Abstract
Despite great advances in protein structure analysis, label-free and ultrasensitive methods to obtain the natural and dynamic three-dimensional (3D) structures are still urgently needed. Surface-enhanced Raman spectroscopy (SERS) can be a good candidate, whereas the complexity originated from the interactions between the protein and the gradient surface electric field makes it extremely challenging to determine the protein structure. Here, we propose a deciphering strategy for accurate determination of 3D protein structure from experimental SERS spectra in seconds by simply summing SERS spectra of isolated amino acids in electric fields of different strength with their orientations in protein. The 3D protein structure can be reconstructed by comparing the experimental spectra obtained in a well-defined gap-mode SERS configuration with the simulated spectra. The gradient electric field endows SERS with a unique advantage to section biomolecules with atomic precision, which makes SERS a competent tool for monitoring biomolecular events under physiological conditions.
An effective spectroscopic strategy enables label-free determination of protein structure in physiological environment in seconds.
INTRODUCTION
The arrangement and combination of 20 amino acids create abundant proteins with complex structures and diverse functions. Accurate identification of proteins and deciphering of their structures are vital to drug development, clinical diagnosis, immunology, bioengineering, and so forth (1–5). Among various characterization techniques, traditional structural biology methods [nuclear magnetic resonance, cryo–electron microscopy (cryo-EM), and x-ray] can provide the accurate structural information for a protein of interest (POI). The developing trajectory of these techniques indicates that they require not only delicate experimental procedures but also deciphering strategies to enable a fast and accurate reconstruction of three-dimensional (3D) structure of a POI by comparing the calculated spectra or images of them with clear configurations with the experimental results (6, 7). One of the examples is the maximum likelihood strategy used for single-particle analysis in cryo-EM, and cryo–transmission electron microscopy had not been widely used in structural biology until the deciphering strategies became matured (8). Although the above techniques have facilitated the identification of the structure-function relationship of POIs, the measurements still have to be performed either in high concentrations, at ultralow temperatures, or in crystalline form (9–11), which are vastly different from the natural environment of POI. It is therefore imperative to develop label-free and ultrasensitive methods that can obtain the natural and dynamic structural information of a POI in physiological environments, particularly at low concentration (12, 13).
Surface-enhanced Raman spectroscopy (SERS) can provide fingerprint information of the molecule of interest as a vibrational spectroscopy and high sensitivity down to single molecule benefiting from the strongly enhanced electric field by the surface plasmon resonance effect of noble metal nanostructures (14, 15). SERS is able to directly analyze biomolecules under physiological conditions (16, 17). In particular, the local enhanced field exponentially decreases with the increasing distance from the nanostructure surface, forming a gradient field at the molecular scale (18). As a result, different functional groups or vibrational modes of surface species may experience substantially different field strengths, offering SERS a great potential to extract the atomic positions and determine the 3D structure of POI with a high precision, just like a molecular-scale computed tomography. Although SERS has been extensively applied in protein analysis (19–22), this advantage has not been fully taken because of the great challenges in obtaining reliable and reproducible SERS spectra. The gap-mode SERS offers a well-defined configuration for generating highly reproducible and reliable SERS spectra by placing small molecules in the gap of plasmonic nanoparticle (NP) and flat surface of noble metals (23–26). To make gap-mode SERS a powerful deciphering tool for protein structural determination (as illustrated in Fig. 1A), both the experimental and data interpretation issues need to be solved. The former includes the denaturation issue as a result of direct interactions between proteins and the metal surfaces and the diverse orientations of big proteins on the surface.
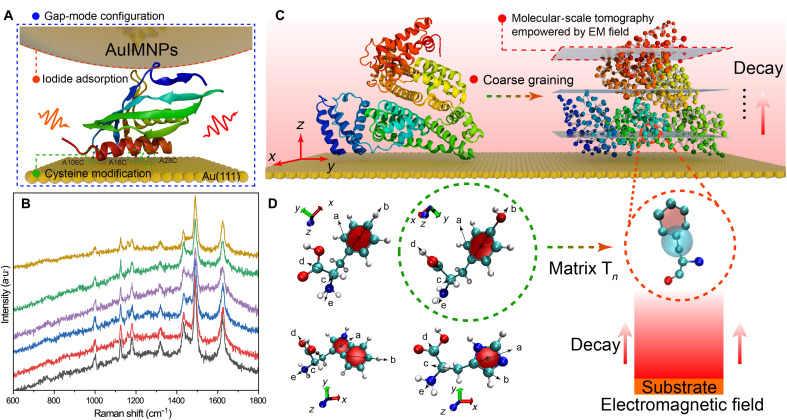
Fig. 1. Scheme of the proposed strategy for deciphering 3D protein structure.
(A) Schematic diagram of protein in the nanocavity of the gap-mode configuration constructed by Au(111) surface and AuIMNPs, where every protein in the gap is subjected to the highly localized plasmonic field. Ketosteroid isomerase (KSI) was used as a model system (Protein Data Bank: 1buq), in which the positions of 16, 28, and 106 are replaced by cysteine. These cysteines can bind to the Au surface through thiol-Au interaction, allowing for orientation-controlled immobilization. (B) SERS spectra of KSI underneath six different NPs on a same single-crystal surface, indicating high reproducibility of the gap-mode configuration. a.u., arbitrary units. (C) On the basis of the coarse-grained model, the POI (bovine serum albumin as a model) on the surface can be sectioned into different layers, where each amino acid would experience different strengths in the decay electromagnetic field. The gray planes, indicating different field strengths, are similar to the sectioning plane in computed tomography but at the molecular scale, giving SERS the additional power to extract the amino acids position with atomic precision. (D) Every two vectors could define a bond rotation in amino acids. Thus, amino acids require at least three rotation matrices (Tn) describing the dynamic structure of amino acids, which would then be used to transform PDs under specific orientation to real orientation.
The latter requires an efficient strategy for interpreting SERS spectra of proteins. Although many elegant theoretical approaches have been proposed to simulate SERS spectra of simple molecules with the aid of quantum mechanics (QM) and machine learning (27–34), it is still challenging to accurately simulate Raman spectra for proteins with large molecular weight and flexible and dynamic structures under real physiological conditions (35). For example, the most important parameter to determine the SERS intensity is the polarizability derivatives (PDs), which can be obtained by performing all-electron calculation. However, calculation of PDs of all vibrational modes of proteins requires enormous computation power scaling with N3 (N is number of electrons) (36). The placement of PDs in the gradient local field further increases the computation costs, making it almost impossible to accomplish with the state-of-the-art computational resources.
The combination of the above two challenges constitutes a substantial obstacle for SERS to be applied in protein structure determination. Here, we proposed a deciphering strategy on the basis of well-defined SERS detecting configuration and a separable PD-based algorithm with rotational coarse-grained model (SPARC) to bridge the experimental results and SERS theories for protein structure determination. On one hand, we constructed a gap-mode configuration with well-defined local gradient field on the basis of iodide-modified NPs over single-crystal surface. The strongly adsorbed monolayer iodides not only prevent strong chemical interactions between the protein and metal surfaces but also remove the impurities on the surface, enabling highly reproducible and reliable SERS spectra to be obtained. On the other hand, we innovatively introduced a coarse-grained model to isolate and obtain the PDs of each amino acid, as depicted in Fig. 1C. Following this divide-and-conquer strategy, the expensive QM and electrodynamics calculations of the complex interactions between protein and the electric field can be simplified into simple matrix computation of PDs, rotation matrix, and gradient local electric field. Such simplification enables a fast calculation of SERS spectra for proteins with the aid of full-atom molecular dynamics (MD) simulation (fig. S1). As a result, the protein structure can be determined in seconds with a high accuracy by analyzing the similarity between the calculated spectra for various configurations and the experimental spectrum. The feasibility of SPARC as a universal tool for scrutinizing interfacial molecular structures is verified with molecules of different sizes, ranging from small molecules to peptides and proteins, on surface. Such an accurate and cost-effective deciphering strategy disruptively enables rapid analysis of the 3D structures of complex biomolecules via SERS. It can be a promising tool to explore the dynamics of interfacial proteins in physiological environments.
