Abstract
Osteosarcoma is a highly aggressive cancer and lacks effective therapeutic targets. We found that L3MBTL2 acts as a tumor suppressor by transcriptionally repressing IFIT2 in osteosarcoma. L3MBTL2 recruits the components of Polycomb repressive complex 1.6 to form condensates via both Pho-binding pockets and polybasic regions within carboxyl-terminal intrinsically disordered regions; the L3MBTL2-induced condensates are required for its tumor suppression. Multi-monoubiquitination of L3MBTL2 by UBE2O results in its proteasomal degradation, and the UBE2O/L3MBTL2 axis was crucial for osteosarcoma growth. There is a reverse correlation between L3MBTL2 and UBE2O in osteosarcoma tissues, and higher UBE2O and lower L3MBTL2 are associated with poorer prognosis in osteosarcoma. Pharmacological blockage of UBE2O by arsenic trioxide can enhance L3MBTL2-induced condensates and consequently suppress osteosarcoma growth. Our findings unveil a crucial biological function of L3MBTL2-induced condensates in mediating tumor suppression, proposing the UBE2O-L3MBTL2 axis as a potential cancer therapeutic target in osteosarcoma.
Blockage of UBE2O by arsenic trioxide enhances L3MBTL2-induced condensates, consequently repressing tumor growth in osteosarcoma.
INTRODUCTION
Osteosarcoma is the most common malignant bone tumor in children and young adults (1). Treatment regimens, which consist of surgical resection and neoadjuvant chemotherapy, has remained largely unchanged for more than 40 years (2), underscoring an urgent need to gain a better understanding of the molecular mechanism underlying osteosarcoma progression in order to develop new therapeutic strategies.
Aberrant changes in the transcriptional and epigenetic landscape have long been implicated in the development and progression of osteosarcoma. For instance, inactivation of TP53 tumor suppressor genes (3) and amplification of MYC (4) are the most common recurrent molecular alterations, which are associated with unfavorable prognosis (5, 6). However, the significance of such alterations in osteosarcoma remains unexplored. CRISPR functional screens have been well developed as the unbiased and efficient tool for revealing genes that play critical roles in cancer progression, including cell proliferation/viability (7, 8), drug sensitivity (9), metastasis (10), and tumor immunity (11). We hypothesized that a CRISPR-Cas9 screen via single guide RNA (sgRNA) library targeting of both transcription factors and epigenetic regulators would identify key genes involved in progression of osteosarcoma, providing new targets for treating patients with osteosarcoma.
Phase-separated biomolecular condensates have been implicated in control of gene transcription, as the components of transcription machinery, including transcription factors, coactivators, and RNA polymerase II, are often assembled in such condensates (12), and dysregulation of both active and repressive transcriptional condensates contributes to the malignant state (13–15). Therefore, it is possible that understanding the mechanisms of condensate formation may provide a promising avenue for developing effective therapies for cancers. It has recently been reported that targeting the core transcription regulatory circuitry (CRC), which is critical for osteosarcoma progression, seems to benefit patients with osteosarcoma, and that disruption of CRC phase separation diminishes the chromatin accessibility and suppresses osteosarcoma chemoresistance and metastasis by targeting master transcription factors of the CRC, such as homeobox B8 (HOXB8) and fos like 1 (FOSL1) (16). Here, using sgRNA library targeting both transcription factors and epigenetic regulators, we performed a CRISPR-Cas9 screening for cell viability and found that lethal (3) malignant brain tumor-like protein 2 (L3MBTL2), a putative Polycomb group protein, is a tumor suppressor of osteosarcoma. L3MBTL2 suppresses tumor growth by transcriptionally repressing interferon induced protein with tetratricopeptide repeats 2 (IFIT2) via recruitment of the components of Polycomb repressive complex 1.6 (PRC1.6) to form nuclear condensates. The blockage of E3 ligase Ubiquitin-conjugating enzyme E2O (UBE2O) by arsenic trioxide (ATO) may enhance the L3MBTL2-induced condensates and consequently impair tumor growth in osteosarcoma.
RESULTS
CRISPR-Cas9 screening identifies L3MBTL2 as the tumor suppressor in osteosarcoma
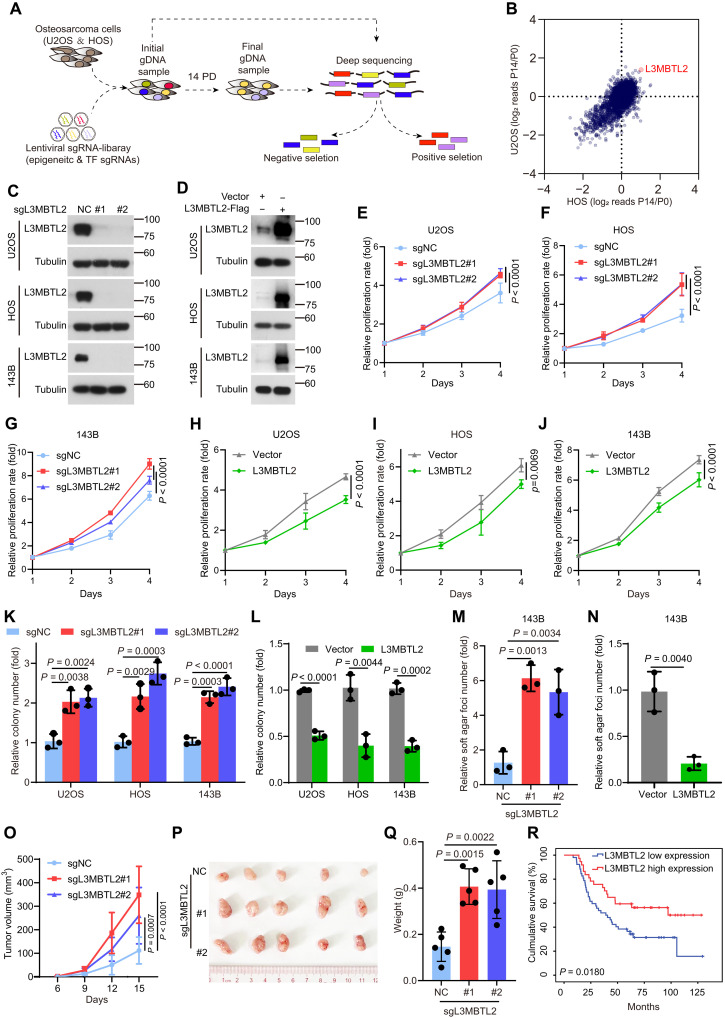
To identify whether genes had tumor-promoting or tumor-suppressing activity relevant to osteosarcoma, we constructed a pooled sgRNA library targeting a total of 2669 genes including transcription factors and epigenetic regulators in the Cas9-expressing vector, and there were 10 sgRNAs for each targeted gene and 500 nontargeted sgRNA controls in this library. Using U2OS (TP53 wild type) and HOS (TP53-R156P mutant) cells, cell viability screens with this library were performed (Fig. 1A). Overall, we identified 224 drop-out genes in this screening using a 5% false discovery rate (FDR) as the cutoff (fig. S1A), and well-known oncogenes in osteosarcoma, such as MYC, CDK1, CDK9, or AURKB (4), were shown in this list as well (tables S1 and S2). In addition, 29 genes were enriched in this screening (fig. S1B), and well-defined tumor suppressors of osteosarcoma, including TP53 (17) or KEAP1 (18), were included in this list (tables S1 and S2). These results indicated that our screening was successful. Among 29 enriched genes, sgRNAs targeting L3MBTL2 was the most significantly enriched in this screening (Fig. 1B) and shown in the top 10 most-enriched genes in both cell lines (fig. S1, B to E). As a crucial subunit of noncanonical PRC1.6, L3MBTL2 has transcriptional repression activity (19–21) and plays critical roles in various cellular processes, including mammalian embryonic development (22), spermatogenesis (23), DNA double-strand break (DSB) repair (24), and endoplasmic reticulum (ER) stress response (25). However, the role of L3MBTL2 in cancer remains poorly defined, leading us to further investigate L3MBTL2 in osteosarcoma.
Fig. 1. CRISPR-Cas9 screening of cell viability identifies L3MBTL2 as the tumor suppressor in osteosarcoma.
(A) Schematic of CRISPR screening strategy. (B) Scatter plot comparison of CRISPR screening in U2OS and HOS cells. Plotted is the average log2 fold change in abundance of all sgRNAs targeting a given gene at final (P14) versus initial reference (P0). (C and D) The indicated stable cells were analyzed by Western blotting. Data are representative of n = 3 biologically independent experiments. (E to J) Cell viability of the indicated stable cells was measured by MTT assay at the indicated time points. (K to N) Colony formation (K and L) and anchorage-independent (M and N) assays were performed for the indicated stable cells. The colony numbers per field were counted. Data in (E to N) are means ± SD of n = 3 biologically independent experiments. P values are shown. (O to Q) The indicated 143B stable cells were subcutaneously injected into mice. (O) Tumor size was measured at the indicated time points, and tumors were dissected at the end point. (P) Representative images of subcutaneous xenograft tumors. (Q) Tumor weight was measured at the end point. n = 5 biologically independent mice. Data are means ± SD. P values are shown. (R) Overall survival curves were analyzed on the basis of L3MBTL2 protein levels in patients with osteosarcoma by Kaplan-Meier method. Fifty-two and 37 cases with low and high expression of L3MBTL2, designed as L3MBTL2 low expression and L3MBTL2 high expression, respectively, were plotted, and P values are shown.
To determine the function of L3MBTL2 in osteosarcoma, we generated stable depletion or overexpression of L3MBTL2 in U2OS, HOS, U2OS/MTX300, and 143B cell lines (Fig. 1, C and D, and fig. S1, F and G). L3MBTL2 depletion and overexpression was able to increase and impede cell proliferation, colony formation, and anchorage-independent growth, respectively (Fig. 1, E to N, and fig. S1, H and I). Consistently, tumor growth in vivo was enhanced and impaired by depleting and overexpressing, respectively, L3MBTL2 in an orthotopic osteosarcoma model using 143B cells (fig. S1, J to O). Likewise, L3MBTL2 depletion was able to promote tumor growth in vivo using a subcutaneous xenograft mouse model, whereas ectopic L3MBTL2 had a reverse effect (Fig. 1, O to Q, and fig. S1, P to U). Moreover, higher protein level of L3MBTL2 was associated with better overall survival in patients with osteosarcoma (Fig. 1R), as determined by immunohistochemical staining using specific anti-L3MBTL2 antibody in osteosarcoma tissues (fig. S1, V and W). Consistently, we confirmed the correlation between L3MBTL2 expression and prognosis in pan-cancer by using a publicly accessible online tool KM Plotter (http://kmplot.com; fig S2). Together, our results indicate that L3MBTL2 acted as a tumor suppressor in osteosarcoma.
L3MBTL2 suppresses tumor growth by transcriptionally repressing IFIT2, which depends on its Pho-binding pocket
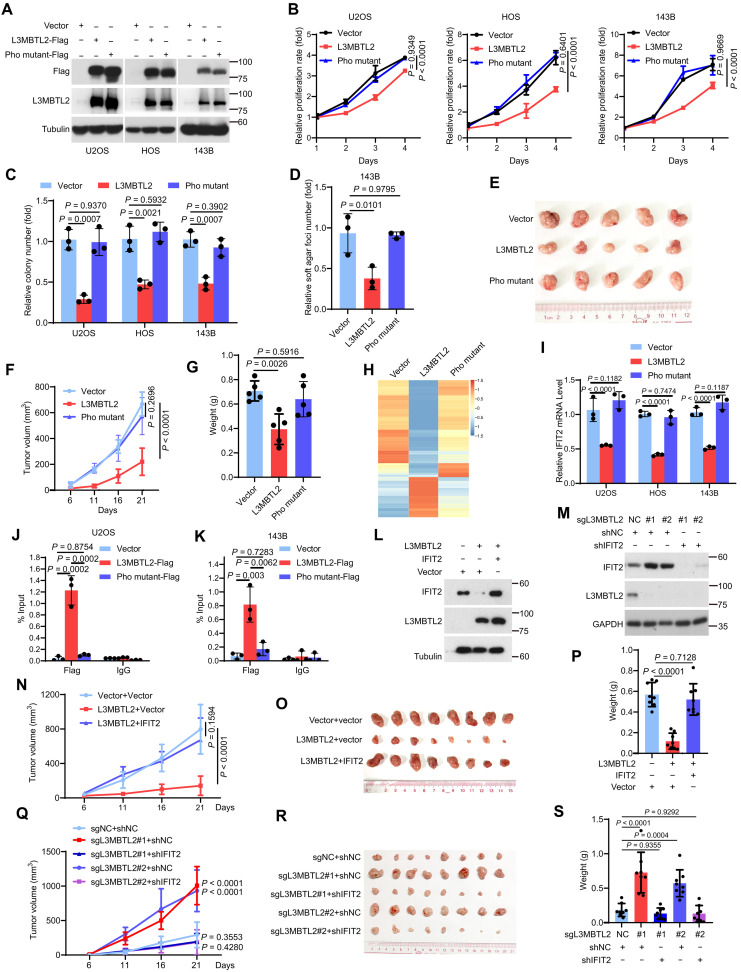
Previous studies have unveiled that L3MBTL2 regulates self-renewal of embryonic stem cells (ESCs) through its atypical C2/C2-type Zinc finger and MBT domains (22), and sumoylation of L3MBTL2 facilitates repression of its target genes (26). In addition, the Pho-binding pocket, an evolutionarily conserved hydrophobic groove from Drosophila Sfmbt (27), is critical for its direct binding to chromatin and the interaction with other subunits of the PRC1.6 complex in ESCs (28). To decipher which domain was critical for the tumor suppression of L3MBTL2 in osteosarcoma, as shown in fig. S3A, we generated four L3MBTL2 mutants: Pho mutant [five key amino acid residues were mutated in the Pho-binding pocket: G271K/A274E/L280A/L297A/L301A, critical for the binding of other subunits in PRC1.6 (28)], Cage mutant [four key amino acid residues were mutated in methyl-lysine binding aromatic cage: D546A/F570A/W573A/Y577A, critical for the binding of methylated histones (29)], ΔFCS [deletion of the atypical C2C2 zinc finger domain, Δ81-116, responsible for binding of regulatory RNAs (30)], and Sumo mutant [sumoylation-defective mutant, K215R/K433R/K541R/K675R/K700R, critical for the repression of a subset of endogenous target genes(26)]. Their functions were assessed in osteosarcoma cell lines stably expressing each mutant (Fig. 2A and fig. S3B). The effects of the Cage mutant, ΔFCS, and Sumo mutant on colony formation were similar compared to wild-type L3MBTL2 (fig. S3C), whereas the Pho mutant did not inhibit cell proliferation, colony formation, or anchorage-independent growth, as well as on tumor growth in vivo using a subcutaneous xenograft mouse model (Fig. 2, B to G). Consistently, the enhancement of cell proliferation and colony formation by depleting L3MBTL2 could be rescued by wild-type L3MBTL2, not the Pho mutant (fig. S3, D to F). Collectively, these results indicated that the Pho-binding pocket is required for the function of L3MBTL2 in osteosarcoma.
