Abstract
Chromatin regulation plays a pivotal role in establishing and maintaining cellular identity and is one of the top pathways disrupted in autism spectrum disorder (ASD). The hippocampus, composed of distinct cell types, is often affected in patients with ASD. However, the specific hippocampal cell types and their transcriptional programs that are dysregulated in ASD are unknown. Using single-nucleus RNA sequencing, we show that the ASD gene, lysine demethylase 5A (KDM5A), regulates the development of specific subtypes of excitatory and inhibitory neurons. We found that KDM5A is essential for establishing hippocampal cell identity by controlling a differentiation switch early in development. Our findings define a role for the chromatin regulator KDM5A in establishing hippocampal cell identity and contribute to the emerging convergent mechanisms across ASD.
The autism gene and chromatin regulator KDM5A mediates hippocampal cell identity by controlling a differentiation switch.
INTRODUCTION
Epigenetic chromatin remodeling is an essential step in cellular differentiation, and it is believed to dictate and maintain cellular identity (1). Studies have shown that undifferentiated cells have a globally “open” chromatin state (2, 3), which transitions to a more compact state as cells differentiate and acquire their identity (4). Chromatin regulation is required for proper gene expression and for brain development (5), with critical functions in establishing cell identities and the wiring of neuronal circuits (1, 6). It is also one of the top pathways disrupted in autism spectrum disorder (ASD) with causative mutations identified in several genes encoding chromatin remodelers (e.g., ARID1B, CHD8, and SETD5) (7–9).
Recent studies have shown that ASD genes are enriched in specific cell types in the brain during development (10, 11) and that changes in gene regulatory networks in ASD occur in a cell type–specific manner in the brain (12). The cellular landscape of the brain is extremely diverse, allowing for complex cognitive functions and behaviors. The diversity of the neuronal and nonneuronal cell types in the brain has been demonstrated at several levels including morphological (13), transcriptomic (14–17), epigenomic (18), as well as circuitry and electrophysiological properties (19). The cellular complexity of the brain increases within specific brain regions and reflects the specific functions of each region. The hippocampus, known for its role in memory formation (20), is characterized by a distinct cellular diversity mediating specific hippocampal functions (21, 22). It is composed of different regions that include the dentate gyrus and the cornu ammonis 1 (CA1), CA2, and CA3. Each of these regions has its own identity, unique properties, and specific cell types expressing region-specific markers. Single-cell sequencing studies have elucidated the different cellular subtypes in the hippocampus and identified their specific markers (16, 23, 24).
Multiple brain regions have been implicated in the pathogenesis of ASD, including the hippocampus (25). It has been shown that the hippocampus is impaired in individuals with ASD, both structurally (26, 27) and functionally (28, 29), including an abnormal pattern of hippocampal development with larger hippocampal volume and abnormal functional connectivity during learning in individuals with ASD compared to controls (26–29). However, the specific cell types in the hippocampus that are particularly affected in ASD are not known. We recently identified a disease gene, lysine demethylase 5A (KDM5A), with pathogenic variants in patients with ASD, lack of speech, and intellectual disability, among other phenotypes. In addition, we showed that complete loss of KDM5A in the constitutive knockout mouse model (Kdm5a−/−) results in transcriptional dysregulation in the hippocampus and in neurobehavioral abnormalities, including severe deficits in social behavior, vocalization, learning, and memory, and increased repetitive behaviors, in addition to abnormal dendritic morphogenesis (30). KDM5A is a chromatin regulator that belongs to the KDM5 family of lysine-specific histone H3 demethylases. Evidence from our studies demonstrates that KDM5A is critical for brain development and is essential for mediating complex behaviors including learning and memory (30). Murine Kdm5a is first detected at embryonic day 9.5 (E9.5) in the auditory pit (31) and at E10.5 in the brain (31). KDM5A is ubiquitously expressed across cell types in the brain (32), its expression pattern throughout development is dynamic, and it is differentially expressed at different stages of cell lineage birth and maturation (33). To investigate whether KDM5A has a cell type–specific function in the brain, we performed single-nucleus RNA sequencing (snRNA-seq) on hippocampal tissue from wild-type (WT) and Kdm5a knockout (Kdm5a−/−) mice. We found a role for KDM5A in establishing identities of specific subtypes of hippocampal cells. Furthermore, we found that loss of KDM5A alters the developmental trajectory of hippocampal neurons and their transcriptional networks by altering the expression of common and unique sets of genes in the different cellular subtypes.
RESULTS
Loss of KDM5A alters hippocampal cell composition
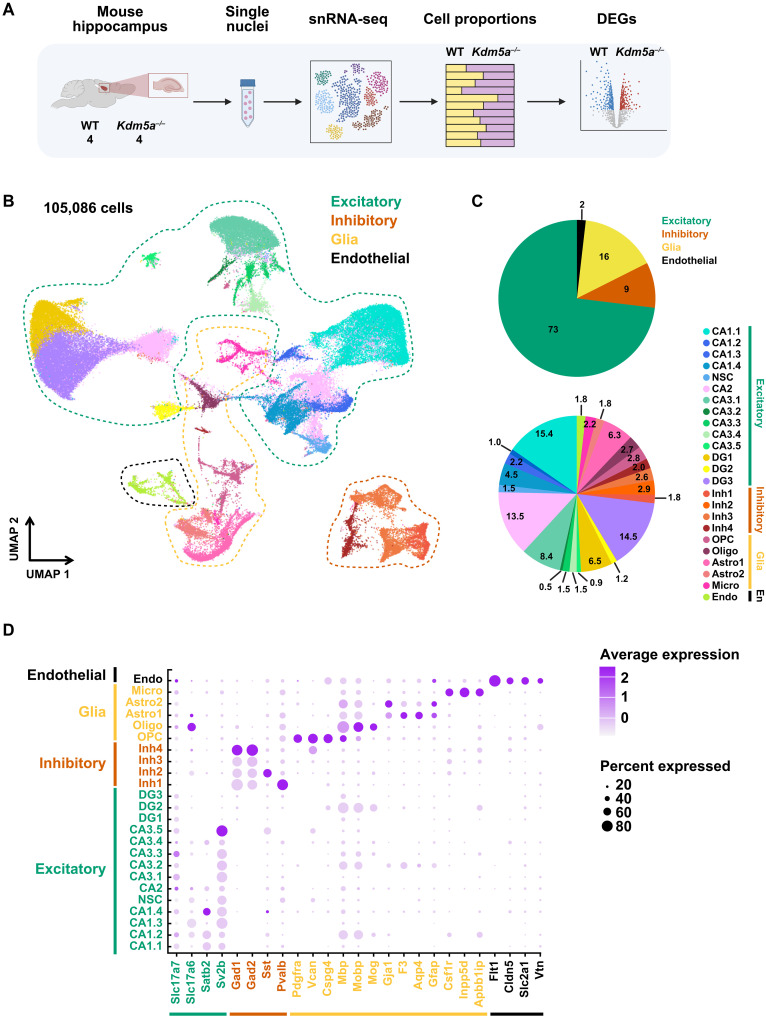
To investigate the effect of losing the chromatin regulator and ASD gene KDM5A on the development of the hippocampus, we profiled hippocampi from 20-week-old WT and Kdm5a knockout (Kdm5a−/−) mice using snRNA-seq (Fig. 1A). We had previously generated and characterized the Kdm5a constitutive knockout mouse model (Kdm5a−/−) and confirmed the complete loss of KDM5A transcript and protein in the Kdm5a−/− (30). We sequenced a total of 105,086 nuclei that met our quality control measures (fig. S1), and we recapitulated all known major cell types in the hippocampus (Fig. 1, B and C). We assigned identities to the cell clusters by assessing the expression of known markers of the different hippocampal cell types (16, 23, 24, 34) and identified subclusters of hippocampal CA1, CA2, CA3, and dentate gyrus excitatory neurons, as well as inhibitory neurons, glia, neural stem cells, and endothelial clusters (Fig. 1D and fig. S2, A and C). We assessed the cellular composition of the sequenced cells and determined the percentage of each cell type. As expected, excitatory neurons represented the predominant cell type (73%) of all the sequenced cells (Fig. 1C). In addition, we confirmed that Kdm5a is ubiquitously expressed across all cell types in the hippocampus (fig. S2B).
Fig. 1. snRNA-seq from WT and Kdm5a−/− mice identifies 24 clusters of hippocampal cell types.
(A) Schematic summary of the experimental and analytical workflow. The figure was created using BioRender. (B) UMAP plot of 105,086 nuclei from hippocampi of four WT and four Kdm5a−/− mice (20 weeks old) colored by cluster identity and annotated according to known cell types. (C) Cell type composition across all cells from WT and Kdm5a−/− hippocampi. Pie chart depicting the percentage of all cells belonging to each major cell type group (top). Pie chart depicting the percentage of all cells belonging to each cell type cluster (bottom). (D) Dot plot showing the relative expression of marker genes for major cell types: excitatory, inhibitory, glia, and endothelial. The dot size represents the percentage of nuclei expressing each marker gene, and the dot color intensity represents the average expression of the gene (light color, low expression; dark color, high expression).
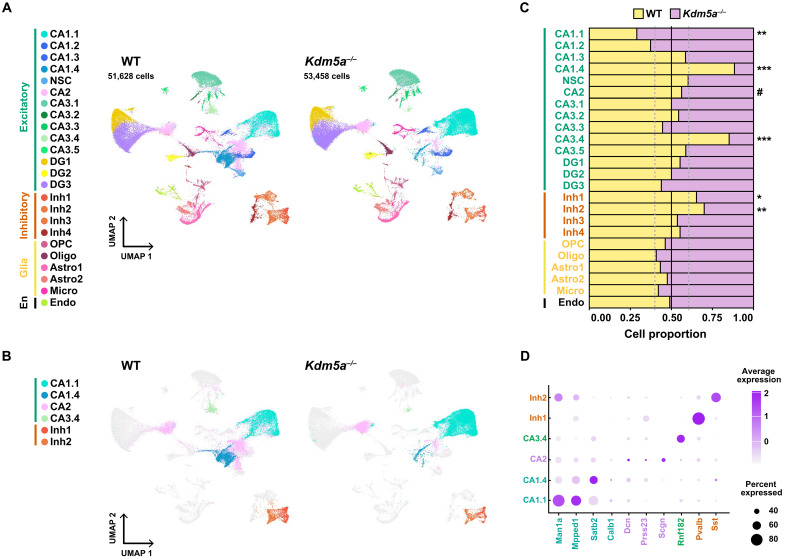
Next, we compared the cell clusters between WT and Kdm5a−/− hippocampi. We sequenced similar numbers of nuclei from WT (51,628) and Kdm5a−/− (53,458) (P = 0.9159) hippocampi and found that the cell proportions of the major cell types (excitatory, inhibitory, glia, and endothelial) were similar among all replicates of the same genotype: four WT and four Kdm5a−/− samples (Fig. 2A and fig. S3, A and B). We analyzed the proportions of WT and Kdm5a−/− nuclei for each cluster and identified six distinct clusters that are most vulnerable to the loss of KDM5A: two inhibitory subclusters (Inh1 Pvalb positive and Inh2 Sst positive) and four excitatory subclusters (CA1.1, CA1.4, CA2, and CA3.4) (Fig. 2, B and C, and fig. S3C). We found that the two inhibitory subclusters (Inh1 and Inh2) and excitatory subclusters CA1.4 and CA3.4 have lower cell proportions from Kdm5a−/− compared to WT. On the other hand, we found that subcluster CA1.1 has more cells from Kdm5a−/− compared to WT (Fig. 2C and fig. S3C). Subcluster CA2 is composed of two spatially separated populations on the uniform manifold approximation and projection (UMAP) plot (Fig. 2B). Although we did not find a change in cell proportions for CA2 between genotypes (Fig. 2C and fig. S3C), the composition of the subcluster shifts in the Kdm5a−/− compared to WT (Fig. 2, A and B). The six clusters that are vulnerable to the loss of KDM5A each express known markers of hippocampal excitatory neurons in the CA1 (Man1a, Mpped1, Satb2, and Calb1), CA2 (Dcn, Prss23, and Scgn), or CA3 (Rnf182) or inhibitory neurons (Pvalb and Sst) confirming their assigned identities (Fig. 2D).
