Abstract
FDG parametric Ki images show great advantage over static SUV images, due to the higher contrast and better accuracy in tracer uptake rate estimation. In this study, we explored the feasibility of generating synthetic Ki images from static SUV ratio (SUVR) images using three configurations of U-Nets with different sets of input and output image patches, which were the U-Nets with single input and single output (SISO), multiple inputs and single output (MISO), and single input and multiple outputs (SIMO). SUVR images were generated by averaging three 5-min dynamic SUV frames starting at 60 minutes post-injection, and then normalized by the mean SUV values in the blood pool. The corresponding ground truth Ki images were derived using Patlak graphical analysis with input functions from measurement of arterial blood samples. Even though the synthetic Ki values were not quantitatively accurate compared with ground truth, the linear regression analysis of joint histograms in the voxels of body regions showed that the mean R2 values were higher between U-Net prediction and ground truth (0.596, 0.580, 0.576 in SISO, MISO and SIMO), than that between SUVR and ground truth Ki (0.571). In terms of similarity metrics, the synthetic Ki images were closer to the ground truth Ki images (mean SSIM = 0.729, 0.704, 0.704 in SISO, MISO and MISO) than the input SUVR images (mean SSIM = 0.691). Therefore, it is feasible to use deep learning networks to estimate surrogate map of parametric Ki images from static SUVR images.
Index Terms—: parametric Ki image, FDG PET, deep learning
I. Introduction
18F-fluorodeoxyglucose (FDG) PET imaging has been widely used for detection, staging, and therapeutic response assessment in oncology [1]. Tumors typically have higher activities of glucose metabolism, leading to higher uptake rates of glucose analogy FDG in tumors than in normal tissues [2–6]. For cancer diagnosis and staging, whole-body PET imaging is typically performed to scan from the base of the skull to mid-thigh, covering most relevant organs [6]. Combined with other imaging modalities, such as CT and MRI, FDG PET imaging help to localize suspected tumor regions and provide more functional information about tumors [7, 8].
Standard uptake value (SUV) is widely used in clinics for PET quantification and is generated from static FDG PET images, acquired typically at 60 minutes post tracer injection [6]. In comparison, parametric Ki value derived from dynamic PET frames is a potentially more quantitative measurement for the FDG metabolic rate than the semi-quantitative measurement of SUV [9, 10]. The derivation of Ki value is based on the theory that the FDG uptake process can be approximated using two-tissue irreversible compartmental model [11, 12]. Through the acquisition of dynamic FDG PET images after tracer injection and the measurement of the arterial tracer activity as the input function, Ki value can be calculated using Patlak graphical analysis for each voxel [12, 13]. As a common physiological metric, the glucose metabolic rate can be converted from the Ki value with the value of the glucose concentration and a correction term of lumped constant [14, 15]. The two-tissue irreversible compartmental model is applicable for many tissue regions over the body, but is not valid in organ regions with dual blood supply, such as liver [16]. Kinetic parameters in these regions could be estimated by more complex models. For example, based on the two-tissue compartmental model, the generalized Patlak analysis estimated Kloss in additional to Ki, with considerations of FDG dephosphorylation. The generalized Patlak analysis can reduce the estimation bias, but encounter higher noise than the standard linear Patlak analysis [17]. The voxel-wise model selection was also implemented to apply different compartmental models in whole-body PET images [18].
Both dynamic PET image acquisition and input function measurement are time-consuming and cause discomfort to the patients [19, 20]. To simplify the process, a dual-time-point Ki calculation method was implemented to reduce the number of frames in linear regression calculation of Patlak analysis [21]. Image-derived and population-based input functions can also replace the input function measurement through arterial blood sampling to reduce discomfort of patients [22, 23]. To further simplify the process, it would be appealing to generate Ki images from single timepoint static PET images.
Deep neural networks have become increasingly popular in medical imaging research, such as image denoising [24], segmentation [25], and detection and classification of tumors [26]. In tasks of denoising low dose PET images, 3D U-Net could achieve superior performance than conventional denoising techniques, such as Gaussian filter [27]. Generative adversarial networks (GAN) [28] are also widely used to denoise images of various modalities, such as low dose CT [29], PET [30] and fast MRI [31]. Various kinds of deep neural networks were also used in image synthesis across different imaging modalities [32]. For example, U-Net, which was originally designed for segmentation tasks of biomedical images, such as segmenting neuronal structures in microscopy images [33], was also developed to generate attenuation maps of PET images from input MRI images [34]. GANs have been used to train SPECT emission images to generate attenuation maps, which can help attenuation correction process without CT [35]. Deep learning networks have been used to predict PET images between different tracers. Based on the correlation between glucose metabolism and synaptic vesicle glycoprotein 2A (SV2A) in the brain, PET images of 11C-UCB-J could be generated from FDG PET images using 3D U-Net [36].
Inspired by the deep learning architecture used for image synthesis between different imaging modalities and tracers, we applied various configurations of 3D U-Nets to synthesize parametric Ki images from static FDG PET images in this study. One recent study used a 3D U-Net structures to generate dynamic PET frames from MR images, where the output dynamic PET frames were further converted to Ki images using Patlak analysis [37]. However, the process of generating Ki images was isolated from the 3D U-Net, and this work focused more on the correlation between MR and dynamic PET images instead of absolute quantification. Another study used 3D U-Net to generate parametric Ki images from static SUV images without input function measurement [38]. The SUV images in their study were acquired by a total-body PET/CT scanner, thus had higher counts than the whole-body PET images from typically clinical PET scanners using continuous bed motion (CBM) or step-and-shoot techniques [38]. A More recent study used an improved cycle-GAN, in additional to U-Net, to generate parametric Ki images from SUV images in the chest regions [39]. Our work implemented similar deep learning networks to generate parametric Ki images in the body trunk without input function measurement. However, we used SUV ratio (SUVR) images, instead of SUV images, as input to training the U-Nets, based on the findings that static SUVR values are better correlated with parametric Ki values than SUV in tumor regions [40]. The static PET images in our study were acquired by a Siemens mCT PET/CT scanner using CBM acquisition and had lower counts than those from total-body scan with long axial FOV. In addition to the U-Net configuration used in [38], we explored U-Net configurations with different sets of input and output image patches to explore if additional information, such Vb values, helped to train U-Nets. Currently, the PET/CT scanners with CBM imaging techniques are still widely used in clinics and our deep learning workflow could be used to generate parametric Ki images from the static PET images acquired by these scanners.
II. Materials and methods
A. Data Generation
There were 25 whole-body FDG dynamic PET scan datasets included in this study. The subjects were scanned on a Siemens Biograph mCT scanner using the multi-pass CBM mode. After the subject’s FDG injection ranging from 256 to 373 MBq, a 6-min single bed scan on the heart was performed to capture the initial phase of input function. Then the whole-body scanning process was performed with 19 CBM passes, with each pass taking 120 seconds for the initial 4 frames from 6 min to 15 min post tracer injection, and 300 seconds for the remaining frames after 15 min post tracer injection [21]. Images were reconstructed into dynamic frames each with the voxel size of 2.036mm×2.036mm×2.027mm. During the acquisition of dynamic frames, blood samples were collected through arterial cannulation after tracer injection and were centrifuged to obtain plasma. The plasma radioactivity of the samples was measured by gamma counters to obtain the arterial input function after decay correction [21].
The static SUV image of each subject was generated as the average of three dynamic SUV frames from 60 minutes to 75 minutes post tracer injection, with the frame index from 14 to 16 [21]. Left ventricle blood pool regions were segmented manually in static SUV images for all subjects, and static SUV images were then normalized by the mean values in the blood pool regions to generate SUVR images. One U-Net configuration was implemented to read three input SUVR patches generated from individual dynamic SUV frames from 60 minutes to 75 minutes, and then normalized by mean values of blood pool in the middle frames. Ground truth Ki images were generated with images of intercept Vb values from all dynamic SUV frames and plasma input functions of the tracer, using Patlak analysis with t* = 20 min [21, 41]. The output Ki images in the training dataset were preprocessed by multiplying the Ki values by 100, so that most output values of U-Nets were in the range of 0 to 10, to improve training efficiency. Both SUVR and Ki images were cropped to only include the subject’s body trunk with a matrix size of 160×288×256. We used patch-based approach in the training and random patches of smaller regions, with a matrix size of 64×64×64, were created from both the SUVR and the Ki images.
