which is a group of ELPs that are rarely observed in bacterial genomes. Midasin AAA ATPase was found to have 23 copies in the region from 0.966 to 1.23 Mbps (Fig. 2 and fig. S1C), suggesting that it may be functionally involved in ribosome maturation. It is not unexpected that Endozoicomonas contained ELPs, but through gene annotation analysis using conserved domain search and EffectiveDB, we found that the ELPs occupy up to 8.73% of the total 8E genome (E. acroporae, 5.51%; Endozoicomonas montiporae, 3.76%) (fig. S3). The unexpectedly high percentage of ELPs in the genome is also found in another Acropora coral isolate Echinolittorina marisrubri sp. 6C (14). However, the biological, mechanical, and original eukaryotic sources of these high proportions of ELPs in Endozoicomonas are still unclear (see Supplementary Materials for detailed functional annotation and genomic features of 8E). No signs of genome reduction were found in 8E, but some signs (gene duplication and TRs) pointed toward genome expansion. We found multiple neighboring copies of chromosome segregation ATPases (33 copies), chromosome segregation proteins (16 copies), and glycosyltransferases (6 copies). However, there was a high density of variously sized DNA TRs (1126 TRs), ranging from 30 to 2000 bp, that were scattered between 4.94 to 5.07Mbp (74 TRs) and 5.84 to 6.31Mbp (361 TRs). 8E is the second known species of large-genome Endozoicomonas (>7 MB), and with technological advancements and widespread use of sequencing, we anticipate that more species of large-genome Endozoicomonas will be sequenced. It is worth thoroughly investigating the genome evolution mechanism in animal-associated bacteria with large genomes.
Fig. 2. The genome comparison atlas of 8E.
The genome of 8E was restarted from dnaA, and the map was generated by CGView. Each lane corresponds to an Endozoicomonas genome, and BLAST comparisons were performed by CGView BLAST (BLAST+ 2.12) with 0.001 expected value cutoff for DNA comparison. From center to exterior: GC skew + (green); GC skew − (purple); GC content; 8E (1); E. montiporae CL-33T (2); E. numazuensis HC50T (3); E. arenosclerae CBAS572T (4); E. atrinae WP70T (5); Endozoicomonas elysicola MKT110T (6); E. acroporae Acr-14T (7); Endozoicomonas ascidiicola AVMART05T (8). WD, WD40; AN, ankyrin-related protein; Mi, midasin AAA ATPase; Chro, chromosome segregation ATPase; Gly, glycotransferase family.
dddD gene–mediated DMSP lyases operon
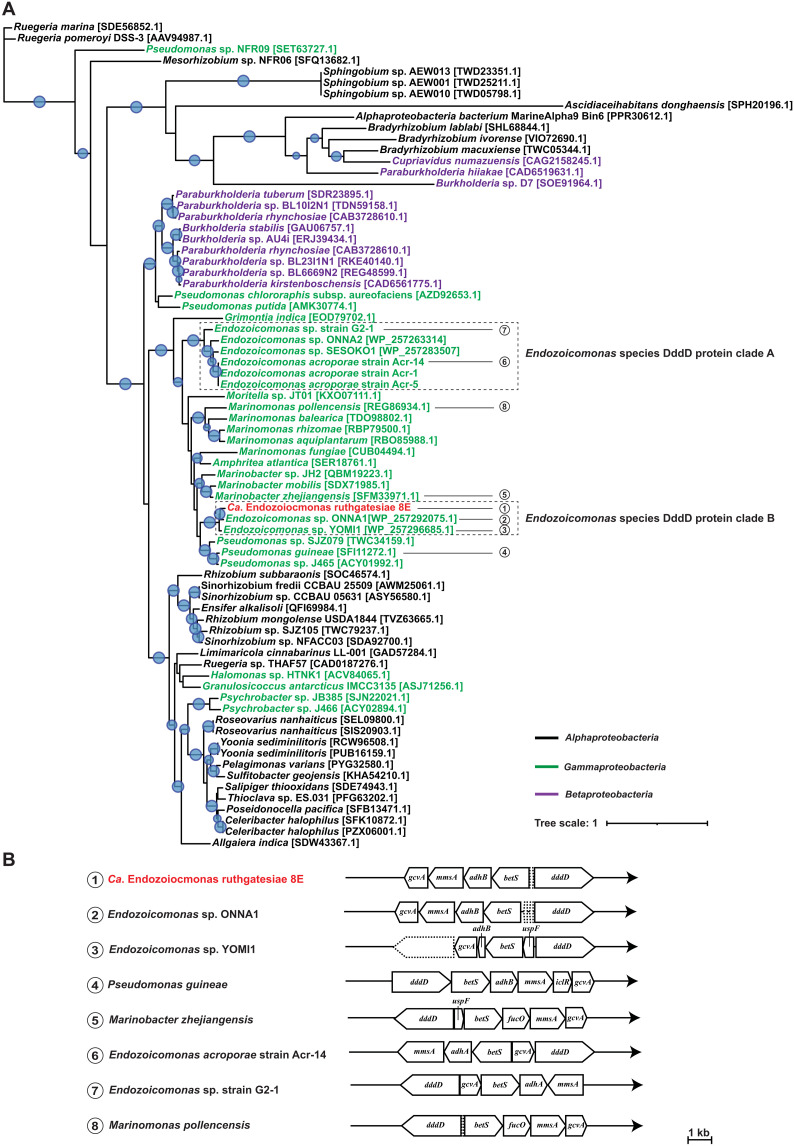
Since 8E was isolated from the DMSP-rich coral genus Acropora, we focused on functions related to DMSP metabolism, as DMSP is an important molecule in the coral holobiont relative to scavenging free radicals in stressful conditions (38). Notably, the gene dddD encodes CoA-transferase lyase, which is a key participant in the DMSP cleavage pathways. dddD was found in the 8E genome along with a complete DMSP lyase operon. We downloaded and annotated all Endozoicomonas genomic assembly on National Center for Biotechnology Information (NCBI) for the DddD protein sequence similarity comparison (table S5). We built a DddD protein phylogenetic tree for Proteobacteria to locate the position of 8E in other DddD-harboring Proteobacteria (Fig. 3A). All seven Endozoicomonas DddD sequences were clustered with other Gammaproteobacteria (Fig. 3A). Among them, seven Endozoicomonas species separated into two different clades. Clade A contained four Endozoicomonas species, including E. acroporae, and clade B contained three Endozoicomonas species, including 8E (Fig. 3A). The DddD protein of 8E had a lower similarity with clade B Endozoicomonas (>67.9% identity) than with Marinobacter zhenjiangenesis and Pseudomonas sp. SJZ079 (>75.5% identity) (table S3), suggesting that the two clades of Endozoicomonas might have different horizontal transfer sources. On the other hand, the gene arrangement and gene content of the dddD-mediated DMSP cleavage operon in the two clades are different (Fig. 3B). In clade A, E. acroporae and Endozoicomonas sp. G2_1 have the same gene arrangement, and both strains contain adhA (long-chain alcohol dehydrogenase α). Clade B Endozoicomonas species contain adhB (long-chain alcohol dehydrogenase β), and their gene arrangements are phylogenetically close to the Endozoicomonas species in clade A but are otherwise more similar to that of Pseudomonas guineae (Fig. 3B). Although most of Endozoicomonas strains included in the analysis are unculturable, we speculated that species placed in two different clades might display similar DMSP metabolic features as E. acroporae or 8E.
Fig. 3. DddD protein phylogenetic tree and DMSP cleavage operons.