Fig. 2. L3MBTL2 suppresses tumor growth by transcriptionally repressing IFIT2, which depends on its Pho-binding pocket.
(A) The indicated stable cells were analyzed by Western blotting. (B) Cell viability of the indicated stable cells was measured by MTT assay at the indicated time points. (C and D) Colony formation (C) and anchorage-independent assay (D) were performed for the indicated stable cells. The colony numbers per field were counted. (E to G) The indicated 143B stable cells were subcutaneously injected into mice. (E) Representative images of subcutaneous xenograft tumors. (F) Tumor size was measured at the indicated time points. (G) Tumor weight was measured at the end point. n = 5 biologically independent mice. Data are means ± SD. P values are shown. (H) The heatmap shows relative expression levels of down- or up-regulated genes in the indicated 143B stable cells. (I) The relative IFIT2 mRNA levels were normalized to the GAPDH levels in the indicated stable cells as determined by RT-qPCR. (J and K) ChIP-qPCR analysis of L3MBTL2-Flag or Pho mutant-Flag occupancy at IFIT2 promoter region in the indicated stable cells. (L and M) The indicated 143B stable cells were analyzed by Western blotting. (N to S) The indicated 143B stable cells were subcutaneously injected into mice. (N and Q) Tumor volumes from mice subcutaneously injected with the indicated stable 143B cells were measured at the indicated days, and tumors were dissected at the end point. (O and R) Representative images of subcutaneous xenograft tumors. (P and S) Tumor weight was measured at the end point. n = 8 biologically independent mice. Data are means ± SD. P values are shown. Data in (A, L and M) are representative of n = 3 biologically independent experiments. Data in (B to D and I to K) are means ± SD of n = 3 biologically independent experiments. P values are shown.
We then performed RNA sequencing (RNA-seq) analyses in 143B cells overexpressing vector, L3MBTL2, or Pho-mutant of L3MBTL2 (Fig. 2H and tables S3 to S5). We found that the repression genes by L3MBTL2 were enriched in the tumor necrosis factor (TNF)/nuclear factor κB (NF-κB) pathway (fig. S3, G and H), such as IL1A, IL1B, IL6, CCL2, BIRC3, IFIT1, IFIT2, IFIT3, ICAM, and CXCL11 (fig. S3I), which is known to be crucial for tumor progression in osteosarcoma (31). Among these genes, the IFIT family including IFIT1, 2, 3 caught our attention, as their function in cancers relates to cell content (32). L3MBTL2, but not its Pho mutant, efficiently repressed the IFIT family (Fig. 2I and fig. S3J). IFIT2, neither IFIT1 nor IFIT3, was overexpressed in osteosarcoma tissues compared to normal tissues in our previously published osteosarcoma RNA-seq data (33), as shown in fig. S3 (K to M), and IFIT2 was also overexpressed in osteosarcoma using 33 paired tumor and normal tissues (fig. S3N). Notably, knockdown of IFIT2 decreased cell proliferation and colony formation (fig. S3, O to Q), and the decrease of colony formation in 143B cells stably expressing L3MBTL2 was abolished by introducing ectopic IFIT2, but not IFIT1 and IFIT3, into those cells (fig. S3R). Consistently, mRNA level of IFIT2 was increased by knockout of L3MBTL2 in 143B, U2OS, and HOS cells (fig. S3S). Chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP-qPCR) showed that endogenous L3MBTL2 was able to bind to IFIT2 promoter (fig. S3, T to U), and the enrichment of L3MBTL2, but not its Pho mutant, on IFIT2 promoter was detected in both 143B and U2OS cells (Fig. 2, J and K). Furthermore, when using a subcutaneous xenograft mouse model in vivo, overexpression of IFIT2 reversed the suppression of tumor growth induced by ectopic L3MBTL2, while knockdown of IFIT2 abolished the enhancement of tumor growth induced by knockout of L3MBTL2 (Fig. 2, L to S). Collectively, these results demonstrate that L3MBTL2 suppressed tumor growth by transcriptionally repressing IFIT2 in osteosarcoma, which depends on its Pho-binding pocket.
L3MBTL2 protein forms condensates via both polybasic regions and Pho-binding pocket
We observed droplet-like condensates, which were negative for the lipid marker dye DiD, in the nuclei of cells transfected with L3MBTL2-EGFP (enhanced green fluorescent protein), but not in those with Pho mutant L3MBTL2-GFP (fig. S4, A and B), which abolished tumor-suppressive activity of L3MBTL2, indicating that the formation of nuclear membrane-less condensates may be crucial for the suppression of IFIT2 transcription and tumor growth by L3MBTL2.
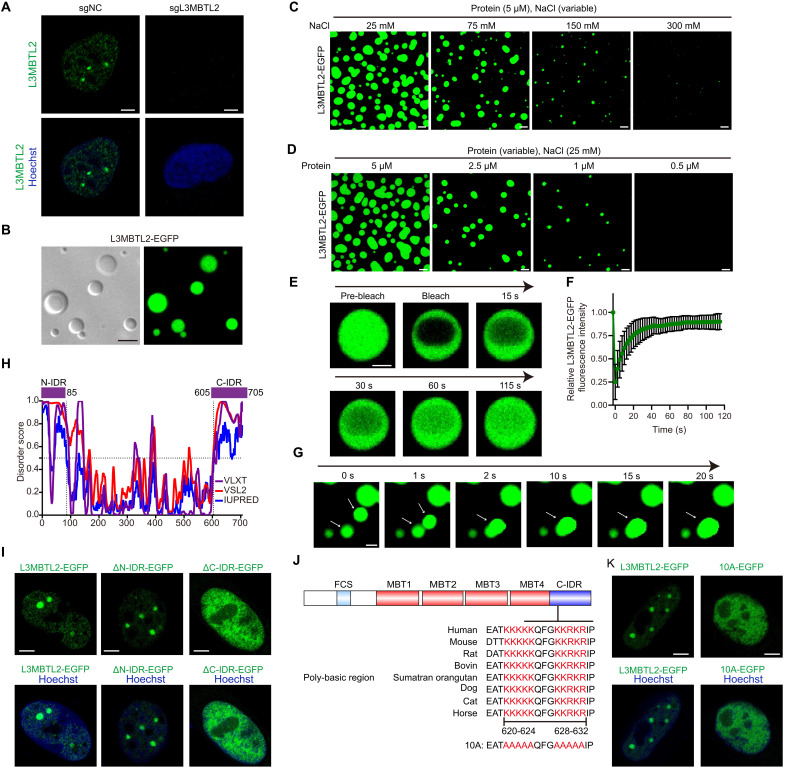
Many membrane-less biomolecular condensates such as nucleoli (34), stress granules (35), Polycomb bodies (36), Cajal bodies (37), etc., are often assembled via liquid-liquid phase separation (LLPS). Phase-separated condensates are involved in a wide range of cellular processes, and dysregulation of LLPS is implicated in the development of many diseases including cancer (14, 15, 38–40). Previous studies have found that canonical PRC1 Polycomb subunits can promote the formation of multicomponent condensates via LLPS in the nucleus, which augments activities of Polycomb complex and enhances chromatin compaction (41–44). Because L3MBTL2 was able to form condensates at its endogenous level (Fig. 3A and fig. S4, C to E), we questioned whether L3MBTL2 has the ability to undergo phase separation. First, purified L3MBTL2-EGFP exhibited remarkable turbidity, with an optical density at 600 nm, which was dependent on concentrations of both crowding agent poly(ethylene glycol) (PEG) and sodium chloride (NaCl; fig. S4, F and G). Second, confocal fluorescence microscopy also showed that purified L3MBTL2-EGFP, not EGFP alone, formed numerous, micrometer-sized spherical droplets in the absence of a crowding agent (Fig. 3B and fig. S4, H and I), which were disrupted by certain concentrations of NaCl (Fig. 3, C and D). This suggests that electrostatic interactions were critical for L3MBTL2-EGFP LLPS in vitro. Third, L3MBTL2-EGFP droplets were dynamic with rapid fluorescence recovery after photobleaching (FRAP) and experienced fusion in vitro and in cells (Fig. 3, E to G, and fig. S4, J to L). Collectively, L3MBTL2 formed nuclear condensates via LLPS.
Fig. 3. L3MBTL2 protein forms condensates via both polybasic regions and Pho-binding pocket.
(A) Representative images of immunofluorescence staining for endogenous L3MBTL2 (anti-L3MBTL2, active motif, green) in the indicated U2OS stable cells. (B) Representative differential interference contrast (left) and fluorescence (right) microscopy images of 10 μM purified L3MBTL2-EGFP in the phase separation buffer with 25 mM NaCl for 1 min at 25°C. (C) Representative images of 5 μM purified L3MBTL2-EGFP in the phase separation buffer with the indicated NaCl concentration and 1% (w/v) PEG-6000 for 1 min at 25°C. (D) Representative images of purified L3MBTL2-EGFP at the indicated protein concentration in the phase separation buffer with 25 mM NaCl and 1% (w/v) PEG-6000 for 1 min at 25°C. (E) Representative FRAP recovery images of L3MBTL-EGFP droplets in the phase separation buffer with 50 mM NaCl and 1% PEG-6000 at the indicated times. Scale bar, 2.5 μm. (F) Relative quantification of FRAP recovery kinetics of L3MBTL2-EGFP droplets averaged from n = 12 biologically independent experiments. Data are means ± SD. (G) Representative time-lapse images showing fusion of two droplets formed by 5 μM L3MBTL-EGFP proteins in the phase separation buffer with 50 mM NaCl and 1% (w/v) PEG-6000 for 1 min at 25°C. Scale bar, 2 μm. (H) Analysis of the intrinsically disorder tendency of L3MBTL2 by using PONDR and IUPred2. (I) Representative images of U2OS cells transiently expressing L3MBTL2 wild type (1 to 705 amino acids), ΔN-IDR (86 to 705 amino acids), or ΔC-IDR (1 to 604 amino acids) fused to EGFP. (J) Conservation of PBR sequences (620 to 624 and 628 to 632 amino acids) within L3MBTL2 C-IDR across homologs. 10A: Ten K or R in PBR of L3MBTL2 were mutated to A. (K) Representative images of U2OS cells transiently expressing L3MBTL2 wild type or 10A mutant fused to EGFP. Data in (A to D, G, I, and K) are representative of n = 3 biologically independent experiments. Scale bars, 5 μm (A to D, I, and K).
To explore the structural basis of L3MBTL2 condensates, we analyzed the amino acid sequences of L3MBTL2 using PONDR (45) and IUPred2 (46). As shown in Fig. 3H, the N-terminal region (1 to 85 amino acids) and the C-terminal region (605 to 705 amino acids) of L3MBTL2 were predicted to be intrinsically disordered regions (IDRs), and we thus designated these two IDRs as N-IDR and C-IDR, respectively. Depletion of C-IDR, but not of N-IDR, markedly disrupted L3MBTL2-EGFP condensate formation in vitro and in cells (Fig. 3I and fig. S4, M to O). Polybasic region [poly-lysine or poly-arginine (PBR)] was critical for phase separation (47), and there were two conserved PBRs (620 to 624 and 628 to 632) within the C-IDR of L3MBTL2 (Fig. 3J). To directly test whether these two PBRs contribute to L3MBTL2-EGFP LLPS, we mutated these 10 conserved lysine and arginine residues to alanine, marking them as 10A (Fig. 3J). In contrast to the Pho mutant of L3MBTL2, the PBR mutant 10A of L3MBTL2 retained the ability to bind to components of PRC1.6 (fig. S4P). However, cells transfected with 10A-EGFP mutant failed to form condensates in nuclei (Fig. 3K and fig. S4Q), and droplet formation in vitro was also markedly reduced when using the purified 10A-EGFP (fig. S4, R to U). Notably, purified Pho mutant-EGFP protein exhibited capacity to form droplet in vitro (fig. S4, T and U), indicating that L3MBTL2 undergoes LLPS predominantly relying on its polybasic regions in vitro, whereas both polybasic regions and the Pho-binding pocket of L3MBTL2 were required for its nuclear condensate formation in vivo.