RESULTS
Deciphering strategy
The first crucial step toward protein structure determination via SERS is the development of a delicate experimental technique that can provide highly reproducible and reliable data encoding the information of the 3D structure of proteins. As mentioned before, the decay nature of SERS enables the protein to be sectioned at the molecular scale by the local gradient field (similar to computed tomography, as illustrated in Fig. 1C), allowing the SERS technique to extract the atomic positions of amino acids with high precision and thus reconstruct the protein structure. For this purpose, we proposed to borrow the gap-mode SERS configuration with some modifications to achieve a well-defined distribution of the gradient electromagnetic (EM) field, as illustrated in Fig. 1A. Instead of pure single-crystal surface, we modified the atomically flat Au(111) surface with cysteine (Cys), of which the POI was placed on top. Then, we placed iodide-modified Au NPs (AuIMNPs) over POIs. In this way, POIs are naturally trapped in the gap formed between AuNPs and the Au(111) surface. The distribution of the gradient EM field, including the decay constant of the plasmonic field, can be regulated by the size of AuNPs, the wavelength and incident angle of the excitation laser, as well as the size of POI (37, 38). The AuIMNPs and Cys-modified Au(111) surface prevent the direct interaction of POI with them and can effectively avoid the protein denaturation issue. Iodide modification helps remove the reductants from the surface, which improves the reliability of SERS detection. Furthermore, the Cys layer helps the protein to be immobilized in a bioconjugated way with a defined orientation on the Au(111) surface, which can effectively remove the spectral averaging effect and enable us to obtain highly reproducible and reliable SERS spectra. The well-defined gap-mode configuration provides a unique EM field distribution, which encodes valuable 3D structural information in the experimentally obtained SERS spectra. This, in turn, empowers the great deciphering capability of SERS for determining the 3D structure of protein, which has not been achieved before. To demonstrate reliability and reproducibility of this gap-mode configuration, we engineered ketosteroid isomerase (KSI) protein with three Cys residues on one side (Fig. 1A and fig. S2), so that it can be adsorbed on an atomically flat Au(111) single-crystal surface with the designed orientation. A nanocavity of well-defined size was then constructed by placing 55-nm AuIMNPs on the Au(111) surface (see fig. S3 for the helium ion microscopy image of the sample). The iodide monolayer would prevent the direct interaction between KSI and AuIMNPs surface and help retain the native structure of KSI (20, 39). This configuration provides highly reproducible SERS spectra as shown in Fig. 1B, which enables the reconstruction of KSI structure based on its SERS spectra as will be discussed later.
The next step is to decode the experimentally obtained protein SERS spectra and reconstruct its interfacial structure. For this purpose, it is crucial to obtain the accurate simulated SERS spectrum for the protein with specific structure, which requires an accurate description of the interaction of PDs and gradient local electric field. Basically, the scattering tensor (Sn) of a molecule that determines the SERS intensity can be described as follows
| (1) |
where is the PDs and represents the enhanced local fields along the incidence and scattering directions. SPARC uses a gradient enhanced local field exponentially decreasing with the increase of distance (r, Cα of amino acid in the corresponding layer), i.e., , which implies that the electric field can vary over the dimension of a molecule or vibrational mode (Fig. 1C). In addition, according to the surface selection rules of SERS, only the vibrational modes with a component (α′zz) parallel to the electromagnetic (EM) field can be effectively enhanced, giving the orientation of amino acids. Eventually, the combination of distance-dependent field strength and the surface selection rules results in a unique spectrum for every conformation of protein on the surface, which makes it possible to determine 3D structure of proteins with SERS. Then, SPARC uses PDs of amino acids with a specific orientation to obtain α′zz. The orientation of amino acids in a protein can be defined by several geometrical vectors, and it requires two vectors to define rotation matrices (Tn) to describe the dynamic structure of amino acids. The rotation matrix is the most important operation for accurately calculating interactions between proteins and gradient local electric fields (Fig. 1D and fig. S4), which can be calculated by the atomic coordinates of POI from MD result. By aligning the orientation of amino acids to their real orientations in POI via rotation transformation, the PDs of each amino acid can be obtained by rotating corresponding empirical PDs (more validations can be found in figs. S5 and S6 and table S1). Therefore, the SERS spectrum of protein (Sprotein) can be the sum of spectra of m amino acids (SAA) accumulated by n vibrational modes () and expressed as follows
| (2) |
where ωs is the frequency of scattered light and Mi and Ms represent the enhancement factors for incident light and scattered light, respectively. Ω represents Euler angles that determine the molecular orientation relative to the local fields. Fp is the Purcell factor. c, ħ, kB, and T are the speed of light, reduced Planck constant, Boltzmann constant, and kelvin temperature, respectively (18). By using Eq. 2, SERS spectra can be acquired with a high calculation speed via performing matrix computation with an averaged scaling factor kscale (more details can be found in the “Correction of algorithm and correlated coefficient” section), which was introduced to compensate for the overestimation coming from high-order polarizability (as indicated in fig. S7). This issue can be addressed by considering the interactions between the quantified EM field with the general integrated polarizability (αn, including contribution of all orders of polarizability) obtained with time-dependent perturbation Hamiltonian (33). As a result, SERS spectra of small molecules can be accurately calculated with the optimized structure in a localized field by overlapped integrals for corresponding orbitals and EM field. However, it is too expensive to directly use this method to track dynamic protein structure and calculate its SERS spectra. All elements needed in the calculation are orbital-dependent. It means that we can further follow our idea to rotate the orbitals under the specific orientation and then calculate the overlapped integrals (see details in Materials and Methods). Note that it takes high-order polarizability into account and eradicates the so-called gauge problem, due to integrals for Gaussian functions in Cartesian coordinates. As a result, the calculation scarifies little speed but markedly improves the accuracy by considering high-order terms of polarizability and EM field.
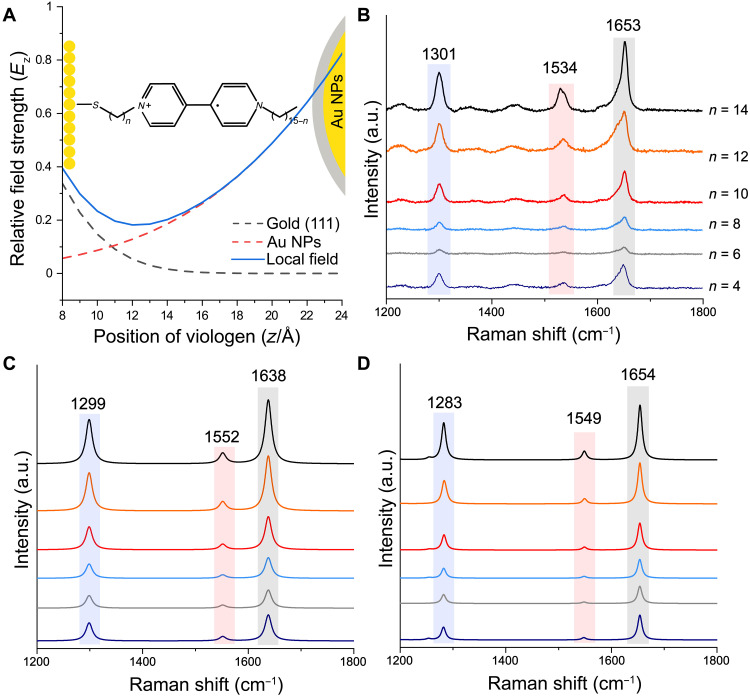
To testify the performance of SPARC, we revisited a simple, elegant, structurally well-defined system by Li et al. (18), which is composed of a set of viologen-based monolayers on Au(111) single-crystal surface with silica-coated Au NPs on the top, so-called gap-mode configuration (Fig. 2A). They demonstrated that the plasmonic field in the gap can be treated as a combination of fields from the Au(111) surface and the Au NPs. Thus, the distribution of the field in the longitudinal direction can be expressed as
| (3) |
where A (B) and α (β) are the maximum intensity and decay constant of a plasmonic field by the Au surface (the Au NPs), respectively. z is the distance between the Au(111) surface, and γ is the gap size of the nanocavity. The experimental SERS spectra for viologen of different alkane lengths are given in Fig. 2B. Because both conformation of viologen molecules and plasmonic field distribution are given, we can conveniently obtain the integral corrected PDs of viologen molecules and calculate their SERS spectra by SPARC. As shown in Fig. 2C, the calculation results obtained by SPARC agree well with both the experimental results (Fig. 2B) and reported theoretical results (Fig. 2D), indicating the reliability of our SPARC method.