Fig. 2. Loss of KDM5A alters the composition of specific subclusters of hippocampal CA1, CA2, CA3, and inhibitory cell types.
(A) UMAP plots of 51,628 nuclei from WT and 53,458 nuclei from Kdm5a−/− hippocampi colored by cluster identity and annotated according to known cell identity markers. (B) UMAP plots of nuclei from WT and Kdm5a−/− hippocampi colored by cluster identity and annotated according to known cell identity markers. Clusters with changing cell proportions in WT and Kdm5a−/− hippocampi are colored. Clusters not changing between WT and Kdm5a−/− hippocampi are in gray. (C) Proportion of cells in each cluster by genotype (CA1.1, **P = 0.0023; CA1.4, ***P < 0.0001; CA2, #P = 0.3216; CA3.4, ***P < 0.0001; Inh1, *P = 0.0223; Inh2, **P = 0.004). Data were analyzed using chi-square test. (D) Dot plot showing the relative expression of marker genes for specific neuronal subtypes. The dot size represents the percentage of nuclei expressing each marker gene, and the dot color intensity represents the average expression of the gene (light color, low expression; dark color, high expression).
KDM5A regulates common and unique genes in the vulnerable hippocampal cells
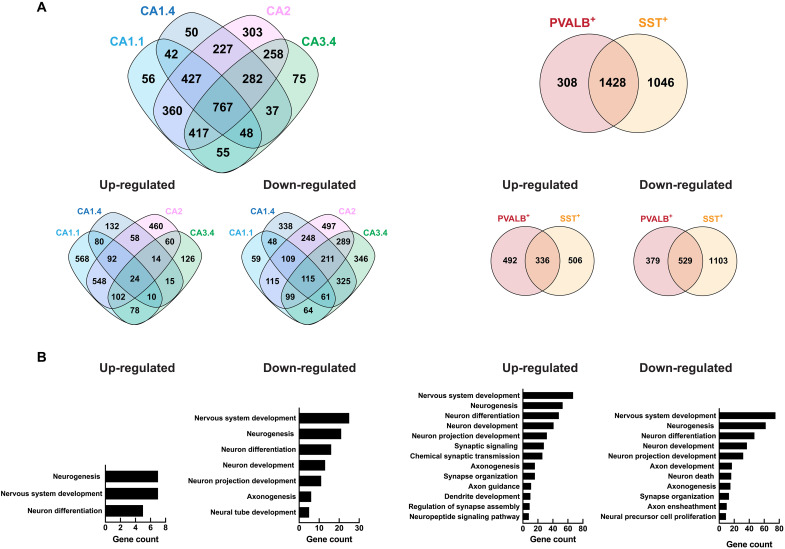
We assessed the transcriptional changes between WT and Kdm5a−/− in the six affected clusters to understand the unique sensitivity of these clusters to the loss of KDM5A. Thousands of differentially expressed genes (DEGs) were identified in the six clusters of interest [table S1; false discovery rate (FDR)–corrected P ≤ 0.05 and log2 fold change ≥ |0.3|], some of which overlap between clusters indicating a common function of KDM5A in the different hippocampal cell types (table S1). We defined a signature for KDM5A that is common across affected cell types, by combining the expression in the four affected excitatory clusters together (CA1.1, CA1.4, CA2, and CA3.4) and the two affected inhibitory clusters together (Inh1 and Inh2) and then comparing each to the expression of the combined unaffected excitatory and inhibitory clusters, respectively. We found that of the 2688 DEGs in the combined affected excitatory cluster, 196 are mouse homologs of known ASD genes [Fisher’s exact test, P < 2.2 × 10−16, odds ratio (OR) = 2.48], and of the 2385 DEGs in the combined affected inhibitory cluster, 177 are mouse homologs of known ASD genes (Fisher’s exact test, P < 2.2 ×10−16, OR = 2.50) (table S2). We identified 767 DEGs that are common among all four excitatory clusters irrespective of their direction of dysregulation (Fig. 3A, top). Of these, 24 genes are commonly up-regulated in all four excitatory clusters, 115 are commonly down-regulated in all four excitatory clusters, and the remaining 628 genes are dysregulated in opposite directions in at least one of the four clusters (Fig. 3A, bottom). We also identified 1428 DEGs that overlap between the two inhibitory clusters irrespective of their direction of dysregulation (Fig. 3A, top). Of these, 336 genes are commonly up-regulated in both inhibitory clusters, 529 genes are commonly down-regulated in both inhibitory clusters, and the remaining 563 genes are dysregulated in opposite directions in both clusters (Fig. 3A, bottom). Several DEGs in the six clusters of interest are mouse homologs of known ASD genes, many of which are common among all six clusters (e.g., Astn2, Cntn4, and Gfap) (table S1). Gene ontology (GO) analysis showed that these common DEGs are enriched for neurogenesis, neuron differentiation, axon guidance, dendritic development, and synaptic signaling (Fig. 3B). The data suggest a common role for KDM5A in regulating pathways underlying neuronal development in all six clusters of excitatory and inhibitory neurons. In addition to the common DEGs, we identified DEGs that are unique to each of the six clusters irrespective of their direction of dysregulation; there are 56 exclusive DEGs for CA1.1 that do not overlap with DEGs from any of the other clusters, 50 for CA1.4, 303 for CA2, and 75 for CA3.4. For the inhibitory clusters, 308 DEGs are unique to the parvalbumin positive (PVALB+) cluster, and 1046 are unique to the somatostatin positive (SST+) cluster (Fig. 3A, top). Considering directionality, we identified 568 genes that are exclusively up-regulated in CA1.1, 132 in CA1.4, 460 in CA2, 126 in CA3.4, 492 in PVALB+, and 506 in SST+ (Fig. 3A, bottom). In addition, we found 59 genes that are exclusively down-regulated in CA1.1, 338 in CA1.4, 497 in CA2, 346 in CA3.4, 379 in PVALB+, and 1103 in SST+. We performed GO analysis on the DEGs that are exclusive per cluster and found similar GO terms between the unique and the common DEGs (fig. S4). Thus, KDM5A regulates common and unique genes in the six subtypes of neurons that are most vulnerable to its loss. Although some of the dysregulated genes may be different in each of the cell types, the molecular pathways they regulate converge on essential neurodevelopmental processes, highlighting the importance of KDM5A in mediating neuronal development and function.
Fig. 3. Differential gene expression analysis reveals common function of KDM5A in the six vulnerable clusters.
(A) Venn diagrams showing overlap of DEGs between WT and Kdm5a−/− in excitatory (left) and inhibitory (right) clusters, irrespective of their direction of dysregulation (top). Overlap between commonly up-regulated DEGs and commonly down-regulated DEGs in all four excitatory (left) and inhibitory (right) clusters (bottom). (B) GO analysis of the DEGs that are commonly up-regulated or commonly down-regulated in excitatory (left) and inhibitory (right) clusters. Data are from DEGs with an FDR-corrected P ≤ 0.05 and log2 fold change ≥ |0.3|.
We examined the transcriptional changes between WT and Kdm5a−/− in all the cells in order to compare the DEGs with previously published bulk RNA-seq data and publicly available chromatin immunoprecipitation–sequencing (ChIP-seq) data. We identified a total of 3099 DEGs between WT and Kdm5a−/−, including 1881 up-regulated and 1218 down-regulated genes (table S1; FDR-corrected P ≤ 0.05 and log2 fold change ≥ |0.3|). Of these DEGs, 200 are mouse homologs of known ASD genes (Fisher’s exact test, P < 2.2 × 10−16, OR = 2.15) (table S1). We compared the DEGs to data from hippocampal bulk RNA-seq in WT and Kdm5a−/− mice that we had previously generated (30) and found 104 common DEGs irrespective of their direction of dysregulation (Fisher’s exact test, P < 2.2 × 10−16, OR = 2.96). Of these, 15 are commonly up-regulated genes (Fisher’s exact test, P = 0.2078, OR = 1.41), 25 are commonly down-regulated genes (Fisher’s exact test, P = 1.33 × 10−05, OR = 2.82), and the remaining 64 are dysregulated in opposite directions (fig. S5A and table S3). Furthermore, to identify potential direct targets of KDM5A, we compared the hippocampal bulk RNA-seq data to publicly available anti-KDM5A ChIP-seq data generated from mouse embryonic stem cells (35). We found 75 DEGs (59 up-regulated and 16 down-regulated) that overlap with KDM5A ChIP-seq peaks, suggesting that KDM5A directly binds and regulates the expression of these genes (fig. S5B and table S3). Comparing these two datasets to the snRNA-seq dataset generated here identified three genes as high-priority possible direct targets of KDM5A: Hapln1 and Nptx2 are commonly up-regulated in the bulk RNA-seq and snRNA-seq datasets and have KDM5A ChIP-seq peaks; Cmss1 is commonly down-regulated in the bulk RNA-seq and snRNA-seq datasets and has KDM5A ChIP-seq peaks (fig. S5C and table S3).
Kdm5a−/− shows accelerated hippocampal development
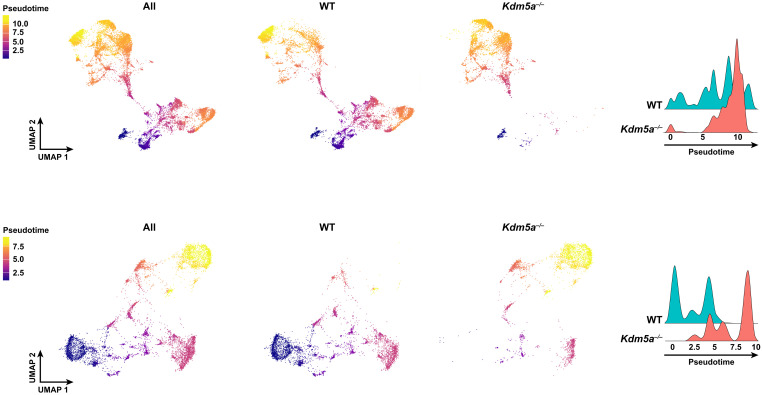
We evaluated the developmental trajectories between WT and Kdm5a−/− hippocampi through pseudotime analysis using Monocle (36). Integrating Monocle clustering with our cell type annotations identified two main clusters (Fig. 4, top and bottom) that had cells with a “neural stem cell” expression profile. The cells in these two clusters were used as starting points of the pseudotime trajectories. In both clusters, the WT cells are distributed throughout the entire trajectory, from early to later time points in development (Fig. 4). However, for the Kdm5a−/− cells, we identified a shift in the distribution of cells toward the end of the pseudotime trajectory compared to WT cells (Fig. 4). The shift of Kdm5a−/− cells and their accumulation at the end of the developmental trajectories suggests an accelerated development and a more mature cellular identity in the Kdm5a−/− hippocampus.