L3MBTL2 condensates execute tumor suppression by selectively enriching components of the PRC1.6 complex
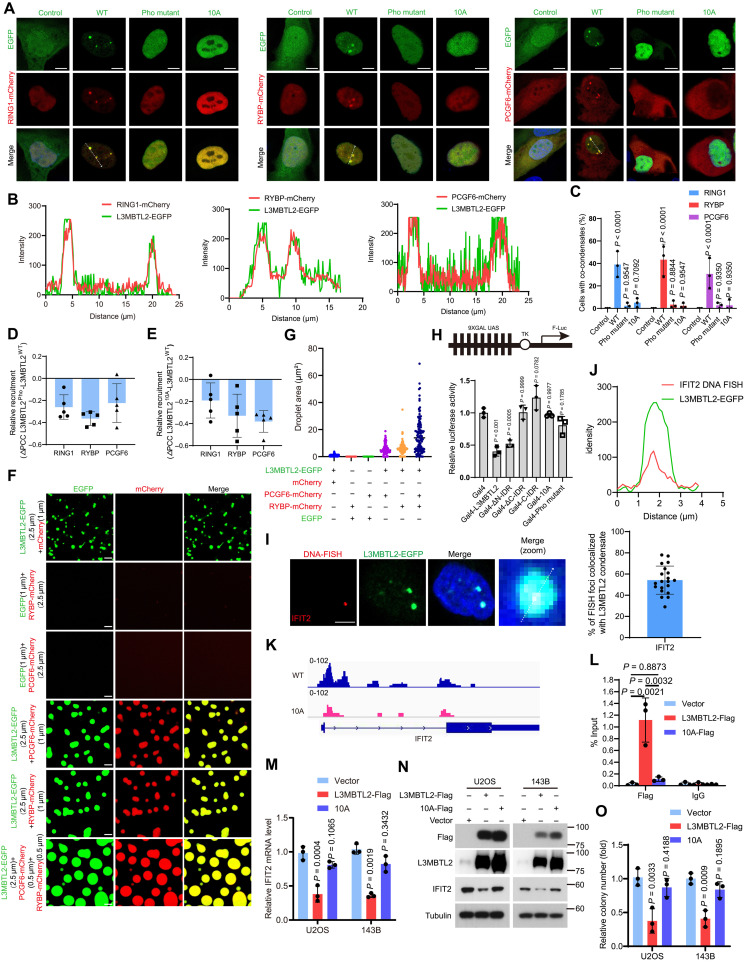
Given that the Pho-binding pocket, which is critical for the binding of other subunits in PRC1.6 (28), is required for both condensate formation and functions of L3MBTL2 in osteosarcoma, we surmised that other subunits of the PRC1.6 should be present in L3MBTL2 condensates. We constructed each component of PRC1.6 tagged with mCherry, and condensate was hardly detected in cells transiently overexpressing each plasmid (fig. S5A). Then, each plasmid was cotransfected with L3MBTL2-EGFP, Pho mutant L3MBTL2-EGFP, or 10A L3MBTL2-EGFP and analyzed their recruitment into L3MBTL2 condensates through calculating the Pearson correlation coefficients (PCCs). Four components (WDR5-, CBX3-, MAX-, and HDAC1-mCherry) were not colocalized with L3MBTL2-EGFP condensates, which were not required for IFIT2 transcriptional repression (fig. S5B-S5G, S5P). Seven components (Ring1-, RYBP-, PCGF6-, YAF2-, MGA-, E2F6-, and TFDP1-mCherry) were colocalized with L3MBTL2-EGFP condensates, but not with Pho mutant L3MBTL2-EGFP or 10A L3MBTL2-EGFP, in nuclei, as indicated by negative ΔPCC (Fig. 4, A to E, and fig. S5, H to P). Furthermore, we observed that both formation of exogenous L3MBTL2-EGFP condensates and recruitment of endogenous PCGF6 or RYBP into L3MBTL2 condensates exhibited a concentration-dependent manner via a doxycycline-inducible expression system (fig. S6, A to G). Using both purified RYBP-mCherry and PCGF6-mCherry as representatives of components of PRC1.6, purified RYBP-mCherry or PCGF6-mCherry alone hardly formed droplets, while mixing purified RYBP-mCherry, PCGF6-mCherry, or both with purified L3MBTL2-EGFP, but not purified Pho mutant L3MBTL2-EGFP, markedly enhanced droplet formation (Fig. 4, F and G, and fig. S6, H and I).
Fig. 4. L3MBTL2 condensates execute its tumor suppression by selectively enriching the components of PRC1.6 complex.
(A) Representative images of U2OS cells cotransfected with the indicated plasmids for 24 hours. Scale bars, 5 μm. (B) Line scan analysis of the fluorescence intensity along with the indicated line in (A). (C) Percentage of n = 100 cells with co-condensates in the indicated cells from (A). (D and E) Relative change in recruitment of the indicated PRC1.6-mCherry for L3MBTL2 wild type, Pho (D), or 10A (E). n = 5. means ± SD. (F) Representative images of the indicated proteins in the phase separation buffer with 25 mM NaCl for 1 min at 25°C. Scale bars, 5 μm. (G) Quantification of the droplets area in (F) was calculated. (H) HEK293T cells were transfected with the indicated constructs, along with the 9xGal-TK-luc reporter and the Renilla control reporter for 24 hours. Then, cells were analyzed for the relative luciferase activity. (I) Left: Confocal images of L3MBTL2-EGFP condensates and IFIT2 loci by DNA FISH in U2OS cells stably expressing pTeton-L3MBTL2-EGFP. Right: Quantification of DNA FISH foci colocalized with L3MBTL2 nuclear condensate (n = 20). Scale bar, 5 μm. (J) Line scan analysis of the fluorescence intensity along with the indicated line in (I). (K) Genome browser screenshots of CUT&Tag tracks illustrating binding of L3MBTL2 wild type and 10A to the IFIT2 promoter in the indicated 143B stable cells. (L) Chip-qPCR analysis of L3MBTL2 occupancy at IFIT2 promoter region in the indicated stable cells. n = 3. means ± SD. (M) The relative IFIT2 mRNA levels were normalized to the GAPDH levels in the indicated stable cells, as determined by RT-qPCR. n = 3. means ± SD. (N) The indicated stable cells were analyzed by Western blotting. (O) Colony formation assays were performed for the indicated stable cells. n = 3. means ± SD. Data in (A, C, F, I, and N) are representative of n = 3 biologically independent experiments.
Next, we explored whether condensate formation was required for L3MBLT2-mediated transcription repression using a Gal4-responsive reporter assay. Gal4-L3MBTL2 and Gal4-ΔN-IDR significantly repressed the reporter activity compared to the Gal4 control group, while Gal4-ΔC-IDR, Gal4-10A, Gal4-pho mutant, and Gal4-C-IDR did not (Fig. 4H). We observed the localization of the target gene IFIT2 within the L3MBTL2 condensate by DNA fluorescence in situ hybridization (FISH) in cells expressing inducible L3MBTL2-EGFP (Fig. 4, I to J). Consistently, CUT&Tag and ChIP-qPCR results revealed a reduced binding capacity of the L3MBTL2 10A mutant to the IFIT2 promoter (Fig. 4, K and L, and table S6), which, in turn, could not repress IFIT2 at both mRNA and protein levels and consequently failed to inhibit colony formation in both U2OS and 143B cells (Fig. 4, M to O). These results demonstrate that L3MBTL2 condensates via phase separation were required for the inhibition of L3MBTL2 on both IFIT2 transcription and colony formation in osteosarcoma cells. Notably, The L3MBTL2 nuclear condensates did not colocalize with the known components of transcription machinery, such as MED1 and BRD4 (fig. S7, A and B), as well as canonical PRC1 subunits, including BMI1 and PHC1 (fig. S7, C and D), indicating that L3MBTL2 condensates executed tumor suppression by selectively enriching the components of PRC1.6 complex in osteosarcoma.
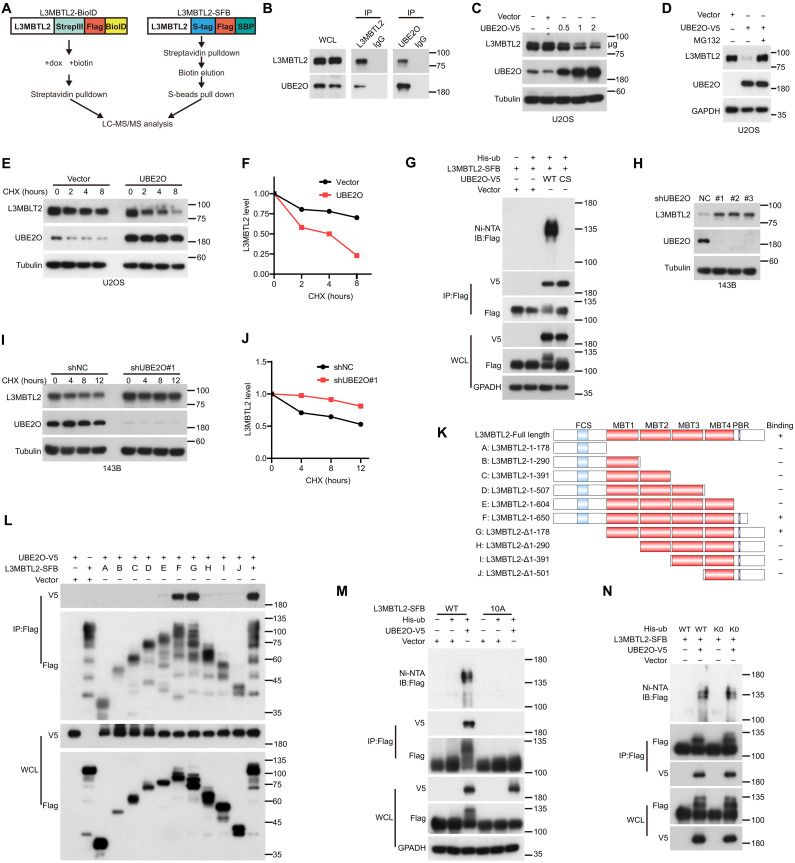
UBE2O multi-monoubiquitinates and degrades L3MBTL2 by binding the polybasic regions of L3MBTL2
Because there was no significant difference in mRNA level of L3MBTL2 between normal and osteosarcoma tissues (fig. S8A), but protein level of L3MBTL2 was decreased in osteosarcoma tissues compared to normal tissues (fig. S8B), and proteasome inhibitor MG132 could increase ectopic and endogenous L3MBTL2 protein (fig. S8, C and D), indicating that the proteasomal degradation of L3MBTL2 may play a key role in osteosarcoma cells. We sought to identify the E3 ligase responsible for degrading L3MBTL2 by using two mass spectrometry (MS)–based strategies, tandem affinity purification (TAP), and BioID-mediated proximity labeling system (Fig. 5A). UBE2O, an atypical E2/E3 hybrid ubiquitin ligase (48), was identified as a L3MBTL2-interacting protein in both MS results (fig. S8E and tables S7 and S8), and the interaction between L3MBTL2 and UBE2O was confirmed by coimmunoprecipitation (co-IP) at endogenous and exogenous levels (Fig. 5B and fig. S8F). There was an inverse correlation between UBE2O and L3MBTL2 at their endogenous protein levels in multiple osteosarcoma cell lines (fig. S8G). Ectopic UBE2O promoted proteasome-mediated L3MBTL2 degradation in a dose-dependent manner and shortened the half-life of L3MBTL2 (Fig. 5, C to F, and fig. S8, H and I). Furthermore, UBE2O, but not its catalytic dead mutant C1040S, enhanced the ubiquitination of L3MBTL2 (Fig. 5G), and knockdown of UBE2O had no effect on L3MBTL2 mRNA level but increased L3MBTL2 protein level and prolonged the half-life of L3MBTL2 in cells (Fig. 5, H to J, and fig. S8, J and K). These results suggest that UBE2O is an E3 ligase responsible for the degradation of L3MBTL2 protein in osteosarcoma.
Fig. 5. UBE2O multi-monoubiquitinates and degrades L3MBTL2 via binding the polybasic regions of L3MBTL2.
(A) Schematic description of mass spectrometry strategies. Left: L3MBTL2 fused to proximity-dependent biotin identification (BioID) protein. Right: L3MBTL2 fused to tandem affinity purification tag (SFB). (B). The co-IPs were performed using U2OS cells with anti-L3MBTL2, anti-UBE2O, or anti-IgG. WCL, whole cell lysate. (C) U2OS cells were transfected with UBE2O-V5 at the indicated concentrations for 48 hours and then were analyzed by Western blotting. (D) U2OS cells transfected with vector or UBE2O-V5 were incubated with or without MG132 for 8 hours and then were analyzed by Western blotting. (E) U2OS cells transfected with vector or UBE2O were incubated with cycloheximide (CHX; 20 μg/ml) for the indicated time points and then were analyzed by Western blotting. (F) Quantitation of L3MBTL2 protein levels in (E). (G, M, and N) HEK293T cells were cotransfected with the indicated plasmids for 48 hours and then were analyzed by immunoprecipitation using Ni-NTA (nitrilotriacetic acid) beads or anti-Flag beads followed by Western blotting. Ni-NTA, Ni2+-NTA. WT: UBE2O wild type (G), L3MBTL2 wild type (M), or ubiquitin wild type (N). CS (G): UBE2O catalytic mutant, C1040S; 10A (M): Ten K or R in PBR of L3MBTL2 were mutated to A; or K0 (N): ubiquitin lysine-deficient mutant. (H) The indicated 143B stable cells were analyzed by Western blotting. (I) The indicated 143B stable cells were treated with CHX (20 μg/ml) for the indicated time points and then were analyzed by Western blotting. (J) Quantitation of L3MBTL2 protein levels in (I). (K) Schematic description of L3MBTL2-SFB truncations used for co-IP assays with UBE2O-V5. (L) HEK293T cells were cotransfected with the indicated plasmids and then were analyzed by immunoprecipitation using anti-Flag beads followed by Western blotting. Data in (B to J and L to N) are representative of n = 3 biologically independent experiments.