B. U-Net Structures
In our work, three deep learning network configurations were implemented using the Pytorch library [42] based on the 3D U-Net structure [36]. All configurations read static SUVR image patches as input and generate their corresponding Ki image patches as output. Fig. 1 shows the first two configurations of U-Nets with different numbers of input SUVR image patches and single output Ki image patch. The U-Net accepting single SUVR patch as input was denoted as single input and single output (SISO). The network with multiple input patches, specifically three SUVR patches corresponding to SUVR images of 60–65 min, 65–70 min, and 70–75 min post-injection, was denoted as multiple inputs and single output (MISO). Three individual SUVR frames provide additional information about the change of tracer activities between 60 minutes and 75 minutes post-injection. We aim to explore if the additional dynamic information could further improve the Ki image prediction. Both U-Net configurations had contraction and expanding paths, with each path having 5 operational layers. Every layer consisted of two 3D convolutional and ReLU operations. The contraction path of the U-Nets encoded the input image patches as smaller features by connecting two consecutive layers with the max-pooling operation, while the expanding path decoded the features with up-sampling operations between two layers. The 3D convolutional operations used 3×3×3 kernels with 1 voxel padding. The first layer had 64 filters, representing 64 features extracted from the input SUVR patches. The number of filters doubled in the next layer, hence there were 128, 256, 512 and 1024 filters in Layers 2, 3, 4 and 5. The output of each layer in the contraction path was concatenated to the layer at the same depth in the expanding path through skip connection. While patches of SUVR and Ki images were used in training process, entire SUVR images of cropped regions in the subjects’ whole body were used as the input of U-Nets in the testing process to generate Ki images.
Fig. 1.
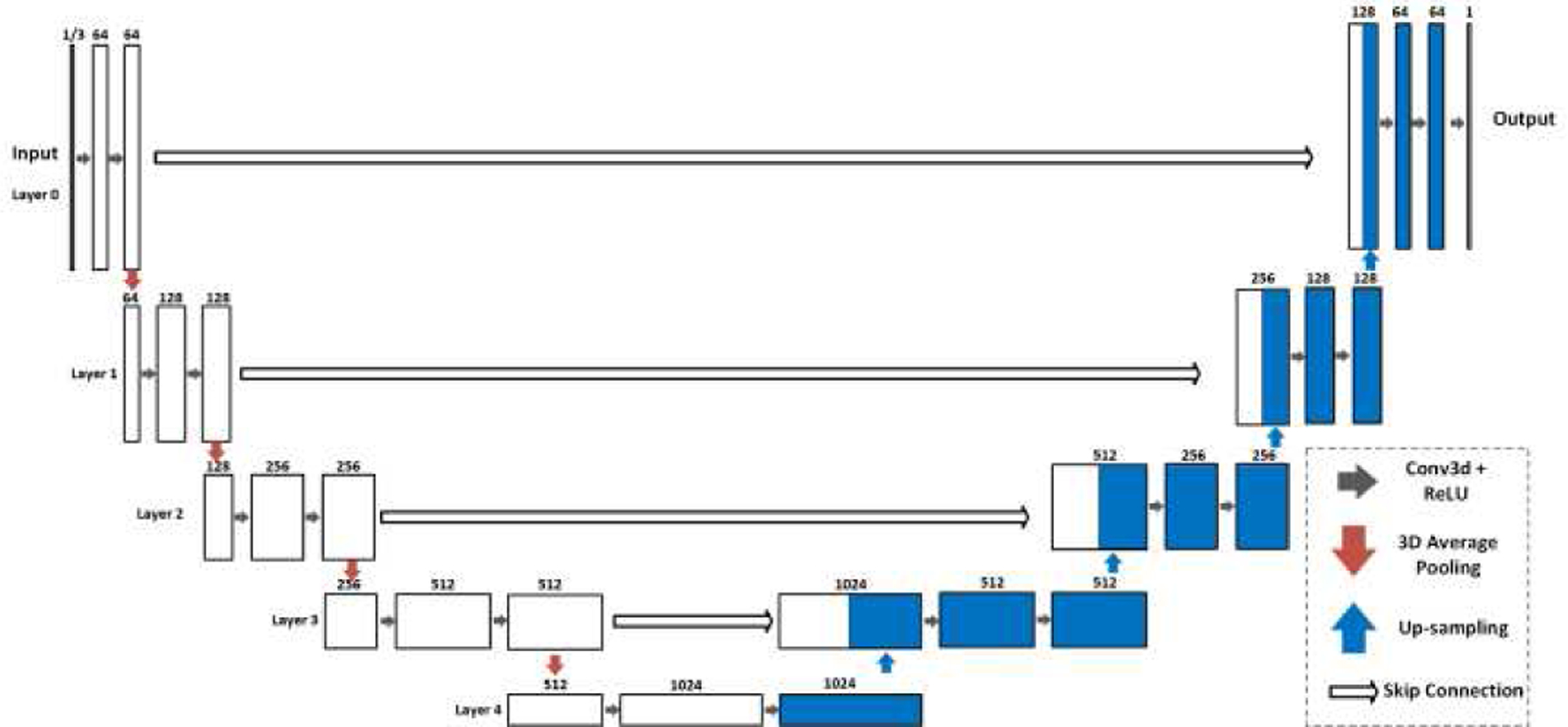
The SISO and MISO U-Nets with 5 operational layers to generate output Ki images from SUVR images. The SISO U-Net reads one SUVR patch as input, while the MISO U-Net has the input of three.
The U-Net can be modified to generate multiple image patches as the multi-task outputs. In Patlak analysis, images of intercept Vb values were generated together with Ki images. In this study, the multi-task U-Net was implemented to incorporate patches of Vb values as the additional output to Ki patches to examine whether Vb values improved the training performance. The multi-task U-Net only accepted single input of SUVR patch, and thus was denoted as single input and multiple outputs (SIMO). This configuration had single contraction path and was divided into two expanding paths after the bottleneck layer between the contraction and the expanding paths. Like the previous two U-Net configurations, the SIMO U-Net used the skip connection to concatenate the output of each contraction path layer to the input of the corresponding expanding path layer.
C. U-Nets Training and Testing
The three configurations of U-Nets were all trained with a NVIDIA Quadro RTX 8000 GPU card. The 25 subjects were partitioned into 5 groups to perform 5-fold cross-validation. In each group, the SUVR and Ki images of 20 subjects were assigned to train the U-Nets, while the SUVR images of the remaining 5 subjects were used as the input of the U-Nets to generate corresponding synthetic Ki images to evaluate the training performance. For every subject, 400 patches of the SUVR and Ki images were randomly generated as the input and output of the U-Nets. All U-Nets were trained with a batch size of 14, and a total of 300 epochs. The SISO and MISO U-Nets used the L1 loss of the scaled Ki patches as the cost function, while in the SIMO U-Net, the cost function was the sum of the L1 losses of scaled Ki and unscaled Vb patches. We did not scale Vb values in the cost function because most of the Vb values were already in the range of 0 to 10. The initial learning rate was set to 10−4, with decay rate of 0.96 for every epoch. The U-Net is a fully convolutional neural network, including operations of 3D convolution, average pooling, up-sampling and skip connection. Therefore, the U-Net can accept input images with variable sizes, and the image size is only limited by the average pooling operations [33]. In the validation and testing process, the images of body trunks (matrix size of 160×288×256) were set as input of the U-Nets to generate corresponding images of Ki distribution over the body trunks. Due to the limitation of GPU memory, the U-Nets used CPU with a large system memory to generate whole-body Ki images.
D. Evaluation
The regions of representative organs, such as liver, heart, kidney, and aorta wall, were first evaluated visually in the coronal view of SUVR, synthetic and ground truth Ki images. Difference images in color map were generated between synthetic and ground truth Ki images, so the regions with high bias between two images can be observed. We also used 2D joint histograms to compare the voxel values in two images. The joint histograms were plotted between synthetic and ground truth Ki images, after removing background voxels, and were compared with the joint histograms between input SUVR and ground truth Ki images.
Two image similarity metrics, structure similarity index (SSIM) and normalized mean square error (NMSE), were used in this study to compare synthetic and ground truth Ki images. The value of SSIM ranges from −1 to 1, and higher values indicate higher similarity between two images [43]. The NMSE is non-negative, and lower values suggest higher similarity. SSIM values were calculated between synthetic and ground truth Ki images and were compared with the values between the corresponding input SUVR and ground truth Ki images to examine whether synthetic Ki images generated from the U-Nets were more similar to ground truth Ki images than SUVR images. The NMSE values were calculated between synthetic images and ground truth Ki images, after removing background voxels, to compare the performance of different U-Nets. Because the definitions of SUVR and Ki are inherently different, thus it is not feasible to calculate the NMSE values between SUVR and Ki images as the reference for evaluation.