(A) DddD protein maximum-likelihood tree. Protein sequences of bacterial CoA-transferase/lyase DddD are obtained from NCBI, except for several Endozoicomonas species without accession numbers. Sequence accession numbers are annotated at the end of the bacteria name. Font color represents bacterial taxonomy based on class: black, Alphaproteobacteria; green, Gammaproteobacteria; red, 8E (classified into Gammaproteobacteria; Fig. 1); and purple, Betaproteobacteria. Maximum-likelihood tree was generated using iqtree and MAFFT v7 to align the selected protein sequences with 1000 replicates. Blue bubbles on the corners of branches indicate a >90 bootstrap value. Bacteria species with DMSP DddD cleavage operons present are listed at the bottom. 1. 8E, 2. Endozoicomonas sp. ONNA1, 3. Endozoicomonas sp. YOMI1, 4. P. guineae, 5. Marinomonas zhejiangensis, 6. E. acroporae Acr-14, 7. Endozoicomonas sp. G2-1, 8. M. pollencensis. (B) dddD gene–mediated DMSP cleavage operon. Gene arrangement in DMSP cleavage operons was distinguished by Prokka and NCBI databases (table S4). dddD, DMSP CoA transferase/lyase; ad, alcohol dehydrogenase; betS, choline/glycine/betaine transporter; adh, long-chain alcohol dehydrogenase; gcvA, LysR family transcriptional regulator; mmsA, malonate-semialdehyde dehydrogenase; iclR, transcriptional repressor; dapA, 4-hydroxy-tetrahyfrodipicolinate synthase; cdhR, HTH-type transcriptional regulator; uspF, universal stress protein F; fucO, lactaldehyde reductase. Dashed line blocks represent hypothetical proteins.
8E as a potent DMS producer
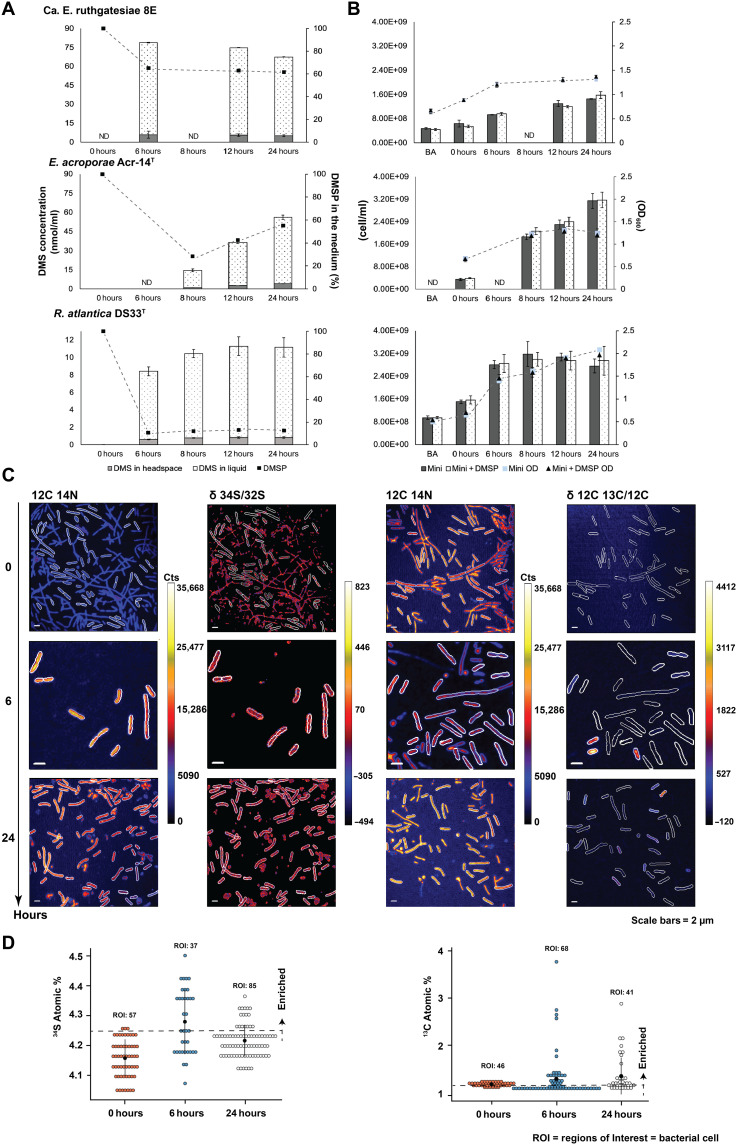
To detect and compare the DMSP metabolic ability in 8E and other bacteria, we measured the production of DMS and consumption of DMSP in 8E, E. acroporae Acr-14T, and a common species of DMSP degrader in the Rosebacter clade, Ruegeria atlantica DSM5823T. The second time points of two Endozoicomonas species represent the earliest detectable time of DMS after the addition of DMSP (Fig. 4A). On the basis of our observations using GC-MS, the overall trend of DMSP metabolism in the three bacteria can be categorized into two types: rapid and gradual emission. At the first sampling point (6 hours), the highest DMS peaks were detected from the culture of 8E with a consumption of 34.8% (from 0.1 to 0.0655 ±0.00309 mM) of the DMSP in the medium (Fig. 4A). After the bacteria entered the stationary phase (Fig. 4B), the DMS(g) concentration gradually decreased (from 5.84 to 5.53 nmol/ml).
Fig. 4. DMS concentration, DMSP consumption, and growth condition of bacterial cultures in minimal medium.
(A) Time courses for production of DMS (nmol/ml) and DMSP utility (%). DMS in headspace: The DMS concentration in treatment and the values were direct measurements of DMS by GC-MS. DMS in liquid: The DMS concentration in the treatment medium and the values were calculated based on Bunsen solubility coefficient. The line chart shows the percentage of DMSP in treatment medium starting from 0.1 mM DMSP. (B) Bacterial cell number and optical density at 600 nm (OD600) value. Bar charts show the cell number measured by flow cytometry. Line charts show the value of OD600 which started from 108 cell/ml. BA, before acclimation; mini, control without DMSP; mini + DMSP, treatment with DMSP; mini OD, control OD600 value; mini + DMSP OD, treatment OD600 value. (C) The DMSP metabolism of 8E were determined using 34S and 13C label stable isotope at the single-cell level. Example parallel images of 12C14N counts, sulfur isotope ratio (34S/32S), and carbon isotope ratio (12C13C/12C). (D) 34S atomic % and 13C atomic % isotopic composition of ROIs in the scatterplot. The nature 34S atomic % is 4.25 and 13C is 1.07; ROIs higher than the value were marked as enriched. ND, not determined.
Meanwhile, a different DMS production profile was observed in E. acroporae and R. atlantica. These species initially consumed more than half (0.0729 and 0.0876 mM, respectively) of the DMSP in the medium but emitted five and nine times lower DMS(g) concentration (1.07 and 0.056 nmol/ml, respectively) than 8E (Fig. 4A). In addition, the DMS concentration in these cultures continued to accumulate over time, reaching 56.1 nmol/ml at 24 hours for E. acroporae and 11.15 nmol/ml at 24 hours for R. atlantica.
To further confirm the DMSP cleavage of the rapid emission type in 8E, we used two different stable isotopes of DMSP (34S-DMSP and 313C -DMSP) (fig. S4) and visualized the bacterial element ratios using NanoSIMS (Fig. 4C). 34S was labeled to monitor the DMS emission, and 313C -DMSP was used to monitor DMSP in the cell. At least 37 bacterial cells [regions of interest (ROIs)] were analyzed in each isotopic treatment. The strong DMS emission caused no obvious 34S enrichment in 8E. The highest ratio of 34S/32S was 4.50 (natural/baseline δ 34S/32S ≈ 4.25) (Fig. 4D) after 6 hours. In contrast, we observed enrichment of 13C at 6 and 24 hours, with 67 ROIs showing δ 13C/12C higher than the natural δ 13C/12C (natural/baseline value of δ 13C/12C ≈ 1.07) and 4 ROIs had two times higher than the natural δ 13C/12C value (Fig. 4D). However, for both 34S and 13C, the enrichment of isotope signals was not equally distributed in every cell. Instead, a few cells appeared brighter and had a stronger signal than the surrounding cells (Fig. 4C). Otherwise, the cell morphology was diverse. Cells with higher δ 13C/12C values were mostly short and rod-shaped (Fig. 4C).