Fig. 4. Developmental trajectory tracing using pseudotime analysis reveals accelerated development and more mature cellular identity in Kdm5a−/− compared to WT hippocampus.
Pseudotime UMAP analysis of all (left), WT (middle), and Kdm5a−/− (right) cells colored by pseudotime (early, blue; late, yellow) in the two Monocle clusters that contain neural stem cells. These cells were used as starting points of the pseudotime trajectory. Top and bottom: The developmental trajectory of cells in each of the two Monocle clusters.
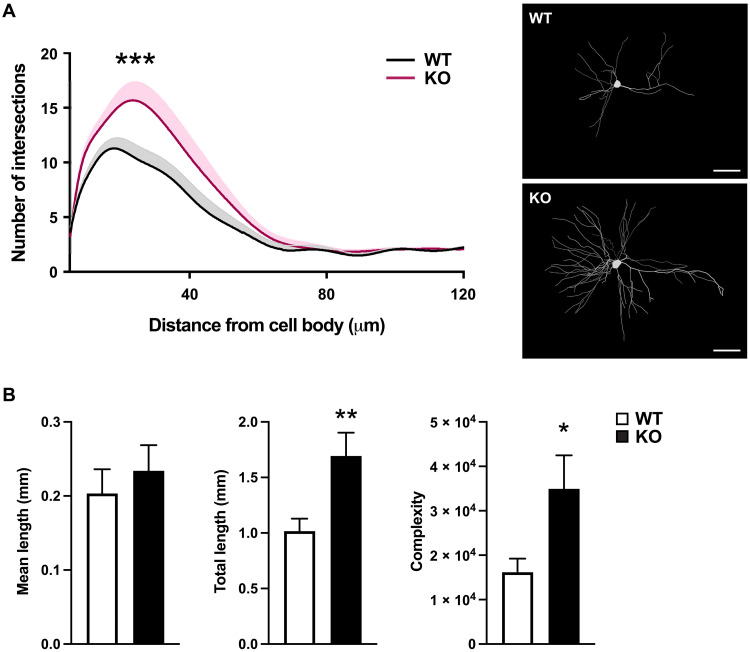
Given the accelerated developmental trajectory identified by pseudotime analysis, we analyzed hippocampal neuronal morphology to determine whether there are any differentiation defects following loss of KDM5A. We measured dendritic complexity and length in vivo by Golgi-Cox staining of brains from WT and Kdm5a−/− mice. Sholl analysis revealed a significant increase in dendritic complexity (~116%) of hippocampal neurons from Kdm5a−/− mice (Fig. 5). Furthermore, hippocampal neurons from Kdm5a−/− mice have a significant increase in total dendritic length (~67%), while the mean dendritic length is unchanged compared with neurons from WT littermates (Fig. 5B). The data suggest an accelerated differentiation of hippocampal neurons upon loss of KDM5A.
Fig. 5. Kdm5a knockout mice have impaired dendritic morphogenesis in the hippocampus.
(A) Sholl analysis from Golgi-Cox–stained neurons revealed an increase in dendritic complexity of hippocampal CA1 neurons from Kdm5a−/− (KO) mice (pink) compared to WT (black) littermates (***P < 0.0001). Data were analyzed using two-way analysis of variance (ANOVA), followed by Tukey’s multiple comparisons test. Representative tracings of Golgi-Cox–stained hippocampal CA1 neurons (right). Scale bars, 20 μm. (B) Golgi-Cox staining showed significantly increased dendritic length (**P = 0.0095) and complexity (*P = 0.0338) of hippocampal CA1 neurons from Kdm5a−/− mice compared to WT littermates. Mean dendritic length is unchanged (P = 0.5255). Data were obtained from mice at 14 to 16 weeks of age. Data were analyzed using unpaired t test. All values are means ± SEM (WT, n = 3; Kdm5a−/−, n = 3; WT neurons, n = 15; Kdm5a−/− neurons, n = 15).
Distinct CA2, CA3, and inhibitory cell populations are sensitive to KDM5A
CA2 is spatially divided into two subclusters on the UMAP (fig. S2A): One of them is marked by the expression of Rps6, encoding the 40S ribosomal protein S6, whose phosphorylation indicates neuronal activity during synaptic plasticity (37, 38); and the other is marked by the expression of Nme2, which encodes a nucleoside diphosphate kinase involved in neurite outgrowth inhibition (39). Loss of KDM5A showed a decrease in the Rps6 cluster accompanied by an increase in the Nme2 cluster (Fig. 2B and fig. S2A), suggesting a role for KDM5A in mediating CA2 cell identity. To validate our finding, we performed in situ hybridization (ISH) on brain sections from WT and Kdm5a−/− mice for Rps6 and Nme2. We found that in the CA2 region of the hippocampus, there is a decrease in the percentage of cells expressing Rps6 (fig. S6, A and B) and an increase in the percentage of cells expressing Nme2 (fig. S6, C and D) in the Kdm5a−/− compared to WT, confirming the shift in CA2 cellular identity revealed by the snRNA-seq data.
Cluster CA3.4 is marked by the expression of Shisa6 and Smad3, both genes encode molecules involved in regulating synaptic transmission in the hippocampus (40, 41). CA3.4 is distinct from other CA3 clusters in that it expresses markers of mossy fibers like Calb2, Thbs2, Drd2, Prrx1, Ttn, Crispld1, and Hk2 (24). In Kdm5a−/−, there is a decrease in the CA3.4 cell proportion (Fig. 2C), suggesting that KDM5A is important in establishing the specific mossy-like CA3 cell identity. To confirm the decrease in Shisa6-marked CA3 cells, we performed ISH on brain sections from WT and Kdm5a−/− mice and found that in the CA3 region of the hippocampus, the percentage of cells expressing Shisa6 is decreased in Kdm5a−/− compared to WT (fig. S6, E and F), validating the decrease in CA3.4 cluster in Kdm5a−/− hippocampus.
The two inhibitory clusters sensitive to KDM5A loss express Pvalb or Sst, markers of parvalbumin and somatostatin interneurons, respectively (Fig. 2, C and D). These two interneuron subtypes play an essential role in regulating network activity in the hippocampus, and it has been recently shown that they undergo long-term inhibitory synaptic plasticity with a specific impact on CA1 pyramidal neurons (42). To validate the decrease in cell proportions of these two inhibitory subtypes in the Kdm5a−/−, we performed single-molecule fluorescence ISH (smFISH) on brain sections from WT and Kdm5a−/− mice. We found that in the hippocampus, the number of cells coexpressing the pan-inhibitory marker Gad1 and Pvalb or coexpressing Gad1 and Sst is decreased in the Kdm5a−/− compared to WT, confirming our snRNA-seq data (fig. S7). This suggests an essential role for KDM5A in mediating the somatostatin and parvalbumin cellular identities in the hippocampus.
KDM5A functions early in development to specify proper CA1 cell identity
The unique increase in cell proportion in subcluster CA1.1 in the Kdm5a−/− prompted us to further investigate the different CA1 subcluster identities. The CA1 layer of the hippocampus is divided into two sublayers, the deep CA1 and the superficial CA1, that are distinct in location (43), density (44), gene expression (45, 46), and functions (47, 48). The deep CA1 neurons develop at E12.5, while the superficial CA1 begins forming at E16.5 (49). To determine whether the two CA1 subclusters CA1.1 and CA1.4, which are sensitive to loss of KDM5A, correspond to either of the deep or superficial CA1 layers, we examined their developmental trajectories with pseudotime analysis. We found that CA1.1 has a more mature cellular identity compared to CA1.4 (fig. S8), suggesting that CA1.1 may correspond to deep CA1 cells, formed earlier in development, whereas CA1.4 may correspond to superficial CA1 cells, formed later in development. The shift in developmental trajectories of Kdm5a−/− cells toward a more mature cell identity compared to WT (Fig. 4 and fig. S8), combined with the identified changes in cell proportions where CA1.1 is increased and CA1.4 is decreased in the Kdm5a−/− compared to WT (Fig. 2C), indicates that the CA1.1 subcluster has a more mature identity and corresponds to deep CA1 cells that are increased in the Kdm5a−/−, while the CA1.4 subcluster has a younger identity and corresponds to superficial CA1 cells that are decreased in the Kdm5a−/−.
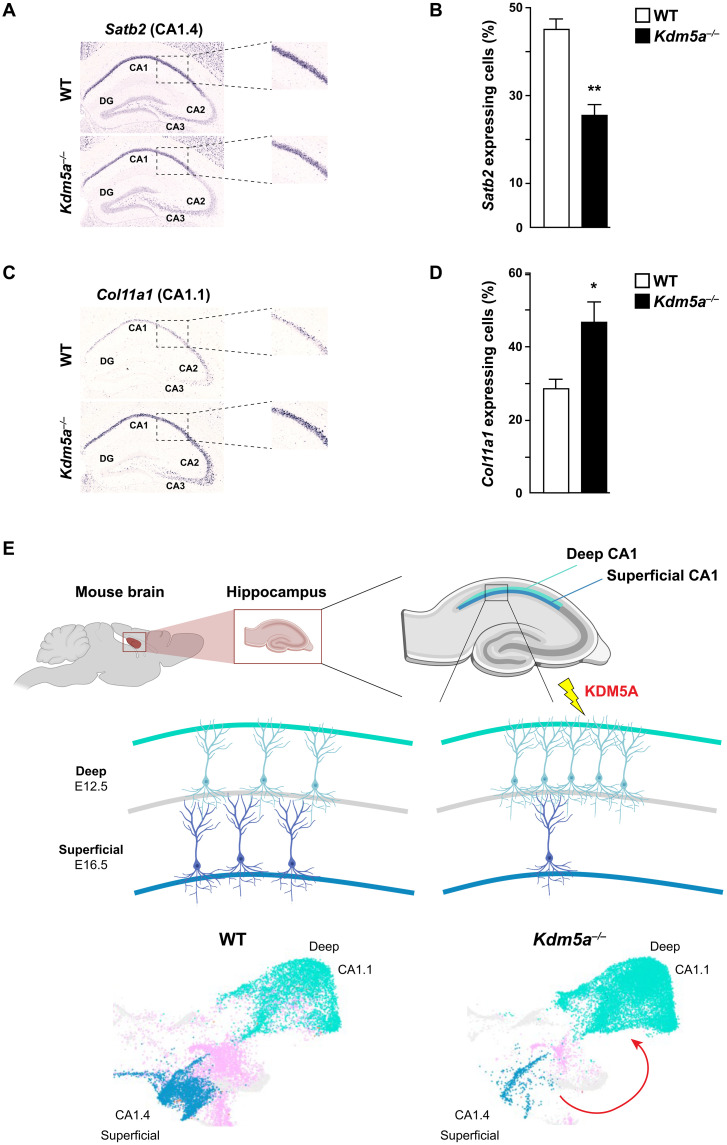
To better understand the changes in deep and superficial CA1 identities upon loss of KDM5A, we assessed the expression of known markers of superficial and deep cells in the two clusters, CA1.4 and CA1.1. Satb2, which encodes the transcriptional regulator special AT-rich sequence binding protein 2 (SATB2), known for its expression in the hippocampal CA1 neurons (50) and for its role in synaptic plasticity and memory formation (51), is also a known marker of superficial CA1 neurons (52). By filtering for genes unique to a cluster, we found that Satb2 is exclusively expressed in the CA1.4 cluster, pointing to a superficial identity for CA1.4. To validate our findings, we performed ISH on brain sections from WT and Kdm5a−/− for Satb2 and found that in the Kdm5a−/−, there is a decrease in the percentage of cells expressing Satb2 in the CA1 compared to WT, confirming the superficial identity of the CA1.4 cluster (Fig. 6, A and B). In addition, we performed Western blot analysis on hippocampal tissue from WT and Kdm5a−/− mice and found that SATB2 expression is decreased in the Kdm5a−/− compared to WT (fig. S9). Hs6st3, which encodes a heparan sulfate sulfotransferase known to be involved in cellular proliferation and differentiation (53), was exclusively expressed in CA1.1. ISH on brain sections from WT and Kdm5a−/− confirmed an increase in the percentage of cells expressing Hs6st3 in the CA1 compared to WT (fig. S6, G and H). We further analyzed DEGs between WT and Kdm5a−/− in cluster CA1.1 and found that markers of deep CA1 cells are up-regulated in Kdm5a−/− compared to WT, including Col11a1, Astn2 (52), and Pvalb (49) (table S1). Col11a1 has been shown to distinctly mark the hippocampal CA1 deep layer (52), so to validate the up-regulation of Col11a1 and confirm the CA1.1 deep cellular identity, we performed ISH on brain sections from WT and Kdm5a−/− for Col11a1. We found an increase in the percentage of cells expressing Col11a1 in the Kdm5a−/− CA1 compared to WT (Fig. 6, C and D), which is in line with its up-regulation as a DEG and with the increased cellular proportion in CA1.1 in the Kdm5a−/− compared to WT.