To investigate how UBE2O degrades L3MBTL2 protein, we mapped the domains necessary for interaction between L3MBTL2 and UBE2O. Using the co-IP assay, we found that the PBRs of L3MBTL2 and the CC domain of UBE2O were critical for this interaction (Fig. 5, K and L, and fig. S9, A and B). The PBR mutant 10A of L3MBTL2 completely eliminated its interaction with UBE2O and its ubiquitination induced by UBE2O (Fig. 5M). Given that UBE2O can process diverse types of ubiquitination, including monoubiquitination (49), multi-monoubiquitination(50), and polyubiquitination (51), we used K0, in which all seven lysine residues in ubiquitin was mutated with arginine, which is unable to form polyubiquitin chain. As shown in Fig. 5N, K0 had similar ubiquitination pattern on L3MBTL2 compared to wild-type ubiquitin (WT) in cells, indicating that UBE2O may multi-monoubiquitinate L3MBTL2.
To further identify the ubiquitination sites of L3MBTL2 by UBE2O, we performed an anti-FLAG affinity purification of L3MBTL2 in U2OS cells cotransfected with UBE2O. MS peptide analysis revealed nine unique ubiquitination sites in L3MBTL2, and almost 60% of ubiquitination events were located in the area of C-IDR domain (table S9). However, single point or combined mutations of these lysine residues had similar ubiquitination on L3MBTL2 by UBE2O (fig. S10, A and B), indicating that multiple lysine residues may be monoubiquitinated in L3MBTL2 by UBE2O and that L3MBTL2 with some mutated lysine residues may become flexible to monoubiquitination by UBE2O. Together, these results demonstrated that UBE2O multi-monoubiquitinates and degrades L3MBTL2 via binding of the polybasic regions of L3MBTL2 in osteosarcoma cells.
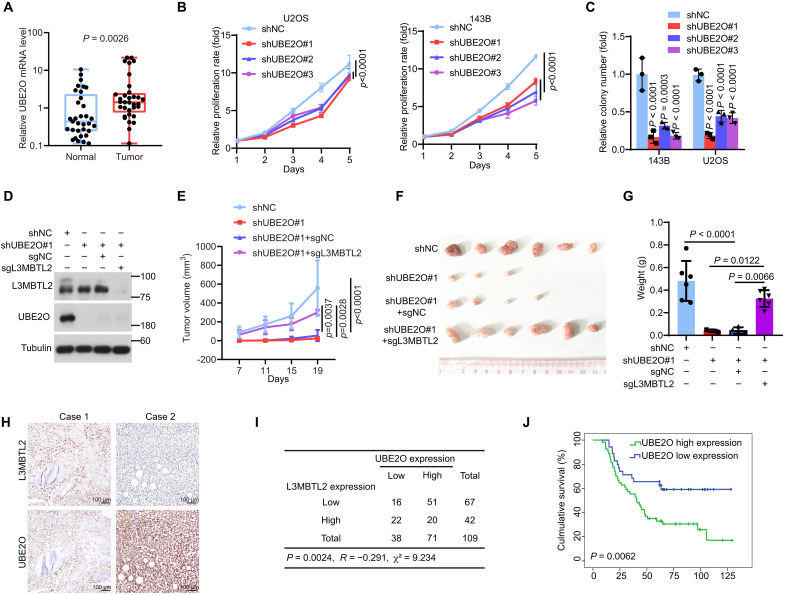
UBE2O promotes tumor growth via mainly degrading L3MBTL2 in osteosarcoma
Next, we sought to determine the function of UBE2O in osteosarcoma, which has not yet been explored. As shown in Fig. 6A, UBE2O mRNA level was up-regulated in osteosarcoma tissues compared to normal tissues. Knockdown of UBE2O inhibited cell proliferation and colony formation in U2OS and 143B cells (Fig. 6, B to C). Knockout of L3MBTL2 reversed the suppression of tumor growth induced by knockdown of UBE2O in a subcutaneous xenograft mouse model (Fig. 6, D to G). Using the clinical osteosarcoma tissues, the protein levels of UBE2O and L3MBTL2 were detected to be inversely correlated (Fig. 6, H to I), and higher UBE2O protein level was associated with poorer overall survival (Fig. 6J). Together, our results reveal that UBE2O promoted tumor growth mainly by degrading L3MBTL2 in osteosarcoma.
Fig. 6. UBE2O promotes tumor growth by mainly degrading L3MBTL2 in osteosarcoma.
(A) UBE2O mRNA levels were normalized to GAPDH levels in the paired osteosarcoma and normal tissues as determined by RT-qPCR. n = 33. Data are means ± SD. P values are shown. (B) Cell viability of the indicated stable cells was measured by MTT assay at the indicated time points. (C) Colony formation assay was performed for the indicated stable cells. The colony numbers per field were counted. Data in (B and C) are means ± SD of n = 3 biologically independent experiments. P values are shown. (D) The indicated 143B stable cells were analyzed by Western blotting. Data are representative of n = 3 biologically independent experiments. (E to G) The indicated 143B stable cells were subcutaneously injected into mice. (E) Tumor growth was measured at the indicated time points. (F) Representative images of xenograft tumors. (G) Tumor weight was measured at the end point. n = 6 biologically independent mice. Data are means ± SD. P values are shown. (H) Representative immunohistochemical images of both L3MBTL2 and UBE2O from 109 osteosarcoma tissues. (I) The correlation between L3MBTL2 and UBE2O protein levels in 109 osteosarcoma tissues from (I). P = 0.002, chi-square tests. R: Spearman correlation coefficient. (J) Overall survival curves were analyzed on the basis of UBE2O protein levels in patients with osteosarcoma by Kaplan-Meier method. Thirty-five and 54 cases with low and high expression of UBE2O, designed as UBE2O low expression and UBE2O high expression, respectively, were plotted, and P values are shown.
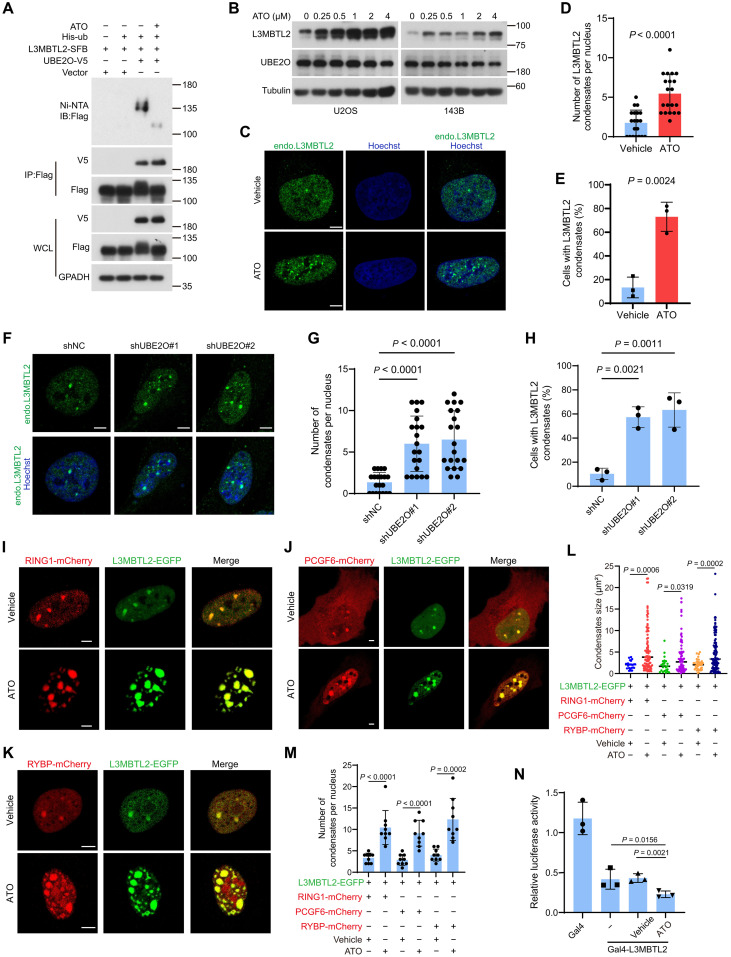
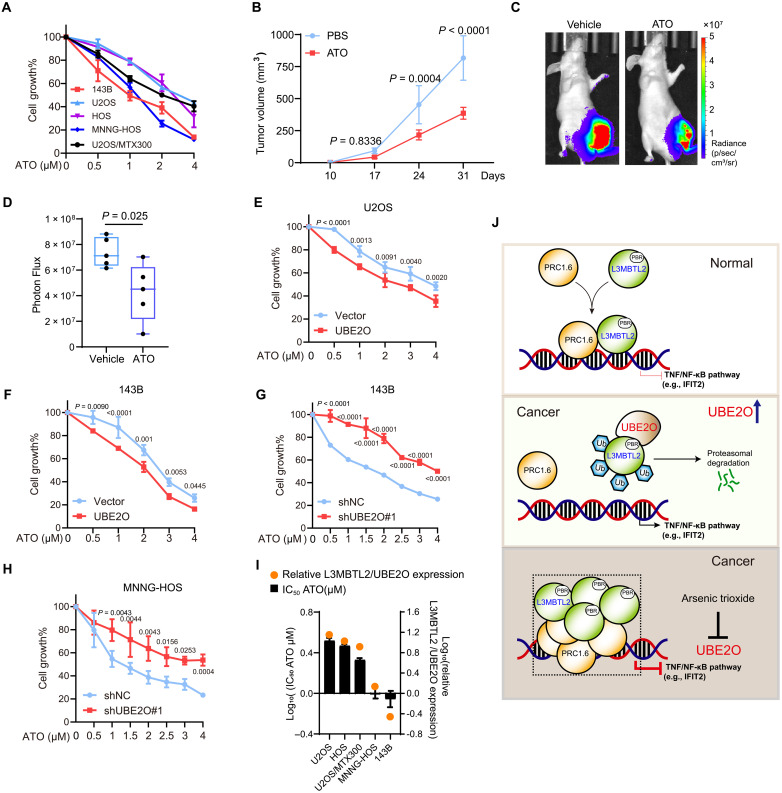
Pharmacological blockage of UBE2O by ATO suppresses osteosarcoma growth through the UBE2O/L3MBTL2 axis
Given that UBE2O-mediated L3MBTL2 multi-monoubiquitination and degradation are critical for cell proliferation and tumor growth, drugs that inhibit UBE2O E3 ligase activity may be beneficial for patients with osteosarcoma. It has been reported that ATO can inhibit UBE2O by cross-linking adjacent cysteines within its catalytic domain (52). Therefore, we assessed whether ATO can have any effect on osteosarcoma. ATO diminished L3MBTL2 multi-monoubiquitination by UBE2O (Fig. 7A), increased protein level of L3MBTL2 (Fig. 7B), and augmented formation of L3MBTL2 condensates in cells (Fig. 7, C to E). Consistently, knockdown of UBE2O showed similar effect (Fig. 7, F to H). The co-condensate of L3MBTL2 with the PRC1.6 components, such as RING1, PCGF6, and RYBP, was also increased in cells treated with ATO (Fig. 7, I to M). Consistently, the Gal4-responsive reporter assay showed that ATO treatment further decreased the L3MBTL2-induced inhibitory luciferase activity in cells (Fig. 7N). Moreover, ATO impeded cell viability in a dose-dependent manner in multiple osteosarcoma cell lines (Fig. 8A) and reduced tumor growth in mice bearing 143B cells at a clinically relevant dose (Fig. 8, B to D). This inhibition of ATO was enhanced and attenuated by overexpression and knockdown of UBE2O in cells, respectively (Fig. 8, E to H). Evaluation of relative protein levels of L3MBTL2 to UBE2O (L3MBTL2/UBE2O; fig. S8G) revealed that the highest and lowest rates of L3MBTL2/UBE2O were correlated with the worst and best inhibition by ATO in cells, respectively (Fig. 8I). Together, these results suggest that the blockage of UBE2O by ATO suppresses osteosarcoma growth through the UBE2O/L3MBTL2 axis.
Fig. 7. ATO stabilizes L3MBTL2 protein and augments the L3MBTL2-induced condensates.