Transcriptional responses of Endozoicomonas species to DMSP
On the basis of the variation of DMSP utilization in the two Endozoicomonas species (Fig. 4A), we hypothesized that these two species might also display different gene regulations, especially in DMSP metabolism. We performed RNA sequencing and collected transcripts at the time points when the DMS signal was first detected (8E at 6 hours; E. acroporae at 8 hours, both with and without addition of DMSP) (Fig. 4A). High-quality (Phred score > 35) sequences were also collected (table S4). More than 10 million reads were obtained for all treatments, and more than 90% of the total reads were mapped to the two bacterial genomes. In total, around 90% of mapped reads were assigned to unique features (table S4).
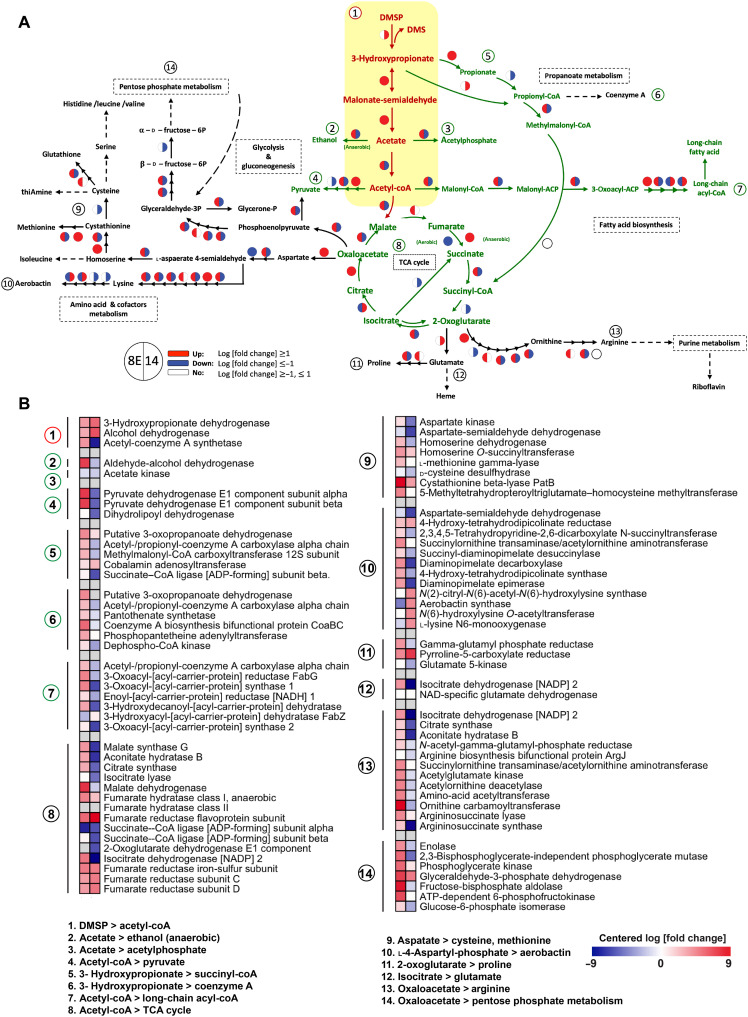
To understand the associated and subsequent responses of DMSP metabolism, we created a map of a putative metabolic scenario derived from all known intermediates of the dddD-centric process and initiated enzymatic reactions (Fig. 5).
Fig. 5. Differential expression of DMSP related pathway in two Endozoicomonas species.
(A) The putative dddD centered pathway. The center pathway of dddD-mediated DMSP is marked in red, and the pathway directly linked with the dddD-mediated DMSP pathway is marked in green. The indirectly linked pathway is marked in black. The circle symbol represents the log [fold change] according to DESeq2, with down-regulation (log [fold change] ≤ 1) in blue, up-regulation (log [fold change] ≥ 1) in red, and no regulation (log [fold change] ≥ −1 and ≤ 1) in white. The color in the left side of the circle belongs to 8E, and the color in the right belongs to E. acroporae Acr-14T. (B) The log fold change heatmap of the putative pathway related with DMSP degradation. The numbers indicate the genes included in the pathway in (A).
The map starts from the dddD-mediated DMSP metabolic pathway and expands with each product. DMSP is a source of both sulfur and carbon in the demethylation pathway and is used only as a carbon source in cleavage enzyme–containing bacteria, including Endozoicomonas. The acetate produced from the dddD-mediated DMSP metabolic operon suggests a direct role of DMSP in bacterial central carbon metabolism. However, two bacteria displayed distinct transcriptional regulation after DMSP consumption, especially in energy and carbohydrate metabolism (Fig. 5 and fig. S3). Most of the genes related to the tricarboxylic acid (TCA) cycle, glycolysis, and gluconeogenesis were up-regulated in 8E but not in E. acroporae, suggesting that DMSP may suppress the energy metabolism in E. acroporae. The up-regulated transcription in amino acid metabolism also differed between the two bacteria; the valine and isoleucine biosynthesis pathway were up-regulated in 8E (fig. S3A), while proline and polyamine biosynthesis increased in E. acroporae. The average normalized count of the genes in the up-regulated/down-regulated pathway mentioned above is 10 times more than the control group (fig. S3B).
The metabolic pathway not directly linked with the DMSP operon also differs between the two bacteria. Pantothenate and aerobactin biosynthesis were notably up-regulated in E. acroporae. The transporter-related genes were also affected by DMSP addition; the reads of genes related to branched-chain and general l–amino acid ABC transporters were 10 times higher in 8E bacteria treated with DMSP than in 8E cultures that received no DMSP treatment (fig. S3A). Further, in E. acroporae, we found that four transporters [molybdate, methyl-galactoside, Fe (III) hydroxamate, and zinc] were up-regulated (fig. S3B). Notably, secretion system structural genes and genes governing cell processes were expressed distinctly between the two bacteria. The increase of normalized read in the type I secretion system, sec-signal recognition particle (SRP), quorum sensing, and flagellar was only observed in 8E (fig. S3B). On the other hand, in E. acroporae, the type III secretion system and twin-arginine targeting were found to be up-regulated (fig. S3B).
DISCUSSION
Endozoicomonas is a core group in the microbiota of coral, and many studies over the past decade have found them to be functionally associated with coral health. However, their ecological functions are mostly unknown. In the present study, we discovered a novel, ubiquitous Endozoicomonas species, 8E, which has potent DMSP degradation activity via the DddD protein–mediated cleavage pathway, compared to E. acroporae (11) and the common oceanic DMSP utilizer, R. atlantica (39). DMSP cleavage activity was previously thought to be rare in Endozoicomonas, but the identification and proof of function in this study revealed that DMSP degradation is a common ecological function of Endozoicomonas species in coral reefs. Using comparative transcriptomic analysis, we uncovered variation in the regulation of dddD gene–mediated DMSP metabolism in Endozoicomonas. This included directly and indirectly related pathways, as well as other bacteria. These findings suggest that the underlying utilization mechanism of DMSP in bacteria is more complex, diverse, and crucial than previously thought.
Differential DMSP metabolic features in two Endozoicomonas species
Within 12 hours of incubation, we clearly observed different profiles of DMSP degradation and uptake between 8E and E. acroporae. 8E displayed rapid and efficient DMSP cleavage, produced the highest DMS concentration (79.92 nmol/ml) within 6 hours of incubation, and consumed less than 35% of the supplied DMSP (Fig. 4A). Contrastingly, E. acroporae consumed more than 70% of the supplied DMSP in the first 8 hours and produced only a small amount of DMS (14.55 nmol/ml) (Fig. 4B). The underlying molecular mechanism causing the metabolic differences is still unclear, but three possibilities based on these results include (i) molecular variations at the gene and enzyme levels (i.e., variations in the dddD gene cluster and differentiations in enzymatic kinetics and activity), (ii) variations in the physiological purpose of DMSP metabolism, and (iii) more than one DMSP lyase gene participated in DMSP degradation.
