Abstract
Genetically encoded voltage indicators (GEVIs) allow the direct visualization of cellular membrane potential at the millisecond time scale. Among these, red-emitting GEVIs have been reported to support multichannel recordings and manipulation of cellular activities with reduced autofluorescence background. However, the limited sensitivity and dimness of existing red GEVIs have restricted their applications in neuroscience. Here, we report a pair of red-shifted opsin-based GEVIs, Cepheid1b and Cepheid1s, with improved dynamic range, brightness, and photostability. The improved dynamic range is achieved by a rational design to raise the electrochromic Förster resonance energy transfer efficiency, and the higher brightness and photostability are approached with separately engineered red fluorescent proteins. With Cepheid1 indicators, we recorded complex firings and subthreshold activities of neurons on acute brain slices and observed heterogeneity in the voltage‑calcium coupling on pancreatic islets. Overall, Cepheid1 indicators provide a strong tool to investigate excitable cells in various sophisticated biological systems.
Genetically encoded red voltage indicators map neuronal and pancreatic action potentials in cultured cells and tissues.
INTRODUCTION
Fluorescent indicators are powerful tools for spatiotemporally resolved mapping of cellular activities. Genetically encoded voltage indicators (GEVIs) allow noninvasive readout of membrane potential changes of large neuronal ensembles at the single-neuron level, thus enabling millisecond–time scale recording of neuronal activities, including subthreshold potentials (1). A common challenge associated with voltage imaging is its high noise level due to limited photon counts, which arises from a combination of the high acquisition frame rate (typically at 0.5 to 2 kHz) and the low copy number of the membrane-embedded sensor protein (1). This high level of imaging noise has a substantial impact on the signal-to-noise ratio (SNR), particularly for voltage imaging in vivo, which is further complicated with the presence of tissue autofluorescence and light scattering (2). For this reason, GEVIs with red-shifted emission spectra are highly sought after, since they avoid much of the autofluorescence window and suffer less from scattering. Moreover, a brighter fluorophore could increase the baseline fluorescence signal, which, in turn, promotes the SNR.
Existing red GEVIs are limited by speed, sensitivity, or brightness. For example, voltage-sensing domain–based GEVIs exhibit response time constants exceeding 5 ms (3, 4); FlicRs (5, 6) have fast kinetics but small dynamic range and dim fluorescence, which limited their performance in tissue. In comparison, rhodopsin-based GEVIs [e.g., QuasArs (7–9) and Archons (10, 11)] feature submillisecond kinetics and high dynamic range [20 to 50% ΔF/F0 per action potential (AP)]; yet, they suffer from low brightness due to their limited fluorescence quantum yield (typically <4%) (10). To improve the molecular brightness, we C-terminally fuse a red fluorescent protein (RFP) donor to a voltage-sensing rhodopsin, which serves as an electrochromic Förster resonance energy transfer (eFRET) quencher. The resulting eFRET GEVIs, including QuasAr2-mRuby2 (12) and voltage-activated red neuronal activity monitors (VARNAMs) (6, 13), exhibit substantially improved brightness while maintaining fast response kinetics and have shown great potential in the in vivo applications despite currently limited in one-photon imaging (14). However, their voltage sensitivities are limited to ~20% ΔF/F0 per 100 mV (6, 12, 13) owing to low FRET efficiencies (15, 16).
Here, we report a pair of red eFRET GEVIs, Cepheid1b and Cepheid1s, with improved voltage response, brightness, and photostability, which enable voltage imaging in mouse brain slices with laser power lower than 2 W/cm2. Both Cepheid indicators support multiplexed imaging with green fluorescent indicators for calcium [GCaMP6s (17)] or glutamate [iGluSnFR (18)]. They are also capable of pairing with CheRiff (7), a spectrally orthogonal blue-shifted optogenetic actuator, to achieve all-optical electrophysiology measurement of neuronal excitability. We further demonstrate that Cepheid indicators report APs and subthreshold potentials ex vivo and in vivo. On acute brain slices, Cepheid1b simultaneously record burst firing and subthreshold depolarization activities in dozens of neurons. On pancreatic islet tissue, we noninvasively observed glucose-stimulated correlations between electrical spiking and calcium oscillations in multiple cells simultaneously by dual-color imaging using Cepheid1b and GCaMP6f.
RESULTS
AlphaFold2-aided design of GEVI scaffolds
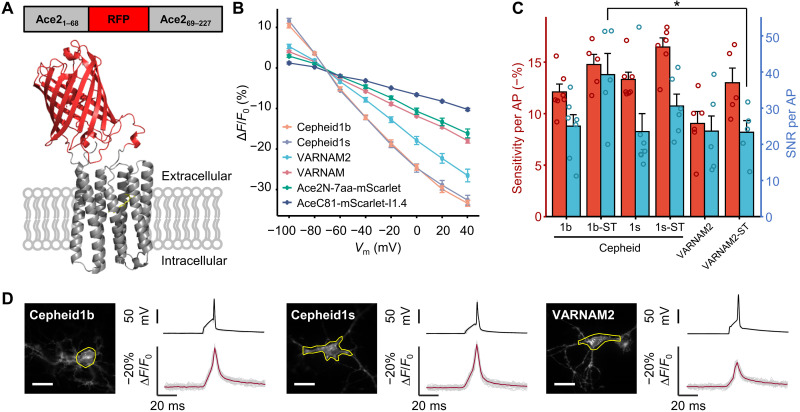
We have shown previously that the voltage responsiveness of eFRET GEVIs depends critically on the baseline FRET efficiency (EFRET) between the fluorescent donor and the retinal acceptor (16, 19). For engineering red eFRET GEVIs, we sought to maximize the EFRET through a combination of approaches. First, we chose the red-shifting Asp81Cys mutation of Ace rhodopsin (AceD81C) as the voltage-sensing module to maximize its spectral overlap with RFP emission (16). Second, we applied AlphaFold2 (20) computational structural modeling to guide our optimization of the donor-acceptor distance R and the orientation factor κ2, which are quantitatively linked to EFRET as described in Eq. 1
| (1) |
Whereas previous eFRET GEVI designs have focused exclusively on the C-terminal fusion of fluorescent protein donors, AlphaFold2 (20) predicts a 4- to 12-Å shorter donor-acceptor distance and substantially higher κ2 when the RFP donor is inserted into the first extracellular loop (ECL1) of the Ace rhodopsin (Fig. 1A, fig. S1, and table S1). Alternatively, insertion into the third intracellular loop (ICL3) is also predicted to improve the overall EFRET. However, with limitations of AlphaFold2 including the inability of predicting non–amino acid fluorophores and the lack of dynamic simulation, the above computational predictions require subsequent experimental evaluations.
Fig. 1. Design and characterization of Cepheid1 indicators in cultured cells.