(A) HEK293T cells were cotransfected the indicated plasmids for 48 hours and then were analyzed by immunoprecipitation using Ni-NTA and anti-FLAG beads followed by Western blotting. WCL, whole cell lysate. (B) U2OS and 143B cells were treated with increasing concentration of ATO for 12 hours and then were analyzed by Western blotting. (C) Representative images of immunofluorescence staining for endogenous L3MBTL2 in U2OS cells treated with ATO (1 μM) or vehicle for 12 hours. Scale bars, 5 μm. (D) Quantification of the numbers of L3MBTL2 condensate in cells from (C). Data are means ± SD of n = 20 cells. (E) Percentage of n = 100 cells with L3MBTL2 condensates in cells from (C). Data are means ± SD of n = 3 biologically independent experiments. (F) Representative images of immunofluorescence staining for endogenous L3MBTL2 in the indicated 143B stable cells. Scale bars, 5 μm. (G) Quantification of the numbers of L3MBTL2 condensate per nucleus in the indicated 143B stable cells (F). Data are means ± SD of n = 20 cells. (H) Percentage of n = 100 cells with L3MBTL2 condensates in the indicated 143B stable cells from (F). Data are means ± SD of n = 3 biologically independent experiments. (I to K) Representative fluorescence microscopy images of condensates formed by L3MBTL2-EGFP with Ring1-mCherry (I), PCGF6-mCherry (J), or RYBP-mCherry (K) in U2OS cells treated with ATO (1 μM) for 12 hours. Scale bars, 5 μm. Data in (A to C, F, and I to K) are representative of n = 3 biologically independent experiments. (L and M) Quantification of the size (L) and number (M) of the condensates from (I to K). Data are means ± SD of n = 10 cells. (N) HEK293T cells were transfected with the indicated constructs, along with the 9xGal-TK-luc reporter and the Renilla control reporter for 24 hours. Then, cells transfected with Gal4-L3MBTL2 were treated with or without ATO for 12 hours and analyzed for luciferase activity.
Fig. 8. ATO suppresses cell proliferation and tumor growth through the UBE2O/L3MBTL2 axis in osteosarcoma.
(A) The indicated osteosarcoma cell lines were treated with the indicated concentration of ATO for 48 hours, and then, cell viability was determined by MTT assay. (B to D) 143B-Luc cells were orthotopically injected into mice. Starting on day 7, mice were treated with ATO (2 mg/kg, daily) by intraperitoneal injection. (B) Tumor growth was measured at the indicated time points. (C) Representative bioluminescence images of mice at the end point. (D) Automated quantification of bioluminescence images. n = 5 biologically independent mice. Data are means ± SD. P values are shown. (E to H) The indicated stable cells were treated with the indicated concentration of ATO for 48 hours, and then, cell viability was determined by MTT assay. (I) Median inhibitory concentration (IC50) values of ATO in the indicated osteosarcoma cell lines as determined by MTT assay, and the log10 both IC50 of ATO and relative protein levels of L3MBTL2 to UBE2O (L3MBTL2/UBE2O) in each cell line were plotted. (J) A proposed model for function and regulation of L3MBTL2 phase separation in osteosarcoma. In normal condition, L3MBTL2 forms nuclear condensates with other components of the PRC1.6 complex to transcriptionally repress TNF/NF-κB pathways (e.g., IFIT2). UBE2O, a E2/E3 hybrid ubiquitin-protein ligase, is highly expressed in osteosarcoma, and multi-monoubiquitinates and degrades L3MBTL2 via specifically binding to polybasic region (PBR) of L3MBTL2, and consequently promotes tumor growth by activating the TNF/NF-κB pathways (e.g., IFIT2). ATO, the inhibitor of UBE2O, stabilizes L3MBTL2 protein and augments the L3MBTL2-induced co-condensate with the components of PRC1.6 complex, inhibiting the TNF/NF-κB pathways (e.g., IFIT2), subsequently suppress osteosarcoma growth.
DISCUSSION
In this report, as proposed in Fig. 8J, under normal condition, L3MBTL2 forms phase-separated condensates with other subunits of the PRC1.6 complex to transcriptionally repress TNF/NF-κB pathways (e.g., IFIT2). However, in osteosarcoma, UBE2O as an E3 ligase responsible for the proteasomal degradation of L3MBTL2 via multiple monoubiquitination is overexpressed, which, in turn, promotes tumor growth. The UBE2O blockage by ATO can augment the L3MBTL2-induced condensates and subsequently suppress tumor growth in osteosarcoma.
Transcriptional aberrations are common molecular characteristics of osteosarcoma (53), which suggests that identifying key transcription regulators may advance therapeutic strategies for patients with osteosarcoma. We report that L3MBTL2, as a key subunit of the PRC1.6 complex, acts as a tumor suppressor in osteosarcoma by repressing IFIT2 (Figs. 1 and 2). This is the first evidence that L3MBTL2 acts as a tumor suppressor in human cancers. L3MBTL2 has been shown to be involved in DNA DSB repair and ER stress response (24, 25), which are linked to tumorigenesis. Huang et al. (54) have recently reported that L3MBTL2 promotes pancreatic cancer progression through transcriptional suppression of CGA (54). However, we found that CGA was undetectable in most osteosarcoma cell lines and tumor tissues in our previously published osteosarcoma RNA-seq data (33, 55). On the other hand, GEPIA database analyses (56) showed that IFIT2, a key downstream target for L3MBTL2 in osteosarcoma, as shown in this report, was significantly up-regulated and correlated with poor prognosis in pancreatic cancer. These results imply that L3MBTL2 may act differently in a variety of cancer types.
Our mass spectrometry analysis has identified UBE2O as the E3 ligase responsible for degrading L3MBTL2 in U2OS cells. UBE2O, an E2/E3 hybrid enzyme, can modulate its substrate through mono-ubiquitination, multisite mono-ubiquitination, or polyubiquitination (50, 57, 58). Previous studies have shown that UBE2O plays an oncogenic role in diverse cancers, such as breast cancer (59), prostate cancer (60), liver cancer (61), and lung cancer (52), etc. However, Xu et al. (62) have recently reported that UBE2O induces apoptosis and inhibits tumor growth by ubiquitinating and degrading c-Maf in multiple myeloma. In our study, we found that UBE2O multi-monoubiquitinates and degrades L3MBTL2 by binding the polybasic regions of L3MBTL2, thereby promoting osteosarcoma growth. Moreover, high expression of UBE2O was related to poor prognosis in patients with osteosarcoma (Figs. 5 and 6), suggesting that UBE2O exerts a tumor-supporting role in osteosarcoma. The KM-plot analysis shows that high mRNA levels of L3MBTL2 are associated with favorable prognosis in multiple cancers, except for sarcoma. This is likely due to UBE2O-mediated specific translational modification of L3MBTL2 in osteosarcoma (fig. S2), and disrupting the interaction between L3MBTL2 and UBE2O may be a potential therapeutic strategy for tumor suppression in osteosarcoma.
As a crucial subunit of the PRC1.6 complex, L3MBTL2 assembles and recruits the PRC1.6 complex to target promoters through its Pho-binding pocket domain (28), and biomolecular condensates are enriched with factors important for transcriptional regulation (63). Here, we found that that L3MBTL2 forms condensates by recruiting the PRC1.6 complex via phase separation, which is dependent on its polybasic region and Pho-binding pocket domain, to execute both transcription inhibition and tumor suppression in osteosarcoma (Figs. 3 and 4). Recent studies have found that canonical PRC1-induced chromatin compaction depends on both PBR of CBX2 (41) and PHC-mediated oligomerization of PRC1 (44), indicating phase separation of canonical PRC1 is driven not only by IDRs-mediated nonspecific multivalent interactions but also by specific molecular interactions. Our results also support this model for phase separation of PRC1, as both mutations of conserved amino acid residues in PBR and Pho-binding pocket could impede the capacity of L3MBTL2 to form condensates in osteosarcoma cells. Disruption of oncogenic condensates seems to be a promising strategy for cancer patients (16, 64, 65). However, it remains unknown how tumor-suppressive condensates are targeted. Our work shows that ATO inhibited UBE2O to stabilize L3MBTL2 protein and subsequently enhanced L3MBTL2 condensates, resulting in the inhibition of cell proliferation and tumor growth in vitro and in vivo, and that the effects of ATO predominantly relied on the UBE2O/L3MBTL2 axis in osteosarcoma (Figs. 7 and 8). We could not exclude the contribution of other targets to the therapeutic effects of ATO in osteosarcoma, as inhibition of cell viability induced by ATO is partially dependent on UBE2O. On the other hand, the PROTAC for UBE2O may generate a good strategy for osteosarcoma treatment.
MATERIALS AND METHODS
Ethics
This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (no. B2022-688). Animal care and experiments were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals and the Principles for the Utilization and Care of Vertebrate Animals and were approved by the Animal Research Committee of SYSUCC (no. L102012020001Z).
Cell culture, stable transfection, and transient transfection
Human cell lines U2OS, HOS, 143B, and human embryonic kidney (HEK) 293T were cultured according to instructions from the American Type Culture Collection. U2OS/MTX300 cells, a methotrexate-resistant derivative of the U2OS cell line, was cultured as previously described (66). 143B-Luc cells stably expressing luciferase were cultured as described previously (67). All cell lines used in this study were authenticated by using short-tandem repeat profiling less than 6 months when this project was initiated and were cultured for no more than 1 month.
Stable cell lines expressing short hairpin RNAs (shRNAs), sgRNAs or cDNA were selected with puromycin (0.5 μg/ml). For transient transfection, HEK293T cells were transfected with plasmids by using polyethylenimine linear. U2OS and 143B cells were transfected with plasmids by using Lipofectamine 3000. U2OS cells were transfected with small interfering RNAs (siRNAs) by using Lipofectamine RNAiMAX. Cell-cultured medium was changed after posttransfection for 6 hours. The sequences of siRNAs, shRNAs, and sgRNAs are listed in table S10.
Plasmid construction and lentivirus production
For mammalian expression plasmids, cDNAs for L3MBTL2, UBE2O, IFIT2, RING1, RNF2, RYBP, YAF2, PCGF6, TFDP1, CBX3, E2F6, WDR5, HDAC1, MGA, MAX, MED1, and BRD4 were amplified by PCR and cloned into the pSIN or pcDNA3.1 vectors with or without a tag (Flag, SFB, V5, EGFP, mCherry, or TagBFP). UBE2O C1040S, L3MBTL2 Pho, or 10A mutant and His-ub K0 mutant were generated from WT UBE2O, L3MBTL2, or ubiquitin with Mut Express II Fast Mutagenesis Kit (Vazyme). L3MBTL2 ΔN-IDR and ΔC-IDR indicates the 86 to 705 amino acids and 1 to 604 amino acids of L3MBTL2, respectively.
For prokaryotic expression plasmids, EGFP, mCherry, L3MBTL2-EGFP, L3MBTL2-ΔN-IDR-EGFP, L3MBTL2-ΔC-IDR-EGFP, L3MBTL2-10A-EGFP, PCGF6-mCherry, and RYBP-mCherry were cloned into pSKB2.
SgRNAs targeting L3MBTL2 were designed by using GUIDES (http://guides.sanjanalab.org/) and inserted into lentiCRISPR v2 vector.
The shRNAs against UBE2O and IFIT2 were inserted into PLKO.1 vector. The sgRNAs and shRNAs are listed in table S10.
For Lentiviral packaging, HEK293T cells were cotransfected with psPAX2, pMD2.G, and plasmids containing sgRNAs, shRNAs, or ectopic expression constructs by using polyethylenimine (PEI). The cell-cultured medium was changed after posttransfection for 8 hours, and then, the supernatants containing lentiviruses were collected after 48 hours, centrifuged at 2000 rpm for 6 min, and aliquots were stored at −80°C.
Western blotting and co-IP
For Western blot analysis, whole-cell lysates were prepared by using radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 5 mM EDTA, 50 mM tris-HCl, and 0.5% NP-40) containing phosphatase inhibitor and protease inhibitor cocktail (Bimake). The cell lysates were cleared by centrifugation at 12,000 rpm for 30 min and then were incubated with antibody beads or agarose overnight at 4°C. Then, the precipitates were washed six times with cold RIPA buffer and eluted with 5× SDS sample buffer. The immunoprecipitates were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% nonfat milk for 1 hour at room temperature and incubated with primary antibodies and horseradish peroxidase–conjugated secondary antibody. The chemiluminescent signaling was detected by using ECL reagents (Tanon). The antibodies used and their concentrations are listed in table S11.
ChIP assays
Approximately 2 × 107 cells were cross-linked for 10 min in 1% formaldehyde, quenched with 0.125 M glycine for 5 min at room temperature, and then lysed using 1 ml of cell lysis buffer [20 mM tris-HCl (pH 8.0), 85 mM KCl, 0.5% NP-40, and 1× protease inhibitor cocktail] for 15 min at 4°C. The nuclei were collected through centrifugation at 1000g for 5 min at 4°C and then were lysed using 1 ml of nuclear lysis buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS, and 1× protease inhibitor cocktail] for 10 min at room temperature. The nuclei lysates were sheared using the Covaris E220 Focused Ultrasonicator (150 PIP. 15% duty factor, 200 cycles, and 5 min) to ~500–base pair (bp) fragments. The sheared chromatin in the supernatant was obtained through centrifugation at 12,000g for 10 min at 4°C. For immunoprecipitation, 100 μl of sheared chromatin was diluted in ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM tris-HCl (pH 8.0), and 167 mM NaCl] to a total volume of 1 ml and then were precleared with protein A/G magnetic beads (ChIP grade) for 1 hour at 4°C. The precleared mixture was immunoprecipitated with antibody against Flag, L3MBTL2, and normal immunoglobulin G (IgG), along with protein A/G magnetic beads overnight at 4°C. Then, beads were washed consecutively with low salt wash buffer [0.1% SDS, 1% Triton-X 100, 2 mM EDTA, 20 mM tris-HCl (pH 8.0), and 150 mM NaCl], high salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.0), and 500 mM NaCl], LiCl wash buffer [0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, and 10 mM tris-HCl (pH 8.0)], and 1× TE buffer (1% SDS and 0.1 M NaHCO3) and resuspended in 100 μl of ChIP Elution Buffer [100 mM tris-HCl (pH 8.0) and 10 mM EDTA] containing ribonuclease A at 37°C for 30 min. Then, 20 μg of proteinase K was added and mixed at 65°C for 2 hours. After incubation at 95°C for 10 min, the DNA was purified and subjected to qPCR analysis. The antibodies used and their concentrations are listed in table S11.