The DddD proteins in the two Endozoicomonas species share relatively low sequence identities with M. zhenjiangenesis and Pseudomonas sp. SJZ079 (table S3), indicating the presence of molecular variations at the gene and enzyme levels. Furthermore, the phylogenetic relationship of the DddD protein and operon arrangement shows incongruence with bacterial taxonomy and phylogeny (Fig. 3A), indicating that the dddD gene clusters in both bacteria were horizontally acquired from different sources. Therefore, the different profiles of DMSP degradation may be caused by primary variations in the metabolic features of uniquely sourced dddD genes or operons. Furthermore, the enzymatic kinetics and activity of the DddD protein between the two bacteria would be variable. We propose that the DddD enzyme might play a key role in the different metabolic profiles of the bacteria. The efficiency of DMSP cleavage activity was high in 8E (confirmed by the NanoSIMS results), which produced a DMS concentration five times higher than that of E. acroporae with only half the DMSP consumption (Fig. 4). This difference in observed efficiency suggests that the DddD lyase in 8E might have a high Vmax value (indicating higher DMSP cleavage activity) and a low Km value (indicating higher affinity to DMSP) than that in E. acroporae. Further investigation is required to confirm enzymatic characters. Cloning the purified DddD protein into a clear system such as Escherichia coli and observing heterogeneous expression may be a direction for future study.
Apart from variations in the operons or genes, we also propose that variations in the physiological purpose of DMSP metabolism could lead to varying profiles of DMS production and DMSP uptake in the two bacteria. The up-regulation of genes involved in the TCA cycle and amino acid metabolism in 8E following the addition of DMSP likely indicates that DMSP is used as a substitute carbon source for energy generation in poor nutrient conditions. On the other hand, the energy-related genes were commonly down-regulated when DMSP was added in E. acroporae, suggesting that DMSP is used for non-energy production metabolisms. Similarly, we found that the growth efficiency of 8E was two times higher than that of E. acroporae with the addition of DMSP concentration under 3 mM in the specific minimal medium (fig. S6). This direct observation strongly implies that 8E exhibits superior efficiency in DMSP degradation and tolerance. On the contrary, we hypothesize that the lower growth rate observed in E. acroporae could be attributed to stress conditions, stemming from a reduced affinity for DMSP as a limited carbon source, or the inhibitory associated with high concentration of DMSP. Hence, E. acroporae likely uses DMSP to cope with stressful conditions. The bacterium might store DMSP and gradually cleave it to produce DMS. This strategy is common for DMSP users. Microorganisms accumulate DMSP for its beneficial antioxidant (22), osmolytic, and cryoprotectant (40) properties, which might help soothe physiological stress. Overall, although DMSP might act as an antioxidant or other beneficial antistress molecule in E. acroporae, it could act as an energy source in 8E. Many studies have discussed how DMSP plays different physiological roles in different organisms (22, 27, 41). However, complex comparative studies on the subject are rare, especially concerning only dddD-mediated DMSP utilization in closely related organisms.
After studies determined that DMSP is not only the precursor of atmospheric trace gas DMS but also plays multiple roles in different marine organisms (19, 40), increasing numbers of DMSP lyase have been identified. To date, nine DMSP lyases (including algal Alma1) have been found and categorized into three different lysis pathways based on their byproducts. There is a possibility of a novel degradation enzyme participating in Endozoicomonas DMSP degradation, as more than half of the genes (~2500 genes) are hypothetical. A future dddD gene knockout experiment could be conducted to clarify the hypothesis despite the lack of a successful gene knockout experiment conducted on Endozoicomonas to date.
Differential gene expression in DMSP pathways and relevant metabolisms in Endozoicomonas
According to the results of the biochemical assay and the distinct gene regulation in two Endozoicomonas species that are associated with energy- and carbohydrate-related metabolic pathways, we believe that these two species use DMSP for different biological purposes. This difference is also found when examining the pathway not directly linked with DMSP metabolism (layer 3, Fig. 5 and fig. S3). Genes for flagella-, chemotaxis-, and quorum sensing–related pathways were up-regulated in 8E. We observed a similar trend of gene regulation when the coral mucus-associated bacteria Roseobacter clades were exposed to DMSP (42–46). A flagella-like structure was observed on 8E during incubation in 0.1 mM DMSP in minimal medium (Fig. 1A), suggesting that flagella-related physiological responses, including chemotaxis and quorum sensing, are coupled with DMSP metabolism. However, a similar coupling response was not observed in E. acroporae, indicating that the gene up-regulation concurrence of flagella, chemotaxis, and quorum sensing only appears in some DMSP bacterial degraders.
In addition, the up-regulated genes involved in aerobactin (a bacterial iron-chelating agent, also called siderophore) biosynthesis and the Fe (III) hydroxamate transporter may increase resistance to oxidative stress in E. acroporae but not in 8E. For example, siderophores might increase resistance to oxidative stress in E. coli (47) and in the human pathogen Yersinia pseudotuberculosis (48). We speculate that aerobactin, a citrate-hydroxamate siderophore, could be another way for E. acroporae to mitigate oxidative stress.
Ecological significance and indispensability of dddD-mediated DMSP metabolism in coral reefs
The dominant DMSP lyase in coastal seawater is DddD protein, which is mainly found in Oceanospirillales (class Gammaproteobacteria). However, the ecological roles and influences of Oceanospirillales in DMSP-abundant coral reefs are still unclear. Endozoicomonas, being one predominant group of Oceanospirillales in coral, has long been thought to participate in coral sulfur cycling via DMSP metabolism (16, 17). With the framework of additional Endozoicomonas genomes sequenced in recent years, we inspected the gene contents of our sample and identified 10 Endozoicomonas species that contain DMSP cleavage genes (table S6), indicating that DMSP degradation is a common eco-physiological function in Endozoicomonas and suggesting that this bacterial group plays a more crucial role in the sulfur cycle of coral holobionts and coral reefs than previously thought.
The DMS concentration produced by both Endozoicomonas species detected in our experiment was higher than that of R. atlantica. In our study, the cleavage ability of Endozoicomonas outcompeted that of the widely documented DMSP degraders Roseobacter. In contrast, Ruegeria can tolerate a high DMSP concentration (5 mM) feeding incubation and displayed higher DMS production at this high DMSP concentration than at a low DMSP concentration (0.1 mM) feeding incubation (49). In most cases, DMSP concentration in coral is less than 0.1 mM (38, 50, 51), and the highest DMSP concentration of 0.177 mM was detected in the coral species Acropora millepora under thermal stress (24). Our observations of the low DMS production rate of R. atlantica matched results from previous studies showing that Ruegeria species can shift the DMSP metabolic pathway from demethylation to cleavage along with an increase in DMSP concentration (from 0.1 to 5 mM DMSP) (49). Hence, we suggest that the Ruegeria in the stressed corals mostly metabolize DMSP via a non-DMS generated demethylation pathway. Unlike Ruegeria, Endozoicomonas cannot tolerate high DMSP concentrations but can cope with 0.1 mM DMSP (fig. S6), suggesting that a species of Endozoicomonas, rather than Ruegeria, could be the main producer of DMS in stressed corals.