(A) Diagram showing the red fluorescent protein (RFP) insertion site (top) and the predicted structure of Cepheid1s. (B) Normalized fluorescence-voltage response curves of red genetically encoded voltage indicators (GEVIs). GEVIs names are listed in descending order of measured voltage sensitivities. (C) Voltage sensitivities of red GEVIs for recording action potentials (APs) in cultured neurons. *P < 0.05, two-sample t test. (D) Optical waveforms of Cepheids and VARNAM2 to APs. Region of interest that yielded the trace is marked by yellow circles. Scale bars, 20 μm. VARNAM, voltage-activated red neuronal activity monitors.
Our list of RFP candidates include mRuby3 (21), mRuby4 (22), mScarlet (23), and mScarlet-I1.4, which is an engineered variant of mScarlet-I featuring better folding (fig. S2) and higher brightness (fig. S3 and table S2). To improve the membrane trafficking, we C-terminally fuse a combination of triple-tandem transport signal and endoplasmic reticulum-exiting (ER2) sequences (24) to the rhodopsin (Fig. 1A). All ICL3 insertions fail to properly traffic to the cytoplasmic membrane and were discarded for further analysis (fig. S4). For ECL1 insertions, all RFP-Ace insertions exhibited good expression and membrane trafficking in human embryonic kidney (HEK) 293T cells except for mScarlet, which has poor membrane trafficking (fig. S4). While mScarlet-I1.4 insertion showed the brightest fluorescence (fig. S5), mRuby4 insertion exhibited superior photostability, with half-photobleach time more than 13 min when illuminated with 561-nm laser at 1.6 W/cm2 (fig. S6). Moreover, a preliminary characterization in cultured HEK293T cells revealed high voltage–dependent response and millisecond-level kinetics in both GEVI (fig. S7). We therefore named mScarlet-I1.4 and mRuby4 insertions as Cepheid1b (for brighter fluorescence) and Cepheid1s (for better photostability), respectively.
Characterization of Cepheid indicators in cultured cells
We then applied whole-cell voltage clamping in HEK293T cells to characterize the response kinetics and the dynamic ranges of Cepheid1 indicators. The voltage responsiveness of Cepheid1b and Cepheid1s compares favorably against several recently published red GEVIs, including VARNAM (6), VARNAM2 (13), and Ace2N-7aa-mScarlet (25), which are constructed by fusing mRuby3 and mScarlet to the C terminus of Ace rhodopsin mutants. When the membrane potential is altered stepwise from −70 to +40 mV, the whole-cell fluorescence of VARNAM, VARNAM2, and Ace2N-7aa-mScarlet change by −17.9 ± 0.6% (n = 6 cells), −26.7 ± 1.7% (n = 7 cells), and −16.0 ± 1.0% (n = 6 cells) ΔF/F0, respectively (Fig. 1B and fig. S8). In comparison, Cepheid1b (−32.9 ± 1.0%, n = 6 cells) and Cepheid1s (−33.6 ± 1.5%, n = 8 cells) are nearly twofold as sensitive as VARNAM and Ace2N-7aa-mScarlet and significantly higher than VARNAM2 (−26.7 ± 1.7%, n = 6 cells) with both P values less than 0.05. When applied with hyperpolarization from −70 to −100 mV, the fluorescence response of Cepheid1b (10.4 ± 0.6%) and Cepheid1s (11.6 ± 0.6%) are about onefold higher than VARNAM (4.0 ± 0.2%) and VARNAM2 (5.3 ± 0.5%). Notably, fusing mScarlet-I1.4 to the C terminus of AceC81 reduces the voltage-dependent response by more than twofold, which supports the AlphaFold2 computational modeling prediction (Fig. 1B).
We applied Cepheid1 indicators to image APs in cultured rat hippocampal neurons. Both indicators exhibited good expression and membrane localization in the soma and neurites (Fig. 1D). The sensitivity of Cepheid1b (−12.1 ± 0.8%) and Cepheid1s (−13.3 ± 0.7%) toward APs (ΔF/F0) are more than 34% higher than VARNAM2 (9.0 ± 1.2%) (Fig. 1C). To improve the imaging quality in brain tissue by reducing dendritic fluorescence, we fused Cepheid1 indicators with a 63–amino acid somatic targeting (ST) domain from the voltage-gated potassium channel Kv2.1, which restricts their expression to the soma and proximal dendrites (Fig. 2A and fig. S9) (26). The resulting soma-targeted Cepheid1b-ST exhibit approximately 40% improved SNR (39.2 ± 5.9) (Fig. 1C), which is substantially higher than that of VARNAM2-ST (23.2 ± 3.2), the soma-targeted variant of VARNAM2.
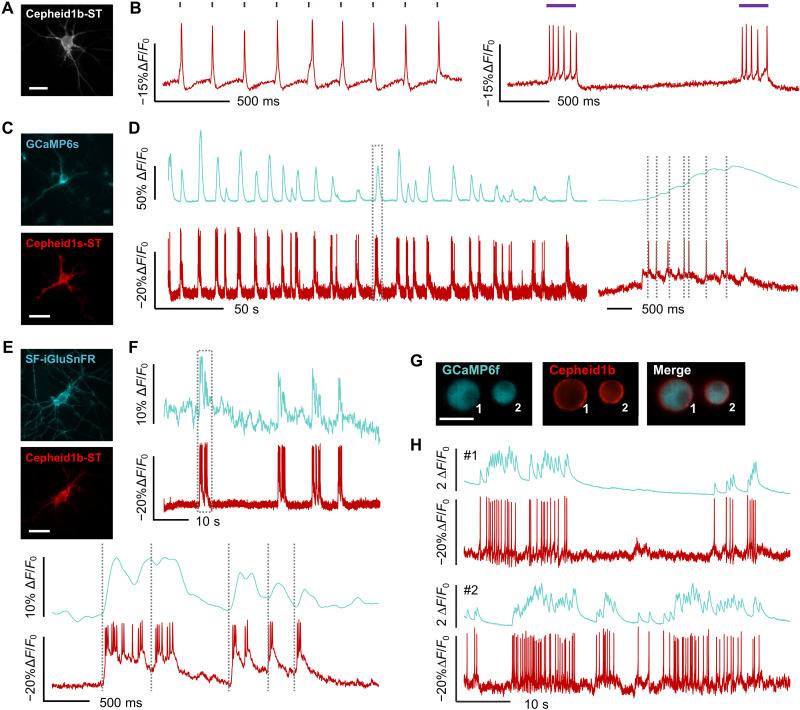
Fig. 2. Multiplexed imaging of Cepheid1b/s with optogenetic tools and blue-shifted biosensors.
(A and B) Epifluorescence image (A) and whole-cell fluorescence response (B) of cultured neuron expressing Cepheid1b-ST-P2A-CheRiff, when optogenetically triggered with 405-nm laser. (C and D) Epifluorescence images (C) and whole-cell fluorescence traces (D) of cultured rat hippocampal neuron expressing GCaMP6s-NES (top) and Cepheid1s-ST (bottom), with zoom-in view of the boxed region shown on the right. (E and F) Epifluorescence images (E) and whole-cell fluorescence traces (F) of cultured rat hippocampal neuron expressing SF-iGluSnFr (top) and Cepheid1b-ST (bottom), with zoom-in view of the boxed region shown at the bottom. (G and H) Epifluorescence images (G) and whole-cell fluorescence traces (H) of mouse pancreatic islet cells expressing GCaMP6f (cyan) and Cepheid1b (red). Scale bars, 20 μm.