RNA extraction and reverse transcription qPCR
Total RNA was purified using an RNAprep pure cell kit (TIANGEN, DP430), and cDNA was synthesized according to the manufacturer’s instructions (Vazyme). Reverse transcription qPCR (RT-qPCR) was performed using a Light Cycler 480 instrument (Roche Diagnostics) with SYBR Green PCR Master Mix (Vazyme). The relative mRNA level of target gene was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The specificity was verified by melting curve analysis. The primers used for RT-qPCR are listed in table S10.
Screening procedure
The CRISPR-Cas9 library was designed for the 2669 genes covering transcriptional and epigenetic factors in the human genome. The sgRNA were designed against the constitutive exons shared by different isoforms of the genes and then filtered for potential off-target sequences, on the basis of similarities to the genome. We used 10 sgRNAs for each gene, and 500 nontargeted sgRNA controls were also included in this library.
To generate the lentivirus, six 10-cm dishes of HEK293T cells were cotransfected with lentivirus library and virus package helper plasmids. For each dish, 4 μg of pMD2G, 8 μg of pSPAX2, and 12 μg of library plasmid were mixed in 1 ml of Opti-MEM; then, 96 μl of PEI was added, mixed, and incubated for 10 min and then added to HEK293T cells. Cell-cultured medium was changed after posttransfection for 6 hours. After 48 hours, the virus was collected and filtered through a 0.45-μm polyvinylidene difluoride membrane. The multiplicity of infection (MOI) of virus for U2OS and HOS was assessed individually according to reference (68). For the cell viability screening, U2OS cell or HOS cells were transduced with virus at a low infection efficiency of 20%, and after 48 hours, cells were treated with puromycin for 72 hours. Then, the sgRNA-transduced cells were obtained, which were donated population doubling 0 (P0). In this step, we separated the cells into two parts. The one part was collected for the genomic DNA; the other part was kept for expanding. Both parts maintained at least 1.4 × 107 transduced cells to preserve a 500-fold library representation. After 14 population doublings, cells were extracted for the genomic DNA and donated population doubling 14 (P14). Last, the sgRNAs were amplified from P0 and P14 with primers listed in table S10. The resulting PCR products were applied to high-throughput sequencing to measure read counts of each sgRNA. Using the Illumina platform and PE150 sequencing, at least 3G clean data were obtained per sample. The data were analyzed with the MAGeCK package, which can assess the statistical significance of sgRNA ranking with the binomial model before identifying the targets; it can perform both negative and positive selection screens simultaneously (69). The full results of the changes are shown in tables S1 and S2.
Cell viability assay
Osteosarcoma stable cells were seeded at the density of 2500 cells (U2OS or HOS stable cells) or 1500 cells (143B stable cells) per well in 96-well microplate, and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was used to determine the cell viability. The cells were incubated with MTT for 4 hours and subjected to detect the optical density value at 490 nm using the microplate reader per day for continuous 4 days.
Colony formation assay
Osteosarcoma stable cells were seeded in six-well plates at the density of 500 per well. After 2 weeks, the cell clones were washed with phosphate-buffered saline (PBS), fixed in methanol, and stained with 0.1% crystal violet. Cell clones containing more than 50 cells were counted.
Anchorage-independent growth assay
The 143B stable cells were resuspended in 2 ml of 0.3% agarose at the density of 1 × 103 and were plated in six-well plates with 0.6% agarose. Cells were cultured for 2 weeks, and the foci formation containing more 50 cells were counted.
Immunohistochemistry staining
Immunohistochemistry (IHC) staining was performed using the standard protocol. The sodium citrate buffer (pH 6.0) was used for antigen retrieval. Slides were blocked and incubated with primary antibodies at 4°C overnight and then followed by secondary antibody and DAB reagents. L3MBTL2 (Bethyl) and UBE2O (GeneTex) were used in this study. IHC staining was evaluated by a pathologist who was blinded to this study. For the evaluation of IHC staining of L3MBTL2 and UBE2O, a semiquantitative scoring criterion was used in which both the staining intensity and positive areas were recorded. A staining index (values 0 to 9), which was obtained as the product of the intensity of positive staining (weak, 1; moderate, 2; and strong, 3) and the proportion of positive cells of interest (0%, 0; <30%, 1; 30 to 60%, 2; and >60%, 3), was calculated. The immunohistochemical cutoff for high or low expression of the indicated molecule was determined on the basis of the ROC curve analysis. The sensitivity and specificity for discriminating dead or alive was plotted as the IHC score, thus generating a ROC curve. The cutoff value was established as the point on the ROC curve where the sum of sensitivity and specificity was maximized. The paraffin-embedded pathologic specimens from 109 patients with osteosarcoma were obtained from the Department of Musculoskeletal Oncology, First Affiliated Hospital, Sun Yat-sen University (table S12).
Immunofluorescence
Cells seed into confocal dish were fixed with 4% paraformaldehyde for 10 min and further permeabilized with 0.5% Triton X-100 for 10 min. Then, cells were blocked with goat serum for 1 hour and followed by incubation with primary antibody overnight at 4°C. Then, cells were washed using PBS and incubated with secondary antibody at room temperature for 1 hour, and then, the nuclei were labeled by using Hoechst 33342 for 4 min and subjected to a confocal microscope. The antibodies are listed in table S11.
For imaging and quantification, at least 15 fields of view per 35-mm glass-bottom dish were randomly chosen by Hoechst nuclear staining and imaged using a Zeiss LSM880 confocal microscope. At least three different dishes were quantified per treatment type. The quantification of the number of condensates per nucleus was performed using ZEN software (version 3.7) in batch analysis mode using the image analysis module with customized parameters.
The degree of colocalization between L3MBTL2 and PRC1.6 subunits was determined by Person’s correlation coefficient, which is defined as
| (1) |
where “i” represents the individual pixel index, “Ch1i” and “Ch2i” denote the values in that pixel for the two channels, and “Ch1aver” and “Ch2aver” represent the sample means for their respective channels.
FRAP experiments
FRAP experiments were performed using a Zeiss LSM880 confocal microscope. For both in vitro and in cells FRAP, the fluorescence signal was bleached using 100% of maximum laser power of the 488-nm laser with 150 interactions for ~2.5 s. Relative recovery was normalized to the initial (before bleaching) fluorescence intensity for each bleached condensate and then used to calculate mean and SD of the recovery time.
Protein expression and purification
6xHis-EGFP, 6xHis-mCherry, 6xHis-ΔN-IDR-EGFP, 6xHis-ΔC-IDR-EGFP, 6xHis-10A-EGFP, 6xHis-Pho-EGFP, 6xHis-PCGF6-mCherry, and 6xHis-RYBP-mCherry were expressed in E. coli BL21(DE3) at 18°C for 18 hours with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG; BioFroxx, 1122GR100). Bacteria lysates were prepared by using lysis buffer [50 mM tris-HCl, 500 mM NaCl, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (pH 7.2)] in an ultrahigh-pressure homogenizer. Supernatant was collected by centrifugation at 12,000g for 1 hour at 4°C and then incubated with Ni Sepharose excel (Cytiva, 17371202) for 2 hours at 4°C. The resin was centrifuged at 1000g for 5 min at 4°C and washed with 10 volumes of lysis buffer using wash buffer [50 mM tris-HCl, 500 mM NaCl, 1 mM DTT, and 30 mM imidazole (pH 7.2)] five times. The purified proteins were eluted using elution buffer [50 mM tris-HCl, 500 mM NaCl, 1 mM DTT, and 500 mM imidazole (pH 7.2)] and concentrated using an Amicon Ultra 15-ml filter with 30-kDa NMWL (Millipore, UFC9030). The concentrated proteins were further loaded onto Superdex 200 increase 10/300 GL column (GE HealthCare) using buffer containing 25 mM tris-HCl (pH 7.2), 150 mM NaCl, and 1 mM DTT. Fractions containing purified proteins were collected according to the results of SDS-PAGE and concentrated, and aliquots were stored at −80°C. The protein concentrations were assessed by ultraviolet absorbance at 280 nm, and the extinction coefficients were calculated using the ProtParam tool.
Phase separation assay
For in vitro phase separation, the purified proteins were incubated in phase separation buffer [25 mM tris-HCl and 1 mM DTT (pH 7.2)] at the indicated NaCl concentrations in the presence or absence of polymeric crowders for 1 min at room temperature, as described in the figure legends, to induce phase separation. Phase separation assays were conducted in a 384-well plate (Cellvis, p384-1.5H-N), and images were acquired using the confocal microscopy (ZEISS LSM880); quantification of the droplets was analyzed by ImageJ.
For nuclear condensate formation analysis in cells, U2OS and 143B cells were transfected with the indicated plasmids and fixed with 4% paraformaldehyde for 15 min; then, the nuclei were stained with Hoechst 33342 and subjected to the confocal microscopy analysis.
Xenograft experiments
This study complies with all relevant ethical regulations regarding animal research. Animal studies were approved by the Animal Research Committee of Sun Yat-sen University Cancer Center and were performed in accordance with established guidelines. Male athymic BALB/C nude mice (4 to 6 weeks old) were obtained from the GemPharmatech.
For subcutaneous transplantation model, U2OS/MTX300(8 × 105) and 143B (8 × 105) cells stably knocking out L3MBTL2, U2OS/MTX300(1.5 × 106), and 143B (8 × 105) cells stably overexpressing L3MBTL2 were subcutaneously injected into the left inguinal area of mice. Tumor sizes were measured, and tumor volume was calculated using following formula: length × width2 × 0.526. The xenograft tumors were dissected and weighed.
For orthotopic mice model, as previously described (70), mice were orthotopically implanted with the stable 143B-Luc cells. Tumor sizes were measured, and tumor volume was calculated using following formula: length × width2 × 0.526. Tumor xenografts were also determined by luciferase-based noninvasive bioluminescence imaging using the platform of IVIS Lumina II (PerkinElmer).
For ATO treatment, 143B-Luc cells were implanted orthotopically into mice. Ten days later, mice were randomly separated into two groups. Mice were treated with intraperitoneal injection of vehicle and ATO (2 mg/kg, daily). On the basis of the current dosing recommendation for ATO in acute promyelocytic leukemia (APL) patients, the recommended dose is 0.15 mg/kg per day. To convert mouse dose in mg/kg to human equivalent doses in mg/kg, the Food and Drug Administration guidelines suggest either dividing the mouse dose by 12.3 or multiplying it by 0.08. With this in mind, we treated mice with 2 mg/kg intraperitoneally three times per week. Measurement of tumor size was performed in orthotopic mice model, and tumor volume was calculated using following formula: length × width2 × 0.526. Tumor xenografts and lung metastatic foci were also analyzed by luciferase-based noninvasive bioluminescence imaging using the platform of IVIS Lumina II (PerkinElmer).
DNA FISH combined with immunofluorescence
U2OS cells stably expressing pTeton-L3MBTL2-EGFP were cultured at a chamber slide (ibidi) and then treated with doxycycline (25 ng/ml) for 24 hours. Then, cells were fixed with 4% paraformaldehyde for 10 min and washed three times with ice-cold PBS for DNA FISH.
Cells were permeabilized in 70% ethanol, 80% ethanol, 90% ethanol, and then 100% ethanol for 5 min at room temperature. Probe hybridization mixture was made mixing 39 μl of FISH Hybridization Buffer (BersinBio, Bes1011), and 1 μl of FISH probes. Ten microliters of mixture was added on the slide, and coverslip was placed on top. The rubber cement was used to seal the coverslip, and once rubber cement solidified, genomic DNA and probes were denatured at 75°C for 8 min, and slides were incubated at 37°C in the dark overnight. Coverslip was removed from slide and then washed once with the prewarmed 2xSCC at 53°C for 5 min, prewarmed 0.1%NP-40/2xSCC for 5 min at 42°C, and 2xSCC for 5 min at room temperature. After washing, the slide was stained with 4′,6-diamidino-2-phenylindole and viewed on a confocal laser scanning microscope (Zeiss). DNA FISH probes were custom-designed and generated by BersinBio; the probe sequences are shown in table S10.
RNA-seq and analysis pipeline
RNA-seq was performed by Novogene using an Illumina X TEN. The 6 GB clean data per sample were collected for RNA-seq.143B cells stably expressing wild-type L3MBTL2, Pho mutant, and Vector were collected, and total RNA was extracted using TRIzol (Life Technologies, 15596026). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and RNA libraries were prepared by using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Libraries were sequenced on an Illumina HiSeq platform, and 150-bp paired-end reads were generated. For RNA-seq data analysis, the read counts were adjusted by edgeR program package through one scaling normalized factor. Differential expression analysis of two conditions was performed using the edgeR R package (3.18.1). The P values were adjusted using the Benjamini and Hochberg method. Corrected P value of 0.05 and absolute fold change of 2 were set as the threshold for significantly differential expression. The results of differential gene expression among vector, L3MBTL2-WT, and Pho mutant are shown in tables S3 to S5.
Proximity labeling assay
Following procedures as previously described (71). To label L3MBTL2-interacting proteins, doxycycline-inducible Flag-Bio-ID-L3MBTL2–expressing U2OS stable cells were treated with doxycycline (200 ng/ml) for 24 hours. Then, 50 μM biotin was added to the culture medium for 2 hours. Afterward, cells were lysed and sonicated in RIPA-SDS buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.125% SDS, 0.125% sodium deoxycholate, and 1% Triton X-100]. The clear lysates were incubated with streptavidin beads at 4°C overnight, and the beads were then washed with buffers in the following order: 1 M KCl buffer, 0.1 M Na2CO3 buffer, 2 M urea in 10 mM tris buffer, and RIPA-SDS buffer twice. The washed beads were suspended in 5× loading buffer containing 10% SDS and boiled for 8 min, and the boiled proteins were subjected to mass spectrometry analysis.