Fig. 2. Testifying the performance of SPARC.
(A) Schematic diagram of the proposed system by Li et al. as well as local electric field distribution for viologen in the gap. Surface-enhanced Raman spectra of viologen with different alkane lengths (n) to the Au(111) surface in the gap of silica coating AuNPs and Au(111) surfaces. (B) Experimental SERS spectra. (C) Simulated SERS spectra by SPARC. (D) Simulated SERS spectra in the literature. (B and D) Reproduced with original data by courtesy of Li et al. (18).
Demonstration of the deciphering strategy
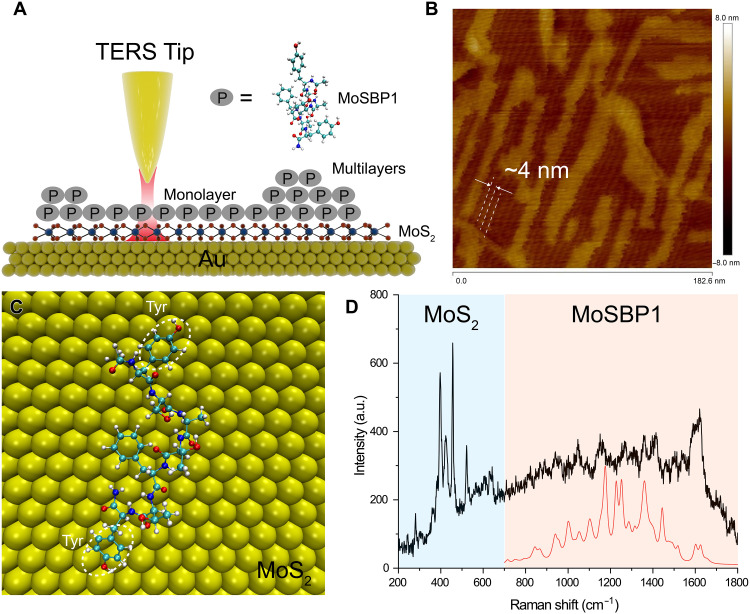
To faithfully demonstrate the feasibility of our strategy for deciphering protein structure, we used the self-assembled peptides (YSATFTTY with acylated N and amidated C termini, named MoSBP1) on MoS2 surface as a model system, which shows well-defined interfacial structure characterized by atomic force microscopy (AFM) (40). If the spectrum reproduced with known structure from AFM is well aligned with the spectrum by tip-enhanced Raman spectroscopy (TERS), then the feasibility of our strategy can be further confirmed. For this purpose, we transferred a monolayer of MoS2 on the Au surface (MoS2-Au; Fig. 3A), and the thin MoS2 layer allows effective plasmonic coupling between the Au surface and Au TERS tip to obtain a strong TERS signal of the peptide. When incubated with the MoS2-Au substrates, MoSBP1 assembled into an elongated layer with three equivalent directions (with 60° angle) as shown in Fig. 3B, which is consistent with the phenomenon observed by Chen et al. (40). The layer is 0.7 nm in height and consists of parallel rows with a periodicity of ca. 4 nm on MoS2-Au substrates (e.g., framed region in Fig. 3B). According to Heinz and colleagues, the two tyrosines (Tyr) at the two ends of the peptide are flat-lying on the MoS2-Au, where the hydrogen bonding between two peptides in an end-to-end way via the Tyr forms the building block of the order assembled structure. These two peptides determine the packing direction(41). In addition, there are several multilayer domains (with a height of 1.4 nm) of MoSBP1 over the monolayer structure, which has not been reported before. Thus, we used SPARC with the known conformation (as indicated in Fig. 3C) to calculate the TERS spectrum of the monolayer of MoSBP1 on MoS2-Au substrates, as the red spectrum shown in Fig. 3D. Note that we used the same approach as outlined in Eq. 3 to calculate the electromagnetic (EM) field along the z direction. The experimental TERS spectrum (black in Fig. 3D) shows several intense peaks in the range of 300 to 700 cm−1 contributed by MoS2 (42), suggesting a strong enhancement in the gap between the Au tip and substrate. The peptide contributes the peaks in the spectral region from 700 to 1800 cm−1. Because of the flat-lying adsorption of Tyr and Phe, their in-plane vibrational modes (e.g., 830 cm−1 for Tyr and 1000 cm−1 for Phe) show low intensity. We evaluate the similarity of our simulated spectra and experimental spectra by Spearman’s rank correlation coefficients (rs; see Materials and Methods for detail). Despite the low intensity and signal-to-noise ratio of the experimental spectrum, our simulated spectrum of monolayer still reveals high similarity (rs = 0.66, normally rs > 0.6 indicates a good theoretical prediction) (43) to the experimental one, which demonstrates the feasibility of SPARC for deciphering protein structure with known sequence.
Fig. 3. Demonstration of the deciphering strategy by TERS.
(A) Schematic diagram of TERS detection of the self-assembled peptide on MoS2. (B) AFM image of the sample, showing a full monolayer of order structure of peptides in the bottom layer and some multilayer features on top. (C) Interfacial repeating unit of peptide on MoS2 in the literature (40). (D) Simulated TERS spectrum (red) by SPARC and experimental TERS spectrum of the monolayer of peptide (black). The peaks in the light blue region are from MoS2, and the light red region is from peptides.
Deciphering SERS spectra of peptides
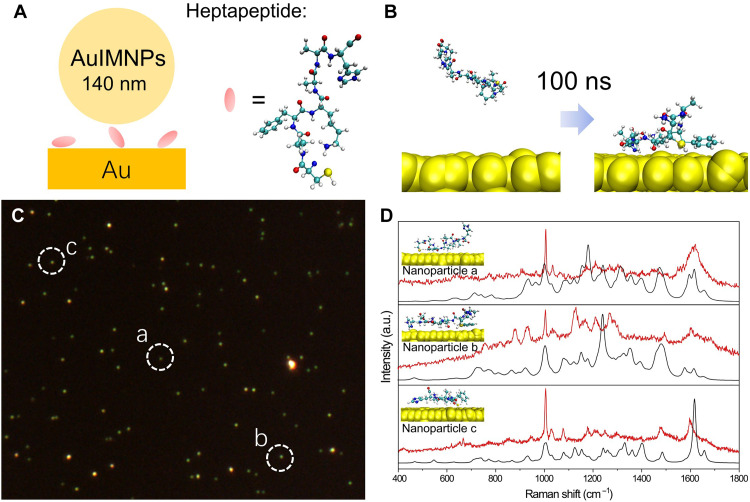
After verifying the reliability of the strategy, it will be interesting to take a step forward to reconstruct the structure of more complex biomolecules from SERS spectra. We assembled a heptapeptide (CAFKAAH; Fig. 4A) with Cys termination on the Au surface via Au─S bond, followed by assembly with 140 nm AuIMNPs. In this way, a nanocavity with a well-defined size was formed to produce SERS signal of peptide in a gap-mode way (Fig. 4A). SPARC was then used to calculate the SERS spectra of POI to decipher protein structure from the experimental SERS spectra. To obtain possible orientations of POIs, we used the GolP-CHARMM force field, which has been accepted as a powerful force field to describe the reasonable protein structure at the interface to simulate the adsorption of peptides on Au (44–46). The heptapeptide is initially placed at least 1 nm away from the surface. After 100 ns, residues like Phe and Cys are successively attached to the Au surface, as shown in Fig. 4B. We performed 35 independent MD simulations (ca. 4 μs) to fully sample possible orientations. According to the free energy of landscapes shown in fig. S8, we extracted the dominant conformations of the peptide. The root mean square deviation (RMSD) between conformations is illustrated in fig. S9A, which also indicates marked structural variations. Experimentally, we combined a dark-field scattering microscope to locate single NPs (Fig. 4C) and acquired the SERS spectra from these NPs. We then compared the similarity of our simulated spectra (based on 40 dominant conformations; fig. S9B) to the experimental spectra of three single NPs in Fig. 4C by Spearman’s rank correlation coefficients (rs) and identified corresponding conformations accordingly. As the peptides right underneath the NPs have less freedom to move, we envisage that one specific orientation may dominate in the final spectra as shown in Fig. 4D. The resulting correlation coefficients are 0.85, 0.81, and 0.77, respectively, indicating good theoretical predictions. In return, the peptide structure can be scrutinized from experimental SERS spectra, as illustrated in Fig. 4D.