Multiplexed recording and manipulation of neuronal activity
The red-shifted spectra of Cepheid1 indicators allow the combination with the photosensitive cation channel CheRiff (7) for all-optical electrophysiology. Patch clamp tests revealed minimal photocurrent of Cepheid1b and Cepheid1s under various illumination conditions (fig. S10), with one exception that when coilluminated with 405 and 561 nm, both GEVIs produce a depolarizing current of ~20 pA. It has been reported that CheRiff can be partially excited by 561-nm illumination at an intensity of 1.5 W/cm2 (6), eliciting a photocurrent of approximately 30 pA. Therefore, in neurons coexpressing Cepheid1b and CheRiff via a self-cleaving 2A peptide, we tested the photocurrent and depolarization caused by 561-nm cross-talk (fig. S11). Under 561-nm illumination at imaging level, we observed a substantially lower photocurrent of ~10 pA and a depolarization of ~2.5 mV. The lower photocurrent may be attributed to the lower expression of CheRiff when coexpressed with Cepheid1b via 2A peptide linkage (27). A leakage at this low level is insufficient to trigger unwanted firing or opening of ion channels and is therefore compatible to all-optically recording neuronal activities.
In cultured rat hippocampal neurons coexpressing CheRiff and Cepheid1b/s-ST, we applied 405-nm laser to stimulate AP firing while simultaneously imaging membrane voltage (Fig. 2A). Under various stimulation patterns, Cepheid1b-ST faithfully recorded tonic APs and burst firing (Fig. 2B). We tested the refractory period of neuronal firing with 10 s of ramped increasing optogenetic stimulation strength. The firing rate during the trial can be overlapped with optogenetic stimulation dosage, while the maximum firing rate is shown as a platform (fig. S12). We then conducted long-term imaging with the photostable Cepheid1s-ST. We found that a few minutes of recovery after continuous recording can notably restore the fluorescence level and reduce phototoxicity. Using this method, Cepheid1s-ST can faithfully report optogenetically induced APs for more than 15 min accumulatively (fig. S13). In some cases, 405-nm light directly causes observable artifact on Cepheid fluorescence level (fig. S11), which explains the fluctuations of the baseline in the ramp experiment (figs. S12 and S13).
We further paired Cepheid1b/s-ST with green-emitting indicators to simultaneously record membrane potential with other neuronal signals such as cytoplasmic calcium and extracellular glutamate. We coexpressed Cepheid1b/s-ST with GCaMP6s (17) in primary rat hippocampal neurons (Fig. 2C and fig. S14). The high photostability of Cepheid1s-ST allowed continuous imaging of time-correlated voltage and calcium spikes for 5 min (Fig. 2D). We also applied Cepheid1b-ST and SF-iGluSnFR (28) to record voltage-induced glutamate release (Fig. 2, E and F, and fig. S15). In addition, the high brightness of Cepheid1b and its spectral orthogonality with GCaMP6f have enabled us to sensitively record membrane potential and cytoplasmic calcium in cultured mouse pancreatic islet cells, which unveils time-correlated spontaneous calcium waves and AP spikes that are reminiscent to neuronal activities (Fig. 2, G and H).
Simultaneous recording of multiple neurons on acute brain slices
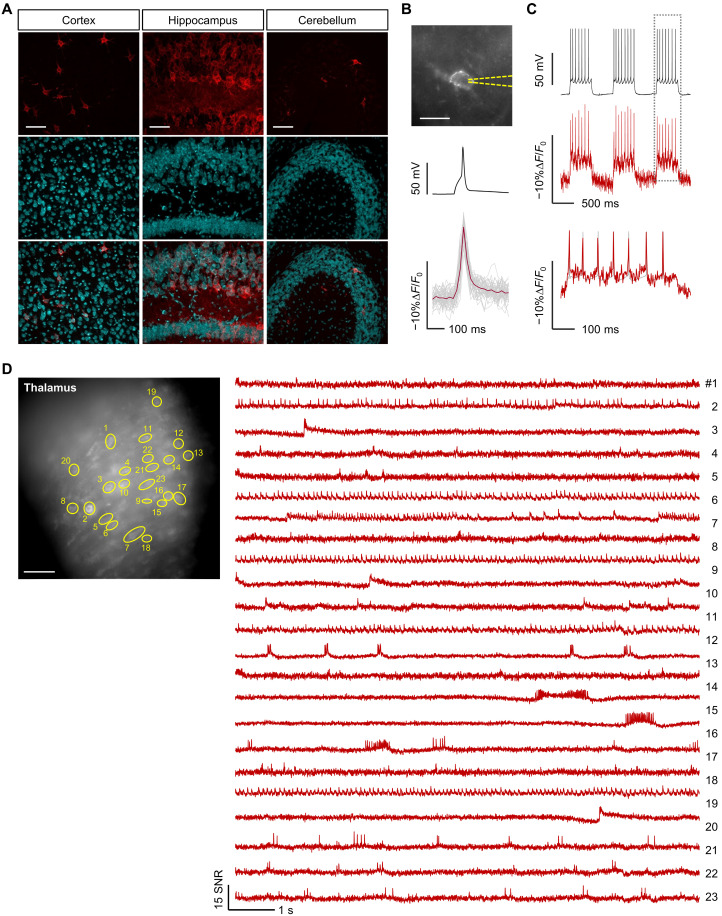
We performed voltage imaging in acute mouse brain slices to assess the ex vivo performance of Cepheid1. We introduced Cepheid1b-ST into mouse brain via adeno-associated virus (AAV) infection, which showed good expression and membrane trafficking across multiple brain regions, including cortex, hippocampus, and cerebellum (Fig. 3A). In acute brain slices, the high sensitivity of Cepheid1b-ST (ΔF/F0 = −9.9 ± 0.3% per AP; Fig. 3B) has allowed faithful report of APs in single trial measurements at the frame rate of 498 Hz. Cepheid1b-ST also resolved current-induced subthreshold and burst APs at 14 Hz with a sensitivity of ~−10% ΔF/F0 (Fig. 3C).
Fig. 3. Voltage imaging with Cepheid1b indicators in acute brain slice.
(A) Confocal images of fixed slices showing expression and localization of Cepheid1b-ST. Scale bars, 50 μm. (B) Fluorescence image (top) and mean electrical (black) and optical (red) waveforms of stimulated action potentials (APs) recorded from a hippocampal neuron expressing Cepheid1b-ST in acute brain slice. Gray and red lines indicate individual and mean optical traces, respectively (n = 48 spikes). Scale bar, 10 μm. (C) Fluorescence response (red) of Cepheid1b-ST to single-trial burst firing (black) in acute brain slice, with zoom-in view of the boxed region shown at the bottom. (D) Representative epifluorescence image of mouse brain slice expressing Cepheid1b-ST in the thalamus (scale bar, 20 μm) (left) and signal-to-noise ratio (SNR) traces indicating spontaneous activity from the labeled cells (right).