Fig. 6. KDM5A regulates the development of hippocampal CA1 superficial and deep neurons.
(A to D) ISH on brain tissue for Satb2 and Col11a1 shows differential expression between WT and Kdm5a−/−. Hippocampal sections from WT (top) and Kdm5a−/− (bottom) mice show decreased expression of Satb2 (A) and increased expression of Col11a1 (C) in Kdm5a−/− compared to WT, with the data quantified in (B) and (D), respectively [WT, n = 3; Kdm5a−/−, n = 3; (B) **P = 0.0028 and (D) *P = 0.0388]. Data were analyzed using unpaired t test. All values are means ± SEM. (E) Schematic model of how loss of KDM5A affects the development and appropriate cell identity specification of hippocampal CA1 superficial and deep neurons. Loss of KDM5A results in an increased cell proportion of deep CA1 neurons (CA1.1) and a decreased cell proportion of superficial CA1 neurons (CA1.4). The figure was created using BioRender.
Together, the data demonstrate an increase in deep CA1 neurons and a decrease in superficial CA1 neurons in Kdm5a−/− hippocampi compared to WT, consistent with the identified cell proportion differences in CA1.1 and CA1.4 between WT and Kdm5a−/− (Fig. 2C) and the more mature and younger identities of CA1.1 and CA1.4, respectively (fig. S8). These findings are further supported by the accelerated differentiation phenotype of hippocampal CA1 neurons from the Kdm5a−/− (Fig. 5). Our results indicate that KDM5A is required for appropriate development of CA1 deep and superficial neurons (Fig. 6E).
DISCUSSION
The hippocampus, known for its essential roles in mediating cognitive abilities, is impaired in individuals with ASD, both structurally (26, 27) and functionally (28, 29), but the underlying defects at the single-cell resolution are unknown. Using snRNA-seq, we unraveled the role of the ASD gene and chromatin regulator KDM5A in mediating the development of specific excitatory and inhibitory neurons essential for proper hippocampal function. Subtypes of excitatory CA1 neurons, marked by the expression of Satb2 (CA1.4), CA3 neurons expressing Shisa6 and Smad3 (CA3.4), and inhibitory neurons expressing Pvalb (Inh1) and Sst (Inh2) are decreased in mice lacking KDM5A. A subtype of CA1 neurons (CA1.1) is increased in the Kdm5a−/− compared to WT, and a subtype of CA2 neurons changes identity from Rps6 positive to Nme2 positive.
Although all six cellular subtypes are sensitive to KDM5A levels and require its function for proper development, DEG analysis identified transcriptomic networks that are unique to each cluster, in addition to ones that are common between these clusters. Thus, the transcriptomic changes that these cells undergo when KDM5A is lost are in part similar indicating a common, broader role for KDM5A in these cell types and in part unique indicating a different specialized role for KDM5A in each of these subtypes. In addition, the DEGs common among these six clusters include homologs of known ASD genes and are enriched for neuron development, maturation, and differentiation terms, which is in line with the pseudotime analysis that suggests a role for KDM5A in neuronal development. By comparing the DEGs from the current snRNA-seq data to those from our previously published hippocampal bulk RNA-seq data and to KDM5A-bound genomic loci from published ChIP-seq data, we identified three genes that are common among these three datasets and that could be potential direct targets of KDM5A: Cmss1, Hapln1, and Nptx2. Cmss1, cms1 ribosomal small subunit homolog, is expressed in the brain and has not been extensively studied, while Hapln1, hyaluronan and proteoglycan link protein 1, has recently been shown, alongside lumican and collagen I, to mediate folding of the developing human neocortex (54). The third high-priority potential KDM5A target is Nptx2, neuronal pentraxin 2, also known as Narp. It is an immediate early gene known for its roles in promoting neurite outgrowth (55), organizing excitatory synapses on hippocampal neurons, clustering AMPA-type glutamate receptors (56), and accumulating at excitatory synapses specifically on gamma-aminobutyric acid (GABA)ergic interneurons expressing parvalbumin (57). The human homolog has been linked to several disorders including epilepsy (55) and Alzheimer’s disease (58). Given these functions, we hypothesize that the increase in dendritic complexity in the Kdm5a−/− hippocampal neurons could be mediated by the up-regulation of Nptx2. Additional studies are needed to demonstrate whether KDM5A mediates a subset of its functions through its direct regulation of Nptx2.
Pseudotime analysis revealed that loss of KDM5A results in a shift toward a more mature cellular identity and a decrease in later developed hippocampal cells compared to WT. The data are in line with the increase in CA1.1 cells (earlier born) and the decrease in the CA1.4 cells (later born) that we identified as deep and superficial cells of the CA1, respectively, indicating a role for KDM5A in regulating neuronal development and differentiation in the hippocampus. In addition, assessing the neuronal arborization of hippocampal neurons showed a remarkable increase in dendritic complexity and branching in the Kdm5a−/− compared to WT, in line with the pseudotime results showing a more mature identity upon loss of KDM5A. Abnormal dendritic maturation has been a recurrent phenotype observed in ASD (59), and changes in either direction—more (60, 61) or less branching (62, 63)—are observed in ASD and can alter proper circuitry and brain function. We previously showed that upon loss of KDM5A, layer II/III pyramidal cortical neurons show a severe reduction in dendritic complexity (30). The two opposing effects induced by the loss of KDM5A in different brain regions suggest that although KDM5A is ubiquitously expressed in the brain, it could have not only common but also distinct functions that are brain region specific. Additional studies investigating the specific role of KDM5A in different brain regions are necessary to pinpoint the region-specific molecular and phenotypic defects observed in this subtype of ASD.
When KDM5A is lost, there is a shift in identity of the CA2 cellular subclusters, with a decrease in CA2 neurons expressing Rps6, a gene known for its function in synaptic plasticity (37, 38), and an increase in CA2 neurons expressing Nme2, a gene known for its role in neurite outgrowth inhibition (39). We also identified a role for KDM5A in mediating the development of a subtype of CA3 neurons; we found a decrease in the Kdm5a−/− of a CA3 subtype that expresses Shisa6 and markers of mossy fibers. This unique CA3 subtype affected by the loss of KDM5A shows a specificity for KDM5A function in the development of hippocampal Shisa6-expressing mossy-like CA3 neurons. CA2 neurons begin forming at around E10, and they are followed later by CA3 neurons that first develop at E11 (64, 65). Our data suggest a role for KDM5A at very early stages in hippocampal development, as early as E10, mediating the development of specific CA2 and CA3 neuronal subtypes. Our data also show a role for KDM5A in regulating the development of Pvalb and Sst expressing interneurons. In the hippocampus, these two subtypes of interneurons originate from the medial ganglionic eminence between E9 and E12, compared to other interneuron subtypes that originate from the caudal ganglionic eminence (66). This places the function of KDM5A at this time window in development because these two specific subtypes of interneurons are sensitive to its loss compared to other subtypes. In addition, synapses formed by these two subtypes of interneurons in the hippocampus are regulated through two distinct pathways, and they each undergo distinct synaptic plasticity mechanisms (42, 67). These interneuron subtype–specific synaptic plasticity mechanisms have a role in regulating output from the CA1 pyramidal neurons (42, 67), so a decrease in the proportion of these cells in the Kdm5a−/− hippocampus can potentially have a damaging impact on hippocampal circuits and their function. Last, changes in cell proportions of excitatory and inhibitory neurons can result in a dysregulation of the excitatory-inhibitory balance, a neurodevelopmental disease mechanism frequently observed in ASD (68).
The CA1 principal neurons (CA1PNs) give rise to the CA1 layer of the hippocampus, which is divided into two functionally distinct sublayers, the deep and the superficial (49). Here, we identified a role for KDM5A in establishing deep and superficial CA1 cellular identities. Our data show that upon loss of KDM5A, the superficial CA1 neurons that are born at E16.5 do not develop properly. We find that in the Kdm5a−/−, there is a decrease in the proportion of superficial CA1 neurons and that more CA1PNs develop into deep CA1 neurons instead. This suggests that KDM5A is essential at E16.5 for the establishment of the superficial CA1 cellular identity. The superficial CA1 neurons are marked by the expression of Satb2 (52), which encodes a transcriptional and chromatin regulator widely expressed in the brain and specifically in CA1 neurons of the hippocampus (50). It has essential roles in long-term memory formation in the adult hippocampus (51), short-term working memory, hippocampus-dependent spatial memory, and synaptic plasticity (69). SATB2 haploinsufficiency results in a neurodevelopmental disorder characterized by intellectual disability, limited to absent speech, and abnormal behaviors (70, 71). It is worth noting that in addition to ASD, patients with pathogenic KDM5A mutations have intellectual disability and absent speech (30). Our previous work has demonstrated that loss of Kdm5a leads to severe impairments in spatial learning and memory (30). The deep and superficial CA1 neurons have distinct functions (47–49, 72, 73), and recent studies have expanded on the unique roles of these sublayers by showing that the CA1 deep neurons are involved in reward configuration, while the CA1 superficial neurons drive memory formation (74). Given all the aforementioned data, the loss of CA1 superficial neurons marked by the expression of Satb2 in Kdm5a−/− is predicted to be one major factor leading to the severe cognitive deficits observed upon loss of KDM5A (30). This finding uncovers a key role for KDM5A in regulating the superficial CA1 neurons and in mediating learning and memory.