Fig. 4. MD simulation and experimental and theoretical SERS spectra of peptides by SPARC.
(A) Schematic diagram of single NP gap-mode SERS. (B) An example of the initial configuration and final configuration of the heptapeptide on a Au surface. (C) Dark-field scattering image of the sample. (D) Theoretical SERS spectra by SPARC (black spectrum) in comparison with experimental SERS spectra (red spectrum) obtained from the NPs as indicated in (C) with white dashed circles. Insets are corresponding configuration scrutinized by our strategy.
Reconstructing protein structure based on its SERS spectra
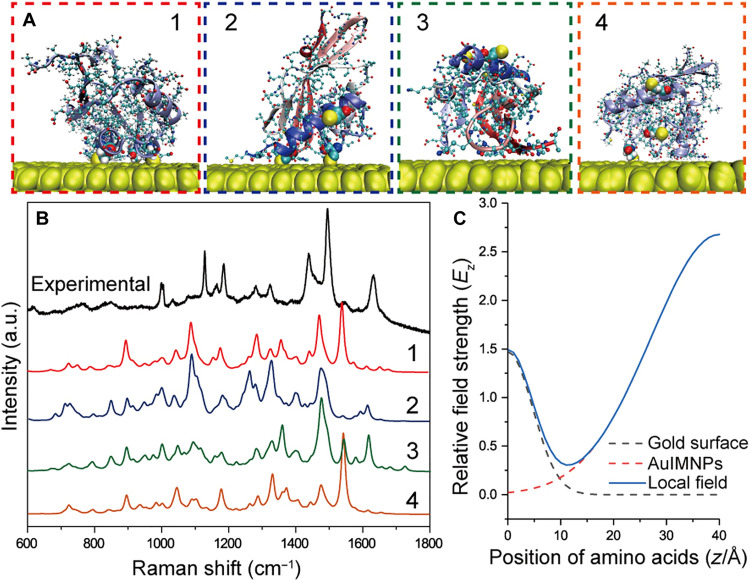
The above shows the results on peptides, and it will be more exciting if proteins with much more complex structure can also be directly deciphered only with SERS spectra. In Fig. 1A, we have shown that we were able to obtain the reproducible spectra of KSI, and we can now use the SPARC to extract its 3D structure. To obtain structural information from the experimental spectrum of this well-defined system, we need to get the atomic coordinates of amino acids and EM field distribution. The atomic coordinates were obtained by over 50 MD simulations (ca. 5 μs) to fully sample all possible stable conformations. The free energy landscapes are shown in fig. S10, from which we extracted the representative orientations of KSI and showed four representative ones in Fig. 5A. We used the same approach as in Eq. 3 to obtain the EM field along the z direction (Fig. 5C). The simulated spectra are given in Fig. 5B, and different orientations result in different spectral features. The correlation coefficient and theoretical spectra of all conformations are illustrated in fig. S11 (A and B). It is interesting to find that conformation 2 in Fig. 5A shows the best correlation with the experimental spectrum of KSI (with rs = 0.75). This conformation agrees well with the intended orientation that would arise by Cys modifications.
Fig. 5. Scrutinization of the KSI structure from the gap-mode SERS spectrum.
(A) Four representative orientations of KSI obtained by MD simulation. (B) Experimental gap-mode SERS spectrum of KSI in comparison with theoretical SERS spectra of four representative conformations calculated by SPARC. (C) Local EM field from contributions of Au surface and AuIMNPs.
From the above result, we can find that our strategy has great potential to obtain the 3D structure of proteins at the surface from its SERS spectrum. Furthermore, the gradient electric field endows SERS with a unique advantage to section protein with atomic precision to reconstruct the 3D protein structure. Hence, with known atomic coordinates and electric field distribution, the SERS spectrum of the POI will be uniquely determined and vice versa. We can also foresee that more structural information can be obtained by tuning the gradient electric field, which promotes spectra deconvolution with enlarged experimental data points. This was demonstrated through SERS measurement of the same system under various laser wavelengths, as depicted in fig. S12. The SERS spectra of KSI displayed various spectral features at different wavelengths, possibly due to the presence of different local electric field distributions within the gap (fig. S13). Because the tensor nature of polarizability and decay of EM involved in SPARC are the most fundamental and universal features of SERS, SPARC should be a general spectrum interpreter that is capable of interpreting SERS spectra of not only protein but also other biomolecules. We further extended SPARC to interpret SERS spectra of DNA (fig. S14). The DNA spectra obtained by SPARC reveal a high correlation with experimental results in the literature (see fig. S15), indicating the potential to analyze the interfacial structure of nuclei acid as well as other biomolecules.
DISCUSSION
In summary, we have reported an accurate and cost-effective deciphering strategy for rapidly reconstructing the 3D protein structure of POI based on SERS. By constructing well-defined detecting configuration, we obtained the reliable and reproducible SERS spectra of protein in the gradient EM field, which contains information of the amino acids position with atomic precision. SPARC can simplify the conformation of proteins at the interface as a spatial arrangement and combination of amino acids. As a result, SERS spectra can be obtained by simple matrix computation of PDs, rotation matrix, and gradient local electric field. Moreover, calculation accuracy can be further improved by introducing interaction Hamiltonian with integrated polarizability. The predicted SERS spectra by SPARC show a high similarity to the experimental spectra of protein in well-defined systems, which, in turn, enable us to reconstruct the 3D structure of proteins with known sequences from the obtained SERS spectra. Because the description of PDs and EM fields are the most fundamental features of SERS, SPARC can be a universal SERS interpreter for any other complex biomolecules, which has also been verified in the DNA system. Our method will open a door for monitoring biomolecular events under physiological conditions, which are important to applications, such as biocatalysis, drug development, and clinical diagnosis. Both experimental and theoretical efforts need to make to address the great challenge. In the experimental aspect, one may tune the EM field to profile protein molecules so that abundant and effective experimental data can be obtained. In the theoretical aspect, more efficient sampling methods should be developed to sample all possible orientations without relying on the less efficient traditional MD simulation. Combined with machine learning, SPARC will be able to consider the coupling effects between amino acids and the influence of the chemical environment in a fast and cost-effective way that is impossible with state-of-the-art QM/ED methods. At that stage, we can foresee that SPARC can be efficient enough to scrutinize the structure of unknown protein in a short time.