Lesion regions were segmented in SUV images by the researchers and verified by a nuclear physician. These regions were evaluated by comparing their mean voxel values in the synthetic and the ground truth Ki images. Among the 25 subjects in the study, 22 of them had hypermetabolic lesions in different parts of the body, such as lung and chest, as displayed in Fig. 7.
Fig. 7.
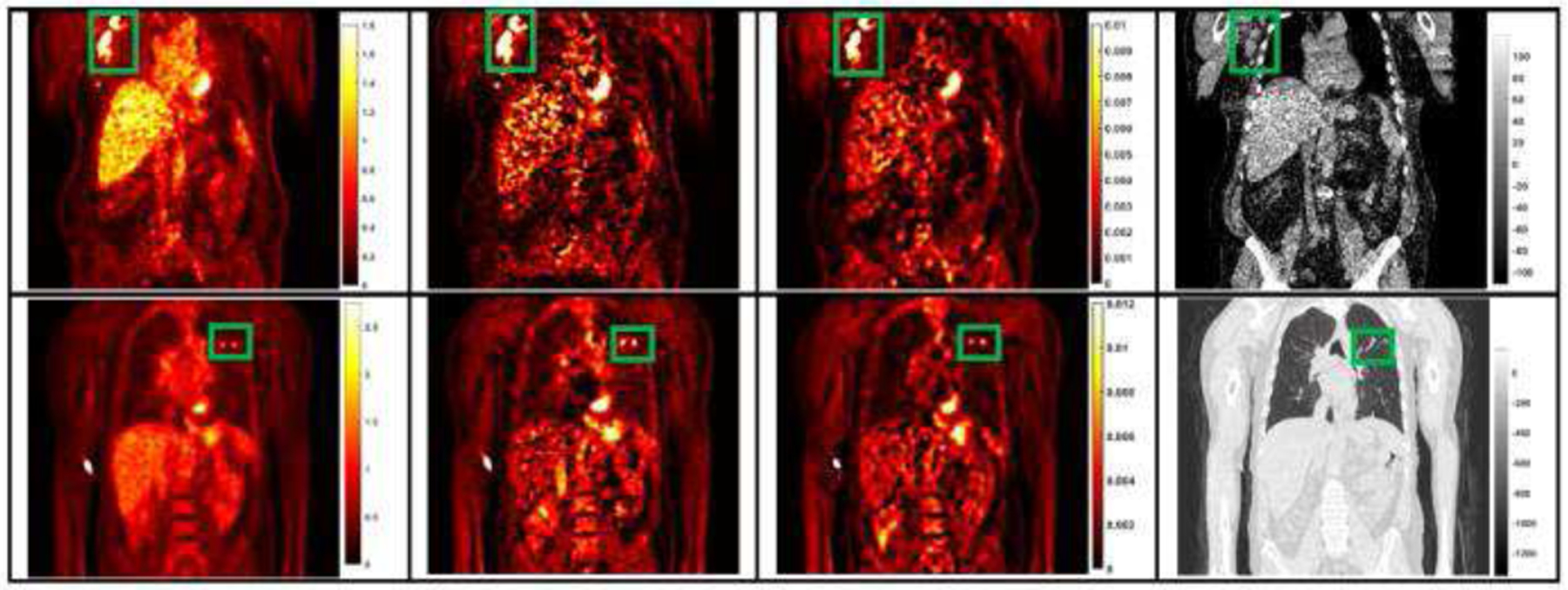
The coronal slices of two sample subject’s SUVR, ground truth Ki, synthetic Ki from SISO, and CT images, with green bounding boxes representing the lesion regions.
III. Results
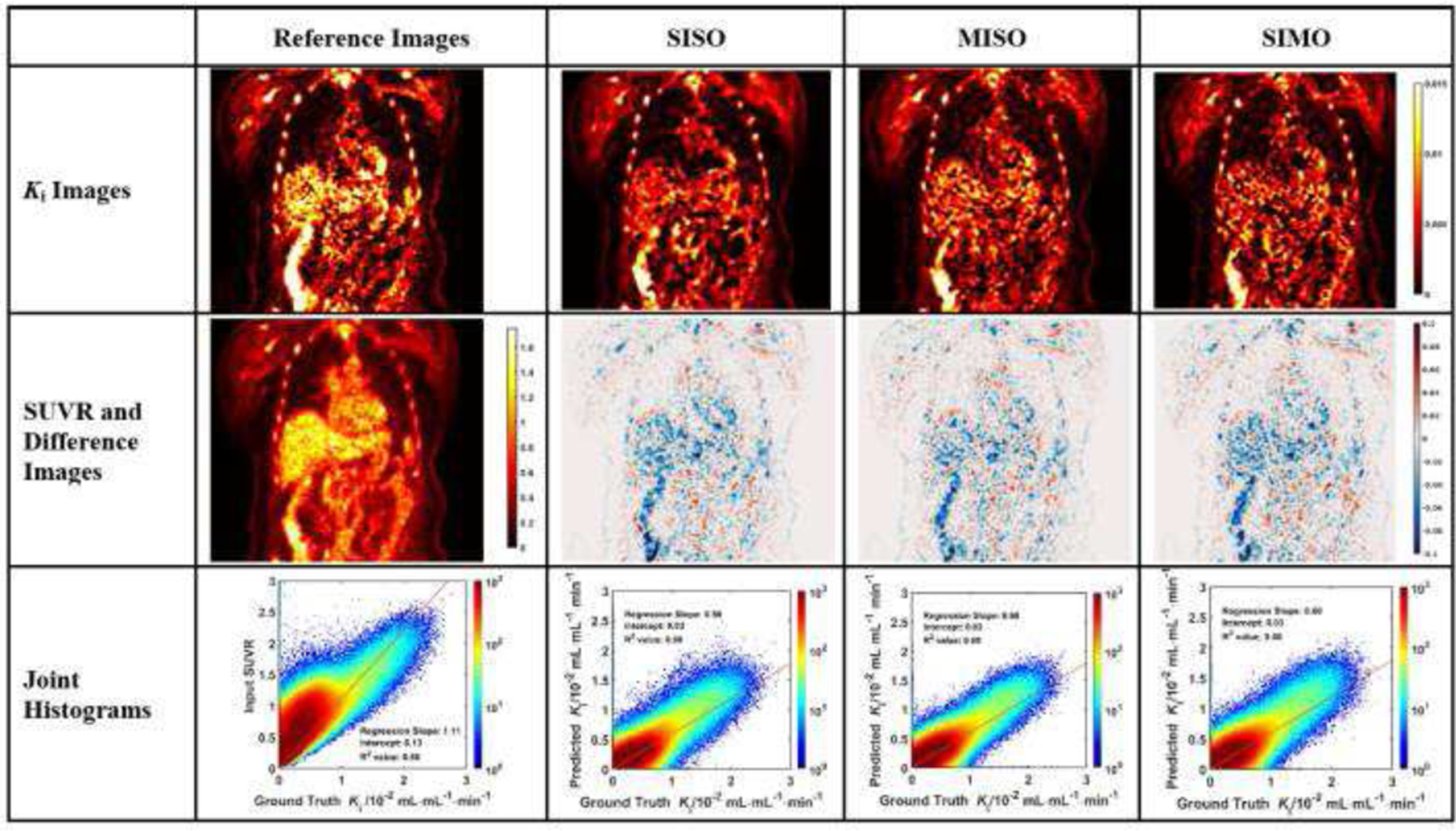
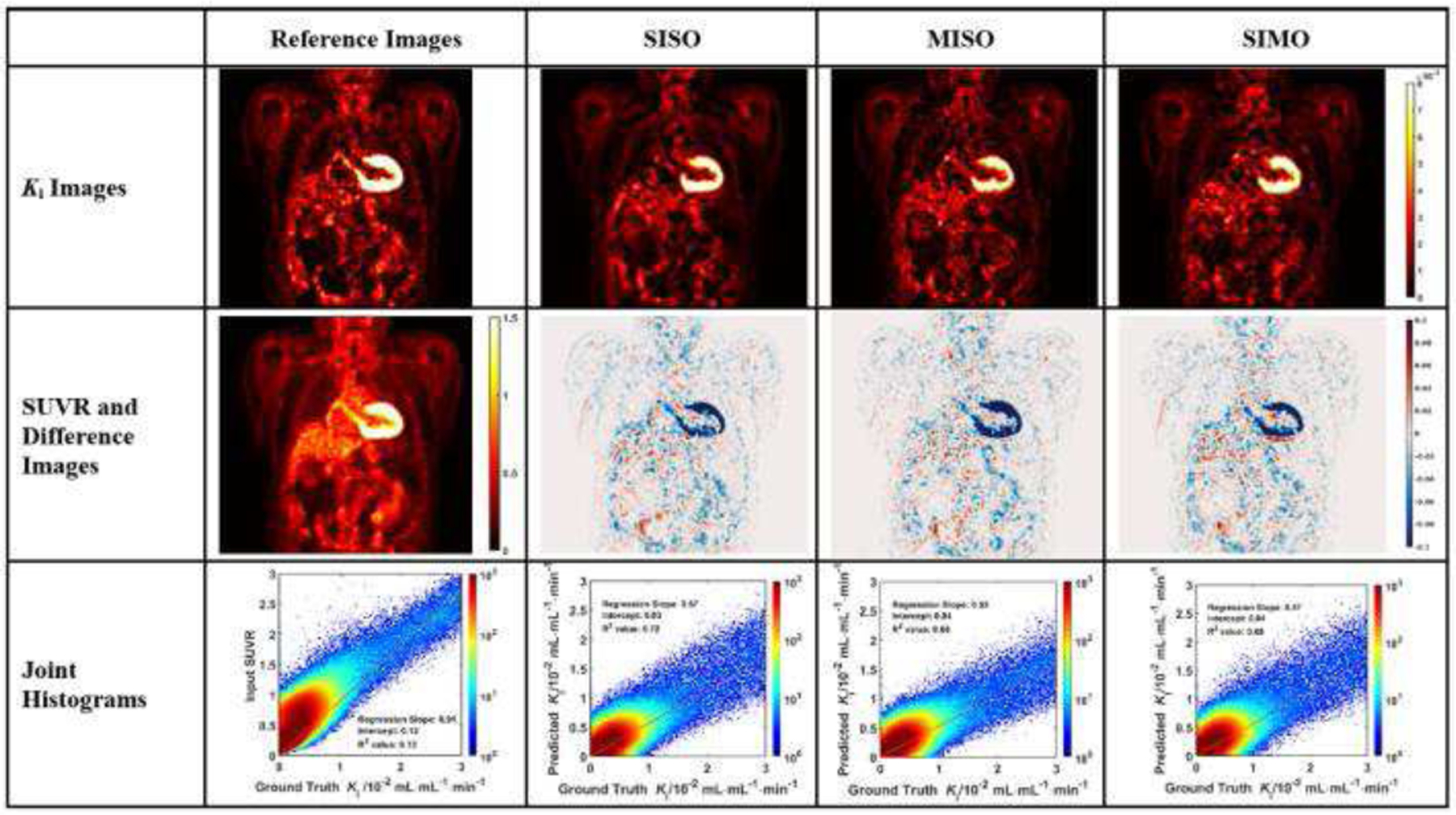
The evaluation of Ki image generation is shown with three sample subjects in Figs. 3 to 5. In each figure, the coronal image slices were chosen to cover the relevant organs, such as liver, heart, and aorta wall. The aorta walls can be visualized more clearly in both synthetic and ground truth Ki images, than in SUVR images. All three networks showed similar difference images, though the myocardial biases appear to be the highest in MISO images.
Fig. 3.
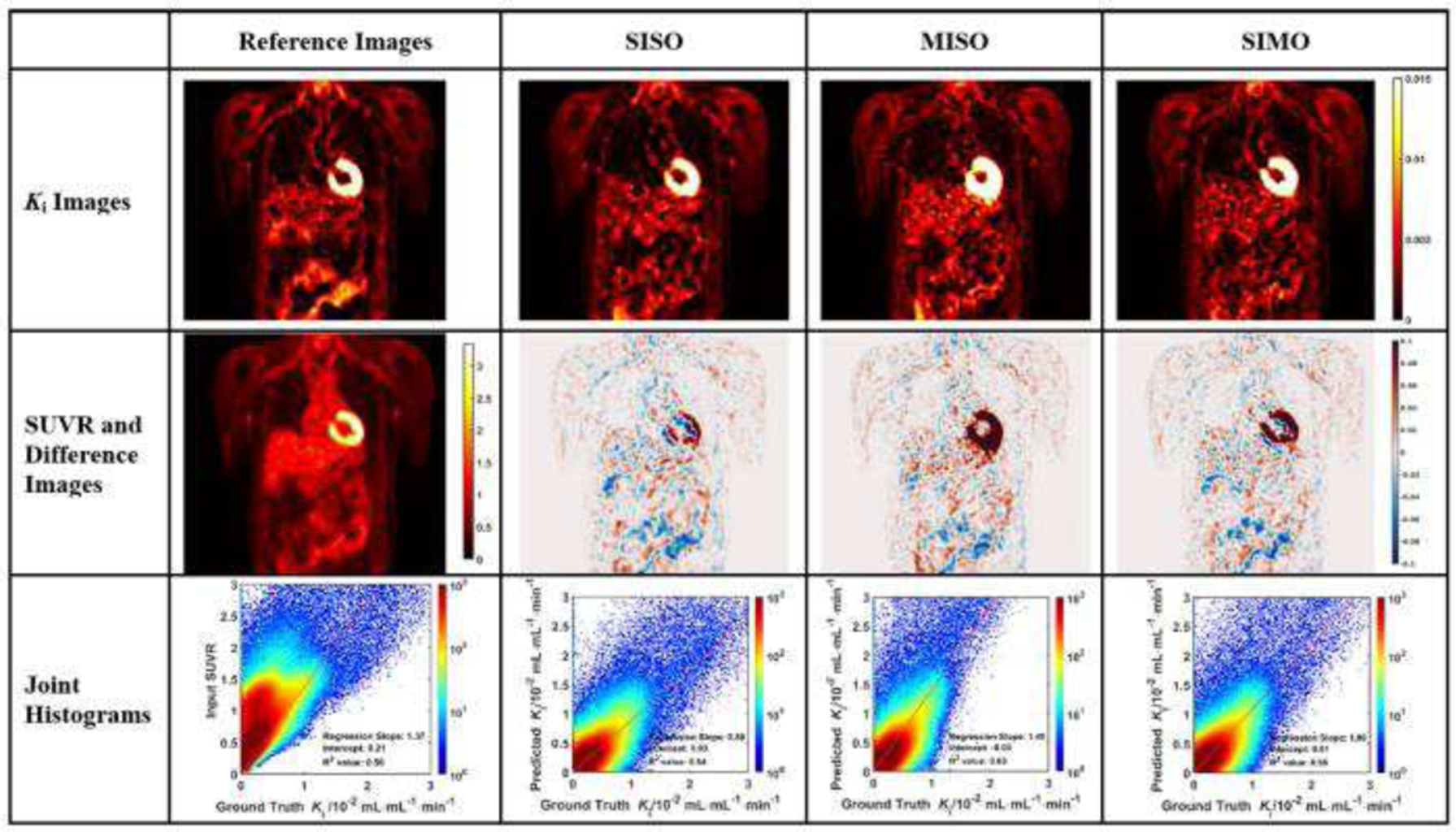
Evaluation analysis of sample subject #1 including the synthetic and reference Ki images, corresponding difference images, SUVR input image, and the joint histograms. In each joint histogram, the regression line was represented by the red straight line, with labels of regression slope, intercept and R2 value.
Fig. 5.
Evaluation analysis of sample subject #3 including the synthetic and reference Ki images, corresponding difference images, SUVR input image, and the joint histograms. In each joint histogram, the regression line was represented by the red straight line, with labels of regression slope, intercept and R2 value.
In each joint histogram, a regression line was derived between synthetic and ground truth Ki values. Certain body regions, including urinary bladder and injection intravenous lines, were excluded from the linear regression analysis. For each subject, we calculated R2 values in the linear regression analysis between the synthetic Ki values generated from various U-Nets and the ground truth Ki values. The R2 values of the prediction in the analysis of the SISO U-Net were higher than the R2 values of SUVR for most subjects (16 out of 25). In the analysis of the other two U-Net configurations, the R2 values of the prediction were generally lower than those of SISO. For the MISO U-Net, 14 out of 25 subjects had higher R2 values than the those of SUVR, while 13 out of 25 subjects had higher R2 values using the SIMO U-Net than those of SUVR.