Our study identifies a critical role for the ASD gene and chromatin regulator KDM5A in regulating hippocampal cell type identity of specific excitatory and inhibitory neurons. Charting the cell type–specific changes that ensue upon loss of this neurodevelopmental disease gene enabled the dissection of molecular changes that underly disease pathogenesis and instructed us on hippocampal development and how chromatin regulation dictates cellular identity. We identify multiple transcriptionally distinct neuronal subtypes of the CA1, CA2, CA3, and inhibitory neurons, further highlighting the rich cellular diversity of these hippocampal regions, in line with recent studies (16, 23, 24, 34). Although much remains to be learned about the different CA1, CA2, and CA3 neuronal subtypes, our results identify the role of an ASD gene in the development of superficial and deep CA1 neurons, in addition to other roles in specific excitatory and inhibitory neuronal populations of the hippocampus. We show that different neuronal subtypes of the CA are differentially affected by loss of KDM5A, which demonstrates that these different subtypes are not only transcriptionally distinct, but that they respond differently to the same genetic defect. This suggests that the different identified subtypes are likely to mediate different functions in the hippocampus, as in the case of the deep and superficial CA1 neurons (47, 48). Further studies to characterize the function of KDM5A in other brain regions will identify how different cellular subtypes respond to the loss of this chromatin regulator and will highlight points of convergence and divergence among the downstream molecular networks it regulates in different cell types.
Loss of KDM5A leads to a more mature cellular identity in the hippocampus, which provides in vivo evidence and complements recent studies showing that chromatin regulators, notably KDM1A, function as an “epigenetic barrier” during neuronal maturation, and inhibiting them leads to an accelerated neuronal maturation (75, 76). Our data suggest that KDM5A may be a player in this epigenetic barrier, regulating transcriptional programs essential for neuronal development and proper maturation. Further studies aimed to identify the molecular function of KDM5A in regulating neuronal maturation will be crucial to our understanding of how chromatin regulators contribute to neuronal differentiation and maturation. In addition, investigating the specific cells, neuronal networks, and molecular pathways that are affected in this genetic subtype of ASD and uncovering the specific cell types that mediate the different ASD phenotype(s) will inform future studies aimed at targeting these cell types for therapies. The information gained will enable the identification of mechanisms that are convergent among different genetic subtypes of ASD where other chromatin regulators and remodelers are mutated.
MATERIALS AND METHODS
Animals
All animal care and use procedures were approved by the University of Texas Southwestern Medical Center (UTSW) Institutional Animal Care and Use Committee (protocol number 2017-102300) and were compliant with U.S. government principles about the care and use of animals, Public Health Service Policy on Humane Care and Use of Laboratory Animals, Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act. Animal husbandry was performed in the UTSW animal facility, accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The Kdm5a constitutive knockout mouse model (Kdm5a−/−) was previously generated and characterized in El Hayek et al. (30). We confirmed the complete loss of KDM5A transcript and protein in the Kdm5a−/− (30).
Tissue collection and nucleus isolation
The WT and Kdm5a−/− hippocampi from 20-week-old mice were processed together and on the same day. Nucleus isolation was adopted and modified from Habib et al. (77). Mice were euthanized with CO2, and the brains were dissected on ice. First, the brain was oriented so that its posterior side is closer to the experimenter. Next, the brain was cut sagittally into its two hemispheres, and the hippocampi were isolated from each hemisphere. The hippocampus was oriented like the brain, where the posterior end was closer to the experimenter. Using a 1-mm-diameter tissue puncher adapted to a syringe and placed at the center of the hippocampus, a tissue punch was collected capturing the entirety of the hippocampus. The punches were immediately stored at −80°C. For each animal, tissue punches from the right and left hippocampi were combined and homogenized using a glass Dounce homogenizer in 500 μl of ice-cold nucleus EZ lysis buffer (NLB) (NUC-101, Sigma-Aldrich) 25 times with pestle A and then 25 times with pestle B. The mixture was transferred to a prechilled Eppendorf tube, and 1000 μl of NLB was added to the tube and incubated on ice for 5 min. The tubes were centrifuged at 800g for 5 min at 4°C. The nuclei were resuspended in 75 μl of nucleus suspension buffer [consisting of 1× phosphate-buffered saline (PBS), 1% bovine serum albumin (AM2618, Thermo Fisher Scientific), and 1% ribonuclease inhibitor (0.2 U/μl; AM2694, Thermo Fisher Scientific)]. The nuclei were then filtered through a 40-μm Flowmi cell strainer (H13680-0040, Bel-Art) to remove cellular debris. Concentration of nuclei for each sample was calculated using Trypan Blue (T8154, Sigma-Aldrich) on a hemocytometer, aiming for a final concentration of ~1000 to 2000 nuclei/μl.
Single-nucleus RNA library preparation and sequencing
Single nuclei were processed using the 10X Genomics Next GEM Single Cell 3′ Reagent Kit v3.1 following the manufacturer’s user guide. All mRNA samples were examined for quantity and quality by NanoDrop and Bioanalyzer 2100 (Agilent). The libraries were constructed following TruSeq Stranded mRNA Sample preparation guide (Illumina), and paired-end sequencing was performed on the Illumina NovaSeq 6000 platform at the McDermott Center Next Generation Sequencing Core at UTSW.
snRNA-seq data analysis
Data were stored, and analyses were performed on the Texas Advanced Computing Center high-performance computing servers, a resource of the University of Texas at Austin (Austin, TX). Raw sequencing data were converted to fastq format using the CellRanger (78) mkfastq command (10X Genomics, v1.1.0). Sequencing reads were aligned to the mm10 mouse reference genome and quantified using CellRanger (78) count (10X Genomics, v1.1.0). We used the R package Seurat (v4.1.0) (79) for further filtering and analysis. Nuclei with greater than 300 genes expressed, greater than or equal to 500 unique molecular identifiers, and less than or equal to 0.4% of counts corresponding to mitochondrial genes were retained for downstream analysis.
SCTransform, with the glmGamPoi package (80), was applied to each sample to normalize and stabilize the variance of molecular count data. Results were saved in a new assay (SCT) with corrected counts, log1p counts, and Pearson residuals. SelectIntegrationFeatures was used to identify features that are repeatedly variable across samples for integration. The number of features requested was 3500. These features were passed to PrepSCTIntegration to prepare for integration of the samples. Anchors between the samples were identified, and these anchors were used to integrate the samples using IntegrateData, creating the “integrated” assay. Principal components analysis dimensionality reduction and UMAP dimensionality reduction were performed on this assay. The top 30 principal components were retained for further analysis. A shared nearest-neighbor graph was constructed using the FindNeighbors function with k = 20. Twenty-four clusters were identified using Louvain clustering in the FindClusters function at a resolution of 0.3. Two of these clusters were merged because they had similar marker genes and no distinctive markers unique to each cluster. One cluster was split into two unique clusters because each exclusively expressed either Pvalb or Sst, markers of parvalbumin and somatostatin cells, respectively.
Differentially expressed genes
FindAllMarkers was used to calculate the expression level in a single cluster versus the average expression in all other clusters, expression level of Kdm5a−/− cells versus WT cells across all clusters, and expression level of Kdm5a−/− cells versus WT cells within each cluster. We used MAST v1.16.0 (81) with a generalized linear model framework and selected genes with a log2 fold change of expression of ≥|0.3| and FDR-corrected P ≤ 0.05 as DEGs. The list of known ASD genes was obtained from the Simons Foundation Autism Research Initiative Gene 2018 database (82) (using the 2022 Q2 release). The genes that were differentially expressed in Kdm5a−/− cells versus WT cells across all cells were overlapped with DEGs identified through bulk RNA-seq on WT and Kdm5a−/− hippocampi from El Hayek et al. (30).
To define a KDM5A signature that is common across affected cell types from the six vulnerable clusters (CA1.1, CA1.4, CA2, CA3.4, Inh1, and Inh2), the expression in all affected excitatory clusters (CA1.1, CA1.4, CA2, and CA3.4) was combined. Similarly, the expression in both affected inhibitory clusters (Inh1 and Inh2) was combined. The FindMarkers function was used to compare the expression level in the combined excitatory clusters versus the average expression in all other excitatory clusters [except the neural stem cell (NSC) cluster]. It was also used to compare the expression level in the combined inhibitory clusters versus the average expression in all other inhibitory clusters. Genes with a log2 fold change of expression of ≥|0.3| and FDR-corrected P ≤ 0.05 were selected as the KDM5A signature genes.
Analysis of candidate KDM5A targets
To nominate possible direct and indirect KDM5A targets, anti-KDM5A ChIP-seq data generated from mouse embryonic stem cells from Beshiri et al. (35) were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (accession number GSE28343). The LiftOver tool from the University of California Santa Cruz Genome Browser (83) was used to convert the peak bed file from the mouse reference genome mm9 to the mm10 assembly. ChIPseeker (84, 85) was used to annotate the genomic region of the peaks. The nearest genes around the peaks were overlapped with DEGs identified through hippocampal bulk RNA-seq from El Hayek et al. (30) and the identified snRNA-seq DEGs from this study. DEGs from the bulk and snRNA-seq data were overlapped with the annotated ChIP-seq peaks to nominate possible direct targets. The remaining DEGs were considered indirect targets. Common genes between the bulk and snRNA-seq data that overlapped with the annotated ChIP-seq peaks were considered high-priority candidate direct targets.
GO analysis
We used the functional annotation tool DAVID (86) to find biological process terms enriched in the DEG sets.
Pseudotime analysis
Monocle3 (v1.2.7) (36) was used for pseudotime analysis. A cell dataset object was created using RNA counts. Cells were clustered using Leiden community detection. Partitions were also calculated using a kNN pruning method. Genes most specifically expressed in each partition were identified, and trajectories were built from the reduced dimension space using reversed graph embedding. Identification of neural stem cells from our Seurat (79) clustering was used to locate neural stem cells in the Monocle partitions. Cells that had an identity of neural stem cells were primarily located in two Monocle clusters (Fig. 4). Cells in these two different clusters were used as trajectory starting points or nodes (Fig. 4). The pseudotime data from one of the Monocle clusters that contained most of the CA1 cells were added back to the Seurat object and visualized on the original UMAP (fig. S8).
Golgi-Cox staining
Mice at 14 to 16 weeks of age were euthanized, and brains were dissected out. Golgi-Cox staining was performed as described in Zaqout and Kaindl (87). The brains were cut into two hemispheres and impregnated with Golgi-Cox solution (1% K2Cr2O7, 1% HgCl2, and 0.8% K2CrO4) at room temperature for 7 days in the dark. Subsequently, they were protected with tissue protectant solution [30% sucrose, 1% PVP40 (polyvinylpyrrolidone), and 30% ethylene glycol in 0.05 M phosphate buffer (pH 7.2)] at 4°C for 24 hours after which the solution was replaced with a fresh one and kept at 4°C for 5 days. Brains were embedded in optimal cutting temperature (OCT) medium. Frozen sagittal sections (150 μm in thickness) were prepared using a microtome (Leica) and loaded on 3% gelatin-coated glass slides. The slides were dried for 5 days at room temperature, dehydrated, and developed as described in Zaqout and Kaindl (87) and mounted with Cytoseal (Thermo Fisher Scientific). Bright-field images were acquired using Nikon Fi3 camera with 60× objective at 0.9-μm steps in z axis. Dendritic and spine analyses were performed using Neurolucida 360 software (MBF Bioscience) at the UTSW Whole Brain Microscopy Facility. For Sholl analysis, images were processed and analyzed in Fiji (ImageJ v2.14.0/1.54f) (88).