MATERIALS AND METHODS
Calculation of amino acids and their PDs
The geometries of amino acids were optimized by density functional theory (DFT) calculations using the basis set B3LYP and adapted the triple splitting valence basis set of 6-311+G*. The structures of amino acids used can be found in fig. S4. All calculations were performed by the Gaussian 09 package (47). The scale factor for vibrational frequency is 0.98 obtained using the peak of phenylalanine at 1000 cm−1. The calculations of nucleic acids are performed at the same level without the scale factor. To obtain PDs of amino acids, we need to transform the atomic PD tensor in the Cartesian coordinate (PDC) to the PD tensor in the normal coordinate frame (PDN) according to the following expression (48)
The Raman scattering factor Sm is calculated from the isotropic polarizability tensor derivative () and the anisotropic PD () of the mth vibrational mode
where
and
After transformation, we can obtain the PDs of each vibrational mode of every amino acid and nucleic acid, which are saved as empirical files to be invoked. To testify the effect of protonation states and side chain on PDs, we took the peak of phenylalanine at 1000 cm−1 as an example, owing to its high sensitivity to the chemical environment change. The five structures are illustrated in fig. S5. We aligned the benzene ring of phenylalanine with the same orientation and compared the PDs values of five structures. As depicted in table S1, the PD value change is modest, and the SD is low. This result indicates that we can obtain reasonable values of PDs based on isolated amino acids. A dipeptide (Ace-phe-ala-nme) with 50 conformations from SPICE dataset was selected to confirm the ability of SPARC for differentiating between different structures (49). The structures and corresponding RMSD are illustrated in fig. S6 (A and B). For DFT-simulated results, only αzz component of PDs was considered to produce the SERS spectra of dipeptide. For SPARC, we ignored the functional group of the dipeptide (i.e., Ace and nme) and calculated the SERS spectra on the basis of two amino acids. As shown in fig. S6C, we showed theoretical spectra of eight conformations of dipeptide simulated by DFT (black curve) and SPARC (red dotted curve). It can be found that the spectra of dipeptide with different orientations and structures can be differentiated. In comparison with DFT-simulated spectra, most of spectral features of the dipeptide can be observed in the spectra simulated by SPARC, which demonstrates the feasibility of SPARC to differentiate between different structures.
MD simulation
The 3D structure of proteins was obtained by homologous modeling using the I-TASSER server, in which full-length atomic models were constructed by iterative template-based fragment assembly simulations (50, 51). Last, five representative protein structures were obtained from the I-TASSER server, for later MD simulation. GROMACS (52) (2019.5) was used for the MD simulation with CHARMM force field (53) and GolP force field (53). A periodic boundary was used in the simulation, where the protein was immersed in a cuboid box of water. The simplified point charge model TIP3P (54) was used to represent water molecules. In the simulation, we first used the steepest descent approach to minimize the energy of the system with a convergence criterion of 1000 kJ mol−1 nm−1 and a step of 0.01 nm. Energy minimization was followed by 100 ns of NVT MD simulation. To visualize the free energy landscape, the anaeig module of GROMACS was used to analyze the trajectories of backbone and project two principle components as horizontal and vertical axes in figs. S8 and S10.
Finite-element method simulation
A commercial finite-element method simulation software (COMSOL Multiphysics package 6.0) was used to model the near-field enhancement of Au NPs on Au film. The perfectly matched layers were used to simulate an open boundary. The permittivity values of Au were from Johnson and Christy (55). The medium over this structure was set to be air. The diameter and size of Au NPs were set on the basis of the helium ion microscopy image (fig. S3).
Algorithm based on Python
Figure S1 schematically shows the processes of SPARC. GROMACS (gmx) was used for MD calculation to obtain the reasonable and dominant conformation of POI on the surface. Multiwfn, a powerful wave function analysis software, was used for calculating SERS spectra (56). Thus, the algorithm has three functions: (i) read coordinates from results provided by gmx, which is based on Bio.PDB.PDBParser; (ii) invoke the parameters to calculate the rotation matrix and calculate the PDs of POI in the real conformation; and (iii) calculate the empirical electromagnetic (EM) field according to coordinates. The SERS activity of each mode can be calculated, and Multiwfn reads the output files. Note that the EM field equation used for gap-mode simulation is adopted from Li et al. (18). The local field distribution of the nanocavity constructed by Au(111) and AuIMNPs in Fig. 5C follows the expression of Ez ∝ 1.469e−0.023z2 + 2.679e−0.003(z−γ)2, with the gap size of the nanocavity (γ) of the size of protein (40 Å). We proposed two methods for accurate calculation of SERS spectra in SPARC. The first method is to use Eq. 2, which is simple matrix computations of empirical PDs, rotation matrix, and EM field by Numpy library. The scaling factors (kscale) are introduced to SERS intensity of each mode to compensate the overestimation to save the calculation resources, which is suitable for calculation of large molecules, like proteins. Another more accurate but time-consuming method is to use Eq. 4 (see the next section for details), in which the overlapped integrals are used for calculating SERS intensity of each mode, which can be used for calculation of small molecules, like viologen. The use of Eq. 2 can also obtain similar results as in Eq. 4 for small molecules as depicted in fig. S7. Then, the SERS activity of all vibrational modes obtained by the above methods can then be merged into one file with three columns (frequency shift, intensity, and width), which can be read by Multiwfn to produce SERS spectrum with Lorentz spectral line.
Correction of algorithm and correlated coefficient
The theoretical SERS spectrum was calculated by SPARC (fig. S7B) and was compared with the theoretical result from Li et al. (18) (fig. S7A). It is obvious that SPARC overestimates the intensity of molecules at a high EM field (when the distance of the molecule to the substrate is less than 1.5 nm), which may be due to the gauge problem (33). To correct this problem, we positioned every amino acid at a different distance (r) to the surface (fig. S7C) and calculated their SERS spectra using SPARC and gauge-invariant method developed by Duan et al. (33). Then, we fitted the intensity of spectra with exponential functions (k = axb). It should be noted that every amino acid under different orientations has its k function due to a unique electronic structure used in the interaction Hamiltonian. As shown in fig. S7D, scaling amino acids separately provides more accurate polarizability. However, it also implies that it is necessary to scale all amino acids with different orientations to fit their k functions to calculate the dynamic SERS protein spectra, which requires enormous computational resources. Instead, we propose to introduce an averaged factor kscale with all the obtained k for all amino acids to avoid the accumulated scaling process. After scaling this algorithm with a general scaling factor (kscale = 0.1 for r < 1 nm), we obtain a theoretical result highly correlated with their result in the literature (Fig. 2D) (18). The SERS spectrum of protein (Sprotein) can be expressed as follows
| (4) |
where ϕg(Ω) and ϕr(Ω) are electronic ground and resonant excited states with respect to the orientation after rotation (Ω), respectively. ∆Erg represents the vertical excitation energy between ϕg(Ω) and ϕr(Ω). and represent the time-dependent operator. Following our idea to integrate Duan’s method into SPARC, all we need is to rotate the orbitals of the molecule. To this end, we extracted molecular orbital (MO) coefficients (C) of amino acids under standard orientation and rotated the orbitals by the Wigner D matrix
where α, β, and γ are Euler’s angle of rotation, which can be calculated from the rotation matrix (T). m, m′, and l are determined by shell type. The reduced Wigner matrices can be written as
In this way, we can construct a diagonal matrix (M), whose matrix elements are in the order of shells. Then, we can obtain ϕg(Ω) and ϕr(Ω) easily with simple matrix multiplication (i.e., C′ = M.T @ C @ M).
Spearman’s rank correlation coefficient was used to evaluate the performance of SPARC (43). The equation of Spearman’s rank correlation coefficient is shown as follows
where σ and cov denote the SD and covariance, respectively. R(x) and R(y) correspond to the ranked Raman intensities of experiment spectra and predicted spectra, respectively. Normally, a high Spearman coefficient, >0.6, is required as a threshold for a good theoretical prediction, in addition to reasonable orientation and relative peak intensity.