Table 1 summarized the statistics of slopes, intercepts and R2 values in the linear regression analysis between the synthetic Ki values obtained from various U-Nets and between the input SUVR and the ground truth Ki values. The statistics were also listed for each validation group in the cross-validation process. The R2 values were labeled as bold if they were higher than the reference R2 values of SUVR. The mean R2 value of all subjects in SISO, MISO and SIMO were all slightly higher than that of SUVR. The statistics of each group generally agreed with the results of all subjects that the R2 values of U-Net predictions are generally higher than the R2 values of SUVR.
TABLE I.
Statistics of Slope, Intercept and R2 in the Linear Regression Analysis in Joint Histograms between Input SUVR, Synthetic Ki and Ground truth Ki Images
| SUVR | SISO | MISO | SIMO | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | R 2 | Slope | Intercept | R 2 | Slope | Intercept | R 2 | Slope | Intercept | R 2 | ||
| Total | Mean | 1.218 | 0.155 | 0.571 | 0.714 | 0.0259 | 0.596 | 0.740 | 0.0366 | 0.580 | 0.747 | 0.0357 | 0.576 |
| Normalized Standard Deviation | 0.235 | 0.215 | 0.211 | 0.366 | 1.769 | 0.227 | 0.423 | 0.676 | 0.216 | 0.367 | 0.6208 | 0.239 | |
| Group 1 | Mean | 1.120 | 0.131 | 0.606 | 0.871 | −0.014 | 0.666 | 0.891 | 0.022 | 0.646 | 0.880 | 0.023 | 0.638 |
| Normalized Standard Deviation | 0.401 | 0.083 | 0.104 | 0.609 | −6.760 | 0.097 | 0.656 | 1.171 | 0.209 | 0.632 | 0.954 | 0.198 | |
| Group 2 | Mean | 1.393 | 0.180 | 0.562 | 0.769 | 0.031 | 0.555 | 0.842 | 0.030 | 0.577 | 0.839 | 0.030 | 0.566 |
| Normalized Standard Deviation | 0.048 | 0.214 | 0.263 | 0.177 | 0.879 | 0.281 | 0.375 | 1.236 | 0.199 | 0.223 | 1.014 | 0.283 | |
| Group 3 | Mean | 1.330 | 0.172 | 0.544 | 0.660 | 0.046 | 0.571 | 0.661 | 0.049 | 0.552 | 0.677 | 0.052 | 0.547 |
| Normalized Standard Deviation | 0.158 | 0.159 | 0.207 | 0.128 | 0.145 | 0.179 | 0.160 | 0.182 | 0.184 | 0.138 | 0.171 | 0.181 | |
| Group 4 | Mean | 0.944 | 0.142 | 0.607 | 0.576 | 0.035 | 0.642 | 0.544 | 0.047 | 0.594 | 0.595 | 0.039 | 0.622 |
| Normalized Standard Deviation | 0.242 | 0.030 | 0.135 | 0.039 | 0.008 | 0.132 | 0.107 | 0.018 | 0.126 | 0.058 | 0.016 | 0.148 | |
| Group 5 | Mean | 1.306 | 0.150 | 0.538 | 0.695 | 0.031 | 0.548 | 0.762 | 0.035 | 0.53 | 0.745 | 0.034 | 0.509 |
| Normalized Standard Deviation | 0.074 | 0.212 | 0.261 | 0.253 | 0.751 | 0.273 | 0.192 | 0.616 | 0.274 | 0.190 | 60.646 | 0.284 | |
Table 2 summarized the statistics of the similarity metrics between synthetic Ki images generated from the U-Nets and ground truth Ki images, compared with the metrics between input SUVR and ground truth Ki images as the reference. The statistics of each validation group were also listed in the cross-validation process. The SSIM values between the synthetic and ground truth Ki images were labeled as bold if they were higher than the reference values. The lowest NMSE values were also labeled in bold among the U-Net predictions. The mean SSIM values between synthetic and ground truth Ki images were higher than that between input SUVR and ground truth Ki images. In the SISO U-Net, 20 out of 25 subjects had higher SSIM scores between synthetic Ki and ground truth Ki images than the scores between input SUVR and ground truth Ki images, while 17 out of 25 in the MISO and 18 out of 25 in the SIMO groups had higher SSIM scores between synthetic and ground truth values. The mean NMSE score between synthetic Ki images from SISO, and ground truth Ki images was lower than those between the synthesis from the other two U-Nets and the ground truth. The Ki images generated from U-Nets were generally more similar to ground truth Ki images than the input SUVR images were, and the SISO U-Net outperformed the other two configurations in terms of SSIM and NMSE.
TABLE II.
Similarity Metrics of SSIM and NMSE between Input SUVR, Synthetic Ki and Ground truth Ki Images
| SUVR | SISO | MISO | SIMO | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SSIM | NMSE | SSIM | NMSE | SSIM | NMSE | SSIM | NMSE | ||
| Total | Mean | 0.691 | 0.729 | 0.139 | 0.704 | 0.165 | 0.704 | 0.153 | |
| Normalized Std | 0.103 | 0.087 | 0.580 | 0.010 | 0.728 | 0.010 | 0.626 | ||
| Group 1 | Mean | 0.653 | 0.675 | 0.014 | 0.643 | 0.016 | 0.638 | 0.016 | |
| Normalized Std | 0.147 | 0.072 | 0.412 | 0.111 | 0.552 | 0.110 | 0.511 | ||
| Group 2 | Mean | 0.727 | 0.771 | 0.008 | 0.759 | 0.010 | 0.757 | 0.009 | |
| Normalized Std | 0.068 | 0.056 | 0.317 | 0.058 | 0.748 | 0.062 | 0.450 | ||
| Group 3 | Mean | 0.672 | 0.735 | 0.007 | 0.712 | 0.008 | 0.713 | 0.008 | |
| Normalized Std | 0.085 | 0.080 | 0.218 | 0.088 | 0.245 | 0.087 | 0.203 | ||
| Group 4 | Mean | 0.717 | 0.713 | 0.016 | 0.693 | 0.020 | 0.695 | 0.016 | |
| Normalized Std | 0.049 | 0.054 | 0.647 | 0.054 | 0.733 | 0.050 | 0.643 | ||
| Group 5 | Mean | 0.689 | 0.750 | 0.007 | 0.713 | 0.009 | 0.718 | 0.009 | |
| Normalized Std | 0.142 | 0.121 | 0.292 | 0.130 | 0.341 | 0.124 | 0.271 | ||
One-tailed pairwise t-tests were conducted to evaluate the difference of the SSIM and R2 values from different U-Nets and the SUVR images. The P values of the t-tested showed that the values of the SSIM and R2 from SISO network are significantly larger than those from SUVR, while the values of SSIM and R2 from the other two networks showed no significant difference from the reference values of SUVR. Therefore, the t-test also indicated that the SISO U-Net outperformed the SUVR and other two U-Nets in terms of SSIM and R2.
The hyperparameters of the U-Nets were optimized using the training and validation datasets. To validate the effectiveness of the hyperparameters, we included a testing dataset with 5 additional subjects to compare their synthetic and ground truth Ki images. The ground truth Ki images were generated using standard Patlak analysis with the image-derived input functions, while the synthetic Ki images were generated using the U-Nets trained in Group 5 in the validation process.
As indicated in Table 4, the U-Nets generally outperformed the input SUVR images in terms of the SSIM and R2 values in the testing dataset, even though the R2 value of MISO was marginally smaller than that of the SUVR. Therefore, the hyperparameters optimized by the training and validation groups did not overfit and were effective to synthesize the Ki images from subjects outside training and validation datasets.
TABLE IV.
Statistics of SSIM and R2 Between the Synthetic and ground truth images in Testing Dataset
| SUVR | SISO | MISO | SIMO | ||
|---|---|---|---|---|---|
| SSIM | Mean | 0.739 | 0.755 | 0.740 | 0.742 |
| Normalized Std | 0.059 | 0.072 | 0.076 | 0.074 | |
| R 2 | Mean | 0.696 | 0.731 | 0.688 | 0.770 |
| Normalized Std | 0.133 | 0.103 | 0.083 | 0.080 |
As shown in Fig. 6, the linear regression analysis of the distributions in lesion regions showed that the R2 values of three U-Nets were similar with the R2 value of SUVR. The R2 value of MISO network was higher than the value of SUVR. Sample lesion regions, labeled by the green bounding boxes, were presented in the coronal image slices in Fig. 7. It can be observed that the synthetic Ki images derived from SISO are visually consistent with the ground truth Ki images and both Ki images have substantially higher lesion contrast than those of SUVR images.