Western blot analysis and antibodies
To detect endogenous proteins, hippocampal tissue was isolated from male and female mice at 18 weeks of age and immediately frozen in liquid nitrogen. From each animal, both hippocampi, the left and right, were isolated and lysed in SDS lysis buffer [250 mM tris (pH 6.8), 4% SDS, 3.2% glycerol, 10 mM N-ethylmaleimide, and 1 mM phenylmethylsulfonyl fluoride] supplemented with 2 mM OPT, and cOmplete mini EDTA-free protease inhibitor and phosSTOP phosphatase inhibitor tablets (Roche). Samples were boiled for 10 min and then centrifuged at 14,000 rpm for 15 min, and the supernatant was collected. Protein concentrations were determined using the DC protein assay (Bio-Rad). Seventy micrograms of protein was loaded per lane, with β-mercaptoethanol and bromophenol blue, onto 8% polyacrylamide gels. Gels were run, and protein was transferred to polyvinylidene difluoride membranes (Millipore) for 2 hours on ice. Membranes were blocked in 5% milk in TBS-T [20 mM tris (pH 7.5), 150 mM NaCl, and 0.1% Tween-20] for 1 hour at room temperature and incubated with primary antibody overnight at 4°C. Membranes were washed in 5% TBS-T, followed by a 1-hour and 30-min incubation in secondary antibody (1:10,000) at room temperature using donkey anti-rabbit (711-035-152, Jackson ImmunoResearch). Following antibody incubation, signal was detected with enhanced chemiluminescence (SuperSignal West Pico chemiluminescent substrate, Thermo Fisher Scientific). Primary antibodies used were SATB2 (ab92446, Abcam; 1:1000) and β-actin (13E5, Cell Signaling Technology; 1:1000). Bands were quantified using Fiji (ImageJ v2.14.0/1.54f) (88).
Single-molecule fluorescent ISH
Mice (30 to 35 weeks of age) were euthanized with a ketamine/xylazine cocktail, followed by perfusion with cold 1× PBS for 3 min and then with cold 4% paraformaldehyde (PFA) in PBS for 15 min at a rate of 8 ml/min. Brains were then dissected out and washed in 10 ml of 1× PBS, followed by an overnight incubation in 4% PFA at 4°C. The next day, the brains were cryoprotected by immersing them in sucrose solutions with gradually increasing concentrations (from 5 to 25%) until they sink. They were next embedded in OCT and allowed to freeze overnight at −80°C. The brains were sectioned at −20°C using a cryostat at 16 μm in thickness by the UTSW Histo Pathology Core. The sections were mounted on Superfrost Plus microscope slides. smFISH was performed using RNAscope Multiplex Fluorescent Reagent Kit v2 (323100) following the manufacturer’s instructions for fixed-frozen tissue samples. The target retrieval was performed manually following the manufacturer’s instructions. Three probes were used: Gad1 mouse 400951-C1, Pvalb mouse 421931-C2, and Sst mouse 404631-C3. Three Opal fluorophores were used: Opal 520 (FP1487001KT, Akoya Biosciences; 1:750) for C1, Opal 570 (FP1488001KT, Akoya Biosciences; 1:750) for C2, and Opal 620 (FP1495001KT, Akoya Biosciences; 1:750) for C3. Mouse-positive (RNAscope 3-plex 320881) and mouse-negative (RNAscope 3-plex 320871) control probes were also used. Slides were imaged at ×20 magnification in z stacks using Zeiss LSM 880 confocal microscope at the UTSW Neuroscience Microscopy Facility. Images of hippocampal sections from n = 3 mice per genotype were analyzed using Fiji (ImageJ v2.14.0/1.54f) (88). For each slice, we quantified the number of fluorescent puncta that are Gad1+, Gad1+Pvalb+, and Gad1+Sst+. The data were plotted as the number of Gad1+Pvalb+ or Gad1+Sst+ puncta over the total number of Gad1+ puncta.
ISH, microscopy, and image analysis
Mice (8 to 10 weeks of age) were euthanized with CO2, and the brains were collected on ice. The brains were immediately placed in freezing blocks filled with OCT and left on dry ice to freeze and then stored in clear plastic wrap at −80°C. The OCT blocks were equilibrated at −20°C before sectioning using Leica CM3050S cryostat. Sagittal sections of 25 μm in thickness were collected on slides then fixed in 4% PFA (15 to 30 min), followed by fresh 0.25% (v/v) acetic anhydride in 0.1 M triethanolamine (pH 8.0) (5 min, ×2), and dehydrated in the Leica autostainer XL in an ethanol sequence reaching 100% ethanol and then air-dried.
ISH was performed at the Baylor College of Medicine RNA ISH Core facility as previously described (89). Sequences for the ISH probes used (Col11a1, Hs6st3, Nme2, Rps6, Satb2, and Shisa6) are available through the Allen Brain Atlas Mouse Brain Data Portal (https://mouse.brain-map.org/). Images were collected using the slide scanner Zeiss Axioscan.Z1, 20× 0.8 numerical aperture lens, and automated quantification was performed as previously described (90). Briefly, a Python script was used to identify the location and signal strength of each gene within the area of interest. Total cells and cells expressing the gene of interest were quantified, and for each gene, the percentage of cells expressing the gene out of the total cells within the corresponding hippocampal region was plotted.
Acknowledgments
The ISH experiments were supported by the RNA In Situ Hybridization Core Facility at Baylor College of Medicine with the expert assistance of C. Ljungberg and funding from a Shared Instrumentation grant from the NIH (S10 OD016167). We thank J. Shelton at the UTSW Histo Pathology Core for the cryostat thin sections used in the smFISH experiment. We thank C. Burroughs for assistance in preparing the figures. Some schematics in the figures were created with BioRender.com.
Funding: This work was supported by UTSW and by grants from the Welch Foundation (I-1946-20210327), the Walter and Lillian Cantor Foundation, and Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD099162) to M.H.C. and by the Peter O’Donnell Jr. Brain Institute Sprouts Program grant to L.E.H.
Author contributions: M.H.C. conceived the study, acquired funds, and oversaw the project. L.E.H., D.D., A.G., A.A., K.K., and M.H.C. designed and performed experiments and analyzed data. L.E.H. and M.H.C. wrote the manuscript. All authors participated in reviewing and editing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The snRNA-seq data generated in this study have been deposited and are available in the NCBI GEO repository with accession no. GSE235490. The code used for data analysis in this study is described in Materials and Methods and is publicly available. This study did not generate new unique reagents.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.T. Chen, S. Y. R. Dent, Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat. Rev. Genet. 15, 93–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V. Azuara, P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, A. G. Fisher, Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538 (2006). [DOI] [PubMed] [Google Scholar]
- 3.M. G. Guenther, S. S. Levine, L. A. Boyer, R. Jaenisch, R. A. Young, A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. Ho, G. R. Crabtree, Chromatin remodelling during development. Nature 463, 474–484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.H. Shen, W. Xu, F. Lan, Histone lysine demethylases in mammalian embryonic development. Exp. Mol. Med. 49, e325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. Mews, E. S. Calipari, J. Day, M. K. Lobo, T. Bredy, T. Abel, From circuits to chromatin: The emerging role of epigenetics in mental health. J. Neurosci. 41, 873–882 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. De Rubeis, X. He, A. P. Goldberg, C. S. Poultney, K. Samocha, A. E. Cicek, Y. Kou, L. Liu, M. Fromer, S. Walker, T. Singh, L. Klei, J. Kosmicki, F. Shih-Chen, B. Aleksic, M. Biscaldi, P. F. Bolton, J. M. Brownfeld, J. Cai, N. G. Campbell, A. Carracedo, M. H. Chahrour, A. G. Chiocchetti, H. Coon, E. L. Crawford, S. R. Curran, G. Dawson, E. Duketis, B. A. Fernandez, L. Gallagher, E. Geller, S. J. Guter, R. S. Hill, J. Ionita-Laza, P. J. Gonzalez, H. Kilpinen, S. M. Klauck, A. Kolevzon, I. Lee, I. Lei, J. Lei, T. Lehtimaki, C. F. Lin, A. Ma'ayan, C. R. Marshall, A. L. McInnes, B. Neale, M. J. Owen, N. Ozaki, M. Parellada, J. R. Parr, S. Purcell, K. Puura, D. Rajagopalan, K. Rehnstrom, A. Reichenberg, A. Sabo, M. Sachse, S. J. Sanders, C. Schafer, M. Schulte-Ruther, D. Skuse, C. Stevens, P. Szatmari, K. Tammimies, O. Valladares, A. Voran, W. Li-San, L. A. Weiss, A. J. Willsey, T. W. Yu, R. K. Yuen; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10k Consortium, E. H. Cook, C. M. Freitag, M. Gill, C. M. Hultman, T. Lehner, A. Palotie, G. D. Schellenberg, P. Sklar, M. W. State, J. S. Sutcliffe, C. A. Walsh, S. W. Scherer, M. E. Zwick, J. C. Barett, D. J. Cutler, K. Roeder, B. Devlin, M. J. Daly, J. D. Buxbaum, Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. J. O'Roak, L. Vives, S. Girirajan, E. Karakoc, N. Krumm, B. P. Coe, R. Levy, A. Ko, C. Lee, J. D. Smith, E. H. Turner, I. B. Stanaway, B. Vernot, M. Malig, C. Baker, B. Reilly, J. M. Akey, E. Borenstein, M. J. Rieder, D. A. Nickerson, R. Bernier, J. Shendure, E. E. Eichler, Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.T. N. Turner, F. Hormozdiari, M. H. Duyzend, S. A. McClymont, P. W. Hook, I. Iossifov, A. Raja, C. Baker, K. Hoekzema, H. A. Stessman, M. C. Zody, B. J. Nelson, J. Huddleston, R. Sandstrom, J. D. Smith, D. Hanna, J. M. Swanson, E. M. Faustman, M. J. Bamshad, J. Stamatoyannopoulos, D. A. Nickerson, A. S. McCallion, R. Darnell, E. E. Eichler, Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 98, 