Purification of protein
Primer STAR Max DNA Polymerase was purchased from Takara (Shiga, Japan). The plasmid mini kit I was provided from Omega Bio-Tek (USA). Escherichia coli BL21 (DE3) competent cell, DMT chemically competent cell, DMT enzyme, isopropyl-β-d-thiogalactopyranoside (IPTG), and kanamycin were purchased from TransGen (Beijing, China). SDS–polyacrylamide gel electrophoresis (PAGE) was purchased from GenScript Biotech Corp. (Nanjing, China). Modified Bradford Protein Assay Kit and LB were purchased from Sangon Biotech (Shanghai, China).
Using E. coli as the expression host, the amino acid sequence of KSI from Pseudomonas putida (UniProtKB: P00947) was optimized and then synthesized by GenScript Biotech Corp. (Nanjing, China). The gene of KSI was ligated with pET-28a (+) under the restriction sites Nde I and Xho I to construct the recombinant plasmid pET-28a (+)–KSI, which was accompanied by His6-tagged in the N terminus of KSI. Mutagenic forward primers and reverse primers (table S1) were used to construct a site-directed mutation plasmid that was called pET-28a (+)–KSI–A16C/A28C/A106C. The plasmids were lastly transformed into E. coli BL21(DE3).
The enzymes were expressed from recombinant plasmids containing the KSI or mutation gene under the control of the T7 promoter. Typically, cultures of recombinant E. coli BL21 (DE3) in LB medium containing kanamycin (40 mg/liter) were grown at 37°C (200 rpm) until optical density at 600 nm reached 0.6 to 0.8 and then induced with 0.2 mM IPTG at 25°C for 16 hours. After induction, cells were harvested by centrifugation at 7000 rpm for 5 min at 4°C, the supernatant was discarded, and the cell pellet was resuspended with phosphate-buffered saline (PBS) buffer. Then, the cells were disrupted by ultrasonication and removed by centrifugation at 9000 rpm for 20 min under 4°C. Obtained crude cell extract was then added to His-Trap column (His-Trap HP 5 ml, GE Healthcare Corp., Piscataway, NJ, USA) that has been pre-equilibrated with binding buffer [20 mM sodium phosphate, 0.5 M NaCl, and 20 mM imidazole (pH 7.4)]. The enzyme-modified column was further equilibrated with binding buffer, followed by elution with buffer containing 20 mM sodium phosphate, 0.5 M NaCl, and 0.5 M imidazole (pH 7.4). The enzymes were filled in dialysis membranes (MD44, 3500D, Solarbio) and dialyzed with PBS buffer overnight. The purity of the obtained enzymes was tested by 10% (w/v) SDS-PAGE, as shown in fig. S2. All the protein concentrations were determined using the modified Bradford Protein Assay Kit with bovine serum albumin as a standard. Amino acid sequence of KSI (Protein Data Bank: 1BUQ): MNTPEHMTAVVQRYVAALNAGDLDGIVALFADDATVEDPVGSEPRSGTAAIREFYANSLKLPLAVELTQEVRAVANEAAFAFTVSFEYQGRKTVVAPIDHFRFNGAGKVVSMRALFGEKNIHAGA. Gene sequence of KSI: ATGAATACTCCCGAACACATGACAGCTGTAGTGCAGCGTTACGTGGCGGCTTTGAATGCAGGTGATCTGGATGGTATTGTAGCGCTCTTCGCCGACGACGCGACCGTGGAGGACCCGGTTGGTAGCGAACCGCGTAGCGGCACCGCAGCGATTCGCGAGTTCTACGCGAACAGCCTGAAGCTGCCACTGGCGGTGGAGCTGACGCAAGAAGTTCGCGCGGTTGCCAATGAAGCTGCGTTCGCTTTCACTGTCTCCTTTGAGTATCAGGGTCGTAAAACCGTCGTGGCTCCGATCGACCACTTTCGTTTTAACGGTGCCGGCAAAGTTGTTTCTATGAGAGCGTTGTTTGGCGAAAAGAACATCCATGCAGGCGCATGA.
Raman instrument
SERS spectra were obtained on a confocal Raman microscope (Renishaw Invia, UK) equipped with multiple lasers. Three wavelengths, including 632.8, 691, and 785 nm, were used as the excitation source in this work. The objective was of ×50 magnification and 0.85 numerical aperture to provide a high incident and collection angle. The laser power used was 0.4 mW, and the typical exposure time for each measurement was 10 s without accumulation unless otherwise stated.
Gap-mode SERS experiments
The Au NPs (with a diameter of 55 and 140 nm) AuIMNPs were prepared according to our previous methods (39, 57). A high-quality Au(111) single-crystal surface was prepared by flame annealing according to the Clavilier method (58). Then, the fresh Au(111) single crystal was immersed in a KSI (or DNA origami) solution overnight to form monolayer biomolecules. Afterward, it was dried under a nitrogen gas flow. AuIMNPs were cleaned by centrifugation and then dropped on the single-crystal surface to form a well-defined gap-mode configuration after evaporation of water (fig. S3). In this way, high-quality and highly reproducible gap-mode SERS signals can be obtained.
AFM imaging
Newly prepared MoS2-Au was immersed in 3 μM MoSBP1 aqueous solution for 1 hour. Afterward, the sample was dried with nitrogen gas. The AFM images were obtained on Dimension FastScan (Bruker). The images were captured using silicon nitride probes (SCANASYST-FLUID+, Bruker) in peak-force mode at room temperature under the protection of argon atmosphere. The preparation of MoS2-Au was based on the previous method (59). MoS2 samples were purchased from Nanjing MKNANO Tech. Co. Ltd. Au(111) substrates were bought from Phasis, which were 200-nm-thick films on mica. Instead of thermal release tape, we used polydimethylsiloxane tape to perform mechanical exfoliation.
TERS measurement
AFM-TERS measurements were performed on a LabRAM HR Evo Nano system (HORIBA Scientific) with side illumination. A 100× objective with a long working distance and a numerical aperture of 0.7 (Mitutoyo) was used for both excitation and collection. A 632.8-nm laser was used for TERS measurements with a p-polarization. The laser power used was 0.1 mW, and the exposure time was 1 s. Au-coated AFM TERS probes used in the TERS measurements were produced by electrochemical deposition of Au on commercial Si AFM tips (ACCESS SNC, Applied Nanostructures Inc.) (60). TERS spectra were obtained in contact mode.
Interpreting SERS spectra of DNA
Following a similar idea, we simplified the DNA or RNA structure into the coarse-grained model with several vectors as shown in fig. S14A. With the given structure of DNA, the algorithm can calculate the SERS spectra according to Eq. 2. Furthermore, unexpectedly, up to now, there are still no calculation results of deoxyadenylic monophosphate (dAMP), deoxyguanosine monophosphate (dGMP), deoxycytidine monophosphate (dCMP), and deoxythymidine monophosphate (dTMP) in the literature. We performed DFT calculation of their Raman spectra as well as that of A, G, C, and T. It can be found in fig. S14 (B to E) that peaks at about 800 cm−1 emerged, which are assigned to O-P-O stretching vibration (61). The ring breathing mode of C and T may also appear at this position; therefore, the overlaps should be carefully considered. To demonstrate its feasibility in predicting SERS spectra of DNA, we first revisited a well-defined system by Halas and colleagues as shown in fig. S15A (62). The correlation coefficient of the two spectra in fig. S15 (A and B) is 0.68, which indicates good performance of SPARC. We then constructed another well-defined system by orientation-controlled immobilization of DNA origami as indicated in fig. S15C. The DNA-origami contains a series of tetrahedral DNA nanostructures (TDNs) with three thiol groups on vertices to anchor firmly to the Au(111) surface (63). Following this method, we used TDN-7 (whose edges contain 7 base pairs giving a length of the edge to be 2.4 nm) to construct this system. As shown in fig. S15D, the averaged spectrum reveals a good agreement with the theoretical spectrum by SPARC, showing a Spearman’s rank correlation coefficient of 0.64.
Acknowledgments
We thank J. Li and S. Duan for helpful discussion and kindness in providing the original data to reproduce Fig. 2 (B and D). We also thank H. Heinz for providing their MD simulation results about MoSBP1 on MoS2.