Tandem affinity purification
Following procedures as previously described (55), U2OS cells were transfected with plasmids encoding C-terminal SFB-tagged L3MBTL2, and stable cells were selected with puromycin (0.5 μg/ml). The cells were lysed in NETN buffer [100 mM NaCl, 20 mM tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM DTT, and 0.5% NP-40] containing 50 mM β-glycerophosphate, 10 mM NaF, and 1 mg/ml/1 each of pepstatin A and aprotinin. The lysates were centrifuged at 12,000 rpm to remove debris and then incubated with streptavidin-conjugated beads for 4 hours at 4°C. The beads were washed five times with NETN buffer, followed by elution with NETN buffer containing biotin (2 mg/ml). The eluates were incubated with S-protein beads for 4 hours. After five washes, the beads were suspended in 5× loading buffer containing 10% SDS and boiled for 8 min, and the boiled proteins were subjected to mass spectrometry analysis.
Mass spectrometry
Following procedures as previously described (72). Mass spectrometry was performed by APTBIO. Each fraction was injected for nanoLC-MS/MS (liquid chromatography–tandem mass spectrometry) analysis. The peptide mixture was loaded onto a reverse-phase trap column (Thermo Scientific Acclaim PepMap100; 100 μm by 2 cm; nanoViper C18) connected to the C18 reversed-phase analytical column (Thermo Scientific Easy Column; 10 cm long; inner diameter, 75 μm; 3-μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min controlled by IntelliFlow technology. The 1-hour gradient was used: zero to 60% buffer B for 50 min, 60 to 90% buffer B for 4 min, and hold in 90% buffer B for 6 min.
LC-MS/MS analysis
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60 min. The mass spectrometer was operated in positive ion mode. MS data were acquired using a data-dependent top 20 method dynamically choosing the most abundant precursor ions from the survey scan [300 to 1800 mass/charge ratio (m/z)] for high energy collision dissociation (HCD) fragmentation. Automatic gain control target was set to 1 × 106; maximum inject time was set to 50 ms, and number of scan ranges was set to 1. Dynamic exclusion duration was 30.0 s. Survey scans were acquired at a resolution of 70,000 at m/z of 100, and resolution for HCD spectra was set to 17,500 at m/z of 100, Automatic gain control target was set to 1 × 105. Isolation width was set to 1.5 m/z, microscans were set to 1, and maximum inject time was set to 50 ms. Normalized collision energy was 27 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled. L3MBTL2 binding partners and ubiquitinated sites identified by mass spectrum are listed in tables S7 to S9.
CUT&Tag assays
Cell (105) samples were washed twice gently with wash buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, and 1× protease inhibitor cocktail] then were treated with 10 μl of concanavalin A–coated magnetic beads (Bangs Laboratories) for 10 min at room temperature. Bead-bounded cells were resuspended with dig wash buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, 1× protease inhibitor cocktail, 0.05% digitonin, and 2 mM EDTA] and a 1:50 dilution of antibody against L3MBTL2 (Active motif, 39569) and incubated on a rotating platform at 4°C overnight.
After removal of primary antibody using magnet stand, cells were incubated with secondary antibody (1:100; Anti-Rabbit IgG antibody, Goat monoclonal: Millipore AP132) for 60 min and then incubated with pA-Tn5 adapter complex for 1 hour. After three washes with Dig-med buffer [0.01% digitonin, 20 mM Hepes (pH 7.5), 300 mM NaCl, 0.5 mM spermidine, and 1× protease inhibitor cocktail], cells were resuspended in Tagmentation buffer (10 mM MgCl2 in Dig-med Buffer) and incubated at 37°C for 1 hour. To amplify libraries, 21 μl of DNA was mixed with 2 μl of a universal i5 and a uniquely barcoded i7 primer. A volume of 25 μl of NEBNext HiFi 2×PCR Master Mix was added and mixed. The sample was placed in a thermocycler with a heated lid using the following cycling conditions: 72°C for 5 min (gap filling), 98°C for 30 s, 14 cycles of 98°C for 10 s, and 63°C for 30 s; final extension at 72°C for 1 min; and hold at 8°C. Libraried clean-up was performed with XP beads (Beckman Counter). Sequencing was performed in the Illumina NovaSeq 6000 using 150-bp paired-end following the manufacturer’s instructions.
Analysis of CUT&Tag data
Raw data (raw reads) of fastq format were firstly processed through in-house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N, and low quality reads from raw data. At the same time, Q20, Q30, and GC content, the clean data were calculated. All the downstream analyses were based on the clean data with high quality. The bam file generated by the unique mapped reads as an input file, using macs2 software for callpeak with cutoff q value <0.05. For differential analysis, first merge the peak files of each sample using the bedtools software. Second, the counts of the reads over the bed were determined for each sample using bedtools multicov. Last, differential accessible peak was assessed using DESeq2. Regions were called differentially accessible if the absolute value of the log2 fold change was 1 at a P value <0.05. The results of differential peaks between L3MBTL2-WT and 10A mutant are shown in table S6.
Statistical analysis
All quantitative data are presented as means ± SD. The error bars indicate the SD. GraphPad Prism 8 and SPSS software were used for the statistical analysis. Unpaired t test was used to compare differences between two groups. One-way analysis of variance (ANOVA) tests were performed to compare differences between subgroups. Survival analysis was carried out by Kaplan-Meier analysis and compared using a log-rank test.
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation for Distinguished Young Scholars of Guangdong Province, China (grant no. 2021B1515020033 to D.Li.); the National Nature Science Foundation in China (NSFC) 82341015 and 82030090 to T.K.; 82322053, 32370819, and 32070765 to D.Li.; 82373026 to L.Zho.; and 82073243 to X.W.; Shenzhen Science and Technology Program (JCYJ20220530144607017) to L.Zho.; Guangdong Basic and Applied Basic Research Foundation (2022A1515220110) to L.Zho.; and Research Start-up Fund of the Seventh Affiliated Hospital, Sun Yat-sen University (ZSQYRSF0012) to L.Zho.
Author contributions: T.K., D.Li., and L.Zho. directed and conceived the project. L.Zho. led this project, performed most of the experiments, and analyzed data. J.W. performed phase separation and animal experiments. S.W. and Y.W. generated CRISPR-Cas9 knockout pooled library. J.Y. collected clinical samples. W.C.,C.Z., R.Z., X.W., L.Zhe., X.H., Y.G., D. Lv, and M.L. provided experimental support. L.Zho., T.K., and D.Li. wrote the manuscript, and the other authors helped revise and proofread the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive for Humans with accession code HRA005341.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legends for tables S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S12
REFERENCES AND NOTES
- 1.L. Mirabello, R. J. Troisi, S. A. Savage, International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 125, 229–234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. S. Meltzer, L. J. Helman, New horizons in the treatment of osteosarcoma. N. Engl. J. Med. 385, 2066–2076 (2021). [DOI] [PubMed] [Google Scholar]
- 3.X. Chen, A. Bahrami, A. Pappo, J. Easton, J. Dalton, E. Hedlund, D. Ellison, S. Shurtleff, G. Wu, L. Wei, M. Parker, M. Rusch, P. Nagahawatte, J. Wu, S. Mao, K. Boggs, H. Mulder, D. Yergeau, C. Lu, L. Ding, M. Edmonson, C. Qu, J. Wang, Y. Li, F. Navid, N. C. Daw, E. R. Mardis, R. K. Wilson, J. R. Downing, J. Zhang, M. A. Dyer; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project , Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 7, 104–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. C. Sayles, M. R. Breese, A. L. Koehne, S. G. Leung, A. G. Lee, H. Y. Liu, A. Spillinger, A. T. Shah, B. Tanasa, K. Straessler, F. K. Hazard, S. L. Spunt, N. Marina, G. E. Kim, S. J. Cho, R. S. Avedian, D. G. Mohler, M. O. Kim, S. G. DuBois, D. S. Hawkins, E. A. Sweet-Cordero, Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 9, 46–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. Feng, D. C. Dean, F. J. Hornicek, D. Spentzos, R. M. Hoffman, H. Shi, Z. Duan, Myc is a prognostic biomarker and potential therapeutic target in osteosarcoma. Ther. Adv. Med. Oncol. 12, 1758835920922055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.E. E. Pakos, P. A. Kyzas, J. P. Ioannidis, Prognostic significance of TP53 tumor suppressor gene expression and mutations in human osteosarcoma: A meta-analysis. Clin. Cancer Res. 10, 6208–6214 (2004). [DOI] [PubMed] [Google Scholar]
- 7.A. Augert, H. Mathsyaraja, A. H. Ibrahim, B. Freie, M. J. Geuenich, P. F. Cheng, S. P. Alibeckoff, N. Wu, J. B. Hiatt, R. Basom, A. Gazdar, L. B. Sullivan, R. N. Eisenman, D. MacPherson, MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38, 97–114.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. Inoue, G. L. Chew, B. Liu, B. C. Michel, J. Pangallo, A. R. D’Avino, T. Hitchman, K. North, S. C. W. Lee, L. Bitner, A. Block, A. R. Moore, A. Yoshimi, L. Escobar-Hoyos, H. Cho, A. Penson, S. X. Lu, J. Taylor, Y. Chen, C. Kadoch, O. Abdel-Wahab, R. K. Bradley, Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.T. Ringel, N. Frey, F. Ringnalda, S. Janjuha, S. Cherkaoui, S. Butz, S. Srivatsa, M. Pirkl, G. Russo, L. Villiger, G. Rogler, H. Clevers, N. Beerenwinkel, N. Zamboni, T. Baubec, G. Schwank, Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell 26, 431–440.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 10.L. van der Weyden, M. J. Arends, A. D. Campbell, T. Bald, H. Wardle-Jones, N. Griggs, M. D. C. Velasco-Herrera, T. Tüting, O. J. Sansom, N. A. Karp, S. Clare, D. Gleeson, E. Ryder, A. Galli, E. Tuck, E. L. Cambridge, T. Voet, I. C. Macaulay, K. Wong; Sanger Mouse Genetics Project, S. Spiegel, A. O. Speak, D. J. Adams, Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature 541, 233–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.F. Li, Q. Huang, T. A. Luster, H. Hu, H. Zhang, W. L. Ng, A. Khodadadi-Jamayran, W. Wang, T. Chen, J. Deng, M. Ranieri, Z. Fang, V. Pyon, C. M. Dowling, E. Bagdatlioglu, C. Almonte, K. Labbe, H. Silver, A. R. Rabin, K. Jani, A. Tsirigos, T. Papagiannakopoulos, P. S. Hammerman, V. Velcheti, G. J. Freeman, J. Qi, G. Miller, K. K. Wong, In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in kras-mutant lung Adenocarcinoma. Cancer Discov. 10, 270–287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Lu, T. Wu, O. Gutman, H. Lu, Q. Zhou, Y. I. Henis, K. Luo, Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 22, 453–464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Yu, Z. Peng, M. Qin, Y. Liu, J. Wang, C. Zhang, J. Lin, T. Dong, L. Wang, S. Li, Y. Yang, S. Xu, W. Guo, X. Zhang, M. Shi, H. Peng, X. Luo, H. Zhang, L. Zhang, Y. Li, X. P. Yang, S. Sun, Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell 81, 1216–1230.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 14.J. H. Ahn, E. S. Davis, T. A. Daugird, S. Zhao, I. Y. Quiroga, H. Uryu, J. Li, A. J. Storey, Y. H. Tsai, D. P. Keeley, S. G. Mackintosh, R. D. Edmondson, S. D. Byrum, L. Cai, A. J. Tackett, D. Zheng, W. R. Legant, D. H. Phanstiel, G. G. Wang, Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. Shi, W. Li, Y. Song, Z. Wang, R. Ju, A. Ulman, J. Hu, F. Palomba, Y. Zhao, J. P. le, W. Jarrard, D. Dimoff, M. A. Digman, E. Gratton, C. Zang, H. Jiang, UTX condensation underlies its tumour-suppressive activity. Nature 597, 726–731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.B. Lu, C. Zou, M. Yang, Y. He, J. He, C. Zhang, S. Chen, J. Yu, K. Y. Liu, Q. Cao, W. Zhao, Pharmacological inhibition of core regulatory circuitry liquid-liquid phase separation suppresses metastasis and chemoresistance in osteosarcoma. Adv. Sci. 8, e2101895 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Kansara, M. W. Teng, M. J. Smyth, D. M. Thomas, Translational biology of osteosarcoma. Nat. Rev. Cancer 14, 722–735 (2014). [DOI] [PubMed] [Google Scholar]