Fig. 6.
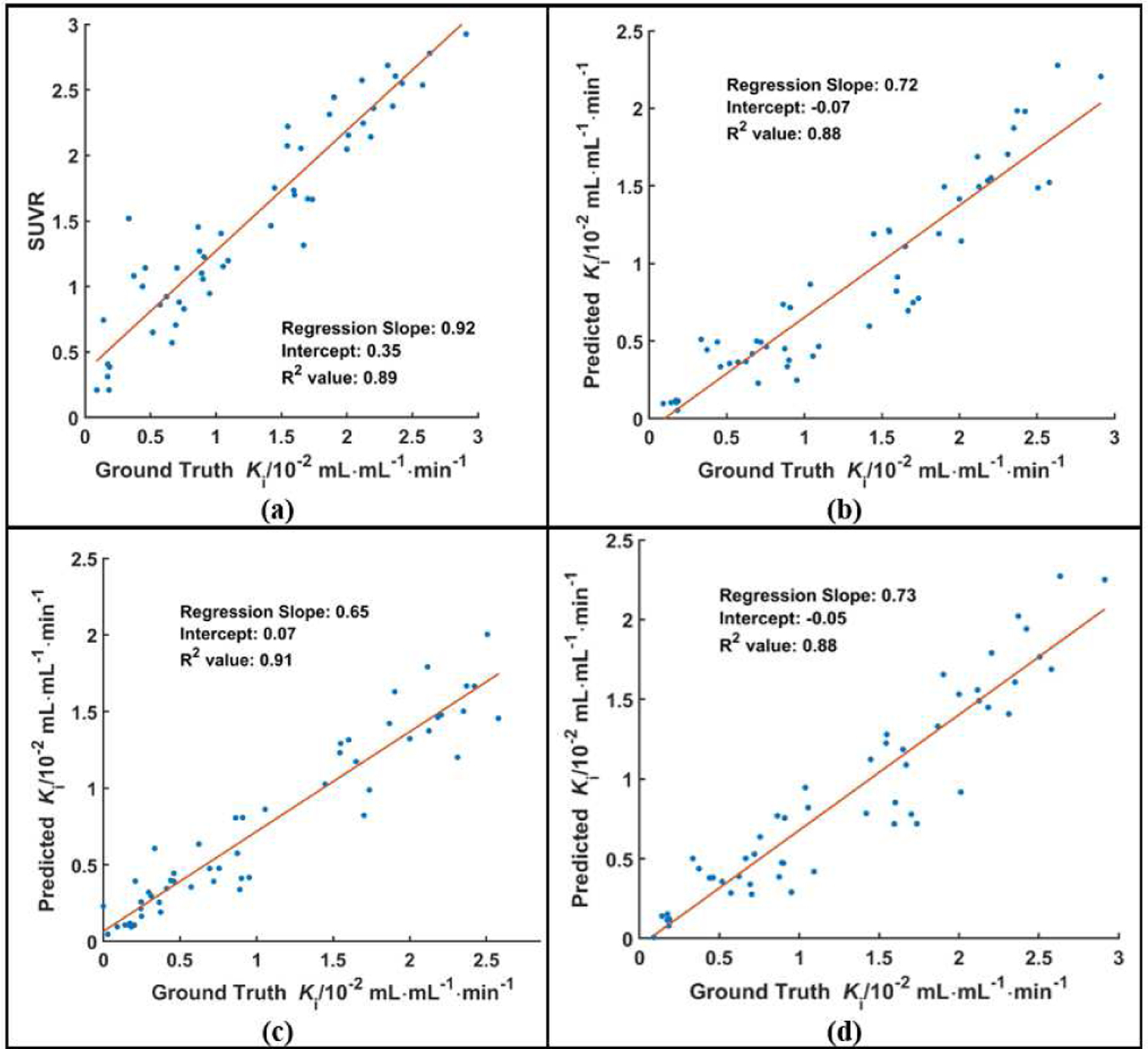
The distributions of the lesion ROI mean values in SUVR (a), synthetic Ki, and ground truth Ki images. The synthetic Ki values were generated from three configurations of U-Nets, which were SISO (b), MISO (c) and SIMO (d).
IV. Discussion
In this study, we developed deep learning methods to generate Ki images from static SUV images using the U-Nets in three configurations. As shown in Figs. 3 to 5, the coronal slices of the synthetic Ki images were visually more consistent with the ground truth Ki slices than the SUVR slices. The aorta walls had higher contrast in both the synthetic and the ground truth Ki slices, than in the SUVR slices. The synthetic Ki images generated from all three U-Nets were visually smoother than ground truth Ki images, likely because SUVR images have lower noise and the subsequent Ki images generated from SUVR images could inherit such low noise level. The difference images showed that the bias between synthetic and ground truth Ki images were small in most regions. Even though higher biases were observed in the regions with high tracer uptake, such as heart and the kidneys, the normalized differences were small.
The linear regression analysis of mean Ki and SUVR values in the body and lesion regions indicated that the synthetic Ki values generated from the U-Nets could correlate slightly better with ground truth Ki values than input SUVR values, in terms of higher R2 values, but such differences are not significant. In addition to the linear regression analysis, the comparison of the SSIM values showed that synthetic Ki images were more consistent with ground truth Ki images than input SUVR images. The statistics of each group in the cross-validation process agreed with the statistics from all subjects. Even though in Group 2, the mean R2 value of SISO is lower than that of SUVR, SISO network still performed better than SUVR in terms of R2 value from all subjects. The statistics in the testing dataset also agree with those in the validation dataset that the U-Nets outperformed the SUVR in terms of mean SSIM and R2 values.
Although three configurations were all based on the U-Net architecture, the SISO U-Net generally outperformed the other two configurations. Visually, the synthetic Ki images generated from SISO were less noisy than the images generated from the other two networks, as shown from Fig. 3 to Fig. 5. In the linear regression analysis of the joint histograms, the mean R2 value of the prediction in SISO was higher than those of the two other configurations. The mean SSIM value in SISO U-Net was also the highest. However, in the evaluation of the lesion regions, the R2 values of U-Net predictions did not have clear differences among all three configurations. The MISO network incorporated more information of the input static SUVR frame than the SISO network. However, individual input frame of MISO was noisier than the single input frame of SISO, which may negatively affect the training performance of MISO network. Therefore, our data suggested that incorporating additional input SUVR or output images of Vb values did not benefit the training performance of U-Nets.
Our work demonstrated that it is feasible to generate parametric Ki images from static SUVR images. The synthetic Ki images generated from various configurations of U-Nets were generally more consistent with ground truth Ki images than input SUVR images, with stronger linear correlations and higher SSIM values. However, the improvement of correlations from the U-Nets over those of SUVR was small and there were still differences between synthetic and ground truth Ki images. As shown in joint histogram analysis from Fig.3 to Fig. 5, the regression slopes between the synthetic and ground truth Ki values were not close to 1 for many subjects. The normalized standard deviation values in Table 1 also showed difference was not minor between the regression slopes across all subjects. Therefore, the synthetic Ki values were not quantitatively accurate compared with ground truth Ki values. In this work, we chose SUVR images instead of SUV images as the network input with the considerations explained in the Appendix that Ki is better correlated with SUVR as compared to SUV. Since input functions vary with subjects, even though population-based input functions can be used [22], values of Θ(T) in the Appendix differ among subjects due to variations of static image acquisition times and input functions. Therefore, the relationship between SUVR and Ki values also varies, and we hope our proposed deep learning methods could recover such underlying relationship. The patient’s motion might also affect the Ki image quality. For example, in Fig. 3, the contour of myocardium is smaller in three synthetic Ki images than in the ground-truth image, probably because this patient’s motion across dynamic frames produced more blurring effect than the static images. In the future, the information of patient’s motion, acquisition times and input functions could be incorporated into deep learning networks to further improve the performance in generating parametric Ki images.