Funding: This work was supported by the National Natural Science Foundation (grant nos. 22021001, 22227802, 22104125, 22032004, 21633005, 21790354, and 92061118) of China, China postdoctoral science foundation (2021M691870), and the Fundamental Research Funds for the Central Universities (20720220018).
Author contributions: H.M., X.W., and B.R. conceived the idea and designed the experiments. H.M. and S.Y. performed the experiments, COMSOL simulations, and conducted the data analysis. H.M. and X.L. developed the Python code of SPARC. H.M. and D.-Y.W. performed DFT calculations and calculated PDs. H.M., J.L., and B.W. performed MD simulations. M.C., X.Z., Y.-F.B., and X.P. prepared the MoS2/Au sample. H.Y. and H.-L.W. conducted the AFM mapping of MoSBP1/MoS2/Au. L.L. prepared the engineered KSI. Y.-F.B., N.C., L.Z., and H.F. prepared Au(111) single-crystal surface and gold NPs. K.D., G.Y., and C.F. prepared the sample of DNA origami. H.M., X.W., D.-Y.W., and B.R. supervised the project. All authors participated in the result discussion and the manuscript preparation.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. A full version of SPARC is available from the Zenodo repository at https://doi.org/10.5281/zenodo.8261122. In addition, a light version without overlapped integrals function can be obtained from the GitHub repository at https://github.com/Oaham725/SPARC.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Tables S1 and S2
REFERENCES AND NOTES
- 1.Panda P. K., Arul M. N., Patel P., Verma S. K., Luo W., Rubahn H.-G., Mishra Y. K., Suar M., Ahuja R., Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 6, eabb8097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo J. P. K., Liu M.-S., Hibshman G. N., Dangerfield T. L., Jung K., McCool R. S., Johnson K. A., Taylor D. W., Structural basis for mismatch surveillance by CRISPR–Cas9. Nature 603, 343–347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao L., Coventry B., Goreshnik I., Huang B., Sheffler W., Park J. S., Jude K. M., Marković I., Kadam R. U., Verschueren K. H. G., Verstraete K., Walsh S. T. R., Bennett N., Phal A., Yang A., Kozodoy L., DeWitt M., Picton L., Miller L., Strauch E.-M., DeBouver N. D., Pires A., Bera A. K., Halabiya S., Hammerson B., Yang W., Bernard S., Stewart L., Wilson I. A., Ruohola-Baker H., Schlessinger J., Lee S., Savvides S. N., Garcia K. C., Baker D., Design of protein-binding proteins from the target structure alone. Nature 605, 551–560 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Simoska O., Lim K., Grattieri M., Yuan M., Dong F., Lee Y. S., Beaver K., Weliwatte S., Gaffney E. M., Minteer S. D., Fundamentals, applications, and future directions of Bioelectrocatalysis. Chem. Rev. 120, 12903–12993 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Sesterhenn F., Yang C., Bonet J., Cramer J. T., Wen X., Wang Y., Chiang C.-I., Abriata L. A., Kucharska I., Castoro G., Vollers S. S., Galloux M., Dheilly E., Rosset S., Corthésy P., Georgeon S., Villard M., Richard C.-A., Descamps D., Delgado T., Oricchio E., Rameix-Welti M.-A., Más V., Ervin S., Eléouët J.-F., Riffault S., Bates J. T., Julien J.-P., Li Y., Jardetzky T., Krey T., Correia B. E., De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y., Liu J., Zhu L., Zhang B., He J., 3D reconstruction from cryo-EM projection images using two spherical embeddings. Commun. Biol. 5, 304 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrmann T., Güntert P., Wüthrich K., Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24, 171–189 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Stiller J. B., Otten R., Häussinger D., Rieder P. S., Theobald D. L., Kern D., Structure determination of high-energy states in a dynamic protein ensemble. Nature 603, 528–535 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerner E., Cordes T., Ingargiola A., Alhadid Y., Chung S., Michalet X., Weiss S., Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L., Liu N., Gao X., Zhu W., Liu K., Wu C., Yan R., Zhang J., Gao X., Yao Y., Deng B., Xu J., Lu Y., Liu Z., Li M., Wei X., Wang H.-W., Peng H., Uniform thin ice on ultraflat graphene for high-resolution cryo-EM. Nat. Methods 20, 123–130 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buhrke D., Hildebrandt P., Probing structure and reaction dynamics of proteins using time-resolved resonance raman spectroscopy. Chem. Rev. 120, 3577–3630 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Langer J., Jimenez de Aberasturi D., Aizpurua J., Alvarez-Puebla R. A., Auguié B., Baumberg J. J., Bazan G. C., Bell S. E. J., Boisen A., Brolo A. G., Choo J., Cialla-May D., Deckert V., Fabris L., Faulds K., García de Abajo F. J., Goodacre R., Graham D., Haes A. J., Haynes C. L., Huck C., Itoh T., Käll M., Kneipp J., Kotov N. A., Kuang H., Le Ru E. C., Lee H. K., Li J.-F., Ling X. Y., Maier S. A., Mayerhöfer T., Moskovits M., Murakoshi K., Nam J.-M., Nie S., Ozaki Y., Pastoriza-Santos I., Perez-Juste J., Popp J., Pucci A., Reich S., Ren B., Schatz G. C., Shegai T., Schlücker S., Tay L.-L., Thomas K. G., Tian Z.-Q., Van Duyne R. P., Vo-Dinh T., Wang Y., Willets K. A., Xu C., Xu H., Xu Y., Yamamoto Y. S., Zhao B., Liz-Marzán L. M., Present and future of surface-enhanced raman scattering. ACS Nano 14, 28–117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang W., Jia S., Chao J., Wang L., Duan X., Liu H., Li Q., Zuo X., Wang L., Wang L., Liu N., Fan C., Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Sci. Adv. 5, eaau4506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong C., Xu M., Xu L.-J., Wei T., Ma X., Zheng X.-S., Hu R., Ren B., Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 118, 4946–4980 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Feng E., Zheng T., He X., Chen J., Tian Y., A novel ternary heterostructure with dramatic SERS activity for evaluation of PD-L1 expression at the single-cell level. Sci. Adv. 4, eaau3494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C. Y., Duan S., Wen B. Y., Li S. B., Kathiresan M., Xie L. Q., Chen S., Anema J. R., Mao B. W., Luo Y., Tian Z. Q., Li J. F., Observation of inhomogeneous plasmonic field distribution in a nanocavity. Nat. Nanotechnol. 15, 922–926 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Han X. X., Huang G. G., Zhao B., Ozaki Y., Label-free highly sensitive detection of proteins in aqueous solutions using surface-enhanced raman scattering. Anal. Chem. 81, 3329–3333 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Xu L.-J., Zong C., Zheng X.-S., Hu P., Feng J.-M., Ren B., Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal. Chem. 86, 2238–2245 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Park H. J., Cho S., Kim M., Jung Y. S., Carboxylic acid-functionalized, graphitic layer-coated three-dimensional sers substrate for label-free analysis of alzheimer's disease biomarkers. Nano Lett. 20, 2576–2584 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Dai X., Fu W., Chi H., Mesias V. S. D., Zhu H., Leung C. W., Liu W., Huang J., Optical tweezers-controlled hotspot for sensitive and reproducible surface-enhanced Raman spectroscopy characterization of native protein structures. Nat. Commun. 12, 1292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda K., Fujimoto N., Uehara H., Uosaki K., Raman scattering of aryl isocyanide monolayers on atomically flat Au(111) single crystal surfaces enhanced by gap-mode plasmon excitation. Chem. Phys. Lett. 460, 205–208 (2008). [Google Scholar]