To simultaneously resolve the electrophysiology across a large neuronal population, we expressed Cepheid1b-ST in the whole brain through delivery with AAV-PHP.eB vector that can cross the blood-brain barrier. In the thalamus, Cepheid1b-ST enables the simultaneous recording of spontaneous neuronal AP dynamics in 23 cells across a large field of view measuring 504 μm by 504 μm (Fig. 3D). The expression of Cepheid1b-ST throughout the brain facilitates the collection of signals from multiple brain areas, such as thalamus (Fig. 3D), habenula (fig. S16), and hippocampus (fig. S17) in a single trail. The resting state and excitability of neurons both were captured, and the neuronal spikings including tonic and burst APs were obtained with an SNR of ~10.
Imaging of electric‑calcium coupling in pancreatic islets
In mammalian pancreatic islets, elevated glucose concentration initiates electrical activities in β cells via the concerted actions of Glut2 transporter, KATP channels and voltage-gated Ca2+ channels (29). The opening of voltage-gated Ca2+ channels underlies the basis for AP firing, increasing intracellular calcium levels to trigger the exocytosis of insulin granules (30). Notably, glucose-induced oscillations of intracellular Ca2+ concentration have been observed in mammalian pancreatic islets (31–34), and patch clamp has been used to study the relationship between membrane potential and [Ca2+]i (35). However, simultaneous noninvasive recording of membrane potential and [Ca2+]i in multiple cells has remained challenging. Furthermore, while electrical coupling between β cells in mammalian islets (36) and oscillatory membrane potential in individual pancreatic β cells (37) has been measured using patch clamp, the spatial patterns of these coupling and oscillation have not been recorded because of the lack of high-throughput detection methods. Moreover, since patch clamp can only access cells on the islet surface, which are mainly β cells (38), cells deeper in the islet have eluded from the previous analysis.
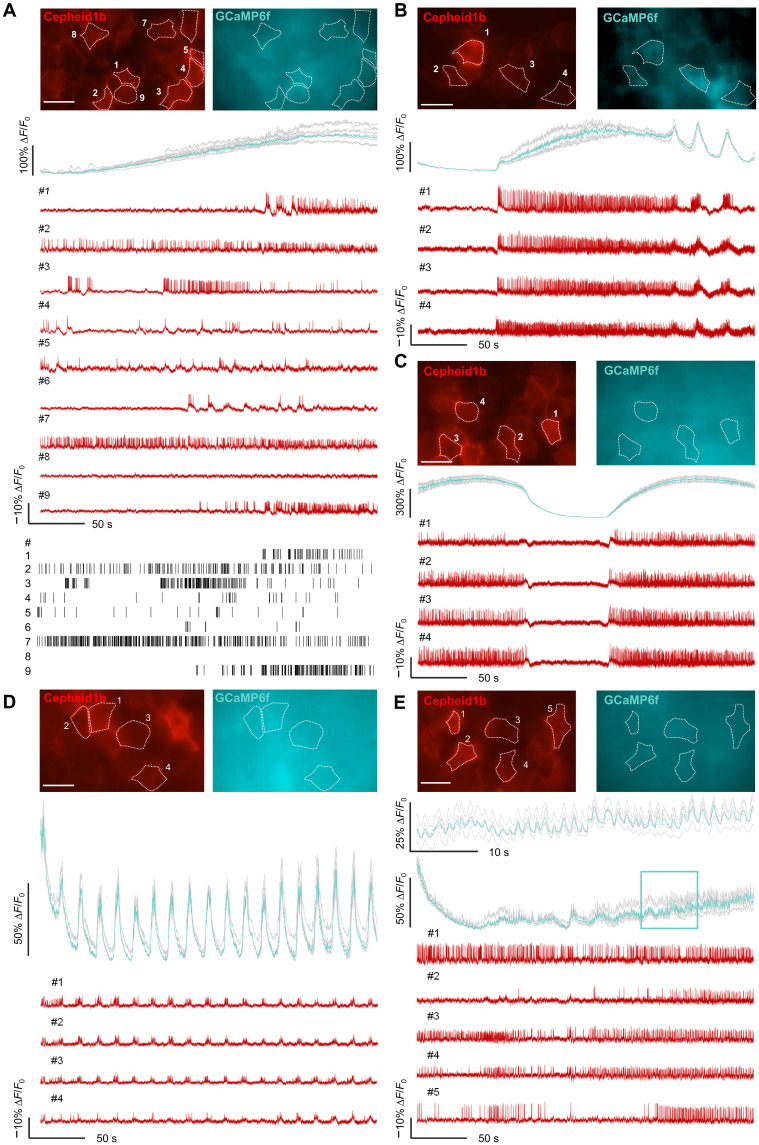
The high brightness of Cepheid1b and its spectral orthogonality with GCaMP6f enables simultaneous recording of membrane potential and cytoplasmic calcium with higher throughput and better access to cells deeper in the islet. We thus infected GCaMP6f+/+ mice with adenovirus encoding Cepheid1b and performed dual-color imaging in isolated pancreatic islets. Upon switching from low (3 mM) to high (10 mM) levels of extracellular glucose, we observed highly heterogeneous patterns of electrical activities in individual islet cells, whereas their calcium signal increased gradually and synchronously over the time course of minutes (Fig. 4A). The electrical activities in individual cells appear uncorrelated with the calcium signal. Following 30 to 60 min of incubation in high-glucose medium, three types of calcium oscillations emerged: fast (~20-s cycle), slow (>100-s cycle), and mixed (20- to 300-s cycle), which were consistent with the previous observations (31–34). The electrical activities of islet cells are spontaneously tuned to be highly synchronized and time-correlated with all three types of calcium oscillations (Fig. 4, B to D). In addition, in a few cases of very fast calcium oscillations (1- to 2-s cycle), electrical activity appeared highly heterogeneous, indicating relatively weak electrical coupling in this type of calcium oscillations (Fig. 4E). Together, the above data demonstrate the power of simultaneously recording electrical and calcium activities in multiple cells in pancreatic islets.
Fig. 4. Voltage imaging with Cepheid1b indicators in pancreatic islets ex vivo.
(A to E) Representative dual-color imaging of calcium (mean, cyan; individual, gray) and voltage (red) in isolated islets at 200-Hz frame rate. At the onset of imaging, extracellular glucose is switched from 3 to 10 mM (A). Following 30 to 60 min of incubation at 10 mM glucose, both mixed (B), slow (C), and fast (D) calcium oscillations are observed together with time-correlated electrical spikes, while weaker electrical coupling is observed with very fast (E) calcium oscillation (zoom-in view of framed region is shown at the top). Scale bars, 20 μm.