58–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N. N. Parikshak, R. Luo, A. Zhang, H. Won, J. K. Lowe, V. Chandran, S. Horvath, D. H. Geschwind, Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. J. Willsey, S. J. Sanders, M. Li, S. Dong, A. T. Tebbenkamp, R. A. Muhle, S. K. Reilly, L. Lin, S. Fertuzinhos, J. A. Miller, M. T. Murtha, C. Bichsel, W. Niu, J. Cotney, A. G. Ercan-Sencicek, J. Gockley, A. R. Gupta, W. Han, X. He, E. J. Hoffman, L. Klei, J. Lei, W. Liu, L. Liu, C. Lu, X. Xu, Y. Zhu, S. M. Mane, E. S. Lein, L. Wei, J. P. Noonan, K. Roeder, B. Devlin, N. Sestan, M. W. State, Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D. Velmeshev, L. Schirmer, D. Jung, M. Haeussler, Y. Perez, S. Mayer, A. Bhaduri, N. Goyal, D. H. Rowitch, A. R. Kriegstein, Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.X. Jiang, S. Shen, C. R. Cadwell, P. Berens, F. Sinz, A. S. Ecker, S. Patel, A. S. Tolias, Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350, aac9462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Tasic, V. Menon, T. N. Nguyen, T. K. Kim, T. Jarsky, Z. Yao, B. Levi, L. T. Gray, S. A. Sorensen, T. Dolbeare, D. Bertagnolli, J. Goldy, N. Shapovalova, S. Parry, C. Lee, K. Smith, A. Bernard, L. Madisen, S. M. Sunkin, M. Hawrylycz, C. Koch, H. Zeng, Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. Tasic, Z. Yao, L. T. Graybuck, K. A. Smith, T. N. Nguyen, D. Bertagnolli, J. Goldy, E. Garren, M. N. Economo, S. Viswanathan, O. Penn, T. Bakken, V. Menon, J. Miller, O. Fong, K. E. Hirokawa, K. Lathia, C. Rimorin, M. Tieu, R. Larsen, T. Casper, E. Barkan, M. Kroll, S. Parry, N. V. Shapovalova, D. Hirschstein, J. Pendergraft, H. A. Sullivan, T. K. Kim, A. Szafer, N. Dee, P. Groblewski, I. Wickersham, A. Cetin, J. A. Harris, B. P. Levi, S. M. Sunkin, L. Madisen, T. L. Daigle, L. Looger, A. Bernard, J. Phillips, E. Lein, M. Hawrylycz, K. Svoboda, A. R. Jones, C. Koch, H. Zeng, Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Zeisel, A. B. Munoz-Manchado, S. Codeluppi, P. Lonnerberg, G. La Manno, A. Jureus, S. Marques, H. Munguba, L. He, C. Betsholtz, C. Rolny, G. Castelo-Branco, J. Hjerling-Leffler, S. Linnarsson, Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 17.A. Zeisel, H. Hochgerner, P. Lonnerberg, A. Johnsson, F. Memic, J. van der Zwan, M. Haring, E. Braun, L. E. Borm, G. La Manno, S. Codeluppi, A. Furlan, K. Lee, N. Skene, K. D. Harris, J. Hjerling-Leffler, E. Arenas, P. Ernfors, U. Marklund, S. Linnarsson, Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Z. Yao, H. Liu, F. Xie, S. Fischer, R. S. Adkins, A. I. Aldridge, S. A. Ament, A. Bartlett, M. M. Behrens, K. Van den Berge, D. Bertagnolli, H. R. de Bezieux, T. Biancalani, A. S. Booeshaghi, H. C. Bravo, T. Casper, C. Colantuoni, J. Crabtree, H. Creasy, K. Crichton, M. Crow, N. Dee, E. L. Dougherty, W. I. Doyle, S. Dudoit, R. Fang, V. Felix, O. Fong, M. Giglio, J. Goldy, M. Hawrylycz, B. R. Herb, R. Hertzano, X. Hou, Q. Hu, J. Kancherla, M. Kroll, K. Lathia, Y. E. Li, J. D. Lucero, C. Luo, A. Mahurkar, D. McMillen, N. M. Nadaf, J. R. Nery, T. N. Nguyen, S. Y. Niu, V. Ntranos, J. Orvis, J. K. Osteen, T. Pham, A. Pinto-Duarte, O. Poirion, S. Preissl, E. Purdom, C. Rimorin, D. Risso, A. C. Rivkin, K. Smith, K. Street, J. Sulc, V. Svensson, M. Tieu, A. Torkelson, H. Tung, E. D. Vaishnav, C. R. Vanderburg, C. van Velthoven, X. Wang, O. R. White, Z. J. Huang, P. V. Kharchenko, L. Pachter, J. Ngai, A. Regev, B. Tasic, J. D. Welch, J. Gillis, E. Z. Macosko, B. Ren, J. R. Ecker, H. Zeng, E. A. Mukamel, A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.H. Markram, E. Muller, S. Ramaswamy, M. W. Reimann, M. Abdellah, C. A. Sanchez, A. Ailamaki, L. Alonso-Nanclares, N. Antille, S. Arsever, G. A. Kahou, T. K. Berger, A. Bilgili, N. Buncic, A. Chalimourda, G. Chindemi, J. D. Courcol, F. Delalondre, V. Delattre, S. Druckmann, R. Dumusc, J. Dynes, S. Eilemann, E. Gal, M. E. Gevaert, J. P. Ghobril, A. Gidon, J. W. Graham, A. Gupta, V. Haenel, E. Hay, T. Heinis, J. B. Hernando, M. Hines, L. Kanari, D. Keller, J. Kenyon, G. Khazen, Y. Kim, J. G. King, Z. Kisvarday, P. Kumbhar, S. Lasserre, J. V. Le Be, B. R. Magalhaes, A. Merchan-Perez, J. Meystre, B. R. Morrice, J. Muller, A. Munoz-Cespedes, S. Muralidhar, K. Muthurasa, D. Nachbaur, T. H. Newton, M. Nolte, A. Ovcharenko, J. Palacios, L. Pastor, R. Perin, R. Ranjan, I. Riachi, J. R. Rodriguez, J. L. Riquelme, C. Rossert, K. Sfyrakis, Y. Shi, J. C. Shillcock, G. Silberberg, R. Silva, F. Tauheed, M. Telefont, M. Toledo-Rodriguez, T. Trankler, W. Van Geit, J. V. Diaz, R. Walker, Y. Wang, S. M. Zaninetta, J. DeFelipe, S. L. Hill, I. Segev, F. Schurmann, Reconstruction and simulation of neocortical microcircuitry. Cell 163, 456–492 (2015). [DOI] [PubMed] [Google Scholar]
- 20.H. Eichenbaum, T. Otto, N. J. Cohen, The hippocampus–What does it do? Behav. Neural Biol. 57, 2–36 (1992). [DOI] [PubMed] [Google Scholar]
- 21.R. Khalaf-Nazzal, F. Francis, Hippocampal development–Old and new findings. Neuroscience 248, 225–242 (2013). [DOI] [PubMed] [Google Scholar]
- 22.P. Andersen, R. Morris, D. Amaral, T. Bliss, J. O'Keefe, Eds., The Hippocampus Book (Oxford University Press, 2006). [Google Scholar]
- 23.F. Ayhan, A. Kulkarni, S. Berto, K. Sivaprakasam, C. Douglas, B. C. Lega, G. Konopka, Resolving cellular and molecular diversity along the hippocampal anterior-to-posterior axis in humans. Neuron 109, 2091–2105.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. S. Cembrowski, L. Wang, K. Sugino, B. C. Shields, N. Spruston, Hipposeq: A comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5, e14997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.F. L. Hitti, S. A. Siegelbaum, The hippocampal CA2 region is essential for social memory. Nature 508, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. M. Schumann, J. Hamstra, B. L. Goodlin-Jones, L. J. Lotspeich, H. Kwon, M. H. Buonocore, C. R. Lammers, A. L. Reiss, D. G. Amaral, The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J. Neurosci. 24, 6392–6401 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.V. P. Reinhardt, A. M. Iosif, L. Libero, B. Heath, S. J. Rogers, E. Ferrer, C. Nordahl, S. Ghetti, D. Amaral, M. Solomon, Understanding hippocampal development in young children with autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry 59, 1069–1079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. Solomon, J. D. Ragland, T. A. Niendam, T. A. Lesh, J. S. Beck, J. C. Matter, M. J. Frank, C. S. Carter, Atypical learning in autism spectrum disorders: A functional magnetic resonance imaging study of transitive inference. J. Am. Acad. Child Adolesc. Psychiatry 54, 947–955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.S. M. Banker, X. Gu, D. Schiller, J. H. Foss-Feig, Hippocampal contributions to social and cognitive deficits in autism spectrum disorder. Trends Neurosci. 44, 793–807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. El Hayek, I. O. Tuncay, N. Nijem, J. Russell, S. Ludwig, K. Kaur, X. Li, P. Anderton, M. Tang, A. Gerard, A. Heinze, P. Zacher, H. S. Alsaif, A. Rad, K. Hassanpour, M. R. Abbaszadegan, C. Washington, B. R. DuPont, R. J. Louie; CAUSES Study, M. Couse, M. Faden, R. C. Rogers, R. A. Jamra, E. R. Elias, R. Maroofian, H. Houlden, A. Lehman, B. Beutler, M. H. Chahrour, KDM5A mutations identified in autism spectrum disorder using forward genetics. eLife 9, e56883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Yokoyama, Y. Ito, H. Ueno-Kudoh, H. Shimizu, K. Uchibe, S. Albini, K. Mitsuoka, S. Miyaki, M. Kiso, A. Nagai, T. Hikata, T. Osada, N. Fukuda, S. Yamashita, D. Harada, V. Mezzano, M. Kasai, P. L. Puri, Y. Hayashizaki, H. Okado, M. Hashimoto, H. Asahara, A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev. Cell 17, 836–848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Z. Yao, C. T. J. van Velthoven, T. N. Nguyen, J. Goldy, A. E. Sedeno-Cortes, F. Baftizadeh, D. Bertagnolli, T. Casper, M. Chiang, K. Crichton, S. L. Ding, O. Fong, E. Garren, A. Glandon, N. W. Gouwens, J. Gray, L. T. Graybuck, M. J. Hawrylycz, D. Hirschstein, M. Kroll, K. Lathia, C. Lee, B. Levi, D. McMillen, S. Mok, T. Pham, Q. Ren, C. Rimorin, N. Shapovalova, J. Sulc, S. M. Sunkin, M. Tieu, A. Torkelson, H. Tung, K. Ward, N. Dee, K. A. Smith, B. Tasic, H. Zeng, A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Cardoso-Moreira, J. Halbert, D. Valloton, B. Velten, C. Chen, Y. Shao, A. Liechti, K. Ascencao, C. Rummel, S. Ovchinnikova, P. V. Mazin, I. Xenarios, K. Harshman, M. Mort, D. N. Cooper, C. Sandi, M. J. Soares, P. G. Ferreira, S. Afonso, M. Carneiro, J. M. A. Turner, J. L. VandeBerg, A. Fallahshahroudi, P. Jensen, R. Behr, S. Lisgo, S. Lindsay, P. Khaitovich, W. Huber, J. Baker, S. Anders, Y. E. Zhang, H. Kaessmann, Gene expression across mammalian organ development. Nature 571, 505–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.N. Habib, Y. Li, M. Heidenreich, L. Swiech, I. Avraham-Davidi, J. J. Trombetta, C. Hession, F. Zhang, A. Regev, Div-Seq: Single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. L. Beshiri, K. B. Holmes, W. F. Richter, S. Hess, A. B. M. M. K. Islam, Q. Yan, L. Plante, L. Litovchick, N. Gevry, N. Lopez-Bigas, W. G. Kaelin Jr., E. V. Benevolenskaya, Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 18499–18504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Cao, M. Spielmann, X. Qiu, X. Huang, D. M. Ibrahim, A. J. Hill, F. Zhang, S. Mundlos, L. Christiansen, F. J. Steemers, C. Trapnell, J. Shendure, The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Z. A. Knight, K. Tan, K. Birsoy, S. Schmidt, J. L. Garrison, R. W. Wysocki, A. Emiliano, M. I. Ekstrand, J. M. Friedman, Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. Klann, T. E. Dever, Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 5, 931–942 (2004). [DOI] [PubMed] [Google Scholar]
- 39.H. Hamasaki, M. Fujitani, T. Yamashita, NME2 associates with PTPσ to transduce signals from chondroitin sulfate proteoglycans. Biochem. Biophys. Res. Commun. 471, 522–527 (2016). [DOI] [PubMed] [Google Scholar]
- 40.R. V. Klaassen, J. Stroeder, F. Coussen, A.-S. Hafner, J. D. Petersen, C. Renancio, L. J. M. Schmitz, E. Normand, J. C. Lodder, D. C. Rotaru, P. Rao-Ruiz, S. Spijker, H. D. Mansvelder, D. Choquet, A. B. Smit, Shisa6 traps AMPA receptors at postsynaptic sites and prevents their desensitization during synaptic activity. Nat. Commun. 7, 10682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. Tapia-Gonzalez, M. D. Munoz, M. I. Cuartero, A. Sanchez-Capelo, Smad3 is required for the survival of proliferative intermediate progenitor cells in the dentate gyrus of adult mice. Cell Commun. Signal 11, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. Udakis, V. Pedrosa, S. E. L. Chamberlain, C. Clopath, J. R. Mellor, Interneuron-specific plasticity at parvalbumin and somatostatin inhibitory synapses onto CA1 pyramidal neurons shapes hippocampal output. Nat. Commun. 11, 4395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.N. J. Bannister, A. U. Larkman, Dendritic morphology of CA1 pyramidal neurones from the rat hippocampus: I. Branching patterns. Branching patterns. J. Comp. Neurol. 360, 150–160 (1995). [DOI] [PubMed] [Google Scholar]