We noticed that most Endozoicomonas genomes (9 of 10) accommodate the dddD gene (table S6) rather than other DMSP cleavage genes. Compared to other DMSP cleavage pathways, dddD is unique and advantageous in the DMSP cleavage pathway as it produces 3-hydroxiopropionate instead of cytotoxic compound acrylate (28). DMSP cleavage does not accumulate harmful compounds in the cell and releases the potent antioxidant DMS, indicating that the selection of the dddD gene in Endozoicomonas may be associated with the establishment of a bacteria-coral mutualism. We performed a simple distribution analysis of 8E across the Indo-Pacific Ocean (fig. S7) and found that 8E are widely distributed and have a similar distribution with E. acroporae (11). We suspect that the ability to cleave DMSP, enabled by the dddD gene, might be one of the reasons for the widespread appearance of Endozoicomonas, especially in the Acropora coral.
MATERIALS AND METHODS
De novo Endozoicomonas isolation, genome sequencing, and assembly
The remarkable DMSP-producing coral genus, Acropora sp., (52) in Kenting, Taiwan, was selected to isolate a new species of Endozoicomonas. The methods used to isolate and culture this species have been previously described (6). A comprehensive description of the morphological, physiological, and biochemical characteristics of strain 8E is provided in the Supplementary Materials. The same method for bacterial genomic DNA isolation as described in (11) was used in this study. High-quality genomic DNA was subjected to two different sequencing techniques. Illumina MiSeq paired-end sequencing was conducted at Yourgene Biosciences, Taiwan, while Nanopore sequencing was performed at the next generation sequencing (NGS) High Throughput Genomics Core, Biodiversity Research Center, Academia Sinica, Taiwan. 8E genome assembly was obtained by combining high-accuracy Illumina short reads and Nanopore sequencing long reads. After conducting a quality check using nanoQC (53), the nanopore reads were assembled using Flye v2.9 (54). The resulting draft contigs were polished using Illumina reads through the utilization of the software Pilon (55). After four rounds of polishing, we achieved 0 corrected single-nucleotide polymorphisms (SNPs), 0 ambiguous bases, 0 small insertions, and 0 deletions. This generated a high-quality contig with a length of 7,377,917 bp. CheckM V1.1.3 (56) was used to assess completeness, contamination, and heterogeneity. The method used to annotate the 8E genome is described in detail in the Supplementary Materials.
Phylogenetically comparing the dddD gene within Endozoicomonas to that of other bacteria
All Endozoicomonas dddD genes and operons used in this study were annotated using Prokka and a web-based CD-Search (57). To compare the position of the DddD protein within Endozoicomonas with that of other Proteobacteria, 67 DddD protein sequences were downloaded from the NCBI (keyword: dddD CoA-transferase/lyase) for phylogenetic tree construction. The collected DddD protein sequences were initially aligned using MAFFT v7. Subsequently, a maximum-likelihood tree was constructed using IQ-TREE (58) with 1000 replicates. Meanwhile, for the comparison of gene arrangement within the operon, we selected eight bacterial strains that contained the DddD protein from two different clades which were close to Endozoicomonas acroporae Acr-14T and 8E, respectively. These bacterial strains included three from the class Gammaproteobacteria (P. guineae, Marinobacter shejiangensis, and Marinomonas pollencensis) and four species from the same genus, Endozoicomonas (Endozoicomonas sp. ONNA1, Endozoicomonas sp. YOMI1, Endozoicomonas sp. G2-1, and E. acroporae Acr-14T). The genomes used in this study were downloaded from the NCBI database and are listed in table S5.
DMSP consumption and DMS-quantified assay
To ensure an equal initial bacterial count at the start of the experiment across different treatments, the initial bacterial population was standardized [optical density at 600 nm (OD600) = 0.6]. Bacterial cultures were initially enriched in mmbv4 medium supplemented with 0.1% glucose for 24 hours. After that, the residual medium was removed via centrifuge, and the bacteria were then resuspended twice using the fresh medium. After medium replacement, the bacteria were cultured in sterile 300-ml gas-tight vials, with rubber stoppers on the tops for sample collection (the bacterial concentration was measured and marked as BA; before acclimation). Before the addition of DMSP, the bacteria were acclimated in the fresh medium for 2 hours. We began collecting gas from the headspace of the bottles after DMSP was added (first measurement at 0 hours).
Two different mediums, with and without subtract combination, were applied. As Endozoicomonas cannot grow in minimal medium with only DMSP, 0.2% casamino acid was added to all minimal medium treatments. The only difference between the control group (labeled as mini) and the DMSP treatment group (labeled as mini + DMSP) is the presence of 0.1 mM DMSP. The second medium used in this study was a rich medium, mmbv4. Unlike the minimal medium, no additional casamino acid was used in any of the mmbv4 medium treatment groups.
DMS(g) measurement used in the assay followed the protocol described in (11). Collection time points for the two Endozoicomonas species were chosen on the basis of the initial emission time of DMS determined through our preliminary test (E. acroporae 8 hours and 8E 6 hours). We sampled R. atlantica DSS-3T at both the 6- and 8-hour time points for DMSP cleavage trend comparison.
DMSP concentration in the medium was determined via alkaline lysis. At each time point, 1.2 ml of the bacterial culture was drawn out from the gas-tight vials using a sterile syringe. The cultures were then centrifuged at 4500 rpm for 5 min, after which the resulting supernatant was collected and mixed with 100 ml of 10 M NaOH in a 20-ml gas-tight vial. Afterward, the vials were incubated at 200 rpm, 25°C for 30 min to ensure the full lysis of DMSP to DMS. The target peak areas from both bacterial cleavage DMS in the headspace and alkaline lysis DMS from DMSP in the medium were quantified using a standard DMS/DMSP prepared calibration curve (detailed information can be found in the Supplementary Materials). Bacterial growth was monitored using OD600 (Analytik Jena, ScanDrop 250, Germany) and flow cytometry.
Stable-isotopic DMSP synthesis
To track the products derived from DMSP, a three-carbon 13 labeled [1,2,3-13C3] stable-isotopic DMSP was essential (fig. S4B). However, no such product was commercially available. We synthesized the stable isotopes by following a synthesis protocol of [1,2,3-13C3] DMSP described in (59, 60). Briefly, DMS was converted from DMSP hydrochloride (Supelco, USA) by alkaline hydrolysis. The resulting DMS was converted to [1,2,3-13C3] DMSP hydrochloride via a Michael addition to [13C3] acrylic acid (99 atomic %, Sigma-Aldrich) under acidic conditions and washed with methylene chloride to remove any excess reactants. The structure and accurate mass of [1,2,3-13C3] DMSP hydrochloride was confirmed by nuclear magnetic resonance and high-resolution mass spectrometry (HRMS) (fig. S5). [34S] DMSP (fig. S4A) was provided by W. B. Whitman from the Department of Microbiology, University of Georgia.
Stable-isotopic labeled DMSP assay and NanoSIMS analysis
In the stable-isotopic labeled DMSP assay, the sampling time points and bacterial incubation conditions were the same as those described for the DMSP consumption and DMS quantification assay. The two different stable isotopes labeled DMSP (fig. S4) generated above were used as the DMSP source for further NanoSIMS visualization and analysis.
We filtered 106 bacterial cells through an Au-Pd (80:20)–coated polycarbonate filter (pore size, 0.2 μm; Millipore). Filters were washed by a series of ethanol solutions from 0, 25, 50, and 75 to 100% (v/v) for dehydration. After that, the isotopic compositions of bacterial cells on these filters were analyzed using a NanoSIMS 50 L (Cameca-Ametek, Gennevilliers, France) housed at Academia Sinica, Taiwan. Secondary ions of 12C2−, 12C13C−, 12C14N−, 32S−, and 34S− were collected simultaneously by multiple electron multipliers. Random areas on the polycarbonate filter were presputtered by 174- to 202-pA Cs+ ion beam for 1 min and analyzed by 12.5- to 13.4-pA Cs+ ion beam for ~45 to 60 min to obtain 50 μm by 50 μm square image data at 512 × 512 pixels resolution. Thirty and 150 mm of entrance slit and aperture slit, respectively, were used to reach a mass resolving power of 5000 for 32S−. The images and isotope ratios of the cells were processed by L’Image software (developed by L. Nittler, School of Earth and Space Exploration, Arizona State University). More than 37 cells were analyzed for each treatment. More than 2 × 105 counts of 12C2 and more than 104 counts of 32S− were collected for each bacterial cell.