DISCUSSION
To summarize, we have reported a pair of red GEVIs with improved voltage dynamic range of 33% −ΔF/F0 per 110 mV, which is the highest among red GEVIs reported to date. The high dynamic range of Cepheid1 allows sensitive detection of APs at 500-Hz frame rate in both cultured neurons (ΔF/F0 = −12%) and brain slices (ΔF/F0 = −10%) under mild illumination with 561-nm laser lower than 2 W/cm2. While Cepheid1b is useful for detecting both subthreshold and APs as a brighter indicator, Cepheid1s enables long-term imaging due to its high photostability. Both GEVIs support multiplexed imaging with green fluorescent indicators and all-optical electrophysiology. Currently, applications of Cepheid indicators are likely restricted to one-photon imaging regime, a limitation shared by all opsin-based GEVIs developed to date. However, we demonstrated that Cepheid1b faithfully reports spike firing in acute mouse brain slices and pancreatic islets, showing its capability for tissue imaging and potential in vivo applications.
Unlike previous engineering efforts that focused exclusively on C-terminal RFP fusions, we achieved higher-voltage responsiveness through insertion into ECL1. This design leveraged the predictive power of computational modeling of chimeric protein structures. In recent years, artificial intelligence models such as AlphaFold or Rosetta are beginning to show their usefulness in understanding protein function and assisting protein design (20, 39, 40). In this work, we also harnessed the power of AlphaFold2 in predicting unknown protein structure and thus estimating the performance of our voltage indicators. However, despite the intuition and inspiration we acquired with AlphaFold2, some of the intrinsic properties of the model limited the reliability of the predictions. First, AlphaFold2 can only take amino acid sequence as the input, and non–amino acid molecules such as fluorophores can only be aligned onto the predicted tertiary structure using crystallography data, which undoubtedly lowered the accuracy (20). Second, the stationary output also created a bias on the assessment of FRET efficiency, which, in the real case, is determined by a dynamic ensemble. Therefore, future improvement would likely involve combining structural prediction models with molecular dynamic simulations.
We envision that the sensitivity of Cepheid1 indicators could be further optimized through directed evolution. On the one hand, the mutation we introduced to Ace rhodopsin domain is directly inherited from a previous work where a short peptide is inserted into the ECL1 region (16). This mutation might not be optimal for our scaffold, and we expect a further improved sensitivity in the future library screening; meanwhile, with several published work with plenty of mutation library data of Ace rhodopsin, engineering positive-responding GEVI is also a promising direction (13, 41). On the other hand, a next-step engineering on mScarlet-I1.4 is also vital for a better performance. We inserted the original mScarlet-I1.4 without circular permutation, which might have caused a compromise in fluorescence intensity compared with C-terminal fusion. To improve this, we propose an optimization in the RFP insertion site, flanking linker sequence, and circular permutation of mScarlet-I1.4.
Cepheid indicators have enabled the visualization of electrical coupling in multiple pancreatic islet cells, revealing synchronized oscillatory cellular membrane potentials that are time-correlated with slow and fast calcium oscillations. The synchronization in membrane potential is substantially weaker either in the absence of calcium oscillation or at very fast calcium oscillation. Future investigation into the underlying mechanism of calcium-sensitive electrical coupling could benefit from voltage imaging-guided single-cell sequencing to analyze the heterogeneous response among islet cells. Furthermore, the genetically encoded property of Cepheid indicators allows for the labeling of specific types of pancreatic islet cells, providing a possible way to solve the debate about whether glucose depolarize or hyperpolarize islet α cells (42).
MATERIALS AND METHODS
Materials and reagents
The reagents used in this study are summarized in table S3. All animal procedures were approved by the Animal Center of Peking University, and the experiments were carried out in accordance with the guidelines of Institutional Animal Care and Use Committee of Peking University.
Molecular cloning
Plasmids used in this study were generated by Gibson Assembly ligating polymerase chain reaction amplified inserted DNA fragment and linearized vector with 25–base pair overlap. The primers for polymerase chain reaction are summarized in table S4. DNA fragments and linearized vector were mixed with Gibson Assembly enzyme (Lightening Cloning Kit). The sequences of plasmids were verified through Sanger sequencing.
Computational modeling of FRET efficiency
The structures of Cepheid1b and Cepheid1s are predicted with AlphaFold2. Non–amino acid chromophores are manually added via structural alignment of the predicted structures against the crystallography data of separate protein domains: Acetabularia rhodopsin II [Protein Data Bank (PDB) ID: 3AM6], mScarlet (PDB ID: 5LK4), and mRuby (PDB ID: 3U0L) as proxies of mScarlet-I1.4 and mRuby4, respectively. The coordinates of fluorophores are used for calculating the distance R (defined as the distance between the centers of mass of donor and acceptor) and the orientation factor κ2, which is defined as κ2 = [(D · A) – 3(D · R)(A · R)]2, where R, D, and A are vectors connecting the centers of mass of donor and acceptor, normalized longitudinal axis of donor and acceptor, respectively.
Expression of Cepheid1 in cultured cells
HEK293T cells were seeded in a 24-well plate and incubated in Dulbecco’s modified Eagle medium (Gibco) containing 10% (v/v) fetal bovine serum (Gibco) at 37°C with 5% CO2. Cells were transfected with Lipofectamine 3000 reagent following the manufacturer’s instructions. Primary rat hippocampal neurons were digested from dissected rat brain at postnatal day 0 and plated onto sterile 14-mm glass coverslips precoated with poly-d-lysine (20 μg/ml) and laminin (10 μg/ml). Neurons were incubated at 37°C with 5% CO2 and were transfected on DIV7 (7 days in vitro) to DIV9 with Lipofectamine 3000 following the manufacturer's instructions. Transfected neurons were imaged after 3 to 7 days.
Imaging apparatus
Fluorescence microscopy in cultured cells and pancreatic islets was performed on an inverted microscope (Nikon-TiE) equipped with a 40×, numerical aperture (NA) of 1.3 oil immersion objective lens, three laser lines (Coherent OBIS; 405, 488, and 561 nm), a spinning disk confocal unit (Yokogawa CSU-X1) and two scientific complementary metal-oxide semiconductor cameras (Hamamatsu ORCA-Flash 4.0 v2). A dual-view device (Photometrics DV2) was used to split the emission into green/red fluorescence channels. Fluorescence imaging experiments in acute slices were performed on an upright microscope (Olympus BX51WI) equipped with a 40×, NA 0.8 water immersion objective lens, a 561-nm laser line (Coherent OBIS) and a scientific complementary metal-oxide semiconductor camera (Hamamatsu ORCA-Flash 4.0 v2). The spectra properties of the filters and dichroic mirrors for various fluorescent indicators used in this study are summarized in table S5.
Fluorescence voltage imaging in HEK293T cells and cultured neurons
To measure the dynamic range and kinetics of the voltage indicators in HEK293T cells, the membrane potential was controlled via whole-cell patch clamp (Axopatch 200B, Axon Instruments). Fluorescence imaging was performed at the frame rate of 1058 Hz. To measure the response of indicators to APs, neurons were current-clamped and injected with 200 to 500 pA of current for 5 to 10 ms to stimulate AP firing. Fluorescence imaging was performed at the frame rate of 484 Hz. For simultaneous imaging of membrane voltage and calcium, Cepheid1 and GCaMP6s–nuclear export signal (NES) were cotransfected into neurons at DIV8. On DIV14 to DIV18, neurons were illuminated with 488-nm (2.4 W/cm2) and 561-nm (1.6 W/cm2) lasers and imaged with a dual-view device at a frame rate of 500 Hz. For simultaneous imaging of membrane voltage and glutamate, Cepheid1 and iGluSnFR were cotransfected into neurons on DIV8 and imaged on DIV14 to DIV19 in dual-view mode (2.4 W/cm2 of 488-nm laser and 1.6 W/cm2 of 561-nm laser) at a frame rate of 500 Hz.