- 18.J. Zhang, X. Wang, W. Wu, H. Dang, B. Wang, Expression of the Nrf2 and Keap1 proteins and their clinical significance in osteosarcoma. Biochem. Biophys. Res. Commun. 473, 42–46 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Z. Gao, J. Zhang, R. Bonasio, F. Strino, A. Sawai, F. Parisi, Y. Kluger, D. Reinberg, PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.P. Trojer, A. R. Cao, Z. Gao, Y. Li, J. Zhang, X. Xu, G. Li, R. Losson, H. Erdjument-Bromage, P. Tempst, P. J. Farnham, D. Reinberg, L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B. Stielow, F. Finkernagel, T. Stiewe, A. Nist, G. Suske, MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLOS Genet. 14, e1007193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. Qin, W. A. Whyte, E. Anderssen, E. Apostolou, H. H. Chen, S. Akbarian, R. T. Bronson, K. Hochedlinger, S. Ramaswamy, R. A. Young, H. Hock, The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11, 319–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. Meng, J. Liao, D. Zhao, H. Huang, J. Qin, T. L. Lee, D. Chen, W. Y. Chan, Y. Xia, L3MBTL2 regulates chromatin remodeling during spermatogenesis. Cell Death Differ. 26, 2194–2207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Nowsheen, K. Aziz, A. Aziz, M. Deng, B. Qin, K. Luo, K. B. Jeganathan, H. Zhang, T. Liu, J. Yu, Y. Deng, J. Yuan, W. Ding, J. M. van Deursen, Z. Lou, L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat. Cell Biol. 20, 455–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. A. Panganiban, H. R. Park, M. Sun, M. Shumyatcher, B. E. Himes, Q. Lu, Genome-wide CRISPR screen identifies suppressors of endoplasmic reticulum stress-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 116, 13384–13393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. Stielow, B. Stielow, F. Finkernagel, M. Scharfe, M. Jarek, G. Suske, SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes. Nucleic Acids Res. 42, 3044–3058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. Alfieri, M. C. Gambetta, R. Matos, S. Glatt, P. Sehr, S. Fraterman, M. Wilm, J. Müller, C. W. Müller, Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements. Genes Dev. 27, 2367–2379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y. Huang, W. Zhao, C. Wang, Y. Zhu, M. Liu, H. Tong, Y. Xia, Q. Jiang, J. Qin, Combinatorial control of recruitment of a variant PRC1.6 complex in embryonic stem cells. Cell Rep. 22, 3032–3043 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Y. Guo, N. Nady, C. Qi, A. Allali-Hassani, H. Zhu, P. Pan, M. A. Adams-Cioaba, M. F. Amaya, A. Dong, M. Vedadi, M. Schapira, R. J. Read, C. H. Arrowsmith, J. Min, Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 37, 2204–2210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. C. Lechtenberg, M. D. Allen, T. J. Rutherford, S. M. Freund, M. Bycroft, Solution structure of the FCS zinc finger domain of the human polycomb group protein L(3)mbt-like 2. Protein Sci. 18, 657–661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. Zhang, B. Chen, K. Jiang, L. Lao, H. Shen, Z. Chen, Activation of TNF-α/NF-κB axis enhances CRL4BDCAF11 E3 ligase activity and regulates cell cycle progression in human osteosarcoma cells. Mol. Oncol. 12, 476–494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.X. F. Tan, Q. Chen, S. H. Hua, G. W. Yip, Roles of interferon induced protein with tetratricopeptide repeats (IFIT) family in cancer. Curr. Med. Chem. 28, 5034–5047 (2021). [DOI] [PubMed] [Google Scholar]
- 33.S. Wang, L. Zhong, Y. Li, D. Xiao, R. Zhang, D. Liao, D. Lv, X. Wang, J. Wang, X. Xie, J. Chen, Y. Wu, T. Kang, Up-regulation of PCOLCE by TWIST1 promotes metastasis in osteosarcoma. Theranostics 9, 4342–4353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Feric, N. Vaidya, T. S. Harmon, D. M. Mitrea, L. Zhu, T. M. Richardson, R. W. Kriwacki, R. V. Pappu, C. P. Brangwynne, Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Molliex, J. Temirov, J. Lee, M. Coughlin, A. P. Kanagaraj, H. J. Kim, T. Mittag, J. P. Taylor, Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. W. Hodgson, H. W. Brock, Are polycomb group bodies gene silencing factories? Cell 144, 170–171 (2011). [DOI] [PubMed] [Google Scholar]
- 37.S. F. Shimobayashi, P. Ronceray, D. W. Sanders, M. P. Haataja, C. P. Brangwynne, Nucleation landscape of biomolecular condensates. Nature 599, 503–506 (2021). [DOI] [PubMed] [Google Scholar]
- 38.J. J. Bouchard, J. H. Otero, D. C. Scott, E. Szulc, E. W. Martin, N. Sabri, D. Granata, M. R. Marzahn, K. Lindorff-Larsen, X. Salvatella, B. A. Schulman, T. Mittag, Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Esposito, C. Fang, K. C. Cook, N. Park, Y. Wei, C. Spadazzi, D. Bracha, R. T. Gunaratna, G. Laevsky, C. J. DeCoste, H. Slabodkin, C. P. Brangwynne, I. M. Cristea, Y. Kang, TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat. Cell Biol. 23, 257–267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.W. Li, J. Hu, B. Shi, F. Palomba, M. A. Digman, E. Gratton, H. Jiang, Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 22, 960–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. J. Plys, C. P. Davis, J. Kim, G. Rizki, M. M. Keenen, S. K. Marr, R. E. Kingston, Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33, 799–813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. Tatavosian, S. Kent, K. Brown, T. Yao, H. N. Duc, T. N. Huynh, C. Y. Zhen, B. Ma, H. Wang, X. Ren, Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 294, 1451–1463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. M. Eeftens, M. Kapoor, D. Michieletto, C. P. Brangwynne, Polycomb condensates can promote epigenetic marks but are not required for sustained chromatin compaction. Nat. Commun. 12, 5888 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. Seif, J. J. Kang, C. Sasseville, O. Senkovich, A. Kaltashov, E. L. Boulier, I. Kapur, C. A. Kim, N. J. Francis, Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat. Commun. 11, 5609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. Linding, L. J. Jensen, F. Diella, P. Bork, T. J. Gibson, R. B. Russell, Protein disorder prediction: Implications for structural proteomics. Structure 11, 1453–1459 (2003). [DOI] [PubMed] [Google Scholar]
- 46.B. Mészáros, G. Erdos, Z. Dosztányi, IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Shen, R. Li, R. Negro, J. Cheng, S. M. Vora, T. M. Fu, A. Wang, K. He, L. Andreeva, P. Gao, Z. Tian, R. A. Flavell, S. Zhu, H. Wu, Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell 184, 5759–5774.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.K. Ullah, E. Zubia, M. Narayan, J. Yang, G. Xu, Diverse roles of the E2/E3 hybrid enzyme UBE2O in the regulation of protein ubiquitination, cellular functions, and disease onset. FEBS J. 286, 2018–2034 (2019). [DOI] [PubMed] [Google Scholar]
- 49.X. Zhang, J. Zhang, A. Bauer, L. Zhang, D. W. Selinger, C. X. Lu, P. ten Dijke, Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 32, 996–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. T. Nguyen, M. A. Prado, P. J. Schmidt, A. K. Sendamarai, J. T. Wilson-Grady, M. Min, D. R. Campagna, G. Tian, Y. Shi, V. Dederer, M. Kawan, N. Kuehnle, J. A. Paulo, Y. Yao, M. J. Weiss, M. J. Justice, S. P. Gygi, M. D. Fleming, D. Finley, UBE2O remodels the proteome during terminal erythroid differentiation. Science 357, eaan0218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. K. Vila, Y. Yao, G. Kim, W. Xia, H. Kim, S. J. Kim, M. K. Park, J. P. Hwang, E. González-Billalabeitia, M. C. Hung, S. J. Song, M. S. Song, A UBE2O-AMPKα2 axis that promotes tumor initiation and progression offers opportunities for therapy. Cancer Cell 31, 208–224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Y. Huang, X. Yang, Y. Lu, Y. Zhao, R. Meng, S. Zhang, X. Dong, S. Xu, G. Wu, UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance. Cell Death Differ. 28, 671–684 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. M. Gianferante, L. Mirabello, S. A. Savage, Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 13, 480–491 (2017). [DOI] [PubMed] [Google Scholar]
- 54.H. Huang, R. Pan, Y. Zhao, H. Li, H. Zhu, S. Wang, A. A. Khan, J. Wang, X. Liu, L3MBTL2-mediated CGA transcriptional suppression promotes pancreatic cancer progression through modulating autophagy. iScience 25, 104249 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D. Liao, L. Zhong, J. Yin, C. Zeng, X. Wang, X. Huang, J. Chen, H. Zhang, R. Zhang, X. Y. Guan, X. Shuai, J. Sui, S. Gao, W. Deng, Y. X. Zeng, J. N. Shen, J. Chen, T. Kang, Chromosomal translocation-derived aberrant Rab22a drives metastasis of osteosarcoma. Nat. Cell Biol. 22, 868–881 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Z. Tang, C. Li, B. Kang, G. Gao, C. Li, Z. Zhang, GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.N. Mashtalir, S. Daou, H. Barbour, N. N. Sen, J. Gagnon, I. Hammond-Martel, H. H. Dar, M. Therrien, E. B. Affar, Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 54, 392–406 (2014). [DOI] [PubMed] [Google Scholar]
- 58.K. Yanagitani, S. Juszkiewicz, R. S. Hegde, UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.X. Liu, F. Ma, C. Liu, K. Zhu, W. Li, Y. Xu, G. Li, Z. Niu, J. Liu, D. Chen, Z. Li, Y. Fu, C. Qian, UBE2O promotes the proliferation, EMT and stemness properties of breast cancer cells through the UBE2O/AMPKα2/mTORC1-MYC positive feedback loop. Cell Death Dis. 11, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. H. Kim, H. J. Yang, C. H. Lee, Y. S. Jeon, J. J. Park, K. W. Lee, J. H. Kim, S. Y. Park, S. J. Song, Y. H. Kim, A. R. Moon, J. H. Lee, Y. S. Song, The positive correlations between the expression of histopathological ubiquitin-conjugating enzyme 2O staining and prostate cancer advancement. Pharmaceuticals 14, 778 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Z. Shi, R. Liu, Q. Lu, Z. Zeng, Y. Liu, J. Zhao, X. Liu, L. Li, H. Huang, Y. Yao, D. Huang, Q. Xu, UBE2O promotes hepatocellular carcinoma cell proliferation and invasion by regulating the AMPKα2/mTOR pathway. Int. J. Med. Sci. 18, 3749–3758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Y. Xu, Z. Zhang, J. Li, J. Tong, B. Cao, P. Taylor, X. Tang, D. Wu, M. F. Moran, Y. Zeng, X. Mao, The ubiquitin-conjugating enzyme UBE2O modulates c-Maf stability and induces myeloma cell apoptosis. J. Hematol. Oncol. 10, 132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. L. J. Lafontaine, J. A. Riback, R. Bascetin, C. P. Brangwynne, The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021). [DOI] [PubMed] [Google Scholar]
- 64.J. Liu, Y. Xie, J. Guo, X. Li, J. Wang, H. Jiang, Z. Peng, J. Wang, S. Wang, Q. Li, L. Ye, Y. Zhong, Q. Zhang, X. Liu, D. M. Lonard, J. Wang, B. W. O’Malley, Z. Liu, Targeting NSD2-mediated SRC-3 liquid-liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat. Commun. 12, 1022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.I. A. Klein, A. Boija, L. K. Afeyan, S. W. Hawken, M. Fan, A. Dall'Agnese, O. Oksuz, J. E. Henninger, K. Shrinivas, B. R. Sabari, I. Sagi, V. E. Clark, J. M. Platt, M. Kar, P. M. McCall, A. V. Zamudio, J. C. Manteiga, E. L. Coffey, C. H. Li, N. M. Hannett, Y. E. Guo, T. M. Decker, T. I. Lee, T. Zhang, J. K. Weng, D. J. Taatjes, A. Chakraborty, P. A. Sharp, Y. T. Chang, A. A. Hyman, N. S. Gray, R. A. Young, Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J.-Q. Yin, J.-N. Shen, W.-W. Su, J. Wang, G. Huang, S. Jin, Q.-C. Guo, C.-Y. Zou, H.-M. Li, F.-B. Li, Bufalin induces apoptosis in human osteosarcoma U-2OS and U-2OS methotrexate300-resistant cell lines. Acta Pharmacol. Sin. 28, 712–720 (2007). [DOI] [PubMed] [Google Scholar]
- 67.L. Zhong, D. Liao, J. Li, W. Liu, J. Wang, C. Zeng, X. Wang, Z. Cao, R. Zhang, M. Li, K. Jiang, Y. X. Zeng, J. Sui, T. Kang, Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct. Target. Ther. 6, 59 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O. Shalem, N. E. Sanjana, E. Hartenian, X. Shi, D. A. Scott, T. S. Mikkelsen, D. Heckl, B. L. Ebert, D. E. Root, J. G. Doench, F. Zhang, Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.W. Li, H. Xu, T. Xiao, L. Cong, M. I. Love, F. Zhang, R. A. Irizarry, J. S. Liu, M. Brown, X. S. Liu, MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D. Liao, L. Zhong, T. Duan, R. H. Zhang, X. Wang, G. Wang, K. Hu, X. Lv, T. Kang, Aspirin suppresses the growth and metastasis of osteosarcoma through the NF-κB pathway. Clin. Cancer Res. 21, 5349–5359 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Y. Wu, L. Zhou, Y. Zou, Y. Zhang, M. Zhang, L. Xu, L. Zheng, W. He, K. Yu, T. Li, X. Zhang, Z. Chen, R. Zhang, P. Zhou, N. Zhang, L. Zheng, T. Kang, Disrupting the phase separation of KAT8-IRF1 diminishes PD-L1 expression and promotes antitumor immunity. Nat Cancer 4, 382–400 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Y. Wu, Q. Yang, L. Xie, C. Yin, H. Qu, J. He, Y. Jiang, T. Li, Possible mechanism of contribution of a secreted aspartic proteinase FpOPSB to the virulence of Fusarium proliferatum causing banana crown rot. Food Frontiers 00, 1–12 (2023). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legends for tables S1 to S12
Tables S1 to S12