- 44.L. Slomianka, I. Amrein, I. Knuesel, J. C. Sorensen, D. P. Wolfer, Hippocampal pyramidal cells: The reemergence of cortical lamination. Brain Struct. Funct. 216, 301–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.C. L. Thompson, S. D. Pathak, A. Jeromin, L. L. Ng, C. R. MacPherson, M. T. Mortrud, A. Cusick, Z. L. Riley, S. M. Sunkin, A. Bernard, R. B. Puchalski, F. H. Gage, A. R. Jones, V. B. Bajic, M. J. Hawrylycz, E. S. Lein, Genomic anatomy of the hippocampus. Neuron 60, 1010–1021 (2008). [DOI] [PubMed] [Google Scholar]
- 46.H. W. Dong, L. W. Swanson, L. Chen, M. S. Fanselow, A. W. Toga, Genomic-anatomic evidence for distinct functional domains in hippocampal field CA1. Proc. Natl. Acad. Sci. U.S.A. 106, 11794–11799 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. Mizuseki, K. Diba, E. Pastalkova, G. Buzsaki, Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nat. Neurosci. 14, 1174–1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Valero, E. Cid, R. G. Averkin, J. Aguilar, A. Sanchez-Aguilera, T. J. Viney, D. Gomez-Dominguez, E. Bellistri, L. M. de la Prida, Determinants of different deep and superficial CA1 pyramidal cell dynamics during sharp-wave ripples. Nat. Neurosci. 18, 1281–1290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. Cavalieri, A. Angelova, A. Islah, C. Lopez, M. Bocchio, Y. Bollmann, A. Baude, R. Cossart, CA1 pyramidal cell diversity is rooted in the time of neurogenesis. eLife 10, e69270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. Huang, N.-N. Song, W. Lan, L. Hu, C.-J. Su, Y.-Q. Ding, L. Zhang, Expression of transcription factor Satb2 in adult mouse brain. Anat. Rec. 296, 452–461 (2013). [DOI] [PubMed] [Google Scholar]
- 51.C. Jaitner, C. Reddy, A. Abentung, N. Whittle, D. Rieder, A. Delekate, M. Korte, G. Jain, A. Fischer, F. Sananbenesi, I. Cera, N. Singewald, G. Dechant, G. Apostolova, Satb2 determines miRNA expression and long-term memory in the adult central nervous system. eLife 5, e17361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.I. Soltesz, A. Losonczy, CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nat. Neurosci. 21, 484–493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H. Habuchi, M. Tanaka, O. Habuchi, K. Yoshida, H. Suzuki, K. Ban, K. Kimata, The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J. Biol. Chem. 275, 2859–2868 (2000). [DOI] [PubMed] [Google Scholar]
- 54.K. R. Long, B. Newland, M. Florio, N. Kalebic, B. Langen, A. Kolterer, P. Wimberger, W. B. Huttner, Extracellular matrix components HAPLN1, lumican, and collagen I cause hyaluronic acid-dependent folding of the developing human neocortex. Neuron 99, 702–719.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 55.C. C. Tsui, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, C. Barnes, P. F. Worley, Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J. Neurosci. 16, 2463–2478 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R. J. O'Brien, D. Xu, R. S. Petralia, O. Steward, R. L. Huganir, P. Worley, Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron 23, 309–323 (1999). [DOI] [PubMed] [Google Scholar]
- 57.M. C. Chang, J. M. Park, K. A. Pelkey, H. L. Grabenstatter, D. Xu, D. J. Linden, T. P. Sutula, C. J. McBain, P. F. Worley, Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat. Neurosci. 13, 1090–1097 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M.-F. Xiao, D. Xu, M. T. Craig, K. A. Pelkey, C.-C. Chien, Y. Shi, J. Zhang, S. Resnick, O. Pletnikova, D. Salmon, J. Brewer, S. Edland, J. Wegiel, B. Tycko, A. Savonenko, R. H. Reeves, J. C. Troncoso, C. J. McBain, D. Galasko, P. F. Worley, NPTX2 and cognitive dysfunction in Alzheimer’s disease. eLife 6, e23798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.V. Martinez-Cerdeno, Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol. 77, 393–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.G. V. Raymond, M. L. Bauman, T. L. Kemper, Hippocampus in autism: A Golgi analysis. Acta Neuropathol. 91, 117–119 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Y. Sun, Y. Gao, J. J. Tidei, M. Shen, J. T. Hoang, D. F. Wagner, X. Zhao, Loss of MeCP2 in immature neurons leads to impaired network integration. Hum. Mol. Genet. 28, 245–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.C. H. Kwon, B. W. Luikart, C. M. Powell, J. Zhou, S. A. Matheny, W. Zhang, Y. Li, S. J. Baker, L. F. Parada, Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. Jiang, R. T. Ash, S. A. Baker, B. Suter, A. Ferguson, J. Park, J. Rudy, S. P. Torsky, H. T. Chao, H. Y. Zoghbi, S. M. Smirnakis, Dendritic arborization and spine dynamics are abnormal in the mouse model of MECP2 duplication syndrome. J. Neurosci. 33, 19518–19533 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.J. B. Angevine, Jr, Time of neuron origin in the hippocampal region. An autoradiographic study in the mouse. Exp. Neurol. Suppl. 2 (Suppl. 2), 1–70 (1965). [PubMed] [Google Scholar]
- 65.B. B. Stanfield, W. M. Cowan, The development of the hippocampus and dentate gyrus in normal and reeler mice. J. Comp. Neurol. 185, 423–459 (1979). [DOI] [PubMed] [Google Scholar]
- 66.L. Tricoire, K. A. Pelkey, B. E. Erkkila, B. W. Jeffries, X. Yuan, C. J. McBain, A blueprint for the spatiotemporal origins of mouse hippocampal interneuron diversity. J. Neurosci. 31, 10948–10970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. E. Horn, R. A. Nicoll, Somatostatin and parvalbumin inhibitory synapses onto hippocampal pyramidal neurons are regulated by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 115, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.S. B. Nelson, V. Valakh, Excitatory/inhibitory balance and circuit homeostasis in autism spectrum disorders. Neuron 87, 684–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Y. Li, Q.-L. You, S.-R. Zhang, W.-Y. Huang, W.-J. Zou, W. Jie, S.-J. Li, J.-H. Liu, C.-Y. Lv, J. Cong, Y.-Y. Hu, T.-M. Gao, J.-M. Li, Satb2 ablation impairs hippocampus-based long-term spatial memory and short-term working memory and immediate early genes (IEGs)-mediated hippocampal synaptic plasticity. Mol. Neurobiol. 10.1007/s12035-017-0531-5, (2017). [DOI] [PubMed] [Google Scholar]
- 70.Y. A. Zarate, J. Kaylor, J. Fish, SATB2-associated syndrome, in GeneReviews((R)), M. P. Adam, D. B. Everman, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Gripp, A. Amemiya, Eds.,(University of Washington, Seattle, 1993).
- 71.Y. A. Zarate, J. L. Fish, SATB2-associated syndrome: Mechanisms, phenotype, and practical recommendations. Am. J. Med. Genet. A 173, 327–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.F. Sharif, B. Tayebi, G. Buzsaki, S. Royer, A. Fernandez-Ruiz, Subcircuits of deep and superficial CA1 place cells support efficient spatial coding across heterogeneous environments. Neuron 109, 363–376.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Geiller, M. Fattahi, J. S. Choi, S. Royer, Place cells are more strongly tied to landmarks in deep than in superficial CA1. Nat. Commun. 8, 14531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R. E. Harvey, H. L. Robinson, C. Liu, A. Oliva, A. Fernandez-Ruiz, Hippocampo-cortical circuits for selective memory encoding, routing, and replay. Neuron 111, 2076–2090.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.G. Ciceri, H. Cho, M. Kshirsagar, A. Baggiolini, K. A. Aromolaran, R. M. Walsh, P. A. Goldstein, R. P. Koche, C. S. Leslie, L. Studer, An epigenetic barrier sets the timing of human neuronal maturation. bioRxiv 2022.06.02.490114 [Preprint] (3 June 2022). 10.1101/2022.06.02.490114. [DOI]
- 76.E. Hergenreder, Y. Zorina, Z. Zhao, H. Munguba, E. L. Calder, A. Baggiolini, A. P. Minotti, R. M. Walsh, C. Liston, J. Levitz, R. Garippa, S. Chen, G. Ciceri, L. Studer, Combined small molecule treatment accelerates timing of maturation in human pluripotent stem cell-derived neurons. bioRxiv 10.1101/2022.06.02.494616 [Preprint] (3 June 2022). 10.1101/2022.06.02.494616. [DOI]
- 77.N. Habib, I. Avraham-Davidi, A. Basu, T. Burks, K. Shekhar, M. Hofree, S. R. Choudhury, F. Aguet, E. Gelfand, K. Ardlie, D. A. Weitz, O. Rozenblatt-Rosen, F. Zhang, A. Regev, Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.G. X. Y. Zheng, J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, T. D. Wheeler, G. P. McDermott, J. Zhu, M. T. Gregory, J. Shuga, L. Montesclaros, J. G. Underwood, D. A. Masquelier, S. Y. Nishimura, M. Schnall-Levin, P. W. Wyatt, C. M. Hindson, R. Bharadwaj, A. Wong, K. D. Ness, L. W. Beppu, H. J. Deeg, C. McFarland, K. R. Loeb, W. J. Valente, N. G. Ericson, E. A. Stevens, J. P. Radich, T. S. Mikkelsen, B. J. Hindson, J. H. Bielas, Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.S. Choudhary, R. Satija, Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 23, 27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.G. Finak, A. McDavid, M. Yajima, J. Deng, V. Gersuk, A. K. Shalek, C. K. Slichter, H. W. Miller, M. J. McElrath, M. Prlic, P. S. Linsley, R. Gottardo, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.S. N. Basu, R. Kollu, S. Banerjee-Basu, AutDB: A gene reference resource for autism research. Nucleic Acids Res. 37, D832–D836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.W. J. Kent, C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, D. Haussler, The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Q. Wang, M. Li, T. Wu, L. Zhan, L. Li, M. Chen, W. Xie, Z. Xie, E. Hu, S. Xu, G. Yu, Exploring epigenomic datasets by ChIPseeker. Curr. Protoc. 2, e585 (2022). [DOI] [PubMed] [Google Scholar]
- 85.G. Yu, L. G. Wang, Q. Y. He, ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 86.D. W. Huang, B. T. Sherman, R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 87.S. Zaqout, A. M. Kaindl, Golgi-Cox staining step by step. Front. Neuroanat. 10, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.M. B. Yaylaoglu, A. Titmus, A. Visel, G. Alvarez-Bolado, C. Thaller, G. Eichele, Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev. Dyn. 234, 371–386 (2005). [DOI] [PubMed] [Google Scholar]
- 90.J. P. Carson, G. Eichele, W. Chiu, A method for automated detection of gene expression required for the establishment of a digital transcriptome-wide gene expression atlas. J. Microsc. 217, 275–281 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for tables S1 to S3
Tables S1 to S3