Electrophysiology for cultured cells
Borosilicate glass capillaries (BF150-86-10, Sutter) were pulled with a micropipette puller (P-1000, Sutter) and the pipette resistance was 4 to 8 megohm. The extracellular solution for whole-cell patch clamp contains 125 mM NaCl, 2.5 mM KCl, 15 mM Hepes, and 30 mM glucose (305 to 310 mosmol/kg; pH 7.3). The intracellular solution contains 125 mM potassium gluconate, 8 mM NaCl, 1 mM EGTA, 10 mM Hepes, 0.6 mM MgCl2, and 0.1 mM CaCl2 (295 mosmol/kg; pH 7.3). Whole-cell current was recorded with an Axopatch 200B amplifier (Axon Instruments). Membrane voltage signal recorded from the patch amplifier was filtered with an internal 5-kHz Bessel filter and digitized at 9681.48 Hz with a National Instruments PCIe-6353 data acquisition board (approximately twice the bandwidth of the Bessel filter).
All-optical electrophysiology
Cultured hippocampal neurons were transfected with plasmid encoding CheRiff and Cepheid1b/s-ST mediated by P2A self-cleaving peptide. Transfected neurons were activated with repeated 2-ms (0.5 W/cm2) 405-nm light pulses, repeated 250- to 300-ms (0.1 to 0.3 W/cm2) 405-nm light steps, or gradually enhanced (from 0 to 0.2 W/cm2 in 10 s) 405-nm light. Images were captured at a frame rate of 500 Hz under constant 561-nm illumination (1.6 W/cm2). For each trial, 25 μM d,l-2-amino-5-phosphonovaleric acid (AMPA), 10 μM 2,3-dioxo-6-nitro-7-sulfamoyl-benzo[f]quinoxaline (NBQX), and 20 μM gabazine were added into extracellular solution to block synaptic transmission.
Intracerebroventricular injection and acute slice measurements
C57BL/6N mouse lines were purchased from Charles River. The AAV vector expressing Cepheid1b-ST: AAV2/PHP.eb-hsyn-Cepheid1b-ST was custom-produced by Chinese Institute for Brain Research. For one 6- to 7-week-old mouse (without regard to sex), 3 μl of AAV2/PHP.eb-hsyn-Cepheid1b-ST (4.8 × 1012 genome copies/ml) was injected into the lateral ventricle. The coordinate for intracerebroventricular injection (in millimeters from Bregma: anteroposterior and mediolateral) was −0.58 to −0.59 and 1.35 to 1.4 and dorsoventral of 1.8 to 1.9 mm.
Acute slices were prepared from 9- to 11-week-old mice (at least 3 weeks after AAV injection). Mouse was deeply anesthetized via isoflurane inhalation and rapidly decapitated. The brain was dissected from the skull and placed in ice-cold artificial cerebrospinal fluid (ACSF) containing 26 mM NaHCO3, 1.25 mM NaH2PO4, 125 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM KCl, and 20 mM glucose (295 mosmol/kg; pH 7.3 to 7.4) and saturated with Carbogen (95% O2 and 5% CO2). The brain was sliced into 250-μm sections with a Leica VT1200s vibratome. Slices were incubated for 45 min at 34.5°C in ACSF and maintained at room temperature (22°C). ACSF was continuously bubbled with Carbogen for the duration of the preparation and subsequent experiment.
For fluorescence imaging and electrophysiology recoding, slices were placed on a custom-built chamber and held by a platinum harp net stretched across. Carbogen-bubbled ACSF was perfused at a rate of 4 ml/min with a longer peristaltic pump. Fluorescence images were captured at a frame rate of 400 to 500 Hz, under 561-nm illumination (2 W/cm2). The intracellular solution for whole-cell patch clamp contains 105 mM potassium gluconate, 30 mM KCl, 4 mM Mg adenosine 5′-triphosphate, 0.3 mM Na2 guanosine 5′-triphosphate, 0.3 mM EGTA, 10 mM Hepes, and 10 mM sodium phosphocreatine (295 mosmol/kg; pH 7.3). Electrophysiology measurements were acquired with a MultiClamp 700B amplifier (Molecular Devices).
Isolation of mouse pancreatic islets and infection of Cepheid1b
Islets of Langerhans were isolated from GCaMP6f+/+ mice. After isolation, the islets were cultured overnight in RPMI 1640 medium containing 10% fetal bovine serum, 8 mM d-glucose, penicillin (100 U/ml), and streptomycin (100 mg/ml) for overnight culture at 37°C in a 5% CO2-humidified air atmosphere. The islets were infected with adenoviruses pAdeno-CMV-Cepheid1b by 1.5-hour exposure in 100 μl of culture medium, with approximately 4 × 109 plaque forming units per islet, followed by addition of regular medium and further culture for 16 to 20 hours before use.
Dissociation into islet single cells
Freshly isolated islets were washed with Hanks’ balanced salt solution and subsequently digested with 0.025% trypsin-EDTA for 3 min at 37°C, followed by brief shaking. Digestion was stopped with addition of culture medium, and the solution was centrifuged at 94g for 5 min. The cells were suspended by RPMI 1640 culture medium. The cell suspension was plated on coverslips in the poly-l-lysine–coated glass bottom dish (D35-14-1-N, Cellvis) or a microfluidic chip. The dishes or chips were then kept for 60 min in the culture incubator at 37°C and 5% CO2 to allow cells to adhere. Additional culture medium was then added, and the cells were cultured for 24 hours before the imaging experiments.
Voltage imaging in mouse pancreatic islets and islet single cells
For simultaneous imaging with Cepheid1b and GCaMP6f, mouse pancreatic islets and mouse islet single cells were illuminated with 488- and 561-nm lasers at 0.2 to 0.5 W/cm2 and 0.9 to 1.8 W/cm2, respectively, and continuously imaged for 60 to 300 s at a camera frame rate of 200 Hz. The samples were kept at 37°C on the microscope stage during imaging. For mouse islets, a polydimethylsiloxane microfluidic chip was used to provide a stable and controllable environment for long-term imaging. The reagents were automatically pumped into the microfluidic chip with a flow rate of 800 μl/hour by the TS-1B syringe pump (LongerPump). Before imaging, the chip and all the solution were degassed with a vacuum pump for 5 min to achieve stable hour-long imaging. The microfluidic chip was pre-filled with KRBB solution [125 mM NaCl, 5.9 mM KCl, 2.56 mM CaCl2, 1.2 mM MgCl2, 1 mM l-glutamine, 25 mM Hepes, and 0.1% bovine serum albumin (pH 7.4)] containing 3 mM d-glucose before use.