As mentioned in the Introduction, the Ki estimation using standard Patlak analysis is not valid for the tissue regions with double blood supply and FDG dephosphorylation, such as in liver [16]. In these regions, the two-tissue reversible compartmental model is a better tool to estimate the parametric values [17]. And in vascular, lung and skin regions, zero or one compartmental model is favored to reduce the artifacts in the Ki value estimation [18]. Therefore, the bias of Ki estimation from the standard Patlak analysis could also contribute to the errors in our Ki estimation based on the SUVR values. The generalized Patlak analysis could help to reduce the bias of Ki estimation in the regions of non-negligible FDG dephosphorylation. But it could introduce higher noise level than the standard Patlak analysis [17]. Therefore, there are trade-off between noise and bias to select different compartmental models to estimate parametric values.
V. Conclusion
An image synthesis workflow, including three deep neural network configurations based on the 3D U-Net architecture, was developed to generate parametric Ki images from the static SUVR images, which can serve as the surrogate to the Ki estimation from the standard Patlak analysis. This workflow reduced the scan time and eliminated the need of input function measurements in the conventional Ki image generation using dynamic PET data and Patlak graphical analysis.
Fig. 2.
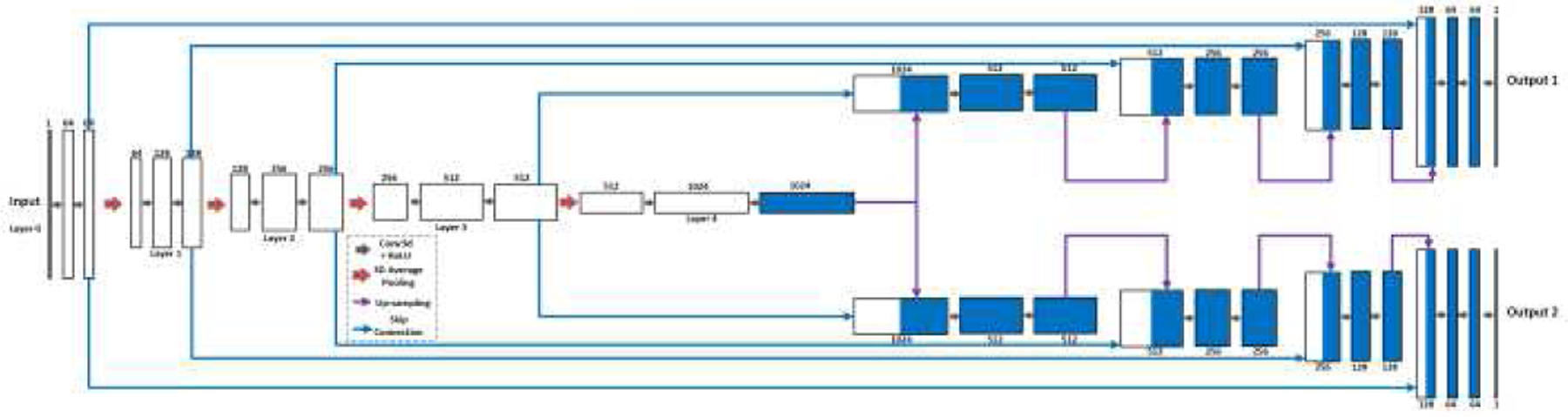
The single input and multiple outputs (SIMO) U-Net with 5 operational layers to generate output Ki and Vb images from SUVR images. The contraction path is shared by both outputs, with the expanding paths split for the outputs.
Fig. 4.
Evaluation analysis of sample subject #2 including the synthetic and reference Ki images, corresponding difference images, SUVR input image, and the joint histograms. In each joint histogram, the regression line was represented by the red straight line, with labels of regression slope, intercept and R2 value.
TABLE III.
Significance Test Results (P-value) of R2 and SSIM Values between Input SUVR, Synthetic Ki and Ground truth Ki Images
| SISO and SUVR | MISO and SUVR | SIMO and SUVR | |
|---|---|---|---|
| SSIM | 0.0014 | 0.1485 | 0.1266 |
| R 2 | 0.0229 | 0.3360 | 0.4137 |
Acknowledgment
This work was supported by National Institute of Health (NIH) grant R03EB027864. All authors declare that they have no known conflicts of interest in terms of competing financial interests or personal relationships that could have an influence or are relevant to the work reported in this paper.
Appendix
This section summarized the finding of J. van den Hoff, et al., regarding the correlation between the SUVR and Ki values [40]. Patlak analysis was the conventional method to generate Ki values from dynamic data [13], based on the equation of
where , representing the tracer net influx rate constant in tissue [11], and Ct(t) and Ca(t) were the tracer concentrations in tissue and plasma at time t post-injection [12]. The term on the left side of the equation could be defined as normalized SUV ratio (SUVR) value. The value of Ki can be well correlated to the value of SUVR for a wide range of subjects, with the assumptions that 1) the plasma input functions of different subjects had the similar shapes, which can be expressed as Ca(t) ≈ N × ba(t); 2) all static SUVR frames were acquired at the approximately the same time post tracer injection; and 3) values of Vb were small compared with SUVR values [40]. Then the correlation can be expressed as . With the fixed time point T, the term is a constant. Therefore, SUVR is linearly proportional to Ki when the assumptions are met and is preferred over SUV as network input in our study to generate Ki.
In clinical scenarios, plasma input functions vary with subjects, and values of Θ(T) differ accordingly, which makes the relationship among Ki, Vb and SUVR vary among subjects. Acquisition times of the static SUV frames also differ with subjects, in the scale of minutes, which makes values of Θ(T) different across subjects. Values of Vb generated by Patlak analysis are also not negligible compared with SUVR values. Thus, the variations in acquisition times and Vb values could contribute to the errors in deriving Ki directly from SUVR and more training data could further improve the network performance.
Footnotes
This work involved human subjects or animals in its research. The authors confirm that all human/animal subject research procedures and protocols are exempt from review board approval.
Contributor Information
Tianshun Miao, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA.
Bo Zhou, Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Juan Liu, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA.
Xueqi Guo, Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Qiong Liu, Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Huidong Xie, Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Xiongchao Chen, Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Ming-Kai Chen, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA.
Jing Wu, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA; Department of Physics, Beijing Normal University, Beijing 100875, China.