Acknowledgments
We thank the two microscopic facilities at Academia Sinica: the IPMB Cell Biology Core EM Division for the technical support in electronic microscopic imaging and the Institute of Astronomy and Astrophysics NanoSIMS lab for stable-isotopic imaging analysis. We thank the NGS Core Facility at the Biodiversity Research Center, Academia Sinica for technical support of the high-throughput sequencing. Last, we thank the research group of Y.-W. Huang from the National Tsing Hua University and W. B. Whitman from the Department of Microbiology, University of Georgia, for providing us with stable isotopes. In recognition of Dr. R. Gates’ commitment to coral research, we have named the coral-isolated bacterium Endozoicomonas ruthgatesiae in her honor.
Funding: S.-L.T. was funded by Academia Sinica, Taipei, Taiwan (investigator award AS-IA-109-L05) and National Science and Technology Council (MOST-110-2611-M-001-004).
Author contributions: Conceptualization: Y.-J.C. and S.-L.T. Performed research: Y.-J.C., Y.-F.C., S.-P.Y., C.-Y.L., P.-W.C., and N.W. Data analysis: Y.-J.C., S.S.-Y.H., and P.-Y.L. Contributed data or analysis tools: Y.-J.C., T.-C.H., W.-N.J., Y.L, D.-C.L., and Y.-W.H. Supervision: S.-L.T. Writing—original draft: Y.-J.C. and S.-L.T. Writing—review and editing: Y.-J.C. and S.-L.T.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The Ca. E. ruthgatesiae 8E genome assembly and the sequencing data of RNA sequencing are available on NCBI under BioProject PRJNA946181. All of the genome sequencing used in this study is listed in table S5. Previously published data used for analyses in this study are listed in the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S8
Tables S1 to S6
References
REFERENCES AND NOTES
- 1.S. Li, T. Young, S. Archer, K. Lee, S. Sharma, A. C. Alfaro, Mapping the green-lipped mussel (Perna canaliculus) Microbiome: A multi-tissue analysis of bacterial and fungal diversity. Curr. Microbiol. 79, 76 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. J. Neave, C. T. Michell, A. Apprill, C. R. Voolstra, Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7, 40579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Nishijima, K. Adachi, A. Katsuta, Y. Shizuri, K. Yamasato, Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. Int. J. Syst. Evol. Microbiol. 63 (Pt 2), 709–714 (2013). [DOI] [PubMed] [Google Scholar]
- 4.R. E. Pike, B. Haltli, R. G. Kerr, Description of Endozoicomonas euniceicola sp nov and Endozoicomonas gorgoniicola sp nov., bacteria isolated from the octocorals Eunicea fusca and Plexaura sp., and an emended description of the genus Endozoicomonas. Int. J. Syst. Evol. Microbiol. 63 (Pt 11), 4294–4302 (2013). [DOI] [PubMed] [Google Scholar]
- 5.L. Schreiber, K. U. Kjeldsen, M. Obst, P. Funch, A. Schramm, Description of Endozoicomonas ascidiicola sp. nov., isolated from Scandinavian ascidians. Syst. Appl. Microbiol. 39, 313–318 (2016). [DOI] [PubMed] [Google Scholar]
- 6.S. Y. Sheu, K. R. Lin, M. Y. Hsu, D. S. Sheu, S. L. Tang, W. M. Chen, Endozoicomonas acroporae sp nov., isolated from Acropora coral. Int. J. Syst. Evol. Microbiol. 67, 3791–3797 (2017). [DOI] [PubMed] [Google Scholar]
- 7.J. L. Meyer, V. J. Paul, M. Teplitski, Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLOS ONE 9, e100316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.K. M. Morrow, D. G. Bourne, C. Humphrey, E. S. Botté, P. Laffy, J. Zaneveld, S. Uthicke, K. E. Fabricius, N. S. Webster, Natural volcanic CO2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. 9, 894–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Z. Qin, K. Yu, S. Chen, B. Chen, Q. Yao, X. Yu, N. Pan, X. Wei, Significant changes in bacterial communities associated with Pocillopora corals ingestion by crown-of-thorns starfish: An important factor affecting the coral’s health. Microorganisms 10, 207 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. J. Neave, C. T. Michell, A. Apprill, C. R. Voolstra, Whole-genome sequences of three symbiotic Endozoicomonas strains. Genome Announc. 2, e00802–e00814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.K. Tandon, C.-Y. Lu, P.-W. Chiang, N. Wada, S.-H. Yang, Y.-F. Chan, P.-Y. Chen, H.-Y. Chang, Y.-J. Chiou, M.-S. Chou, W.-M. Chen, S.-L. Tang, Comparative genomics: Dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J.-Y. Ding, J.-H. Shiu, W.-M. Chen, Y.-R. Chiang, S.-L. Tang, Genomic insight into the host–Endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front. Microbiol. 7, 251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K. Tandon, Y.-J. Chiou, S.-P. Yu, H. J. Hsieh, C.-Y. Lu, M.-T. Hsu, P.-W. Chiang, H.-J. Chen, N. Wada, S.-L. Tang, Microbiome restructuring: Dominant coral bacterium Endozoicomonas species respond differentially to environmental changes. mSystems 7, e0035922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. Pogoreutz, C. A. Oakley, N. Rädecker, A. Cárdenas, G. Perna, N. Xiang, L. Peng, S. K. Davy, D. K. Ngugi, C. R. Voolstra, Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle. ISME J. 16, 1883–1895 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L. M. Fitzgerald, A. M. Szmant, Biosynthesis of ‘essential’ amino acids by Scleractinian corals. Biochem. J. 322 (Pt 1), 213–221 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J.-B. Raina, D. Tapiolas, B. L. Willis, D. G. Bourne, Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.E. Ransome, S. J. Rowley, S. Thomas, K. Tait, C. B. Munn, Disturbance to conserved bacterial communities in the cold-water gorgonian coral Eunicella verrucosa. FEMS Microbiol. Ecol. 90, 404–416 (2014). [DOI] [PubMed] [Google Scholar]
- 18.M. O. Andreae, Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar. Chem. 30, 1–29 (1990). [Google Scholar]
- 19.G. Kirst, Osmotic adjustment in phytoplankton and macroalgae: The use of dimethylsulfoniopropionate (DMSP) In R. P. Kiene, P. T. Visscher, K. D. Maureen, G. Kirst, Eds. Biological and environmental chemistry of DMSP and related sulfonium compounds. (Plenum Press, 1996) pp. 121-129. [Google Scholar]
- 20.G. Kirst, C. Thiel, H. Wolff, J. Nothnagel, M. Wanzek, R. Ulmke, Dimethylsulfoniopropionate (DMSP) in icealgae and its possible biological role. Mar. Chem. 35, 381–388 (1991). [Google Scholar]
- 21.A. M. Theseira, D. A. Nielsen, K. Petrou, Uptake of dimethylsulphoniopropionate (DMSP) reduces free reactive oxygen species (ROS) during late exponential growth in the diatom Thalassiosira weissflogii grown under three salinities. Mar. Biol. 167, 127 (2020). [Google Scholar]
- 22.W. Sunda, D. Kieber, R. Kiene, S. Huntsman, An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320 (2002). [DOI] [PubMed] [Google Scholar]
- 23.C. Aguilar, J.-B. Raina, C. A. Motti, S. Fôret, D. C. Hayward, B. Lapeyre, D. G. Bourne, D. J. Miller, Transcriptomic analysis of the response of Acropora millepora to hypo-osmotic stress provides insights into DMSP biosynthesis by corals. BMC Genomics 18, 612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. G. Gardner, M. R. Nitschke, J. O'Brien, C. A. Motti, J. R. Seymour, P. J. Ralph, K. Petrou, J.-B. Raina, Increased DMSP availability during thermal stress influences DMSP-degrading bacteria in coral mucus. Front. Mar. Sci. 9, 10.3389/fmars.2022.912862, (2022). [Google Scholar]