Data analysis
Most fluorescence images and electrophysiology recordings were analyzed with home-built software written in MATLAB (MathWorks, version R2022a). The fluorescence images obtained in acute slices were preprocessed with a Python-based voltage imaging data analysis package (VolPy). Fluorescence intensities were extracted from the mean values over a manually drawn region of interest around the soma of each labeled cell. Statistical analysis was performed with Origin (version 2019b) and R (version 4.2.0).
Acknowledgments
We thank A. E. Cohen (Harvard University) for discussions. P.Z. is sponsored by Bayer Investigator Award (Peking University).
Funding: This work was supported by the Ministry of Science and Technology (2018YFA0507600 and 2022YFA1304700), the National Natural Science Foundation of China (32088101).
Author contributions: J.Y. and P.Z. conceived the project. Y.H., J.Y., Y.L., Y.C., and P.Z. designed the experiments. Y.H., J.Y., Y.L., Y.C., H.R., R.D., W.Q., K.R., B.X., and M.D. performed the experiments. Y.X. and J.C. engineered and characterized mScarlet-I1.4. Y.H., J.Y., and P.Z. analyzed data and wrote the paper with inputs from other authors.
Competing interests: The authors declare that they no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The amino acid sequences of Cepheid1b/1 s indicators are summarized in table S6. Requests for the plasmids used in this study should be submitted to P.Z.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Tables S1 to S6
REFERENCES AND NOTES
- 1.Y. Bando, C. Grimm, V. H. Cornejo, R. Yuste, Genetic voltage indicators. BMC Biol. 17, 71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Y. Xu, P. Zou, A. E. Cohen, Voltage imaging with genetically encoded indicators. Curr. Opin. Chem. Biol. 39, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.W. Akemann, H. Mutoh, A. Perron, Y. K. Park, Y. Iwamoto, T. Knopfel, Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 108, 2323–2337 (2012). [DOI] [PubMed] [Google Scholar]
- 4.H. Tsutsui, S. Karasawa, Y. Okamura, A. Miyawaki, Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods 5, 683–685 (2008). [DOI] [PubMed] [Google Scholar]
- 5.A. S. Abdelfattah, S. L. Farhi, Y. Zhao, D. Brinks, P. Zou, A. Ruangkittisakul, J. Platisa, V. A. Pieribone, K. Ballanyi, A. E. Cohen, R. E. Campbell, A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 36, 2458–2472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.M. Kannan, G. Vasan, C. Huang, S. Haziza, J. Z. Li, H. Inan, M. J. Schnitzer, V. A. Pieribone, Fast, in vivo voltage imaging using a red fluorescent indicator. Nat. Methods 15, 1108–1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D. R. Hochbaum, Y. Zhao, S. L. Farhi, N. Klapoetke, C. A. Werley, V. Kapoor, P. Zou, J. M. Kralj, D. Maclaurin, N. Smedemark-Margulies, J. L. Saulnier, G. L. Boulting, C. Straub, Y. K. Cho, M. Melkonian, G. K.-S. Wong, D. J. Harrison, V. N. Murthy, B. L. Sabatini, E. S. Boyden, R. E. Campbell, A. E. Cohen, All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Adam, J. J. Kim, S. Lou, Y. Zhao, M. E. Xie, D. Brinks, H. Wu, M. A. Mostajo-Radji, S. Kheifets, V. Parot, S. Chettih, K. J. Williams, B. Gmeiner, S. L. Farhi, L. Madisen, E. K. Buchanan, I. Kinsella, D. Zhou, L. Paninski, C. D. Harvey, H. Zeng, P. Arlotta, R. E. Campbell, A. E. Cohen, Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.H. Tian, H. C. Davis, J. D. Wong-Campos, P. Park, L. Z. Fan, B. Gmeiner, S. Begum, C. A. Werley, G. B. Borja, H. Upadhyay, H. Shah, J. Jacques, Y. Qi, V. Parot, K. Deisseroth, A. E. Cohen, Video-based pooled screening yields improved far-red genetically encoded voltage indicators. 10.1101/2021.11.22.469481Nat. Methods 20, 1082–1094 (2023). [DOI] [PMC free article] [PubMed]
- 10.K. D. Piatkevich, E. E. Jung, C. Straub, C. Linghu, D. Park, H. J. Suk, D. R. Hochbaum, D. Goodwin, E. Pnevmatikakis, N. Pak, T. Kawashima, C. T. Yang, J. L. Rhoades, O. Shemesh, S. Asano, Y. G. Yoon, L. Freifeld, J. L. Saulnier, C. Riegler, F. Engert, T. Hughes, M. Drobizhev, B. Szabo, M. B. Ahrens, S. W. Flavell, B. L. Sabatini, E. S. Boyden, A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.K. D. Piatkevich, S. Bensussen, H. A. Tseng, S. N. Shroff, V. G. Lopez-Huerta, D. Park, E. E. Jung, O. A. Shemesh, C. Straub, H. J. Gritton, M. F. Romano, E. Costa, B. L. Sabatini, Z. Fu, E. S. Boyden, X. Han, Population imaging of neural activity in awake behaving mice. Nature 574, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. Zou, Y. Zhao, A. D. Douglass, D. R. Hochbaum, D. Brinks, C. A. Werley, D. J. Harrison, R. E. Campbell, A. E. Cohen, Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 5, 4625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Kannan, G. Vasan, S. Haziza, C. Huang, R. Chrapkiewicz, J. Luo, J. A. Cardin, M. J. Schnitzer, V. A. Pieribone, Dual-polarity voltage imaging of the concurrent dynamics of multiple neuron types. Science 378, eabm8797 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. Chamberland, H. H. Yang, M. M. Pan, S. W. Evans, S. Guan, M. Chavarha, Y. Yang, C. Salesse, H. Wu, J. C. Wu, T. R. Clandinin, K. Toth, M. Z. Lin, F. St-Pierre, Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6, e25690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Xu, L. Peng, S. Wang, A. Wang, R. Ma, Y. Zhou, J. Yang, D. E. Sun, W. Lin, X. Chen, P. Zou, Hybrid indicators for fast and sensitive voltage imaging. Angew. Chem. Int. Ed. Engl. 57, 3949–3953 (2018). [DOI] [PubMed] [Google Scholar]
- 16.S. Liu, C. Lin, Y. Xu, H. Luo, L. Peng, X. Zeng, H. Zheng, P. R. Chen, P. Zou, A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat. Chem. 13, 472–479 (2021). [DOI] [PubMed] [Google Scholar]
- 17.T. W. Chen, T. J. Wardill, Y. Sun, S. R. Pulver, S. L. Renninger, A. Baohan, E. R. Schreiter, R. A. Kerr, M. B. Orger, V. Jayaraman, L. L. Looger, K. Svoboda, D. S. Kim, Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.J. S. Marvin, B. G. Borghuis, L. Tian, J. Cichon, M. T. Harnett, J. Akerboom, A. Gordus, S. L. Renninger, T. W. Chen, C. I. Bargmann, M. B. Orger, E. R. Schreiter, J. B. Demb, W. B. Gan, S. A. Hires, L. L. Looger, An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Liu, J. Yang, P. Zou, Bringing together the best of chemistry and biology: Hybrid indicators for imaging neuronal membrane potential. J. Neurosci. Methods 363, 109348 (2021). [DOI] [PubMed] [Google Scholar]