- 24.Li J. F., Huang Y. F., Ding Y., Yang Z. L., Li S. B., Zhou X. S., Fan F. R., Zhang W., Zhou Z. Y., Wu D. Y., Ren B., Wang Z. L., Tian Z. Q., Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 464, 392–395 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Wang Y.-H., Zheng S., Yang W.-M., Zhou R.-Y., He Q.-F., Radjenovic P., Dong J.-C., Li S., Zheng J., Yang Z.-L., Attard G., Pan F., Tian Z.-Q., Li J.-F., In situ Raman spectroscopy reveals the structure and dissociation of interfacial water. Nature 600, 81–85 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Benz F., Schmidt M. K., Dreismann A., Chikkaraddy R., Zhang Y., Demetriadou A., Carnegie C., Ohadi H., de Nijs B., Esteban R., Aizpurua J., Baumberg J. J., Single-molecule optomechanics in “picocavities”. Science 354, 726–729 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Banik M., El-Khoury P. Z., Nag A., Rodriguez-Perez A., Guarrottxena N., Bazan G. C., Apkarian V. A., Surface-enhanced raman trajectories on a nano-dumbbell: Transition from field to charge transfer plasmons as the spheres fuse. ACS Nano 6, 10343–10354 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Chulhai D. V., Jensen L., Determining molecular orientation with surface-enhanced raman scattering using inhomogenous electric fields. J. Phys. Chem. C 117, 19622–19631 (2013). [Google Scholar]
- 29.Aprà E., Bhattarai A., El-Khoury P. Z., Gauging molecular orientation through time domain simulations of surface-enhanced Raman scattering. J. Phys. Chem. A 123, 7142–7147 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Jensen L., Aikens C. M., Schatz G. C., Electronic structure methods for studying surface-enhanced Raman scattering. Chem. Soc. Rev. 37, 1061–1073 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Mullin J., Schatz G. C., Combined linear response quantum mechanics and classical electrodynamics (QM/ED) method for the calculation of surface-enhanced Raman spectra. J. Phys. Chem. A 116, 1931–1938 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Duan S., Tian G., Ji Y., Shao J., Dong Z., Luo Y., Theoretical modeling of plasmon-enhanced Raman images of a single molecule with subnanometer resolution. J. Am. Chem. Soc. 137, 9515–9518 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Duan S., Tian G., Xie Z., Luo Y., Gauge invariant theory for super high resolution Raman images. J. Chem. Phys. 146, 194106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu W., Ye S., Zhang Y., Li T., Zhang G., Luo Y., Mukamel S., Jiang J., Machine learning protocol for surface-enhanced Raman spectroscopy. J. Phys. Chem. Lett. 10, 6026–6031 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Bouř P., Sopková J., Bednárová L., Maloň P., Keiderling T. A., Transfer of molecular property tensors in cartesian coordinates: A new algorithm for simulation of vibrational spectra. J. Comput. Chem. 18, 646–659 (1997). [Google Scholar]
- 36.Yang W., Direct calculation of electron density in density-functional theory. Phys. Rev. Lett. 66, 1438–1441 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Xomalis A., Zheng X., Demetriadou A., Martínez A., Chikkaraddy R., Baumberg J. J., Interfering plasmons in coupled nanoresonators to boost light localization and SERS. Nano Lett. 21, 2512–2518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Li M., Meng L., Lin K., Feng J., Huang T., Yang Z., Ren B., Probing the location of hot spots by surface-enhanced Raman spectroscopy: Toward uniform substrates. ACS Nano 8, 528–536 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Xu L.-J., Lei Z.-C., Li J., Zong C., Yang C. J., Ren B., Label-free surface-enhanced Raman spectroscopy detection of DNA with single-base sensitivity. J. Am. Chem. Soc. 137, 5149–5154 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Chen J., Zhu E., Liu J., Zhang S., Lin Z., Duan X., Heinz H., Huang Y., De Yoreo J. J., Building two-dimensional materials one row at a time: Avoiding the nucleation barrier. Science 362, 1135–1139 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Liu J., Zeng J., Zhu C., Miao J., Huang Y., Heinz H., Interpretable molecular models for molybdenum disulfide and insight into selective peptide recognition. Chem. Sci. 11, 8708–8722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang T.-X., Cong X., Wu S.-S., Lin K.-Q., Yao X., He Y.-H., Wu J.-B., Bao Y.-F., Huang S.-C., Wang X., Tan P.-H., Ren B., Probing the edge-related properties of atomically thin MoS2 at nanoscale. Nat. Commun. 10, 5544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye S., Zhong K., Zhang J., Hu W., Hirst J. D., Zhang G., Mukamel S., Jiang J., A machine learning protocol for predicting protein infrared spectra. J. Am. Chem. Soc. 142, 19071–19077 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Wright L. B., Rodger P. M., Corni S., Walsh T. R., GolP-CHARMM: First- principles based force fields for the interaction of proteins with Au(111) and Au(100). J. Chem. Theory. Comput. 9, 1616–1630 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Iori F., Di Felice R., Molinari E., Corni S., GolP: An atomistic force-field to describe the interaction of proteins with Au(111) surfaces in water. J. Comput. Chem. 30, 1465–1476 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Brancolini G., Kokh D. B., Calzolai L., Wade R. C., Corni S., Docking of ubiquitin to gold nanoparticles. ACS Nano 6, 9863–9878 (2012). [DOI] [PubMed] [Google Scholar]
- 47.M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, Williams, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, D. J. Fox. (Wallingford, CT, 2009), vol. Gaussian 09 Rev. D.01. [Google Scholar]
- 48.Pang R., Yu L.-J., Zhang M., Tian Z.-Q., Wu D.-Y., DFT study of hydrogen-bonding interaction, solvation effect, and electric-field effect on Raman spectra of hydrated proton. J. Phys. Chem. A 120, 8273–8284 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Eastman P., Behara P. K., Dotson D. L., Galvelis R., Herr J. E., Horton J. T., Mao Y., Chodera J. D., Pritchard B. P., Wang Y., De Fabritiis G., Markland T. E., SPICE, a dataset of drug-like molecules and peptides for training machine learning potentials. Sci. Data 10, 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C., GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 53.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M., All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 55.Johnson P. B., Christy R. W., Optical constants of the noble metals. Phys. Rev. B 6, 4370–4379 (1972). [Google Scholar]
- 56.Lu T., Chen F., Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 33, 580–592 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Li J. F., Tian X. D., Li S. B., Anema J. R., Yang Z. L., Ding Y., Wu Y. F., Zeng Y. M., Chen Q. Z., Ren B., Wang Z. L., Tian Z. Q., Surface analysis using shell-isolated nanoparticle-enhanced Raman spectroscopy. Nat. Protoc. 8, 52–65 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Clavilier J., Faure R., Guinet G., Durand R., Preparation of monocrystalline Pt microelectrodes and electrochemical study of the plane surfaces cut in the direction of the {111} and {110} planes. J. Electroanal. Chem. Interfacial Electrochem. 107, 205–209 (1980). [Google Scholar]
- 59.Wu F., Liu Z., Hawthorne N., Chandross M., Moore Q., Argibay N., Curry J. F., Batteas J. D., Formation of coherent 1H–1T heterostructures in single-layer MoS2 on Au(111). ACS Nano 14, 16939–16950 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Yang L.-K., Huang T.-X., Zeng Z.-C., Li M.-H., Wang X., Yang F.-Z., Ren B., Rational fabrication of a gold-coated AFM TERS tip by pulsed electrodeposition. Nanoscale 7, 18225–18231 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Rico E., Alvarez-Puebla R. A., Guerrini L., Direct surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids: From fundamental studies to real-life applications. Chem. Soc. Rev. 47, 4909–4923 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Barhoumi A., Zhang D., Halas N. J., Correlation of molecular orientation and packing density in a dsDNA self-assembled monolayer observable with surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 130, 14040–14041 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Lin M., Wang J., Zhou G., Wang J., Wu N., Lu J., Gao J., Chen X., Shi J., Zuo X., Fan C., Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. 54, 2151–2155 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Tables S1 and S2