Richard E. Carson, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
Chi Liu, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT 06511, USA; Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
References
- [1].Reske SN and Kotzerke J, “FDG-PET for clinical use,” Eur. J. Nucl. Med, vol. 28, no. 11, pp. 1707–1723, 2001, 10.1007/s002590100626. [DOI] [PubMed] [Google Scholar]
- [2].Avril N et al. , “Glucose metabolism of breast cancer assessed by 18F-FDG PET: histologic and immunohistochemical tissue analysis,” J. Nucl. Med, vol. 42, no. 1, pp. 9–16, 2001. [PubMed] [Google Scholar]
- [3].Paquet N, Albert A, Foidart J, and Hustinx R, “Within-patient variability of 18F-FDG: standardized uptake values in normal tissues,” J. Nucl. Med, vol. 45, no. 5, pp. 784–788, 2004. [PubMed] [Google Scholar]
- [4].Torizuka T et al. , “In vivo assessment of glucose metabolism in hepatocellular carcinoma with FDG-PET,” J. Nucl. Med, vol. 36, no. 10, pp. 1811–1817, 1995. [PubMed] [Google Scholar]
- [5].Weber WA, Schwaiger M, and Avril N, “Quantitative assessment of tumor metabolism using FDG-PET imaging,” Nucl. Med. Biol, vol. 27, no. 7, pp. 683–687, 2000, 10.1016/S0969-8051(00)00141-4. [DOI] [PubMed] [Google Scholar]
- [6].Boellaard R et al. , “FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0,” Eur. J. Nucl. Med. Mol. Imaging, vol. 37, no. 1, pp. 181–200, 2010, 10.1007/s00259-009-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Townsend DW, Carney JP, Yap JT, and Hall NC, “PET/CT today and tomorrow,” J. Nucl. Med, vol. 45, no. 1 suppl, pp. 4S–14S, 2004. [PubMed] [Google Scholar]
- [8].Judenhofer MS et al. , “Simultaneous PET-MRI: a new approach for functional and morphological imaging,” Nat. Med, vol. 14, no. 4, pp. 459–465, 2008, 10.1038/nm1700. [DOI] [PubMed] [Google Scholar]
- [9].Wang G, Rahmim A, and Gunn RN, “PET parametric imaging: Past, present, and future,” IEEE Trans. Radiat. Plasma. Med. Sci, vol. 4, no. 6, pp. 663–675, 2020, doi: 10.1109/TRPMS.2020.3025086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dimitrakopoulou-Strauss A, Pan L, and Sachpekidis C, “Kinetic modeling and parametric imaging with dynamic PET for oncological applications: General considerations, current clinical applications, and future perspectives,” Eur. J. Nucl. Med. Mol. Imaging, vol. 48, pp. 21–39, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, and Rahmim A, “Dynamic whole-body PET parametric imaging: I. Concept, acquisition protocol optimization and clinical application,” Phys. Med. Biol, vol. 58, no. 20, p. 7391, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carson RE, “Tracer kinetic modeling in PET,” in Positron Emission Tomography: Springer, 2005, pp. 127–159. 10.1007/1-84628-007-9_6 [DOI] [Google Scholar]
- [13].Patlak CS, Blasberg RG, and Fenstermacher JD, “Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data,” J. Cereb. Blood Flow Metab, vol. 3, no. 1, pp. 1–7, 1983. [DOI] [PubMed] [Google Scholar]
- [14].Visser EP et al. , “Comparison of tumor volumes derived from glucose metabolic rate maps and SUV maps in dynamic 18F-FDG PET,” J. Nucl. Med, vol. 49, no. 6, pp. 892–898, 2008. [DOI] [PubMed] [Google Scholar]
- [15].Wienhard K, “Measurement of glucose consumption using [18F] fluorodeoxyglucose,” Methods, vol. 27, no. 3, pp. 218–225, 2002, 10.1016/S1046-2023(02)00077-4. [DOI] [PubMed] [Google Scholar]
- [16].Wang G, Corwin MT, Olson KA, Badawi RD, and Sarkar S, “Dynamic PET of human liver inflammation: impact of kinetic modeling with optimization-derived dual-blood input function,” Phys. Med. Biol, vol. 63, no. 15, p. 155004, 2018, 10.2967/jnumed.121.262668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karakatsanis NA et al. , “Generalized whole-body Patlak parametric imaging for enhanced quantification in clinical PET,” Phys. Med. Biol, vol. 60, no. 22, pp. 8643–8673, 2015, DOI: 10.1088/0031-9155/60/22/8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang G et al. , “Total-Body PET Multiparametric Imaging of Cancer Using a Voxel-wise Strategy of Compartmental Modeling,” J. Nucl. Med, vol. 63, no. 8, pp. 1274–1281, 2021, DOI: 10.2967/jnumed.121.262668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen K et al. , “Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function,” J. Cereb. Blood Flow Metab, vol. 18, no. 7, pp. 716–723, 1998, 10.1097/00004647-199807000-00002. [DOI] [PubMed] [Google Scholar]
- [20].Dimitrakopoulou-Strauss A, Pan L, and Strauss LG, “Quantitative approaches of dynamic FDG-PET and PET/CT studies (dPET/CT) for the evaluation of oncological patients,” Cancer Imaging, vol. 12, no. 1, p. 283, 2012, doi: 10.1102/1470-7330.2012.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu J et al. , “Generation of parametric Ki images for FDG PET using two 5‐min scans,” Med. Phys, vol. 48, no. 9, pp. 5219–5231, 2021, 10.1002/mp.15113. [DOI] [PubMed] [Google Scholar]
- [22].Naganawa M et al. , “Assessment of population-based input functions for Patlak imaging of whole body dynamic 18F-FDG PET,” EJNMMI physics, vol. 7, no. 1, pp. 1–15, 2020, 10.1186/s40658-020-00330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Van Der Weerdt AP, Klein LJ, Boellaard R, Visser CA, Visser FC, and Lammertsma AA, “Image-derived input functions for determination of MRGlu in cardiac 18F-FDG PET scans,” J. Nucl. Med, vol. 42, no. 11, pp. 1622–1629, 2001. [PubMed] [Google Scholar]
- [24].Tian C, Fei L, Zheng W, Xu Y, Zuo W, and Lin C, “Deep learning on image denoising: An overview,” Neural Netw, 2020, 10.1016/j.neunet.2020.07.025. [DOI] [PubMed] [Google Scholar]
- [25].Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, and Ding X, “Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation,” Med. Image Anal, vol. 63, p. 101693, 2020, 10.1016/j.media.2020.101693. [DOI] [PubMed] [Google Scholar]
- [26].Serte S, Serener A, and Al-Turjman F, “Deep learning in medical imaging: A brief review,” Trans. Emerg. Telecommun. Technol, p. e4080, 2020, 10.1002/ett.4080. [DOI] [Google Scholar]
- [27].Lu W et al. , “An investigation of quantitative accuracy for deep learning based denoising in oncological PET,” Phys. Med. Biol, vol. 64, no. 16, p. 165019, 2019. [DOI] [PubMed] [Google Scholar]
- [28].Goodfellow I et al. , “Generative adversarial nets,” Adv. Neural Inf. Process. Syst, vol. 27, 2014. [Google Scholar]
- [29].Yang Q et al. , “Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss,” IEEE Trans. Med. Imaging, vol. 37, no. 6, pp. 1348–1357, 2018, doi: 10.1109/TMI.2018.2827462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y et al. , “3D conditional generative adversarial networks for high-quality PET image estimation at low dose,” Neuroimage, vol. 174, pp. 550–562, 2018, 10.1016/j.neuroimage.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ran M et al. , “Denoising of 3D magnetic resonance images using a residual encoder–decoder Wasserstein generative adversarial network,” Med. Image Anal, vol. 55, pp. 165–180, 2019, 10.1016/j.media.2019.05.001. [DOI] [PubMed] [Google Scholar]
- [32].Nie D et al. , “Medical image synthesis with deep convolutional adversarial networks,” IEEE Trans. Biomed. Eng, vol. 65, no. 12, pp. 2720–2730, 2018, doi: 10.1109/TBME.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015: Springer, pp. 234–241, 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- [34].Gong K, Yang J, Kim K, El Fakhri G, Seo Y, and Li Q, “Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images,” Phys. Med. Biol, vol. 63, no. 12, p. 125011, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shi L, Onofrey JA, Liu H, Liu Y, and Liu C, “Deep learning-based attenuation map generation for myocardial perfusion SPECT,” Eur. J. Nucl. Med. Mol. Imaging, vol. 47, no. 10, 2020, 10.1007/s00259-020-04746-6. [DOI] [PubMed] [Google Scholar]
- [36].Wang R et al. , “Generation of synthetic PET images of synaptic density and amyloid from 18F-FDG images using deep learning,” Med. Phys, vol. 48, no. 9, pp. 5115–5129, 2021, 10.1002/mp.15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gong K, Catana C, Qi J, and Li Q, “Direct patlak reconstruction from dynamic PET using unsupervised deep learning,” in 15th International meeting on fully three-dimensional image reconstruction in radiology and nuclear medicine, 2019, vol. 11072: International Society for Optics and Photonics, p. 110720R, 10.1117/12.2534902. [DOI] [Google Scholar]
- [38].Huang Z et al. , “Parametric image generation with the uEXPLORER total-body PET/CT system through deep learning,” Eur. J. Nucl. Med. Mol. Imaging, pp. 1–11, 2022, 10.1007/s00259-022-05731-x. [DOI] [PubMed] [Google Scholar]
- [39].Wang H et al. , “Deep learning–based dynamic PET parametric Ki image generation from lung static PET,” Eur. Radiol, 2022/November/18 2022, 10.1007/s00330-022-09237-w. [DOI] [PubMed] [Google Scholar]
- [40].van den Hoff J et al. , “The PET-derived tumor-to-blood standard uptake ratio (SUR) is superior to tumor SUV as a surrogate parameter of the metabolic rate of FDG,” EJNMMI research, vol. 3, no. 1, pp. 1–8, 2013, 10.1186/2191-219X-3-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ye Q et al. , “Improved discrimination between benign and malignant LDCT screening-detected lung nodules with dynamic over static 18F-FDG PET as a function of injected dose,” Phys. Med. Biol, vol. 63, no. 17, p. 175015, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen X et al. , “CT-free attenuation correction for dedicated cardiac SPECT using a 3D dual squeeze-and-excitation residual dense network,” J. Nucl. Cardiol, pp. 1–16, 2021, 10.1007/s12350-021-02672-0. [DOI] [PubMed] [Google Scholar]
- [43].Wang Z, Bovik AC, Sheikh HR, and Simoncelli EP, “Image quality assessment: from error visibility to structural similarity,” IEEE Trans. Image Process, vol. 13, no. 4, pp. 600–612, 2004, DOI: 10.1109/TIP.2003.819861. [DOI] [PubMed] [Google Scholar]