- 20.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Zidek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B. T. Bajar, E. S. Wang, A. J. Lam, B. B. Kim, C. L. Jacobs, E. S. Howe, M. W. Davidson, M. Z. Lin, J. Chu, Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Xu, M. Deng, S. Zhang, J. Yang, L. Peng, J. Chu, P. Zou, Imaging neuronal activity with fast and sensitive red-shifted electrochromic FRET indicators. ACS Chem. Nerosci. 10, 4768–4775 (2019). [DOI] [PubMed] [Google Scholar]
- 23.D. S. Bindels, L. Haarbosch, L. van Weeren, M. Postma, K. E. Wiese, M. Mastop, S. Aumonier, G. Gotthard, A. Royant, M. A. Hink, T. W. J. Gadella Jr., MScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2017). [DOI] [PubMed] [Google Scholar]
- 24.V. Gradinaru, F. Zhang, C. Ramakrishnan, J. Mattis, R. Prakash, I. Diester, I. Goshen, K. R. Thompson, K. Deisseroth, Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Beck, Y. Gong, A high-speed, bright, red fluorescent voltage sensor to detect neural activity. Sci. Rep. 9, 15878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. T. Lim, D. E. Antonucci, R. H. Scannevin, J. S. Trimmer, A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron 25, 385–397 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Z. Liu, O. Chen, J. B. J. Wall, M. Zheng, Y. Zhou, L. Wang, H. Ruth Vaseghi, L. Qian, J. Liu, Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7, 2193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. S. Marvin, B. Scholl, D. E. Wilson, K. Podgorski, A. Kazemipour, J. A. Muller, S. Schoch, F. J. U. Quiroz, N. Rebola, H. Bao, J. P. Little, A. N. Tkachuk, E. Cai, A. W. Hantman, S. S. Wang, V. J. DePiero, B. G. Borghuis, E. R. Chapman, D. Dietrich, D. A. DiGregorio, D. Fitzpatrick, L. L. Looger, Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Rorsman, F. M. Ashcroft, Pancreatic β-cell electrical activity and insulin secretion: Of mice and men. Physiol. Rev. 98, 117–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Braun, R. Ramracheya, M. Bengtsson, Q. Zhang, J. Karanauskaite, C. Partridge, P. R. Johnson, P. Rorsman, Voltage-gated ion channels in human pancreatic β-cells: Electrophysiological characterization and role in insulin secretion. Diabetes 57, 1618–1628 (2008). [DOI] [PubMed] [Google Scholar]
- 31.M. Valdeolmillos, R. M. Santos, D. Contreras, B. Soria, L. M. Rosario, Glucose-induced oscillations of intracellular Ca2+ concentration resembling bursting electrical activity in single mouse islets of Langerhans. FEBS Lett. 259, 19–23 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Y.-J. Liu, A. Tengholm, E. Grapengiesser, B. Hellman, E. Gylfe, Origin of slow and fast oscillations of Ca2+ in mouse pancreatic islets. J. Physiol. 508, 471–481 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Zhang, P. Goforth, R. Bertram, A. Sherman, L. Satin, The Ca2+ dynamics of isolated mouse β-cells and islets: Implications for mathematical models. Biophys. J. 84, 2852–2870 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.H. Ren, Y. Li, C. Han, Y. Yu, B. Shi, X. Peng, T. Zhang, S. Wu, X. Yang, S. Kim, L. Chen, C. Tang, Pancreatic α and β cells are globally phase-locked. Nat. Commun. 13, 3721 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. F. Worley 3rd, M. S. McIntyre, B. Spencer, R. J. Mertz, M. W. Roe, I. D. Dukes, Endoplasmic reticulum calcium store regulates membrane potential in mouse islet beta-cells. J. Biol. Chem. 269, 14359–14362 (1994). [PubMed] [Google Scholar]
- 36.Q. Zhang, J. Galvanovskis, F. Abdulkader, C. J. Partridge, S. O. Göpel, L. Eliasson, P. Rorsman, Cell coupling in mouse pancreatic β-cells measured in intact islets of Langerhans. Philos. Trans. A Math Phys. Eng. Sci. 366, 3503–3523 (2008). [DOI] [PubMed] [Google Scholar]
- 37.J. E. Manning Fox, A. V. Gyulkhandanyan, L. S. Satin, M. B. Wheeler, Oscillatory membrane potential response to glucose in islet β-cells: A comparison of islet-cell electrical activity in mouse and rat. Endocrinology 147, 4655–4663 (2006). [DOI] [PubMed] [Google Scholar]
- 38.P. Rorsman, L. Eliasson, T. Kanno, Q. Zhang, S. Gopel, Electrophysiology of pancreatic β-cells in intact mouse islets of Langerhans. Prog. Biophys. Mol. Biol. 107, 224–235 (2011). [DOI] [PubMed] [Google Scholar]
- 39.K. E. Kishi, Y. S. Kim, M. Fukuda, M. Inoue, T. Kusakizako, P. Y. Wang, C. Ramakrishnan, E. F. X. Byrne, E. Thadhani, J. M. Paggi, T. E. Matsui, K. Yamashita, T. Nagata, M. Konno, S. Quirin, M. Lo, T. Benster, T. Uemura, K. Liu, M. Shibata, N. Nomura, S. Iwata, O. Nureki, R. O. Dror, K. Inoue, K. Deisseroth, H. E. Kato, Structural basis for channel conduction in the pump-like channel rhodopsin ChRmine. Cell 185, 672–689.e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Akdel, D. E. V. Pires, E. P. Pardo, J. Janes, A. O. Zalevsky, B. Meszaros, P. Bryant, L. L. Good, R. A. Laskowski, G. Pozzati, A. Shenoy, W. Zhu, P. Kundrotas, V. R. Serra, C. H. M. Rodrigues, A. S. Dunham, D. Burke, N. Borkakoti, S. Velankar, A. Frost, J. Basquin, K. Lindorff-Larsen, A. Bateman, A. V. Kajava, A. Valencia, S. Ovchinnikov, J. Durairaj, D. B. Ascher, J. M. Thornton, N. E. Davey, A. Stein, A. Elofsson, T. I. Croll, P. Beltrao, A structural biology community assessment of AlphaFold2 applications. Nat. Struct. Mol. Biol. 29, 1056–1067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. S. Abdelfattah, R. Valenti, J. Zheng, A. Wong, G. P. Team, K. Podgorski, M. Koyama, D. S. Kim, E. R. Schreiter, A general approach to engineer positive-going eFRET voltage indicators. Nat. Commun. 11, 3444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.E. Gylfe, P. Gilon, Glucose regulation of glucagon secretion. Diabetes Res. Clin. Pract. 103, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Tables S1 to S6