- 25.S. G. Gardner, D. A. Nielsen, O. Laczka, R. Shimmon, V. H. Beltran, P. J. Ralph, K. Petrou, Dimethylsulfoniopropionate, superoxide dismutase and glutathione as stress response indicators in three corals under short-term hyposalinity stress. Proc. R. Soc. B Biol. Sci. 283, 20152418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.E. S. Deschaseaux, G. B. Jones, M. A. Deseo, K. M. Shepherd, R. Kiene, H. Swan, P. L. Harrison, B. D. Eyre, Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol. Oceanogr. 59, 758–768 (2014). [Google Scholar]
- 27.C. R. Reisch, M. A. Moran, W. B. Whitman, Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2, 172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. Yoshii, Cytotoxic effects of acrylates and methacrylates: Relationships of monomer structures and cytotoxicity. J. Biomed. Mater. Res. 37, 517–524 (1997). [DOI] [PubMed] [Google Scholar]
- 29.C.-Y. Li, X.-J. Wang, X.-L. Chen, Q. Sheng, S. Zhang, P. Wang, M. Quareshy, B. Rihtman, X. Shao, C. Gao, F. Li, S. Li, W. Zhang, X.-H. Zhang, G.-P. Yang, J. D. Todd, A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. eLife 10, e64045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.U. Alcolombri, P. Laurino, P. Lara-Astiaso, A. Vardi, D. S. Tawfik, DddD is a CoA-transferase/lyase producing dimethyl sulfide in the marine environment. Biochemistry 53, 5473–5475 (2014). [DOI] [PubMed] [Google Scholar]
- 31.J. D. Todd, R. Rogers, Y. G. Li, M. Wexler, P. L. Bond, L. Sun, A. R. J. Curson, G. Malin, M. Steinke, A. W. B. Johnston, Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Y. Cui, S.-K. Wong, R. Kaneko, A. Mouri, Y. Tada, I. Nagao, S.-J. Chun, H.-G. Lee, C.-Y. Ahn, H.-M. Oh, Y. Sato-Takabe, K. Suzuki, H. Fukuda, T. Nagata, K. Kogure, K. Hamasaki, Distribution of dimethylsulfoniopropionate degradation genes reflects strong water current dependencies in the Sanriku coastal region in Japan: From mesocosm to field study. Front. Microbiol. 11, 1372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Liu, C.-X. Xue, J. Wang, A. T. Crombie, O. Carrión, A. W. Johnston, J. C. Murrell, J. Liu, Y. Zheng, X.-H. Zhang, Oceanospirillales containing the DMSP lyase DddD are key utilisers of carbon from DMSP in coastal seawater. Microbiome 10, 110 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.E. Stackebrandt, B. M. Goebel, Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849 (1994). [Google Scholar]
- 35.M. R. Olm, A. Crits-Christoph, S. Diamond, A. Lavy, P. B. Matheus Carnevali, J. F. Banfield, Consistent metagenome-derived metrics verify and delineate bacterial species boundaries. mSystems 5, e00731-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. Kim, H.-S. Oh, S.-C. Park, J. Chun, Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351 (2014). [DOI] [PubMed] [Google Scholar]
- 37.R. M. Bowers, N. C. Kyrpides, R. Stepanauskas, M. Harmon-Smith, D. Doud, T. Reddy, F. Schulz, J. Jarett, A. R. Rivers, E. A. Eloe-Fadrosh, S. G. Tringe, N. N. Ivanova, A. Copeland, A. Clum, E. D. Becraft, R. R. Malmstrom, B. Birren, M. Podar, P. Bork, G. M. Weinstock, G. M. Garrity, J. A. Dodsworth, S. Yooseph, G. Sutton, F. O. Glöckner, J. A. Gilbert, W. C. Nelson, S. J. Hallam, S. P. Jungbluth, T. J. G. Ettema, S. Tighe, K. T. Konstantinidis, W.-T. Liu, B. J. Baker, T. Rattei, J. A. Eisen, B. Hedlund, K. D. McMahon, N. Fierer, R. Knight, R. Finn, G. Cochrane, I. Karsch-Mizrachi, G. W. Tyson, C. Rinke; Genome Standards Consortium, A. Lapidus, F. Meyer, P. Yilmaz, D. H. Parks, A. M. Eren, L. Schriml, J. F. Banfield, P. Hugenholtz, T. Woyke, Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J.-B. Raina, D. M. Tapiolas, S. Forêt, A. Lutz, D. Abrego, J. Ceh, F. O. Seneca, P. L. Clode, D. G. Bourne, B. L. Willis, C. A. Motti, DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502, 677–680 (2013). [DOI] [PubMed] [Google Scholar]
- 39.J. M. Gonzalez, J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. A. Fuhrman, R. P. Kiene, M. A. Moran, Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53 (Pt 5), 1261–1269 (2003). [DOI] [PubMed] [Google Scholar]
- 40.U. Karsten, K. Kück, C. Vogt, G. Kirst, “Dimethylsulfoniopropionate production in phototrophic organisms and its physiological functions as a cryoprotectant” in Biological and environmental chemistry of DMSP and related sulfonium compounds. (Springer, 1996), pp. 143–153. [Google Scholar]
- 41.U. Karsten, G. Kirst, C. Wiencke, Dimethylsulphoniopropionate (DMSP) accumulation in green macioalgae from polar to temperate regions: Interactive effects of light versus salinity and light versus temperature. Polar Biol. 12, 603–607 (1992). [Google Scholar]
- 42.T. R. Miller, K. Hnilicka, A. Dziedzic, P. Desplats, R. Belas, Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70, 4692–4701 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. R. Miller, R. Belas, Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8, 1648–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 44.M. Landa, A. S. Burns, S. J. Roth, M. A. Moran, Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Li, W. Kuang, L. Long, S. Zhang, Production of quorum-sensing signals by bacteria in the coral mucus layer. Coral Reefs 36, 1235–1241 (2017). [Google Scholar]
- 46.W. M. Johnson, M. C. Kido Soule, E. B. Kujawinski, Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 10, 2304–2316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Adler, N. S. Corbalan, D. R. Peralta, M. F. Pomares, R. E. de Cristóbal, P. A. Vincent, The alternative role of enterobactin as an oxidative stress protector allows Escherichia coli colony development. PLOS ONE 9, e84734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. Li, D. Pan, M. Li, Y. Wang, L. Song, D. Yu, Y. Zuo, K. Wang, Y. Liu, Z. Wei, Z. Lui, L. Zhu, Aerobactin-mediated iron acquisition enhances biofilm formation, oxidative stress resistance, and virulence of Yersinia pseudotuberculosis. Front. Microbiol. 12, 699913 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. S. Wirth, T. Wang, Q. Huang, R. H. White, W. B. Whitman, Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria Species. mBio 11, e00329-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. D. Broadbent, G. B. Jones, R. J. Jones, DMSP in corals and benthic algae from the Great Barrier Reef. Estuar. Coast. Shelf Sci. 55, 547–555 (2002). [Google Scholar]
- 51.D. M. Tapiolas, J.-B. Raina, A. Lutz, B. L. Willis, C. A. Motti, Direct measurement of dimethylsulfoniopropionate (DMSP) in reef-building corals using quantitative nuclear magnetic resonance (qNMR) spectroscopy. J. Exp. Mar. Biol. Ecol. 443, 85–89 (2013). [Google Scholar]
- 52.K. L. Van Alstyne, P. Schupp, M. Slattery, The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs 25, 321–327 (2006). [Google Scholar]
- 53.W. De Coster, S. D’Hert, D. T. Schultz, M. Cruts, C. Van Broeckhoven, NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Kolmogorov, D. M. Bickhart, B. Behsaz, A. Gurevich, M. Rayko, S. B. Shin, K. Kuhn, J. Yuan, E. Polevikov, T. P. L. Smith, P. A. Pevzner, MetaFlye: Scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.B. J. Walker, T. Abeel, T. Shea, M. Priest, A. Abouelliel, S. Sakthikumar, C. A. Cuomo, Q. Zeng, J. Wortman, S. K. Young, A. M. Earl, Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D. H. Parks, M. Imelfort, C. T. Skennerton, P. Hugenholtz, G. W. Tyson, CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. Marchler-Bauer, S. Lu, J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, F. Lu, G. H. Marchler, M. Mullokandov, M. V. Omelchenko, C. L. Robertson, J. S. Song, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, C. Zheng, S. H. Bryant, CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39 (Suppl 1), D225–D229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.B. Q. Minh, H. A. Schmidt, O. Chernomor, D. Schrempf, M. D. Woodhams, A. von Haeseler, R. Lanfear, IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. T. Chambers, C. M. Kunin, D. Miller, A. Hamada, Dimethylthetin can substitute for glycine betaine as an osmoprotectant molecule for Escherichia coli. J. Bacteriol. 169, 4845–4847 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. S. Wirth, W. B. Whitman, An efficient method for synthesizing dimethylsulfonio-34S-propionate hydrochloride from 34S8. J. Label. Compd. Radiopharm. 62, 52–58 (2019). [DOI] [PubMed] [Google Scholar]
- 61.D. Reynolds, T. Thomas, Evolution and function of eukaryotic-like proteins from sponge symbionts. Mol. Ecol. 25, 5242–5253 (2016). [DOI] [PubMed] [Google Scholar]
- 62.M. J. Neave, A. Apprill, C. Ferrier-Pages, C. R. Voolstra, Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.C. S. Yang, M. H. Chen, A. B. Arun, C. A. Chen, J. T. Wang, W. M. Chen, Endozoicomonas montiporae sp nov., isolated from the encrusting pore coral Montipora aequituberculata. Int. J. Syst. Evol. Microbiol. 60 (Pt 5), 1158–1162 (2010). [DOI] [PubMed] [Google Scholar]
- 64.M. T. H. D. Nguyen, M. Liu, T. Thomas, Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23, 1635–1645 (2014). [DOI] [PubMed] [Google Scholar]
- 65.C. Cazalet, C. Rusniok, H. Brüggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, C. Buchrieser, Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Z. Stoytcheva, B. Joshi, J. Spížek, P. Tichý, WD-repeat protein encoding genes among prokaryotes of theStreptomyces genus. Folia Microbiol. 45, 407–413 (2000). [DOI] [PubMed] [Google Scholar]
- 67.M. Hisbergues, C. G. Gaitatzes, F. Joset, S. Bedu, T. F. Smith, A noncanonical WD-repeat protein from the cyanobacterium Synechocystis PCC6803: Structural and functional study. Protein Sci. 10, 293–300 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.X.-J. Hu, T. Li, Y. Wang, Y. Xiong, X.-H. Wu, D.-L. Zhang, Z.-Q. Ye, Y.-D. Wu, Prokaryotic and highly-repetitive WD40 proteins: A systematic study. Sci. Rep. 7, 10585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.E. J. Neer, C. J. Schmidt, R. Nambudripad, T. F. Smith, The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300 (1994). [DOI] [PubMed] [Google Scholar]
- 70.M. Guo, J. Wang, Y. Zhang, L. Zhang, Increased WD40 motifs in Planctomycete bacteria and their evolutionary relevance. Mol. Phylogenet. Evol. 155, 107018 (2021). [DOI] [PubMed] [Google Scholar]
- 71.D. Laslett, B. Canback, ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.T. Seemann, barrnap 0.9: Rapid ribosomal RNA prediction. Google Scholar, (2013).
- 73.T. Seemann, Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 74.R. K. Aziz, D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, O. Zagnitko, The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R. Overbeek, R. Olson, G. D. Pusch, G. J. Olsen, J. J. Davis, T. Disz, R. A. Edwards, S. Gerdes, B. Parrello, M. Shukla, V. Vonstein, A. R. Wattam, F. Xia, R. Stevens, The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D. Arndt, J. R. Grant, A. Marcu, T. Sajed, A. Pon, Y. Liang, D. S. Wishart, PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.S. Lu, J. Wang, F. Chitsaz, M. K. Derbyshire, R. C. Geer, N. R. Gonzales, M. Gwadz, D. I. Hurwitz, G. H. Marchler, J. S. Song, N. Thanki, R. A. Yamashita, M. Yang, D. Zhang, C. Zheng, C. J. Lanczycki, A. Marchler-Bauer, CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.G. Benson, Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.V. Eichinger, T. Nussbaumer, A. Platzer, M.-A. Jehl, R. Arnold, T. Rattei, EffectiveDB—Updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 44, D669–D674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.E. P. Nawrocki, S. R. Eddy, Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.E. P. Nawrocki, “Annotating Functional RNAs in Genomes Using Infernal” in RNA Sequence, Structure, and Function: Computational and Bioinformatic Methods, J. Gorodkin, W. L. Ruzzo, Eds. (Humana Press, 2014), pp. 163–197. [DOI] [PubMed] [Google Scholar]
- 82.S. Kalyaanamoorthy, B. Q. Minh, T. K. F. Wong, A. von Haeseler, L. S. Jermiin, ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.L.-T. Nguyen, H. A. Schmidt, A. Von Haeseler, B. Q. Minh, IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.I. Letunic, P. Bork, Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.S. I. Na, Y. O. Kim, S. H. Yoon, S. M. Ha, I. Baek, J. Chun, UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285 (2018). [DOI] [PubMed] [Google Scholar]
- 86.L. Rodriguez-R, K. Konstantinidis. (PeerJ Preprints, 2016).
- 87.R. C. Team. (2018).
- 88.R. Kolde, M. R. Kolde, Package ‘pheatmap’. R package 1, 790 (2015).
- 89.W. Shen, S. Le, Y. Li, F. Hu, SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLOS ONE 11, e0163962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.J. R. Grant, P. Stothard, The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. C. Darling, B. Mau, F. R. Blattner, N. T. Perna, Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.K. Katoh, J. Rozewicki, K. D. Yamada, MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.S. Andrews. (Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom, 2010).
- 94.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Y. Liao, G. K. Smyth, W. Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 96.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.M. Kanehisa, Y. Sato, K. Morishima, BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 98.S.-H. Yang, C.-H. Tseng, H.-P. Lo, P.-W. Chiang, H.-J. Chen, J.-H. Shiu, H.-C. Lai, K. Tandon, N. Isomura, T. Mezaki, H. Yamamoto, S.-L. Tang, Locality effect of coral-associated bacterial community in the Kuroshio Current from Taiwan to Japan. Front. Ecol. Evol. 8, 569107 (2020). [Google Scholar]
- 99.R. Bernasconi, M. Stat, A. Koenders, A. Paparini, M. Bunce, M. J. Huggett, Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 10, 1529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.K. Damjanovic, L. L. Blackall, L. M. Peplow, M. J. H. van Oppen, Assessment of bacterial community composition within and among Acropora loripes colonies in the wild and in captivity. Coral Reefs 39, 1245–1255 (2020). [Google Scholar]
- 101.E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. D. Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y.-X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. M. Donald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson II, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight, J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.R. C. Team, R Core Team R: A language and environment for statistical computing. Foundation for Statistical Computing., (2020).
- 103.H. Wickham, ggplot2. Wiley interdisciplinary reviews: computational statistics 3, 180–185 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S8
Tables S1 to S6
References